Introduction
The tree shrew (Tupaia belangeri) is widely
distributed in South China and is in the order Scandentia. Based on
several characteristics, including small body size, low cost, easy
maintenance, short reproductive cycle, high brain-to-body mass
ratio and, in particular, its close relation to primates, the tree
shrew is being increasingly used as a valuable animal model and as
an alternative animal model to rodents and non-human primates in
biomedical research, including use as a model in investigations of
viral hepatitis infection (1),
depression (2), myopia (3,4),
tumors (5), aging and learning
behaviors (6).
Interleukins (ILs) are the largest group of
cytokines and are important in the innate and acquired immune
responses of the host. Among them, IL-6 is vital in immune
responses, including hematopoiesis, the acute phase of disease
processes, immune cell differentiation and energy transduction,
through several complex immune signaling pathways (7). Although IL-6 has a protective role
in various bacterial and viral infections, the same activities can
maintain chronic inflammation in conditions, including arthritis
and other autoimmune diseases. Therefore, it is important to define
the sequence features of IL-6 and obtain further insight into the
function of tsIL-6 in the antiviral and antibacterial response to
assist in investigations using tree shrews for models associated
with IL-6.
To date, >89 IL-6 genes have been identified in
mammals, and analyses of the homology of IL-6 among several species
have been reported (8–12). Additionally, a number of important
immune factors and important molecules in the tree shrew have been
investigated in previous studies (13–15). However, as an important cytokine
associated with viral infection, the genomic and mRNA sequence of
tsIL-6 has not been predicted (16) and associated investigations of
expression have not been reported for the tree shrew, restricting
its use in the modeling of human disease.
As the tree shrew is considered to be evolutionarily
closer to humans, than rodents, they may serve as a superior animal
model to investigate human disease mechanisms. The present study
aimed to elucidate the structural characteristics of tsIL-6 and its
expression pattern in response to exogenous challenge, in order to
provide fundamental information on the gene structure and phylogeny
of the tsIL-6 gene. In addition, the tissue-specific distribution
of IL-6 was examined in 14 tissue specimens, and its expression
profiles following lipopolysaccharide (LPS) and
polyinosinic:polycytidylic acid (poly I:C) stimulation in four
tissues associated with the immune system were examined using
reverse transcription-quantitative polymerase chain reaction
(RT-qPCR) analysis.
Materials and methods
Laboratory animals and exogenous
challenge
Healthy 1-year-old, male tree shrews (Tupaia
belangeri; length, 17.89±0.49 cm; weight, 149.80±15.26 cm) were
provided by and maintained at the Center of Tree Shrew Germplasm
Resources, Institute of Medical Biology, The Chinese Academy of
Medical Science and Peking Union Medical College (Kunming, China).
The production certification number was SCXK (Dian) K2013-0001. The
institutional Animal Care and Welfare Committee of the Institute of
Medical Biology, Chinese Academy of Medical Sciences and Peking
Union Medical College approved the present study, and all the
procedures were performed according to ethical standards and
practices.
Immune stimulation was performed by intraperitoneal
injection of 500 μl LPS (1.5 mg/ml; cat. no. L2630) or 500
μl Poly I:C (1.5 mg/ml; cat. no. P9582) (both from
Sigma-Aldrich, St. Louis, MO, USA) in phosphate-buffered saline
(PBS; pH 7.4; HyClone Media, Logan, UT, USA), respectively
(11,12). Tree shrews injected with 500
μl PBS were used as controls. Following injection, all tree
shrews were placed back into their cages and housed individually.
All animals were housed in the same room under a 12 h light/12 h
dark cycle (lights on at 08:00; 150 lux) with 60±10% relative
humidity and an ambient temperature of 25±2°C. The tree shrews had
ad libitum access to water and food. At 0, 3, 6, 12, 24, 48
and 72 h post-injection, two tree shrews in each group were
sacrificed and four samples of tissues associated with the immune
system, the liver, spleen, kidney and intestine, were obtained for
total RNA extraction.
RNA isolation, cDNA synthesis and
cloning, and sequencing of tsIL-6 mRNA
Total RNA was extracted from ~100 mg of tree shrew
splenic tissue with TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) according to the manufacturer's
protocol, and incubated in RNase-free DNase I (Takara Biotechnology
Co., Ltd., Dalian, China) for 20–30 min at 37°C to remove residual
genomic DNA. First-strand cDNA was synthesized from the total RNA
using M-MLV reverse transcriptase (Fermentas; Thermo Fisher
Scientific, Inc., St. Leon-Rot, Germany) with the 5′-rapid
amplification of mRNA ends (RACE) RT primer and
oligo(dT)18.
Based on the published mRNA sequence of tsIL-6
(16), a pair of primers, EXONs
1–5, targeting the two ends of tsIL-6 was designed (Table I). The 5′ and 3′ ends of tsIL-6
were amplified using the RACE approach (17). The 5′ end RACE PCR reaction was
performed using the 5′-RACE 1 adapter forward primer and a
gene-specific 5′-RACE 1 reverse primer, followed by nested PCR with
a 5′-RACE 2 adapter forward primer and a gene-specific 5′-RACE 2
reverse primer. The 3′ end RACE PCR reaction was performed using
the gene-specific 3′-RACE 1 forward primer and 3′-RACE 1 adapter
reverse primer, followed by nested PCR with a gene-specific 3′-RACE
2 forward primer and the 3′-RACE 2 adapter reverse primer.
Approximately 1 μl cDNA synthesized from total RNA from
tissues was used as the template. The reaction was performed in a
volume of 20 μl containing 0.4 μM of each primer, 200
μM dNTPs, 1 unit of LA TaqDNA polymerase (Takara
Biotechnology Co., Ltd.) and 2 μl 10X buffer. We used the
following PCR conditions: one denaturation cycle at 95°C for 2 min,
35 cycles of 94°C for 30 sec, 57°C for 30 sec and 72°C for 30 sec,
followed by one cycle of 72°C for 10 min.
 | Table IPrimer pairs used in polymerase chain
reactions. |
Table I
Primer pairs used in polymerase chain
reactions.
| Primer pair | Forward primer
(5′–3′) | Reverse primer
(5′–3′) |
|---|
| EXONS 1–5 |
CCACAAGCACCTTCAGTCCAG |
CAAAGGAAGGAATGTCCATGTCTA |
| 5′-RACE 1 |
GCCACGCGTCGACTAGTACGGGGGGGGGG |
GCCAGTGCAACCCTGCACTTGTAAA |
| 5′-RACE 2 |
GGCCACGCGTCGACTAGTAC |
TCTTCAGTTTTATCTGTGGAGGTGGGTTCT |
| 3′-RACE 1 |
TCCCACCCCTGACCCAACTTCAAAT |
GCTGTCAACGATACGCTACGTAAC |
| 3′-RACE 2 |
TCAAGAGGAGTGGCTAAAGAAGGTGACAGT |
GCTACGTAACGGCATGACAGTG |
| Genome 1 |
ACGCTTATACTTTTAGTTCTTCATGGA |
CAGCTATGCCAGGCAGTGTTT |
| Genome 2 |
TGTGTCTGAGTTTTAAGCTGCCA |
GGCAGAATGAAGCACATCCAA |
| Genome 3 |
CTGGAGGAGAAGAAATTGTGGAG |
TCCTTCTAGCTCTTTCTTAGGCAA |
| Genome 4 |
CAACCCAGAGGGACCAATTTT |
CAGGCCATTCATTTCCTTTCT |
| Genome 5 |
AGTGCCAAAGTCCAAGGGTC |
TCTGTCCTTGAGCCCACCA |
| GAPDH |
CCATCACCATCTTCCAGGAGCGAG |
CAAAGGTGGAGGAGTGGGTGTCG |
| IL-6 qPCR |
CCTGACCCAACTTCAAATGC |
CACACTACATTAGCCGAGAAGC |
| GAPDH |
CCATCACCATCTTCCAGGAGCGAG |
CAAAGGTGGAGGAGTGGGTGTCG |
All PCR products were gel-purified, cloned into the
pMD19-T vector (Takara Biotechnology Co., Ltd.), and commercially
sequenced by Sangon Biotech Co., Ltd. (Shanghai, China). The
sequences of overlapping clones were assembled to construct the
complete tsIL-6 mRNA sequence.
DNA extraction and cloning of tsIL-6
genomic DNA
Total DNA was isolated from the splenic tissues
using a TIANamp Marine Animals DNA kit (cat. no. DP324; Tiangen
Biotech Co., Ltd., Beijing, China) according to the manufacturer's
protocol. Targeting the conserved regions of mammalian IL-6 genomic
sequences, five pairs of primers (genome 1–5) were designed to
amplify the genome of tsIL-6 (Table
I). All PCR products were gel-purified, cloned into the pMD19-T
vector (Takara Biotechnology Co., Ltd.), and sequenced by Sangon
Biotech Co., Ltd. The sequences of overlapping clones were
assembled to construct the complete tsIL-6 genomic gene sequence.
The tsIL-6 genomic and mRNA sequences were then aligned using
DNAMAN software (version 8.0; Lynno Biosoft, San Ramon, CA, USA) to
verify the intron/exon boundaries.
Amino acid sequences and phylogenetic
analysis
The promoter sequence was analyzed using the JASPAR
database (http://jaspar.genereg.net/cgi-bin/jaspar_db.pl).
Homology sequence searching was performed using the Basic Local
Alignment Search Tool (http://www.ncbi.nlm.nih.gov/blast). Deduced amino acid
sequence comparisons were performed using the Expert Protein
Analysis System (http://www.expasy.org/). The signal peptide and
conserved domains were predicted using the Simple Modular
Architecture Reach Tool (http://smart.embl-heidelberg.de/). Glycosylation sites
were predicted using NetOGlyc 3.1 (http://www.cbs.dtu.dk/services/NetOGlyc).
