Introduction
Chronic hepatitis C virus (CHC) infection is a major
global health problem with ~200 million people infected and 700,000
people dying from hepatitis C-related liver diseases each year
(1). China was considered to have
a particularly high prevalence of 0.43% in the general population
with 29 million hepatitis C virus (HCV) infected individuals
(2,3). Over 70% of acutely infected persons
progress into chronic HCV infection, that consequently causes
progressive liver fibrosis and hepatocellular carcinoma (4,5).
Although successful execution of direct-acting
antiviral therapy (DAA) was recently approved in western countries
according to the World Health Organization (WHO 2017) (6). The combination therapy of pegylated
interferon-α (pegIFN-α) plus ribavirin (RBV) (pegIFN-α/RBV) was
still the most effective treatment for patients with HCV infection
in developing countries, such as China due to economic reasons and
curative concerns (7). Even
though progression to severe liver disease could be prevented in
54–63% of patients through antiviral treatment with pegIFN-α/RBV,
the fact that some patients treated with pegIFN-α/RBV failing to
achieve sustained virological response (SVR) or experienced side
effects should not be neglected (8,9).
Therefore, it is important to identify the factors that may affect
the response to treatment given that interaction between the virus
and host genetics has been theorized to be an important determinant
of treatment response and the natural course of hepatitis C
(10,11).
A genetic association study clarified that the human
leukocyte antigen (HLA) was an essential genetic factor that
regulated immune response and may be one of the tactics used by HCV
to avoid immune clearance (12).
Furthermore, various HLA alleles involved in the immune
response were demonstrated to be linked to spontaneous clearance of
HCV infection and even be potentially predictive for HCV treatment
response (13). Studies to date
about HLA alleles mainly focused on the HLA class I
and II genes regions, by contrast, the study on non-classic genes
located among classic regions was limited, such as LMP2/LMP7
genes, TAP1/TAP2 genes and tapasin genes (14–17). These genes, however, have been
found in a previous study of the authors to be related to HCV
susceptibility and spontaneous clearance.
In the present study, the authors genotyped the
TAP gene, LMP7 gene and tapasin gene to investigate
the possible association of HLA gene polymorphisms with
treatment response to pegIFN-α/RBV in 352 patients with CHC.
Patients and methods
Participants
A total of 352 participants were enrolled in the
study from Jurong People's Hospital (Zhenjiang, China). All
participants were the patients with genotype 1 CHC identified by
the diagnosis of infectious disease in hospital or by doctor
visits, and this infection was suspected to come from their former
remunerated blood donation behaviors. This study protocol was
approved by the institutional review board of Nanjing Medical
University (Nanjing, China). All participants provided written
informed consent. Interviews for donation history and other risk
factors were conducted with signed informed consent from April,
2011 to January, 2016.
Those eligible subjects for the study were included
if they received antiviral treatment of pegIFN-α/RBV for the first
time and HCV antibody presented positive continually for more than
six months. Subjects who were co-infected with hepatitis B virus or
human immunodeficiency virus, or suffered from other types of liver
diseases, alcoholic diseases, metabolic liver diseases and previous
interferon and/or ribavirin therapy during the trial were
excluded.
Investigation
Each participant was interviewed face-to-face using
a structured and standardized questionnaire administered by trained
interviewers. Demographic data, history of common diseases and
therapeutic processes were collected for each subject. After the
interview, a blood sample of ~10 ml was collected as a source of
genomic DNA for serological tests and host DNA genotyping. The
authors followed up these participants and detected viral load at
on-treatment week 0, 4, 12, 24 and 48 and post-treatment week 24.
SVR was defined as HCV RNA below the assay's lower limit of
quantitation at post-treatment week 24. Rapid virological response
(RVR) was defined as HCV RNA down two logarithmic or below the
assay's lower limit of quantitation at post-treatment week 4.
Completed early virological response (cEVR) was defined as HCV RNA
below the assay's lower limit of quantitation at on-treatment week
12.
Finally, 352 cases with HCV infection (anti-HCV
positive) were divided into two groups: 232 sustained virological
response cases (SVR) and 120 non-sustained virological response
cases (non-SVR).