Phosphorylation sites were predicted using the NetPhos2.0 server
(http://www.cbs.dtu.dk/services/NetPhos/).
Multiple alignments of the putative amino acid
sequences of tsIL-6 with IL-6 from other related species were
performed using MUltiple Sequence Comparison by Log-Expectation
(18). Phylogenetic trees were
obtained based on the IL-6 amino acid sequences (data in https://www.ncbi.nlm.nih.gov/nuccore/1051343575?report=fasta)
by Bayesian inference (BI) using MrBayes software v.3.2.6
(http://mrbayes.sourceforge.net/)
(19,20) and by maximum likelihood (ML) using
RAxML v.7.0.4 (https://sco.h-its.org/exelixis/web/software/raxml/)
(21). In BI, two independent
sets of Markov chain Monte Carlo algorithms were run, each with
three heated and one unheated chain, for 1,000,000 generations. The
trees were sampled every 1,000 generations. The first 2,500 trees
were discarded as burn-in. In ML analyses, the 'rapid bootstrap'
option was run from starting random seeds to generate 100
nonparametric bootstrap replicates and the GTR-GAMMA model, the
other parameters were set as default. Cows were used as an outgroup
in both analyses.
Modeling of tsIL-6
The spatial protein structure was predicted using
the SWISS-MODEL workspace (http://www.expasy.ch/swissmod/SWISS-MODEL.html) and
was illustrated using the PyMOL Molecular Graphics system version
1.7.2.1 (DeLano Scientific, San Carlos, CA, USA).
RT-qPCR of tsIL-6
The RT-qPCR reactions were performed using SG Fast
qPCR Master mix (High Rox) in a Stepone Plus system (both from ABI;
Thermo Fisher Scientific, Inc.). For each reaction, a volume of 20
μl containing 0.4 μM of each forward and reverse
primer, 2 μl of cDNA product, and 10 μl of 2X
SYBR-Green Premix Ex Taq II were used for the RT-qPCR reaction. The
primer sequences are provided in Table I (IL-6 qPCR primer). The cycling
profile consisted of an initial denaturation at 95°C for 3 min,
followed by 45 cycles of 95°C for 7 sec, 57°C for 10 sec, and 72°C
for 15 sec, followed by melt curve analysis. The tree shrew
housekeeping gene glyceraldehyde 3-phosphate dehydrogenase (GAPDH)
was used as the reference gene for normalization (22). The normalized mRNA expression
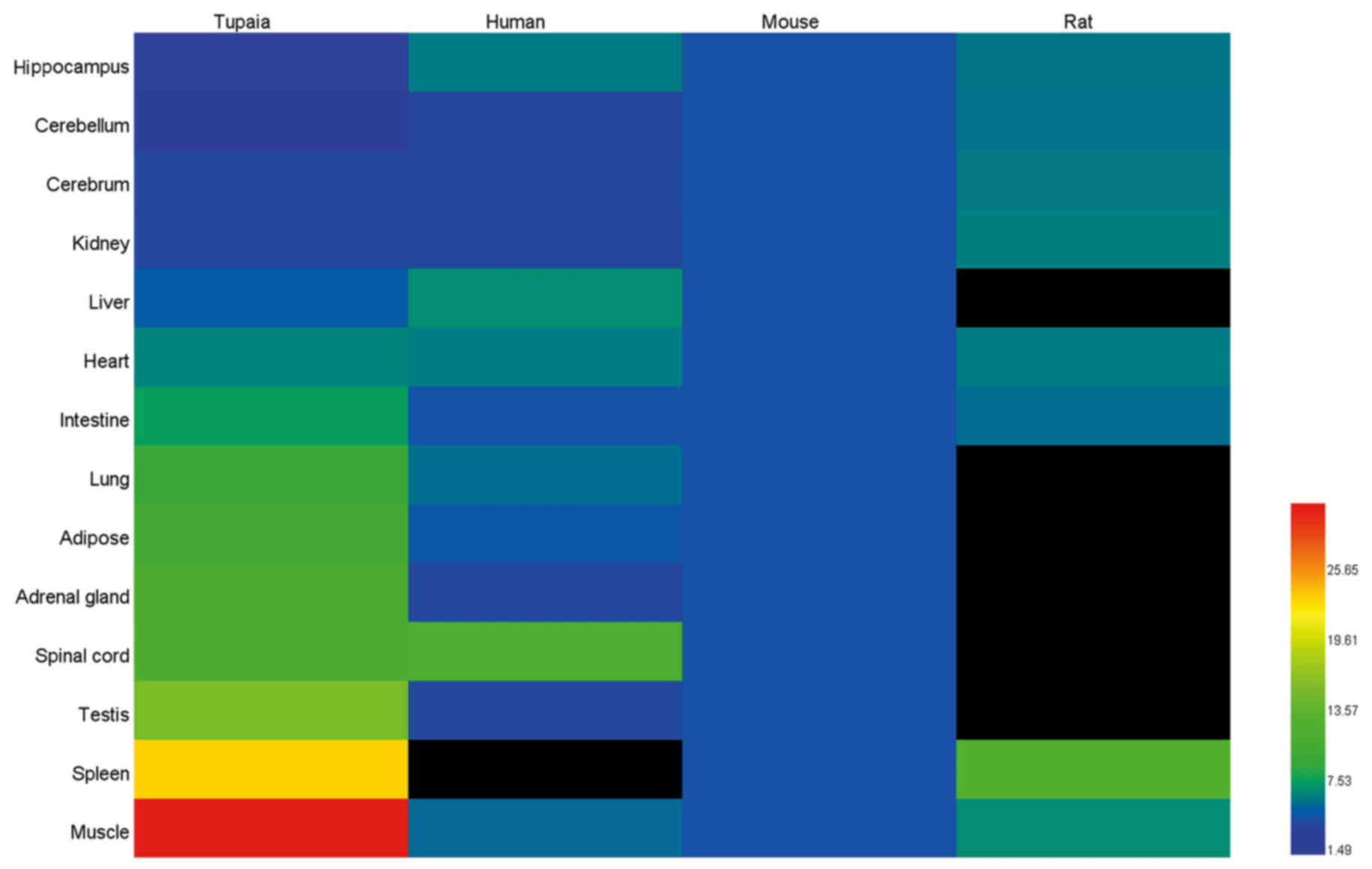
information of tree shrew, human, mouse and rat from BioGPS
(www.biogps.org) was used in order to compare IL-6
gene basal expression in different tissues of tree shrew, human,
mouse and rat.
Statistical analysis
Data were analyzed using one-way analysis of
variance and Student-Newman-Keuls test with SPSS software (version
15; SPSS, Inc., Chicago, IL, USA), The results are expressed as the
mean ± standard error of the mean. P<0.05 was considered to
indicate a statistically significant difference.
Results
Structural features of the tsIL-6
gene
The structural characteristics of the complete
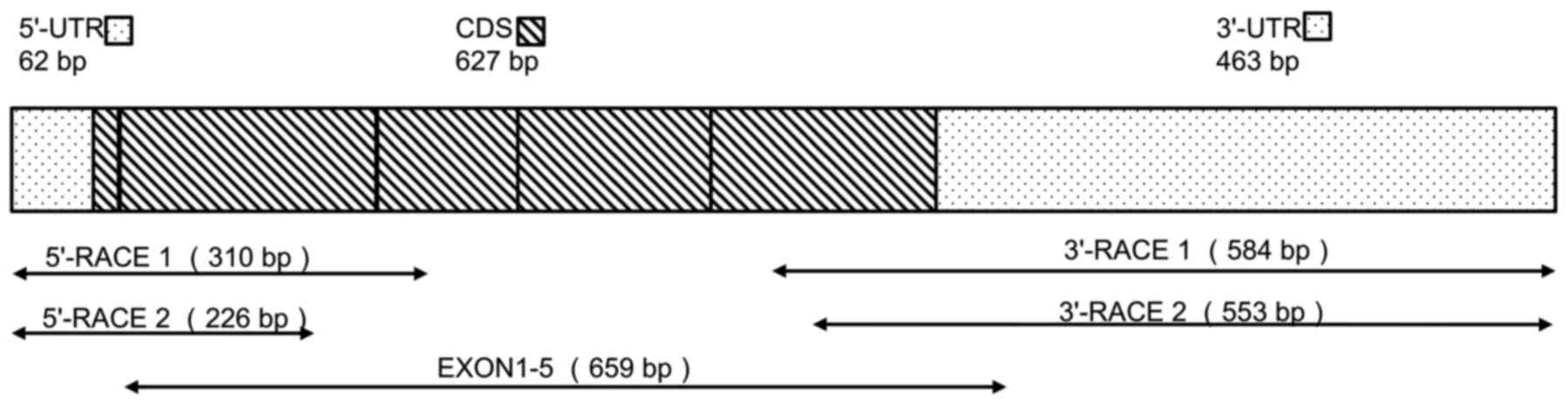
tsIL-6 mRNA are shown in Fig. 1.
An mRNA fragment of 659 bp was amplified using the degenerate
primers, EXONs 1–5. A fragment of 226 bp containing the initial
start codon ATG was then amplified by 5′ RACE, and a 553-bp
fragment containing the poly(A) tail was obtained by 3′ RACE.
Alignment with the DNA sequence produced a 1,152-bp mRNA sequence
with five putative open reading frames (ORFs). Therefore, the
1,152-bp sequence representing the full-length mRNA sequence of
tsIL-6 was obtained (accession no. KX359391), which contained an
ORF of 627 bp, a 62-bp 5′-untranslated region (UTR), a 463-bp
3′-UTR (Fig. 1), and six RNA
instability motifs (ATTTA) in the 3′-UTR (23).
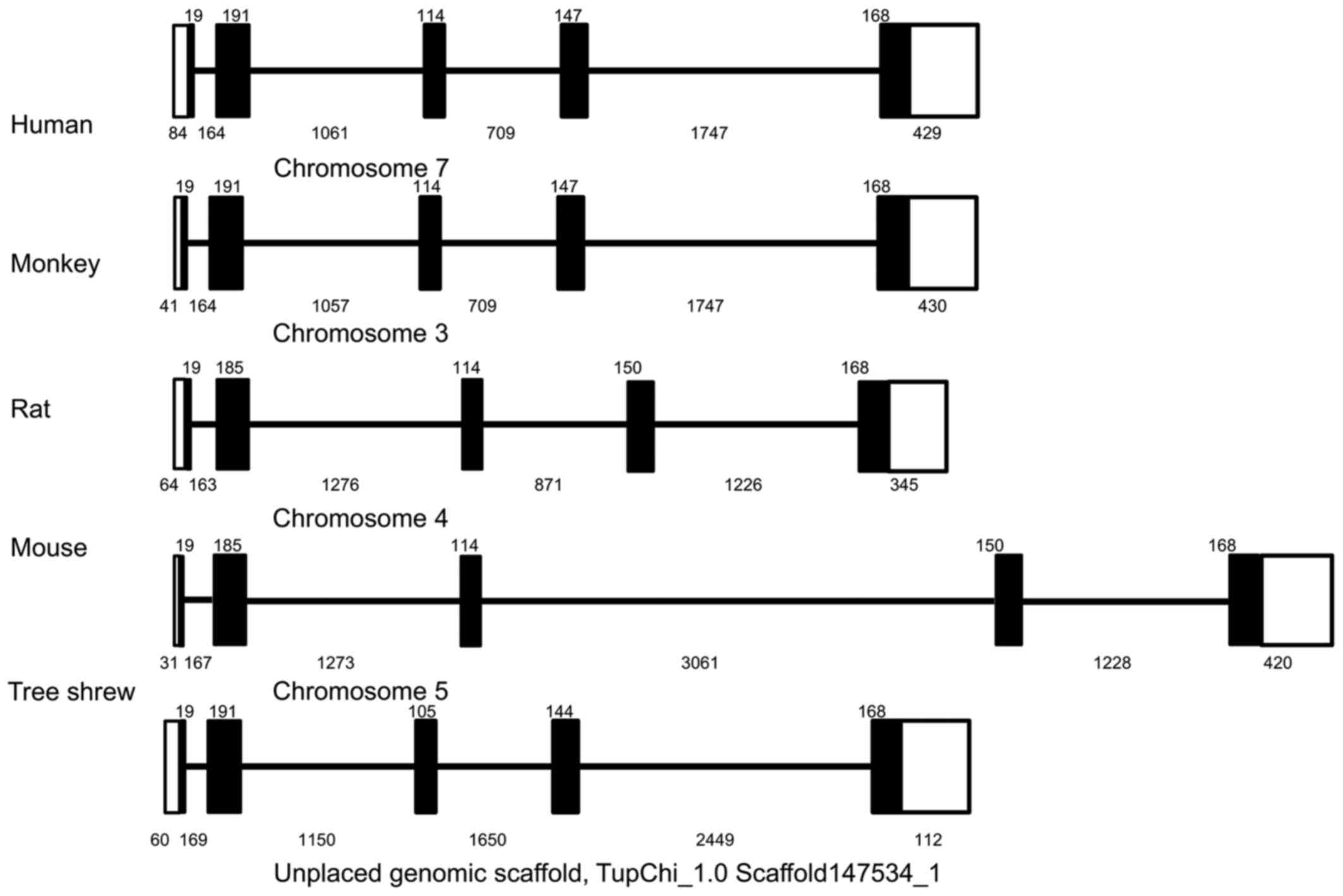
Using a similar strategy, the 5,265-bp genomic
sequence of the tsIL-6 gene was obtained and submitted to GenBank
(accession no. KX359390). In contrast to the structure of the mRNA,
the genomic sequence revealed that the tsIL-6 gene consisted of
five exons and four introns (Fig.
2). All four introns had the typical structure of 'GU-AG', in
which the first two nucleotides were 5′-GU-3′, and the last two
nucleotides were 5′-AG-3′). There was a TATA box with the typical
sequence of TATAATAAT between bases -548 and -539 bp, and a CAAT
box with a typical sequence of GGC/TCAATCT between bases −605 and
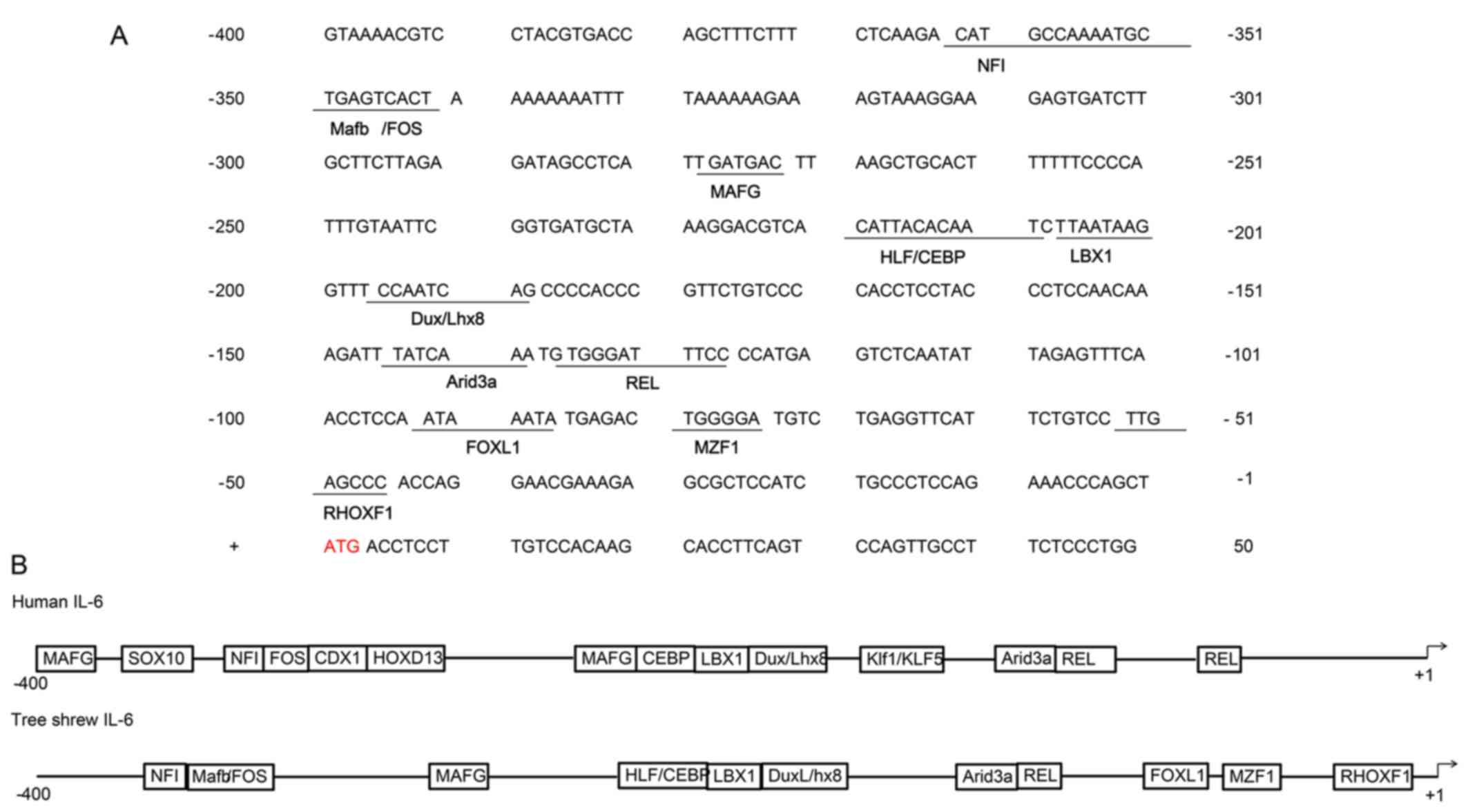
−596 bp. Analysis of the promoter region of tsIL-6 (between −400
and +50 bp from the transcription initiation site) for known
conserved elements using JASPAR revealed several putative
transcription factor binding regions and showed a high degree of
conservation of the mammalian IL-6 gene. The highly conserved NFI,
FOS, MAFG, CEBP, LBX1, DuxLhx8, Arid3a and REL sequences were
identified, shown in Fig. 3A. The
comparison of functional cis-regulatory elements from human and
tsIL-6 promoter regions is shown in Fig. 3B (24).