Laboratory testing
Sera and HCV antibody (anti-HCV) were detected by
ELISA (S20130002; Beijing Wantai Biological Pharmacy Enterprise
Co., Ltd., Beijing, China) under the manufacturers' instructions.
Blood biochemical tests were undertaken by Roche Module P800
Automatic Biochemical Analyzer (Roche Diagnostics GmbH, Basel,
Switzerland). Total RNA was extracted from the serum using TRIzol
LS Reagent, and HCV RNA was detected by RT-PCR with specific
primers using PrimeScript RT-PCR kit (DRR014S; Takara Biotechnology
Co., Ltd., Dalian, China).
Single nucleotide polymorphisms (SNPs)
selection and genotyping assays
The information of SNPs in four candidate genes
(TAP1, TAP2, LMP7 and tapasin) was
obtained from the NCBI dbSNP database (http://www.ncbi.nlm.nih.gov/SNP) and the Chinese Han
population database of HapMap (http://www.hapmap.org). All the SNPs were filtered
with the criteria: MAF (minor allele frequency) ≥0.05. A total of 6
SNPs (TAP1 rs1135216, TAP2 rs1800454, LMP7 rs2071543,
tapasin rs9277972, rs1059288 and rs2282851) were chosen for
genotyping.
DNA extraction was performed by protease K digestion
and phenol-chloroform purification. Genotyping was performed by
using a TaqMan allelic discrimination assay on the ABI PRISM 7900
HT sequence detection system (Applied Biosystems; Thermo Fisher
Scientific, Inc., Waltham, MA, USA). PCR was performed according to
recommended thermal profile: 50°C for 2 min (preheating), 95°C for
10 min (preincubation) followed by 40 cycles at 95°C for 15 sec
(denaturation) and 60°C for 1 min (annealing). Table I presents the TaqMan minor
groove-binding (TaqMan-MGB) probes and specific forward/reverse PCR
primers used in this study (Nanjing BioSteed BioTechnologies Co.,
Ltd., Nanjing, China). Two blank controls and five repeated samples
were assigned to each 384-well format for quality control, and a
100% concordance was achieved.
 | Table IPrimers and probes for TaqMan allelic
discrimination. |
Table I
Primers and probes for TaqMan allelic
discrimination.
| Polymorphism | Sequence
(5′-3′) |
|---|
| TAP1
rs1135216 |
| Primer | F:
CACACATGTGGCTATACCGTTCTC |
| R:
TCGCTGACCCCCTGACA |
| Probe |
FAM-TGCAGAGGTAGGCG-MGB |
|
HEX-TCTGCAGAGGTAGACG-MGB |
| TAP2
rs1800454 |
| Primer | F:
CCTGGAACGCGCCTTGTA |
| R:
CCTTTCACAACCACTCTGGTATCTT |
| Probe |
FAM-TGCTCGTAAGGAGG-MGB |
|
HEX-CTGCTCATAAGGAGG-MGB |
| LMP7
rs2071543 |
| Primer | F:
GCGACCCTCCACTCCTCA |
| R:
GGACACTACAGTTTCTCTATGCGATCT |
| Probe |
FAM-CCGACCTTCATTCC-MGB |
|
HEX-CCGACCTGCATTC-MGB |
| Tapasin
rs9277972 |
| Primer | F:
GTCTAGGTCCTTCAGGTAGAAGTAATCTTT |
| R:
CTAAGTGAAATTGCATACTGTTTTTACTCTAC |
| Probe |
FAM-CCTATAAGGTTAAACTGTTCT-MGB |
|
HEX-CCTATAAGGTTTAACTGTTCT-MGB |
| Tapasin
rs1059288 |
| Primer | F:
TGGGCCTTAGGTCCCTATGC |
| R:
AAGTGATCGTGTGAGTCGTCGTT |
| Probe |
FAM-CAGACAGGCCGGTC-MGB |
|
HEX-ACAGACAGGCCAGTC-MGB |
| Tapasin
rs2282851 |
| Primer | F:
CCTCATTCTTGAATTATCTGCACAGT |
| R:
GCCCAGGAGTCAGAAGCTTTT |
| Probe |
FAM-CCACGTCTCAGCCTA-MGB |
|
HEX-CCACGTCCCAGCCT-MGB |
Statistical analysis
Data were scrutinized and then entered a database
using EpiData 3.1 by two different studies for further analysis.