Features of predicted amino acid
sequences
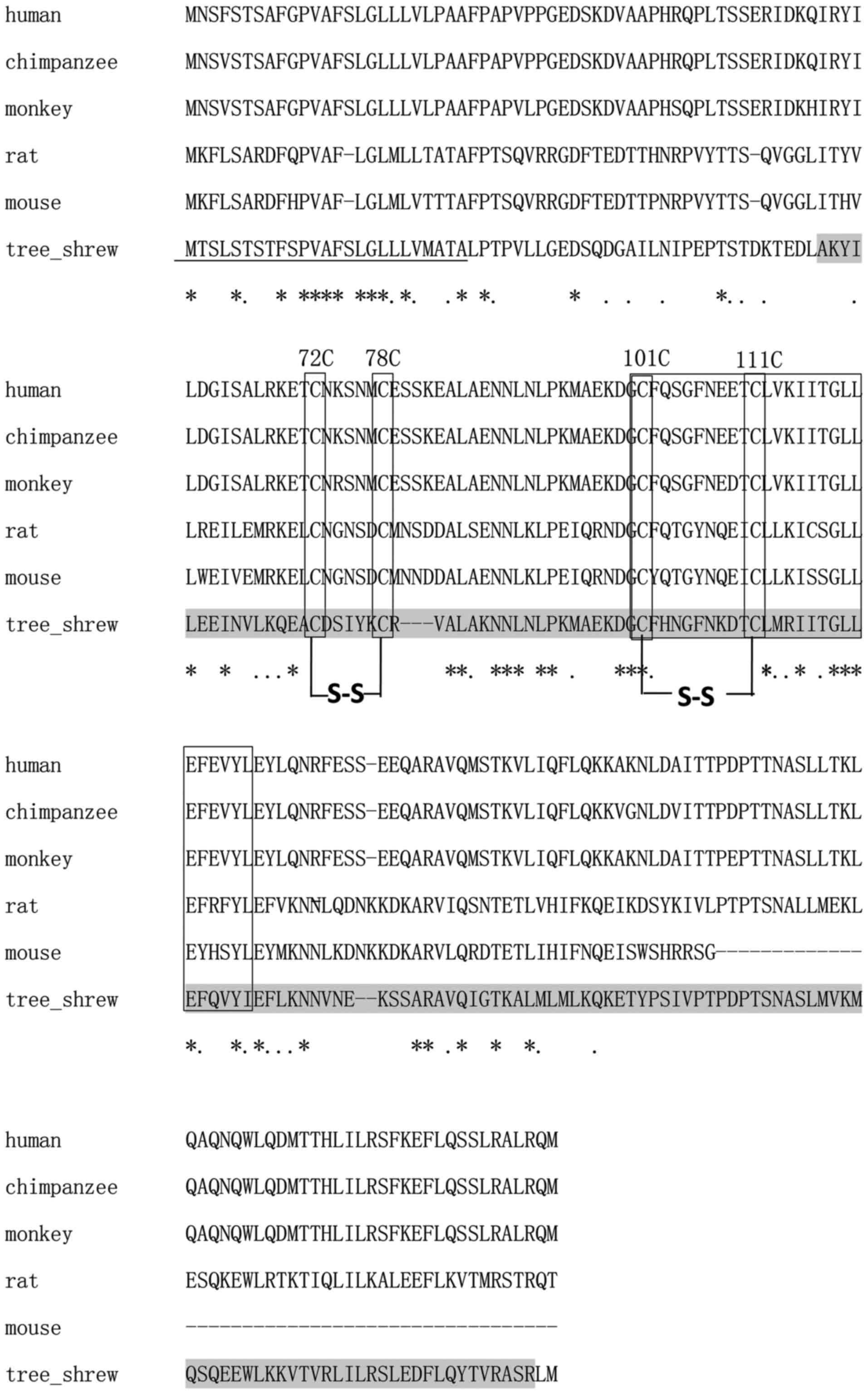
Using the SMART program, the 627-bp ORF of the
putative tsIL-6 was predicted to encode 208 amino acids, consisting
of a signal peptide in the first 25 amino acids of the N-terminus
and an IL-6 domain between amino acid positions 57 and 206, with an
E-value of 1.54e–56 (Fig. 4). The
molecular weight and isoelectric point were theoretically
calculated as 23.46 and 7.97 kDa, respectively. All four conserved
cysteine residues in mammalian IL-6 were observed at amino acid
positions 72, 78, 98 and 108 (Fig.
4). The typical C-X(9)-C-X(6)-G-L-X(2)-[FY]-X(3)-L IL-6 family motif and conserved
disulfide bridges were also identified in the tsIL-6 protein
(Fig. 4). Nine positive positions
were more likely to have O-GalNAc modifications, which were present
at amino acid positions 2, 3, 5, 7, 8, 10, 158, 162 and 177.
The alignment between human IL-6 and tsIL-6 revealed
111 identical amino acid residues. There were 97 amino acid
residues in human IL-6, which were not found in the corresponding
positions in the tree shrew. Among these, 35 amino acid residues
were conserved and 43 amino acid residues were semi-conserved
substitutions, and unlikely to affect protein structure or
function. The remaining 19 residues were non-conserved
substitutions. There were five non-conserved residues located close
to the N-terminus, at a position distant from the residues involved
in receptor binding. There were 14 non-conserved residues located
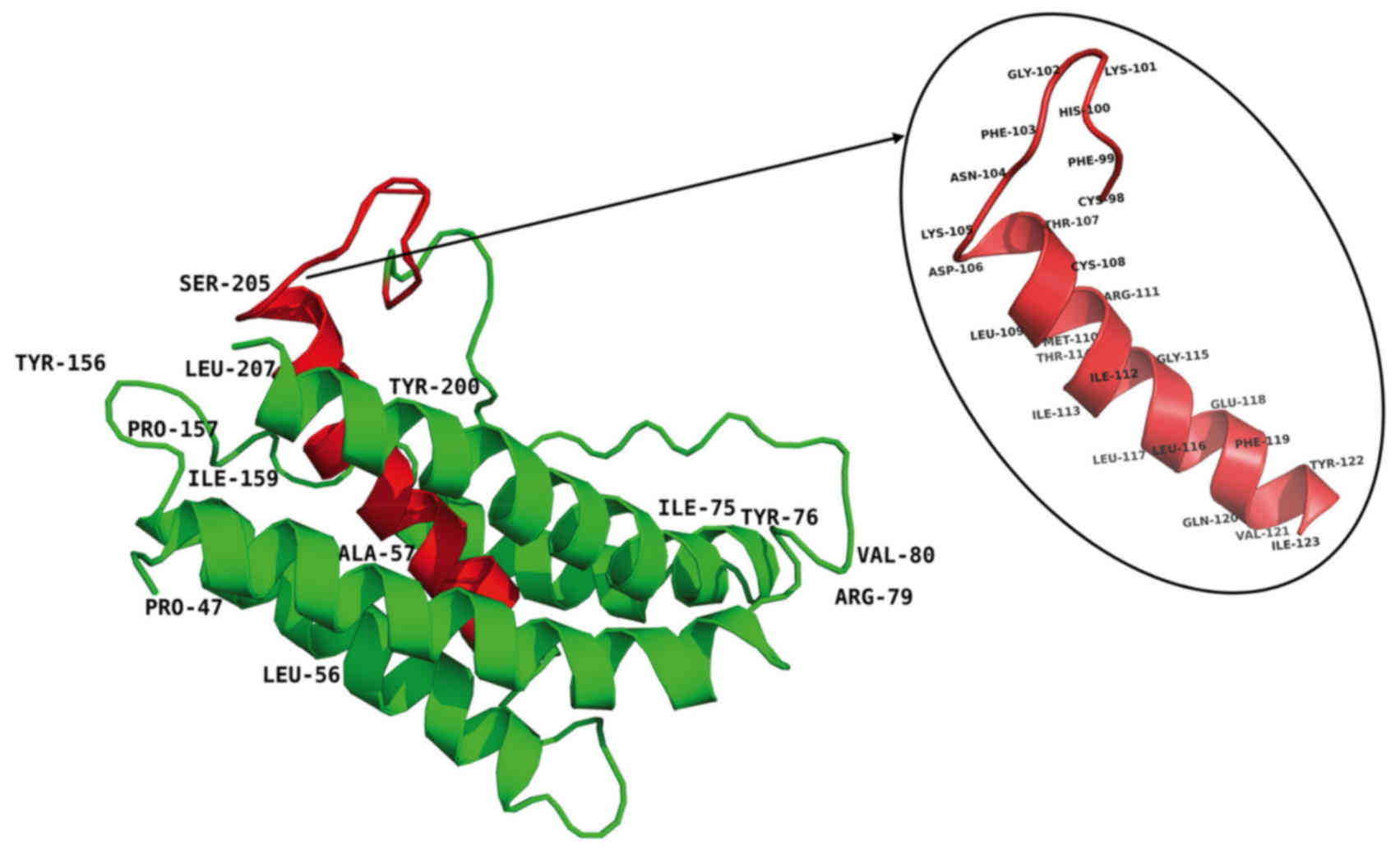
at sites of variable amino acid regions (labeled in Fig. 5). The structural and mutational
analyses of human IL-6 identified residues 49–73, 108–130, 137–157,
169–180 and 184–210 as α-helix A, B, C, D and E, respectively, and
residues 72C–78C and 101C–111C as putative disulfide bond sites
(Fig. 4). Similar α-helical
structure and disulfide bond sites were predicted in the
corresponding sites in tsIL-6. The conserved (C-x(9)-C-x(6)-G-L-x(2)-[FY]-x(3)-L) peptide located at position 98–123
consisted of a partial coil and a partial α-helix B (Fig. 5).