Differences in general demographic characteristics were calculated
by the Student's t-test or one-way analysis of variance and the
Chi-square (χ2) test. Dominant and additive genetic
models were used in the analysis of each SNP. Associations between
SNPs and the treatment response of HCV infection were estimated by
calculating the odds ratios (ORs) with 95% confidence intervals
(CIs). Adjustments for age, sex, baseline viral load, glucose
(GLU), α-fetal protein (AFP), albumin (ALB) and platelets were
conducted with the use of the regression analysis. To evaluate
ability of the genetic and clinical factors to predict HCV
treatment response, the area under the curve (AUC) of the receiver
operating characteristic was calculated. Line and bar charts were
used to present the viral load at each follow-up time-point. All
statistical analyses were two sided, and P<0.05 was considered
to indicate a statistically significant difference. The trend
analysis was calculated with Cochran-Armitage trend test. All the
statistical analyses were performed using the STATA software
(version 12.0; StataCorp LLC, College Station, TX, USA).
Results
Demographic characteristics of the study
populations
The basic characteristics of 232 cases that achieved
an SVR (SVR) and 120 cases who did not achieve an SVR (non-SVR)
were available in the study. The rate of SVR was 65.9%. Patients
with low viral load and high levels of GLU, AFP, TP, ALB and
platelets at baseline were more likely to achieved an SVR, as
presented in Table II.
 | Table IIBaseline characteristics of CHC
patients treated with IFN/RBV. |
Table II
Baseline characteristics of CHC
patients treated with IFN/RBV.
| Variables | N-SVR (n=120) | SVR (n=232) | P-value |
|---|
| Mean age, year | 53.41±8.14 | 53.6±8.50 | 0.783 |
| Age ≥50 | 83 (69.2) | 159 (68.5) | 0.903 |
| Male (%) | 29 (24.2) | 57 (24.6) | 0.934 |
| AST ≥40 U/l
(%) | 70 (58.3) | 121 (52.2) | 0.270 |
| ALT ≥40 U/l
(%) | 79 (65.8) | 138 (59.5) | 0.245 |
| GGT ≥50 (U/l) | 51 (42.5) | 77 (33.2) | 0.085 |
| GLU > 6.01
(mmol/l) | 49 (40.8) | 61 (26.3) | 0.005a |
| AFP >7.02
(ng/ml) | 50 (41.7) | 65 (28.0) | 0.010a |
| T3 (nmol/l) | 1.54±0.49 | 1.50±0.72 | 0.593 |
| T4 (nmol/l) | 128.67±32.96 | 123.50±31.01 | 0.147 |
| Anti-TPO ≥35
I/ml | 14 (11.67) | 35 (15.09) | 0.380 |
| Base HCV-RNA | 6.18±0.75 | 5.82±1.26 | 0.004a |
| TP (g/l) | 77.87±5.92 | 78.51±5.92 | 0.335 |
| ALB (g/l) | 42.56±4.10 | 43.87±4.06 | 0.005a |
| Platelets
(109/l) | 122.51±59.65 | 137.19±52.48 | 0.018a |
| Abnormal | 53 (44.2) | 61 (26.3) | 0.001a |
| Normal | 67 (55.8) | 171 (73.7) | |
| WBC
(109/l) | 4.88±2.62 | 5.02±1.78 | 0.555 |
| Abnormal | 45 (37.5) | 73 (31.5) | 0.256 |
| Normal | 75 (62.5) | 159 (68.5) | |
| Hemoglobin
(g/l) | 132.83±17.50 | 134.07±16.02 | 0.506 |
| Abnormal | 22 (18.3) | 31 (13.4) | 0.216 |
| Normal | 98 (81.7) | 201 (86.6) | |
Association of candidate SNPs with
SVR
The allelic frequencies of candidate genes
(TAP1 rs1135216, TAP2 rs1800454, LMP7
rs2071543, tapasin rs9277972, rs1059288 and rs2282851)
between SVR and non-SVR groups were compared in Table III. The observed genotype
frequencies of these SNPs in the remaining subjects with different
HCV status were all in Hardy-Weinberg equilibrium (all P≥0.05).
LMP7 rs2071543-A variant and TAP2 rs1800454-A
variants were related to a decreased possibility of achieving an
SVR.
 | Table IIIAssociation of SNPs in HLA
with SVR. |
Table III
Association of SNPs in HLA
with SVR.
| Genotype | N-SVR | SVR | SVR rate (%) | OR (95% CI) | P-value |
|---|
| rs1135216 | | | | | |
| AA | 44 (36.7) | 85 (36.6) | 65.9 | 1.00 | – |
| AG | 29 (24.1) | 69 (29.8) | 70.4 | 1.36
(0.74–2.50) | 0.328 |
| GG | 47 (39.2) | 78 (33.6) | 62.4 | 1.14
(0.65–2.01) | 0.654 |
| Dominant | | | | 1.23
(0.74–2.04) | 0.427 |
| Additive | | | | 1.06
(0.80–1.41) | 0.677 |
| rs1800454 | | | | | |
| GG | 78 (65.0) | 176 (75.9) | 69.3 | 1.00 | – |
| GA | 36 (30.0) | 52 (22.4) | 59.1 | 0.52
(0.30–0.90) | 0.020a |
| AA | 6 (5.0) | 4 (1.7) | 40.0 | 0.27
(0.07–1.09) | 0.066 |
| Dominant | | | | 0.49
(0.29–0.82) | 0.007a |
| Additive | | | | 0.52
(0.33–0.82) | 0.005a |
| rs9277972 | | | | | |
| AA | 91 (75.8) | 169 (72.9) | 65.0 | 1.00 | – |
| AT | 21 (17.5) | 45 (19.4) | 68.2 | 1.10
(0.59–2.04) | 0.774 |
| TT | 8 (6.7) | 18 (7.8) | 69.2 | 1.67
(0.67–4.17) | 0.272 |
| Dominant | | | | 1.25
(0.73–2.14) | 0.422 |
| Additive | | | | 1.22
(0.83–1.80) | 0.302 |
| rs1059288 | | | | | |
| TT | 44 (36.7) | 82 (35.3) | 65.1 | 1.00 | – |
| TC | 58 (48.3) | 118 (50.9) | 67.0 | 1.26
(0.75–2.12) | 0.382 |
| CC | 18 (15.0) | 32 (13.8) | 64.0 | 1.02
(0.50–2.12) | 0.949 |
| Dominant | | | | 1.20
(0.73–1.97) | 0.465 |
| Additive | | | | 1.06
(0.75–1.51) | 0.732 |
| rs2282851 | | | | | |
| CC | 73 (60.8) | 134 (57.8) | 43.6 | 1.00 | – |
| CT | 41 (34.2) | 90 (38.8) | 68.7 | 1.27
(0.77–2.08) | 0.352 |
| TT | 6 (5.0) | 8 (3.5) | 57.1 | 0.57
(0.18–1.76) | 0.327 |
| Dominant | | | | 1.16
(0.72–1.88) | 0.533 |
| Additive | | | | 1.03
(0.68–1.55) | 0.891 |
| rs2071543 | | | | | |
| CC | 68 (56.7) | 154 (66.4) | 69.4 | 1.00 | – |
| CA | 43 (35.8) | 70 (30.2) | 61.9 | 0.77
(0.46–1.28) | 0.312 |
| AA | 9 (7.5) | 8 (3.5) | 47.1 | 0.32
(0.11–0.91) | 0.033a |
| Dominant | | | | 0.68
(0.42–1.09) | 0.112 |
| Additive | | | | 0.66
(0.45–0.98) | 0.039a |
In rs1800454 SNPs, the rate of SVR was significantly
higher in patients with the GG genotype compared to those with the
GA and AA allele. The additive model analyses indicated that the
presence of each additional allele was indicated to reduce the
decreased probability of achieving an SVR by ~48% (adjusted OR,
0.52; 95% CI, 0.33–0.82; P=0.005).
In rs2071543 SNPs, for patients with the CC
genotype, a higher SVR rate was observed in comparison to the CA
and AA genotypes. The additive model indicated that each additional
allele contributed to a decreased likelihood of achieving an SVR by
~34% (adjusted OR, 0.66; 95% CI, 0.45–0.98; P=0.039).