Features of the three-dimensional
structure
Analysis of the secondary structure of tsIL-6
revealed that the protein consisted of a bundle of four α-helices
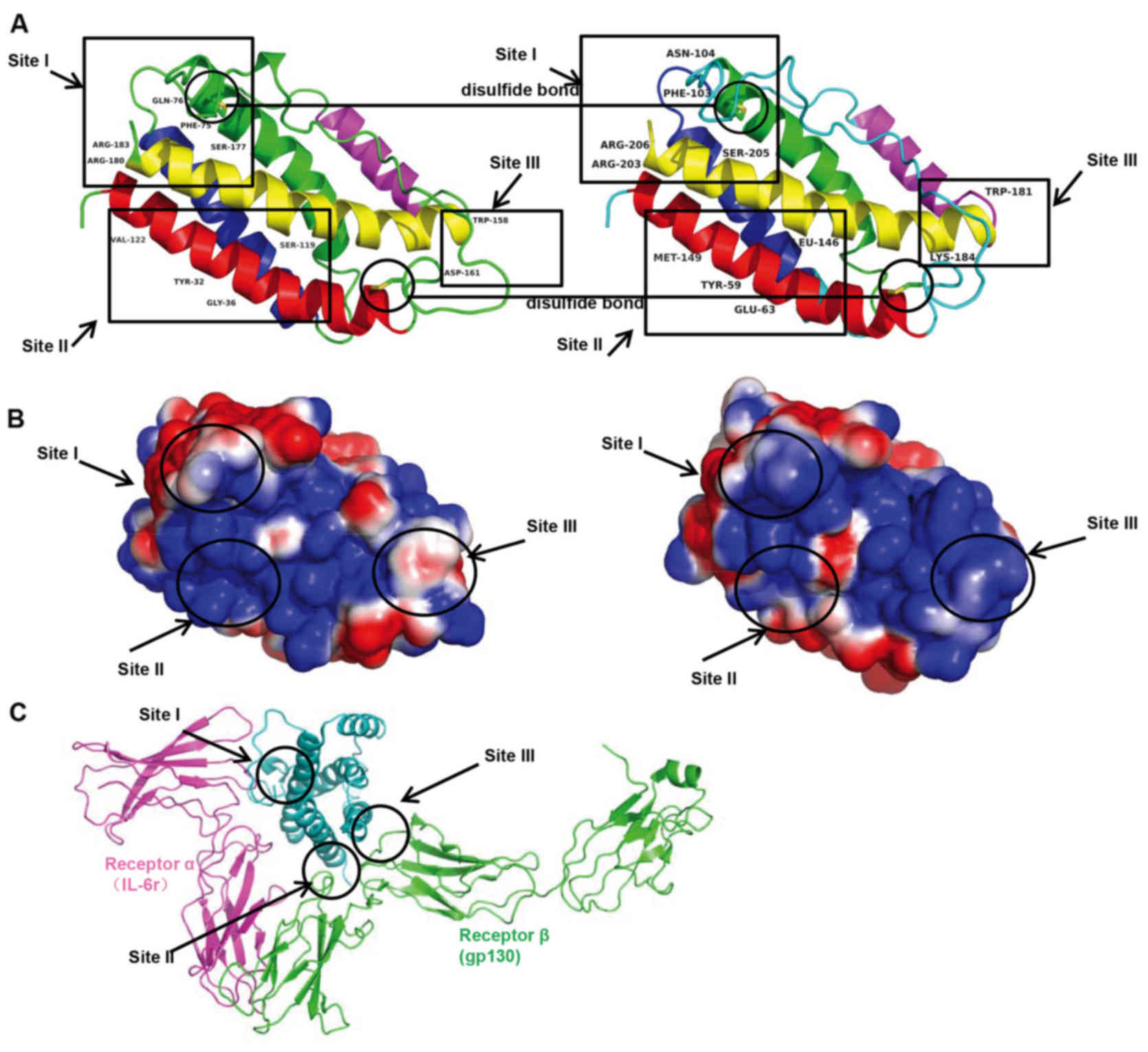
linked by loops and an additional mini-helix (Fig. 5). There were three main IL-6
binding sites: Site I, II and III. Site I was responsible for
IL-6-IL-6 receptor interactions. In the site II region, IL-6
interacted with gp130 in the trimer. Site III was the site of
IL-6-gp130 interactions between trimers. Phe74, Gln175, Ser176,
Arg179 and Arg182 in site I; Tyr31, Gly35, Ser118 and Val121 in
site II; and Trp157 and Asp160 in site III were identified as
important for binding based on mutagenesis investigation (Fig. 6A). The corresponding amino acids
were Phe103, Asn103, Ser205, Arg203 and Arg206 in site I; Tyr59,
Glu35, Leu146 and Met149 in site II; and Trp181 and Lys184 in site
III. The surface charge appeared to be similar in corresponding
site I, and less similar in sites II and III, suggesting
potentially different protein-protein interactions in human and
tsIL-6 (Fig. 6B and C). The
three-dimensional domain structures of human and tsIL-6 were
dimensionally conserved.
Phylogenetic relationship between the
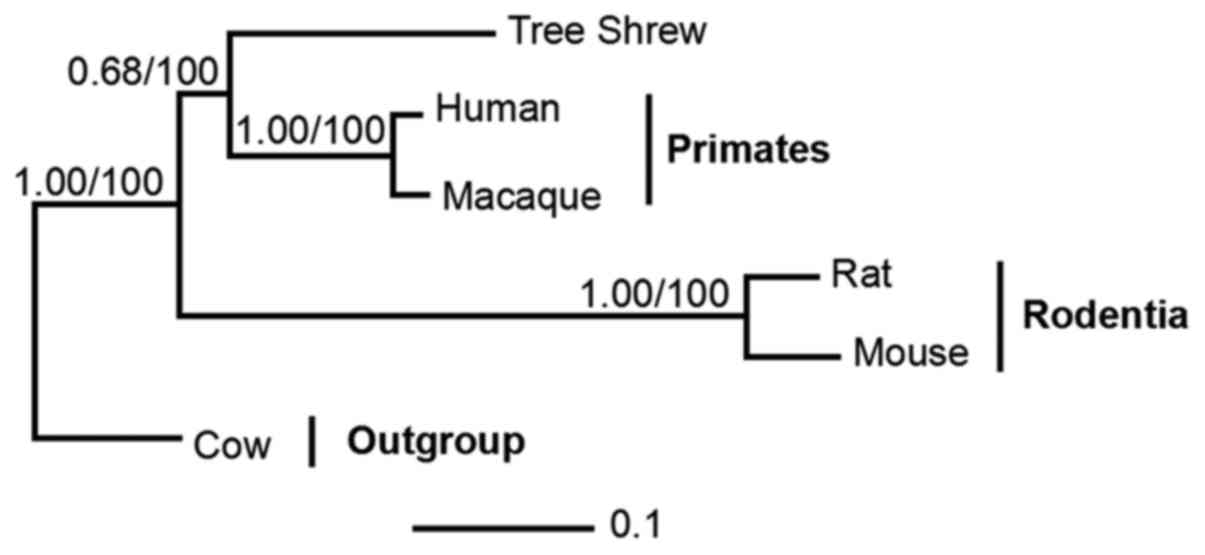
tree shrew and related species
The data comprised orthologs from five taxa,
including representatives of primates (humans and macaques),
Rodentia (mouse and rat) and the Cow outgroup. Phylogenetic trees
were reconstructed using BI and ML methods. The BI and ML analyses
supported the sister clades between tree shrew and primates with
posterior probabilities (PP=0.68) and bootstrap support (BP=100)
(Fig. 7).
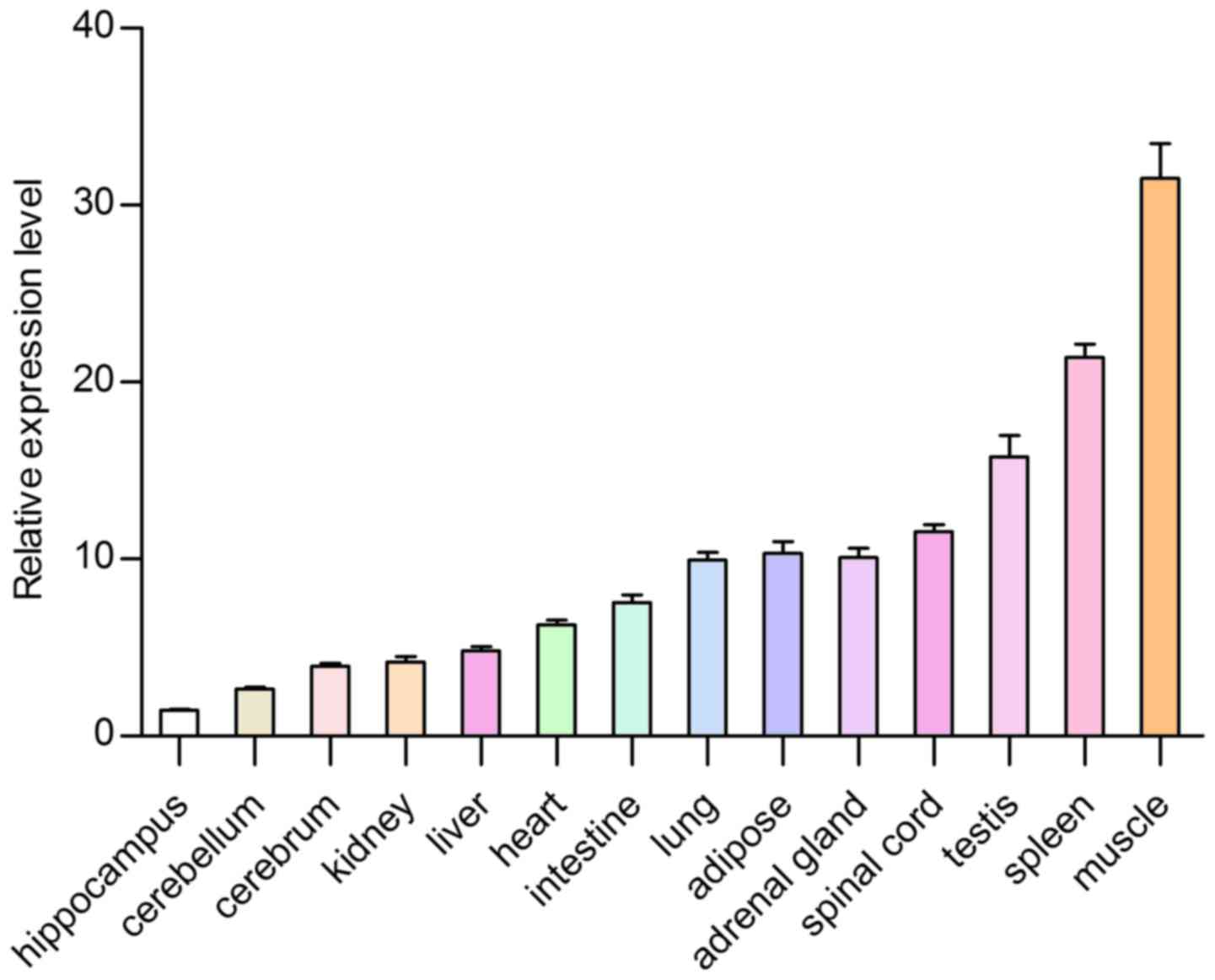
Feature of expression profiles
To further determine the tsIL-6 mRNA expression
distribution in the tree shrew, RT-qPCR was used to measure the
mRNA expression levels of tsIL-6 in 14 tissue specimens from adult
tree shrews. The highest expression levels were found in the muscle
and the spleen, followed by the testis, adipose tissue, spinal
cord, adrenal gland, lung, intestine, heart, liver and kidney,
whereas low expression was found in the cerebrum, cerebellum and
hippocampus (Fig. 8). The present
study further retrieved the normalized mRNA expression information
of humans, mice and rats from BioGPS (www.biogps.org), in order to compare IL-6 gene basal
expression in different tree shrew, human, mouse and rat tissues
(Fig. 9). However, the data from
the human splenic tissue, and the rat liver, lung, adipose, adrenal
gland, spinal cord and testicular tissues were unavailable in
BioGPS. Based on the relative abundance of mRNA expression of IL-6
in human whole blood in BioGPS, it was hypothesized that the
expression level of human IL-6 was likely to be higher in splenic
tissues than the other tissues, such as brain and liver, while the
expression level of rat IL-6 was likely to be similar in all
tissues. It appeared that the expression levels in humans and mice
were similar, whereas the expression of IL-6 in the tree shrew
muscle tissue was more variable and the highest basal mRNA
expression was found here.