Association of rs1800454 and rs2071543
with RVR/cEVR
The association between rs1800454 and rs2071543 with
RVR/cEVR was also analyzed, as presented in Table IV. Carrying rs1800454-A allele
was showed to be a risk factor of both achieving a RVR (dominant
model: OR, 0.41; 95% CI, 0.25–0.70; P=0.001) and a cEVR (additive
model: OR, 0.53; 95% CI, 0.34–0.83; P=0.006). Subjects with
rs2071543-A allele were less prone to achieve a RVR (dominant
model: OR, 0.59; 95% CI, 0.37–0.93; P=0.023). However, no
significant correlation was observed between rs2071543-A allele and
cEVR (all P>0.05).
 | Table IVAssociation of rs1800454 and
rs2071543 in HLA with RVR/cEVR. |
Table IV
Association of rs1800454 and
rs2071543 in HLA with RVR/cEVR.
| Genotype | N-RVR | RVR | OR (95% CI) | P-value | N-cEVR | cEVR | OR (95% CI) | P-value |
|---|
| rs1800454 | | | | | | | | |
| GG | 111 (64.9) | 143 (79.0) | 1.00 | – | 71 (65.1) | 183 (75.3) | 1.00 | – |
| GA | 54 (31.6) | 34 (18.8) | 0.41
(0.24–0.69) | 0.001 | 31(28.4) | 57 (23.5) | 0.64
(0.37–1.10) | 0.107 |
| AA | 6 (3.5) | 4 (2.2) | 0.60
(0.15–2.37) | 0.464 | 7 (6.4) | 3 (1.2) | 0.15
(0.03–0.65) | 0.011a |
| Dominant | | | 0.42
(0.25–0.70) | 0.001 | | | 0.55
(0.33–0.92) | 0.024a |
| Additive | | | 0.50
(0.32–0.79) | 0.003 | | | 0.53
(0.34–0.83) | 0.006a |
| rs2071543 | | | | | | | | |
| CC | 97 (56.7) | 124 (68.5) | 1.00 | – | 62 (56.9) | 159 (65.4) | 1.00 | – |