Expression of tsIL-6 in response to LPS
and poly I:C challenge in vivo
The transcriptional changes of tsIL-6 in four immune
system-associated tissues following injection with LPS and poly I:C
are shown in Fig. 10. All tsIL-6
expression data is available in Tables II and III.
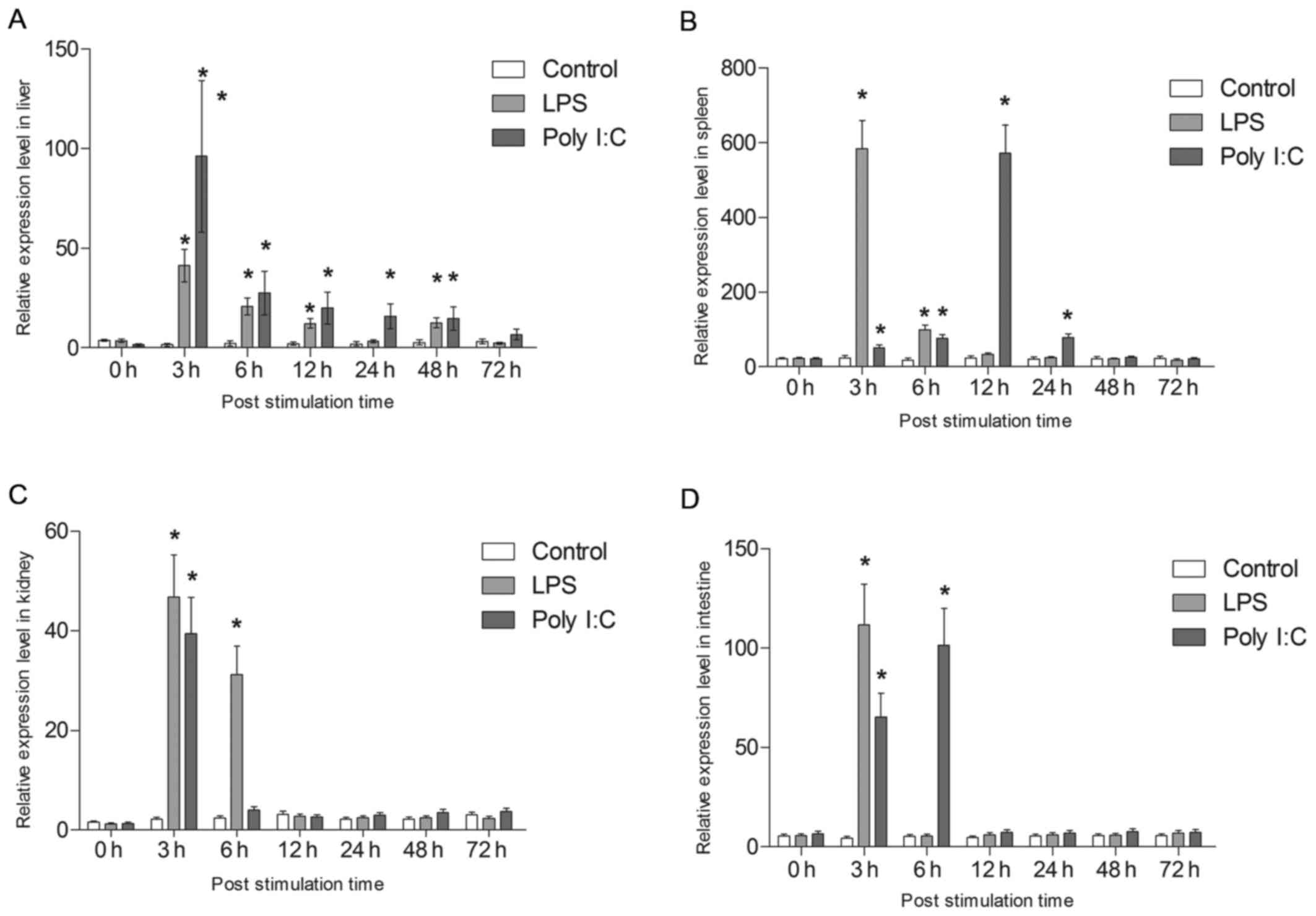 | Figure 10Analysis of the expression of tsIL-6.
Expression of tsIL-6 was determined in the (A) liver, (B) spleen,
(C) kidney and (D) intestinal tissues of the LPS- and poly
I:C-challenged groups and the control group using reverse
transcription-quantitative polymerase chain reaction analysis at 0,
3, 6, 12, 24, 48 and 72 h-post injection. Data are presented as the
mean ± standard deviation (n=6) within the same post-injection
time. *P<0.05 among treatments. tsIL-6, tress shrew
interleukin-6; LPS, lipopolysaccharide; poly I:C,
polyinosinic:polycytidylic acid. |
 | Table IIExpression levels of tsIL-6 in 14
tissues. |
Table II
Expression levels of tsIL-6 in 14
tissues.
| Expression
level | Cerebrum | Cerebellum | Hippocampus | Spinal cord | Heart | Liver | Spleen | Lung | Kidney | Muscle | Adrenal gland | Intestinal | Testis | Adipose |
|---|
| Tree shrew 1 | 4.00 | 2.59 | 1.49 | 11.40 | 6.58 | 5.05 | 23.56 | 9.66 | 4.20 | 31.69 | 11.33 | 7.76 | 15.59 | 10.25 |
| Tree shrew 2 | 4.52 | 2.82 | 1.39 | 11.63 | 7.44 | 4.39 | 20.73 | 10.05 | 3.36 | 37.39 | 8.84 | 5.97 | 13.72 | 11.99 |
| Tree shrew 3 | 4.32 | 2.25 | 1.48 | 11.74 | 5.66 | 5.00 | 20.26 | 9.28 | 4.78 | 35.81 | 8.95 | 8.53 | 15.28 | 12.50 |
| Tree shrew 4 | 3.52 | 2.80 | 1.32 | 13.23 | 6.06 | 4.44 | 18.85 | 8.41 | 5.16 | 23.77 | 10.65 | 7.29 | 12.01 | 8.30 |
| Tree shrew 5 | 3.40 | 2.51 | 1.26 | 11.06 | 6.19 | 4.14 | 23.56 | 11.50 | 4.28 | 30.10 | 11.67 | 8.77 | 17.78 | 9.43 |
| Tree shrew 6 | 3.96 | 2.98 | 1.79 | 10.15 | 5.73 | 5.80 | 21.44 | 10.72 | 3.27 | 30.42 | 9.06 | 7.06 | 20.27 | 9.53 |
 | Table IIIPost-injection expression data. |
Table III
Post-injection expression data.