| CA | 65 (38.0) | 48 (26.5) | 0.55
(0.34–0.89) | 0.015 | 43 (39.5) | 70 (28.8) | 0.67
(0.40–1.11) | 0.117 |
| AA | 9 (5.3) | 9 (5.0) | 0.90
(0.33–2.47) | 0.831 | 4 (3.7) | 14 (5.8) | 1.79
(0.55–5.90) | 0.335 |
| Dominant | | | 0.59
(0.37–0.93) | 0.023 | | | 0.76
(0.47–1.23) | 0.262 |
| Additive | | | 0.71
(0.48–1.03) | 0.073 | | | 0.92
(0.62–1.36) | 0.662 |
Combined effect of rs1800454 and
rs2071543
The evaluation of the combined effects of
rs1800454-A and rs2071543-A was performed to test the association
with SVR. The results showed that SVR rate was lower when patients
carried more unfavorable rs1800454-A and rs2071543-A alleles, and
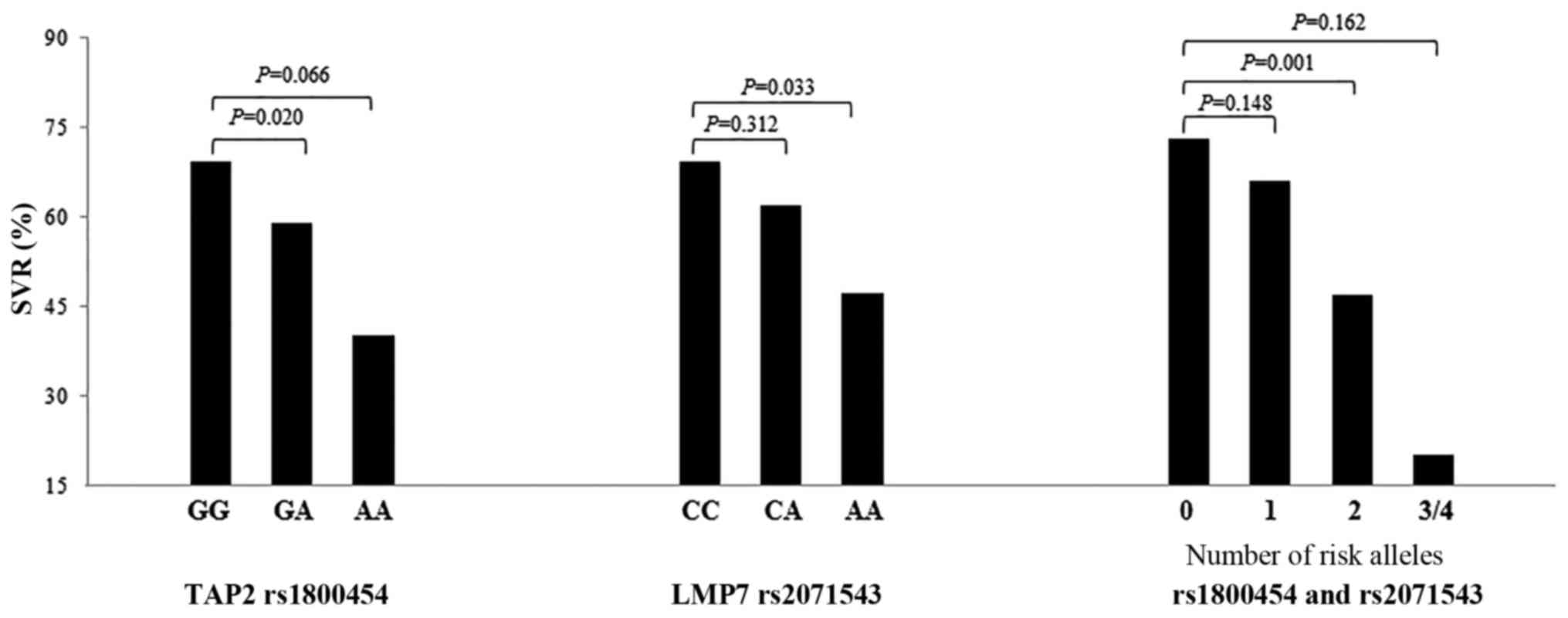
the SVR rate of carrying 3–4 alleles was 20%. Carrying two
unfavorable alleles appeared to have a negative dangerous effect on
the SVR (OR, 0.30; 95% CI, 0.14–0.61; P=0.001), as showed in
Fig. 1 and Table V.
 | Table VCombined effects of rs1800454 and
rs2071543 with SVR. |
Table V
Combined effects of rs1800454 and
rs2071543 with SVR.
| Variable | N-SVR | SVR | SVR rate (%) | OR (95% CI) | P-value |
|---|
| 0 | 41 (34.2) | 112 (48.3) | 73.2 | 1 | – |
| 1 | 50 (41.7) | 97 (41.8) | 66.0 | 0.68
(0.40–1.15) | 0.148 |
| 2 | 25 (20.8) | 22 (9.5) | 46.8 | 0.30
(0.14–0.61) | 0.001a |
| 3–4 | 4 (3.3) | 1 (0.4) | 20.0 | 0.17
(0.01–2.02) | 0.162 |
| Trend | | | | | P<0.001a |
Multivariate stepwise regression
analysis