| Tissue | Post-injection
(h) | Control | LPS | Poly I:C |
|---|
| Liver | 0 | 3.85±0.46 | 3.70±0.73 | 1.54±0.61 |
| 3 | 1.55±0.82 | 41.31±8.18 | 96.13±38.07 |
| 6 | 2.34±1.37 | 20.78±4.11 | 27.58±10.92 |
| 12 | 2.20±0.94 | 12.35±2.45 | 20.00±7.92 |
| 24 | 2.08±1.20 | 3.50±0.69 | 15.90±6.30 |
| 48 | 2.78±1.37 | 12.70±2.51 | 14.71±5.82 |
| 72 | 3.31±1.18 | 2.50±0.49 | 6.80±2.69 |
| Spleen | 0 | 22.14±4.07 | 23.19±4.26 | 22.03±4.05 |
| 3 | 24.67±9.07 | 583.57±107.29 | 51.98±9.57 |
| 6 | 19.13±7.03 | 99.59±18.31 | 76.12±13.99 |
| 12 | 24.06±8.84 | 33.45±6.15 | 572.65±105.28 |
| 24 | 21.33±7.83 | 25.27±4.64 | 78.10±14.36 |
| 48 | 22.18±8.15 | 21.56±3.96 | 25.38±4.66 |
| 72 | 23.00±8.46 | 19.05±3.50 | 22.28±4.11 |
| Kidney | 0 | 1.51±0.39 | 1.25±0.32 | 1.29±0.34 |
| 3 | 2.15±0.56 | 46.75±12.16 | 39.42±10.25 |
| 6 | 2.38±0.62 | 31.20±8.11 | 3.96±1.03 |
| 12 | 3.14±0.82 | 2.70±0.70 | 2.58±0.67 |
| 24 | 2.15±0.56 | 2.40±0.62 | 2.97±0.77 |
| 48 | 2.17±0.57 | 2.40±0.62 | 3.51±0.91 |
| 72 | 3.02±0.79 | 2.30±0.60 | 3.66±0.95 |
| Intestine | 0 | 5.36±1.39 | 5.55±1.44 | 6.56±1.71 |
| 3 | 4.37±1.13 | 111.65±29.03 | 65.25±16.97 |
| 6 | 5.15±1.34 | 5.32±1.38 | 101.25±26.33 |
| 12 | 4.57±1.19 | 5.98±1.55 | 7.21±1.87 |
| 24 | 5.32±1.38 | 5.90±1.53 | 6.90±1.79 |
| 48 | 5.53±1.44 | 5.65±1.47 | 7.65±1.99 |
| 72 | 5.52±1.44 | 6.87±1.79 | 7.32±1.90 |
Following LPS challenge, tsIL-6 mRNA was
significantly increased at 3 h in the liver (P<0.05), with the
highest expression being 16.86-fold higher than that in the
control, following which it reduced gradually. However, the
significantly high expression levels were maintained to 48 h
(P<0.05). In addition, tsIL-6 transcription increased
significantly at 3 h post-poly I:C injection, reaching a peak value
of 39.23-fold higher than that in the control (P<0.05),
following which it showed gradual recovery and returned to control
levels from 48–72 h post-injection (Fig. 10A).
Following stimulation with LPS, the mRNA expression
of tsIL-6 increased significantly between 3 and 6 h post-injection
in the spleen (P<0.05), with the highest value 23.7-fold higher
than that of the control at 3 h. The expression subsequently
returned to the control level at 12 h post-stimulation. The mRNA
expression of tsIL-6 increased gradually between 3 and 12 h
post-injection with poly I:C, with the peak value of 24.3-fold
higher than that in the control at 12 h (P<0.05), following
which it reduced to the control level between 48 and 72 h (Fig. 10B).
A significant upregulation of tsIL-6 was detected at
3 h post-LPS stimulation in the kidney (P<0.05), with athe peak
value being 21.74-fold higher than that in the control, following
which it returned to the control level between 12 and 72 h
post-injection. The mRNA expression of tsIL-6 increased
significantly following poly I:C challenge up to 6 h
post-stimulation (P<0.05), with the peak value being 18.3-fold
higher than that in the control at 6 h (P<0.05), following which
it recovered to the control level between 12 and 72 h (Fig. 10C). A significant upregulation of
tsIL-6 was detected in the intestinal tissues 3 h post-LPS
stimulation (P<0.05), with the peak value being 25.5-fold higher
than that in the control, following which it returned to the
control level between 6 and 72 h post-injection. The mRNA level of
tsIL-6 increased significantly 6 h post poly I:C challenge
(P<0.05), with the peak value being 19.66-fold higher than that
in the control (P<0.05), followed by recovery to the control
level between 12 and 72 h (Fig.
10D).
Discussion
IL-6-type cytokines are important in the
communication between cells of multicellular organisms. They are
involved in the regulation of complex cellular processes, including
proliferation and differentiation, and are key during inflammation
and the immune response. Elucidating the structures, functions and
expression patterns of IL-6 is important for understanding their
essential roles in normal tissue development and diseases. As tree
shrews are increasingly used as novel disease animal models, it is
important to understand the biology of tree shrews at the molecular
level. However, IL-6 in the tree shrew has not been investigated
previously. In the present study, full-length tsIL-6 genomic and
mRNA sequences were first identified, and their evolutionary status
was compared with other species. Subsequently, the conservation of
their functional domains was analyzed, including signal peptides,
conserved cysteine residues and typical IL-6/G-CSF/MGF family
structure. In addition, the expression patterns in different
tissues were analyzed prior to and following LPS and poly I:C
challenge.
To date, three relevant sequences of tsIL-6 have
been recorded. One is in the 2-fold coverage (2X) genomic sequence
assemblies of tree shrews, generated by Lindblad-Toh et al
(25), and the other two are in
the 70-fold coverage (70X) genomic sequence assemblies of tree
shrews, generated by Fan et al (16). According to Lindblad-Toh et
al (25), the partial mRNA of
570 bp encoding a 189-aa peptide has already been documented
(ENSTBET00000004709) in the Ensembl database, although this
incomplete. According to Fan et al (16), the partial mRNA of the 779-bp
sequence encoding a 208-aa peptide has been previously predicted
based on the shotgun sequencing method (Gene ID: 102496137)
(1), which has not been
experimentally identified.
As an important cytokine associated with the immune
system, the present study identified the complete mRNA and genomic
DNA sequence of tsIL-6 for the first time, and comprehensive
analysis and characterization of tsIL-6 was performed. The present
study experimentally confirmed the presence of five exons and four
introns in the tsIL-6 gene, as previously predicted, and filled the
genetic gap in the predicted genomic sequence. It was demonstrated
that all four introns have a typical 'GU-AG' structure (Fig. 2).
In the present study, the full-length tsIL-6 mRNA
was found to consist of 1,152 bp with an ORF of 627 bp encoding 208
amino acids. Six mRNA instability motifs (ATTTA), which are typical
of genes encoding inflammatory mediators, were located in the
tsIL-6 3′-UTR. These motifs suggested that this gene is transiently
transcribed and involved in inflammatory responses. A 25-aa signal
peptide and a conserved IL-6 superfamily domain were predicted in
the tsIL-6 protein, which agreed with the protein characterizations
of IL-6 in humans (Homo sapiens). The majority of reported
mammalian IL-6s contain four conserved cysteine residues, which are
important for IL-6 bioactivity. The tsIL-6 domain also contains the
typical IL-6/G-CSF/MGF family structure of C-X(9)-C-X(6)-G-L-X(2)-[FY]-X(3)-L, indicating that tsIL-6 may share
similar functions with the conservative IL-6 family.
The five exons identified from tsIL-6 genomic
sequences were similar to the IL-6 structure in humans, monkeys and
gorillas. Phylogenetic analysis demonstrated sister clades between
tree shrews and the order of primates, with PP and BP support
(0.68/100). These results partly support the results from a
previous study, with tree shrews constituting a sister clade to
Primatomorpha (Dermoptera and Primates) other than Glires (rodents
and lagomorphs) at the root of the Euarchontoglires (26).
Regarding amino acid length, mice and rats had
missing amino acids, S and E, at positions 15 and 51, respectively,
but an extra amino acid, K, at position 137. Mice also had a
missing 55-aa peptide tail in the C terminus. The tree shrew had
three missing amino acids, SSK, at position 80 and a missing amino
acid, S, at position 136. This suggested that position 136–137 is a
mutation-prone site in IL-6. Regarding the amino acid sequence,
human IL-6 had identities of 98.11, 96.70, 40.65, 30.05 and 52.36%
with chimpanzee, monkey, rat, mouse and tree shrew IL-6,
respectively.
The homology modeling of the predicted tsIL-6
protein (Fig. 5) demonstrated a
high level of structural homology with the crystallographic
structure of human IL-6. Previous human IL-6 studies have
demonstrated a hexameric structure and assembly of the IN-6/IL-6
α-receptor/gp 130 complex (27).
In the three primary IL-6 binding sites, site I, II and III, the
modification of amino acid Gln175 to Asn104 in site I; Gly35 to
Glu35, Ser118 to Leu146, Val121 to Met149 in site II; and Asp160 to
Lys184 in site III altered the surface charge (Fig. 6). The changes in these sites
present an asymmetric pattern of evolution. The protein-protein
interactions in human IL-6 and tsIL-6 may differ as a consequence
of these changes.
The expression levels of IL-6 in different tissues
may indicate their functions in organ development, homeostasis and
diseases. In the tree shrew, the present study determined the
relative mRNA levels of IL-6 in 14 tissues from adult tree shrews.
The highest expression level was found in muscle. Numerous studies
have indicated that active skeletal muscle synthesizes and releases
IL-6, which is important in the adaptation of an organism to
exercise. Investigations in humans have also shown that human
activity results in the production of IL-6 in muscles (28). Strenuous exercise can cause a
marked increase in the plasma concentration of IL-6, which
originates from skeletal muscle. The increase of IL-6 is relevant
to the intensity and duration of exercise. As a result, muscles are
considered to be the site of IL-6 production (29). Therefore, the high expression
level of tsIL-6 may be associated with high locomotor abilities of
tree shrew and tsIL-6 may be important in skeletal muscle. The
second highest expression level was found in the spleen, which is a
tissue associated with the immune system. Moderate expression
levels of IL-6 were observed in the testis, adipose tissue, spinal
cord, adrenal gland, lung, intestine, heart, liver and kidney,
whereas low expression levels were found in the cerebrum,
cerebellum and hippocampus. The expression of tsIL-6 was confirmed
to exist widely in each examined tissue of the tree shrew by
RT-qPCR analysis. The expression level in different tissues may be
due to different species, immunological status, developmental stage
or genetic background. Considering the living environment of tree
shrews in the wild, the relatively high expression level of IL-6
may be associated with chronic infection, neoplasia and autoimmune
diseases (30).
Previous studies have demonstrated that IL-6
transcripts can be induced by LPS or poly I:C stimulation in mice
(31,32), grouper (33) and human corneal fibroblasts
(34). These studies indicated
that IL-6 transcripts were markedly induced under LPS and I:C
stimulation in these models. Similarly, the results of the present
study revealed that tsIL-6 was induced in the liver, spleen, kidney
and intestine following LPS and poly I:C stimulation, suggesting
that IL-6 can be induced in tree shrews as a defense against
bacterial and viral infection.
In conclusion, cloning, sequencing and modeling of
the tsIL-6 gene revealed molecular, structural and functional
conservation between tree shrews and primates. The mRNA and genomic
sequence of tsIL-6 generated in the present study offers an
opportunity to elucidate the genetic basis of its suitability as an
animal model for investigations associated with viral infections.
The availability of these novel gene data provides a valuable basis
and tool for functional gene investigations of tsIL-6.
Acknowledgments
This study was supported by Yunnan Science and
Technology Talent and Platform Program (no. 2017HC019), the
National Science and Technology Support Program (grant no.
2014BAI01B00), the Joint Support for the National Program of Yunnan
Province (grant no. 2015GA009) and the National Nature Science
Foundation of China (grant no. 31301044).
References
|
1
|
Yang EB, Cao J, Su JJ and Chow P: The tree
shrews: Useful animal models for the viral hepatitis and
hepatocellular carcinoma. Hepatogastroenterology. 52:613–616.
2005.PubMed/NCBI
|
|
2
|
Fuchs E: Social stress in tree shrews as
an animal model of depression: An example of a behavioral model of
a CNS disorder. CNS. 10:182–190. 2005.
|
|
3
|
Norton TT and Siegwart JT Jr: Light
levels, refractive development, and myopia - a speculative review.
Exp Eye Res. 114:48–57. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Guo L, Frost MR, Siegwart JT Jr and Norton
TT: Scleral gene expression during recovery from myopia compared
with expression during myopia development in tree shrew. Mol Vis.
20:1643–1659. 2014.PubMed/NCBI
|
|
5
|
Ye L, He M, Huang Y, Zhao G, Lei Y, Zhou Y
and Chen X: Tree shrew as a new animal model for the study of lung
cancer. Oncol Lett. 11:2091–2095. 2016.PubMed/NCBI
|
|
6
|
Lin N, Xiong LL, Zhang RP, Zheng H, Wang
L, Qian ZY, Zhang P, Chen ZW, Gao FB and Wang TH: Injection of
Aβ1-40 into hippocampus induced cognitive lesion associated with
neuronal apoptosis and multiple gene expressions in the tree shrew.
Apoptosis. 21:621–640. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hunter CA and Jones SA: IL-6 as a keystone
cytokine in health and disease. Nat Immunol. 16:448–457. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lohrengel B, Lu M and Roggendorf M:
Molecular cloning of the woodchuck cytokines: TNF-alpha, IFN-gamma,
and IL-6. Immunogenetics. 47:332–335. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Nagarajan G, Swami SK, Ghorui SK, Pathak
KM, Singh RK and Patil NV: Cloning and phylogenetic analysis of
interleukin-6 (IL-6) and tumor necrosis factor-α (TNF-α) from
Indian dromedaries (Camelus dromedarius). Comp Immunol Microbiol
Infect Dis. 34:291–298. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Takakura H, Mori Y and Tatsumi M:
Molecular cloning of caprine IL-6 cDNA and its expression in insect
cells. Int Arch Allergy Immunol. 113:409–416. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhu Q, Li C, Yu ZX, Zou PF, Meng QX and
Yao CL: Molecular and immune response characterizations of IL-6 in
large yellow croaker (Larimichthys crocea). Fish Shellfish Immunol.
50:263–273. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Iliev DB, Castellana B, Mackenzie S,
Planas JV and Goetz FW: Cloning and expression analysis of an IL-6
homolog in rainbow trout (Oncorhynchus mykiss). Mol Immunol.
44:1803–1807. 2007. View Article : Google Scholar
|
|
13
|
Yu D, Wu Y, Xu L, Fan Y, Peng L, Xu M and
Yao YG: Identification and characterization of toll-like receptors
(TLRs) in the Chinese tree shrew (Tupaia belangeri chinensis). Dev
Comp Immunol. 60:127–138. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yu D, Xu L, Liu XH, Fan Y, Lü LB and Yao
YG: Diverse interleukin-7 mRNA transcripts in Chinese tree shrew
(Tupaia belangeri chinensis). PLoS One. 9:e998592014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zheng Y, Yun C, Wang Q, Smith WW and Leng
J: Identification of the full-length β-actin sequence and
expression profiles in the tree shrew (Tupaia belangeri). Int J Mol
Med. 35:519–524. 2015. View Article : Google Scholar
|
|
16
|
Fan Y, Yu D and Yao YG: Tree shrew
database (TreeshrewDB): A genomic knowledge base for the Chinese
tree shrew. Sci Rep. 4:71452014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jain R, Gomer RH and Murtagh JJ Jr:
Increasing specificity from the PCR-RACE technique. Biotechniques.
12:58–59. 1992.PubMed/NCBI
|
|
18
|
Edgar RC: MUSCLE: Multiple sequence
alignment with high accuracy and high throughput. Nucleic Acids
Res. 32:1792–1797. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ronquist F and Huelsenbeck JP: MrBayes 3:
Bayesian phylogenetic inference under mixed models. Bioinformatics.
19:1572–1574. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Meyer X, Chopard B and Salamin N:
Accelerating Bayesian inference for evolutionary biology models.
Bioinformatics. 33:669–676. 2017.
|
|
21
|
Stamatakis A, Ludwig T and Meier H:
RAxML-III: A fast program for maximum likelihood-based inference of
large phylogenetic trees. Bioinformatics. 21:456–463. 2005.
View Article : Google Scholar
|
|
22
|
Remans T, Smeets K, Opdenakker K,
Mathijsen D, Vangronsveld J and Cuypers A: Normalisation of
real-time RT-PCR gene expression measurements in Arabidopsis
thaliana exposed to increased metal concentrations. Planta.
227:1343–1349. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Siemetzki U, Ashok MS, Briese T and Lipkin
WI: Identification of RNA instability elements in Borna disease
virus. Virus Res. 144:27–34. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Fu X, Ding Z, Fan J, Wang H, Zhou F, Cui
L, Boxiang C, Wang W and Liu H: Characterization, promoter analysis
and expression of the interleukin-6 gene in blunt snout bream,
Megalobrama amblycephala. Fish Physiol Biochem. 42:1527–1540. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lindblad-Toh K, Garber M, Zuk O, Lin MF,
Parker BJ, Washietl S, Kheradpour P, Ernst J, Jordan G, Mauceli E,
et al Broad Institute Sequencing Platform and Whole Genome Assembly
Team; Baylor College of Medicine Human Genome Sequencing Center
Sequencing Team; Genome Institute at Washington University: A
high-resolution map of human evolutionary constraint using 29
mammals. Nature. 478:476–482. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhou X, Sun F, Xu S, Yang G and Li M: The
position of tree shrews in the mammalian tree: Comparing multi-gene
analyses with phylogenomic results leaves monophyly of Euarchonta
doubtful. Integr Zool. 10:186–198. 2015. View Article : Google Scholar
|
|
27
|
Boulanger MJ, Chow DC, Brevnova EE and
Garcia KC: Hexameric structure and assembly of the
interleukin-6/IL-6 alpha-receptor/gp130 complex. Science.
300:2101–2104. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ikeda SI, Tamura Y, Kakehi S, Sanada H,
Kawamori R and Watada H: Exercise-induced increase in IL-6 level
enhances GLUT4 expression and insulin sensitivity in mouse skeletal
muscle. Biochem Biophys Res Commun. 473:947–952. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ono T, Maekawa K, Watanabe S, Oka H and
Kuboki T: Muscle contraction accelerates IL-6 mRNA expression in
the rat masseter muscle. Arch Oral Biol. 52:479–486. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gleeson M: Interleukins and exercise. J
Physiol. 529:12000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hacham M, Cristal N, White RM, Segal S and
Apte RN: Complementary organ expression of IL-1 vs. IL-6 and CSF-1
activities in normal and LPS-injected mice. Cytokine. 8:21–31.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Szot P, Franklin A, Figlewicz DP, Beuca
TP, Bullock K, Hansen K, Banks WA, Raskind MA and Peskind ER:
Multiple lipopolysaccharide (LPS) injections alter interleukin 6
(IL-6), IL-7, IL-10 and IL-6 and IL-7 receptor mRNA in CNS and
spleen. Neuroscience. 355:9–21. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Jeong YH, Park JS, Kim DH, Kang JL and Kim
HS: Anti-inflammatory mechanism of lonchocarpine in LPS- or
poly(I:C)-induced neuroinflammation. Pharmacol Res. 119:431–442.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Kimura K, Orita T, Nomi N, Fujitsu Y,
Nishida T and Sonoda KH: Identification of common secreted factors
in human corneal fibroblasts exposed to LPS, poly(I:C), or zymosan.
Exp Eye Res. 96:157–162. 2012. View Article : Google Scholar
|