A stepwise regression model comprising all
statistically significant variables was established. The final
model included the rs1800454, rs2071543, baseline HCV RNA level,
baseline platelet level, baseline GLU level and baseline AFP level
as independent predictors of SVR in Table VI (all P<0.05). Subsequently,
the receiver-operating characteristic analysis for SVR was
performed to estimate the predicted value of the independent
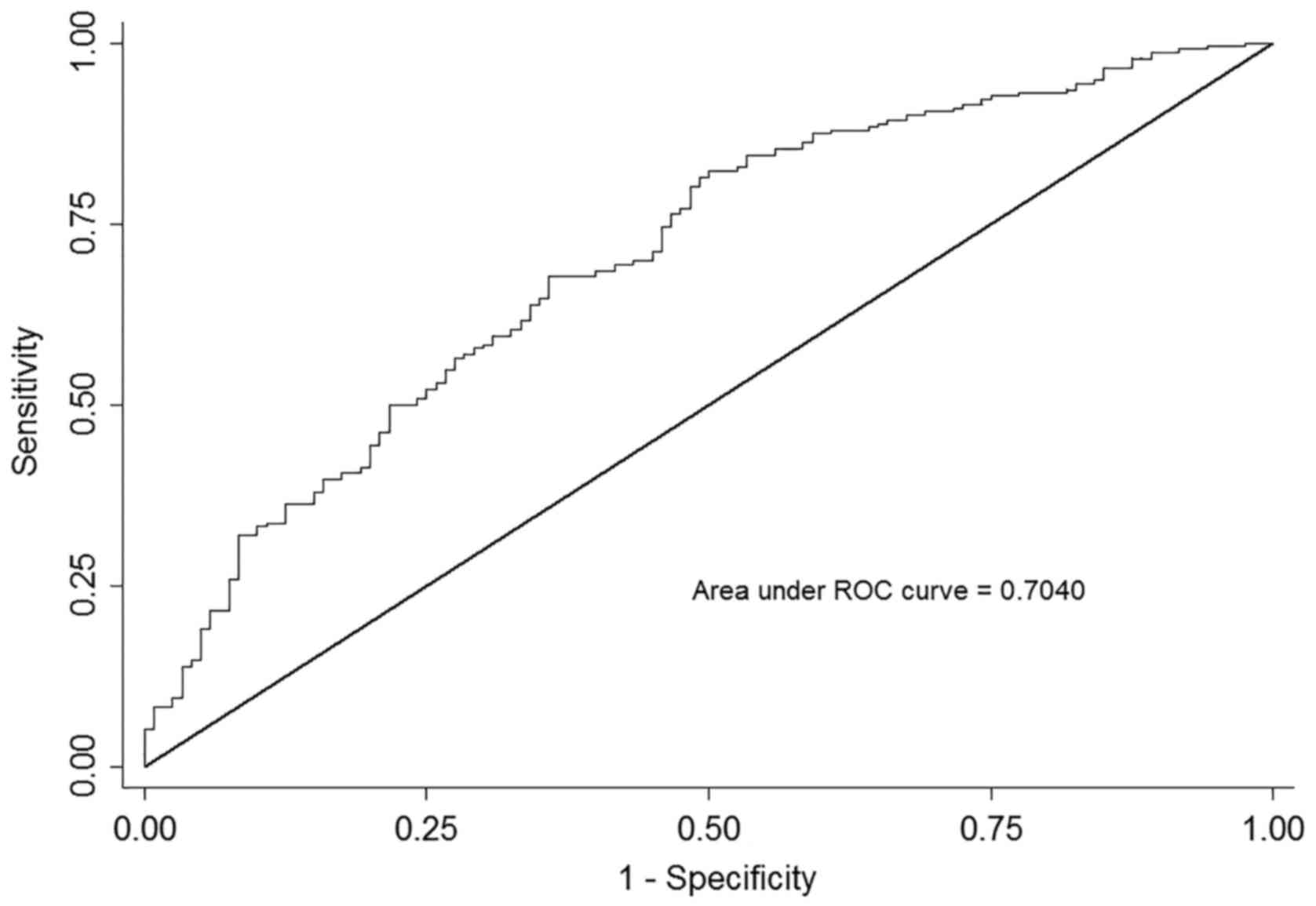
factors. The AUC based on this model, including the above factors,
produced an AUC of 0.704 (95% CI, 0.647–0.761; P<0.001), as
presented in Fig. 2. An
approximately parallel AUC was yielded when adding up one SNP of
rs1800454 or rs2071543, suggesting that the predicted value of
rs1800454 and rs2071543 was at the similar level.
 | Table VIMultivariate Stepwise regression
analysis for independent factors of SVR. |
Table VI
Multivariate Stepwise regression
analysis for independent factors of SVR.
| Variable | Coef. | SE | 95% CI | P-value |
|---|
| rs1800454 | −0.15 | 0.05 | (−0.24, −0.05) | 0.002a |
| rs2071543 | −0.10 | 0.04 | (−0.18, −0.02) | 0.016a |
| GLU | −0.12 | 0.05 | (−0.23, −0.02) | 0.021a |
| RNA level | −0.07 | 0.02 | (−0.11, −0.02) | 0.002a |
| Platelets
(109/l) | −0.14 | 0.05 | (−0.24, −0.03) | 0.009a |
| AFP | −0.11 | 0.05 | (−0.21,
−0.005) | 0.041a |
Association of rs1800454 and rs2071543
with viral kinetics during treatment
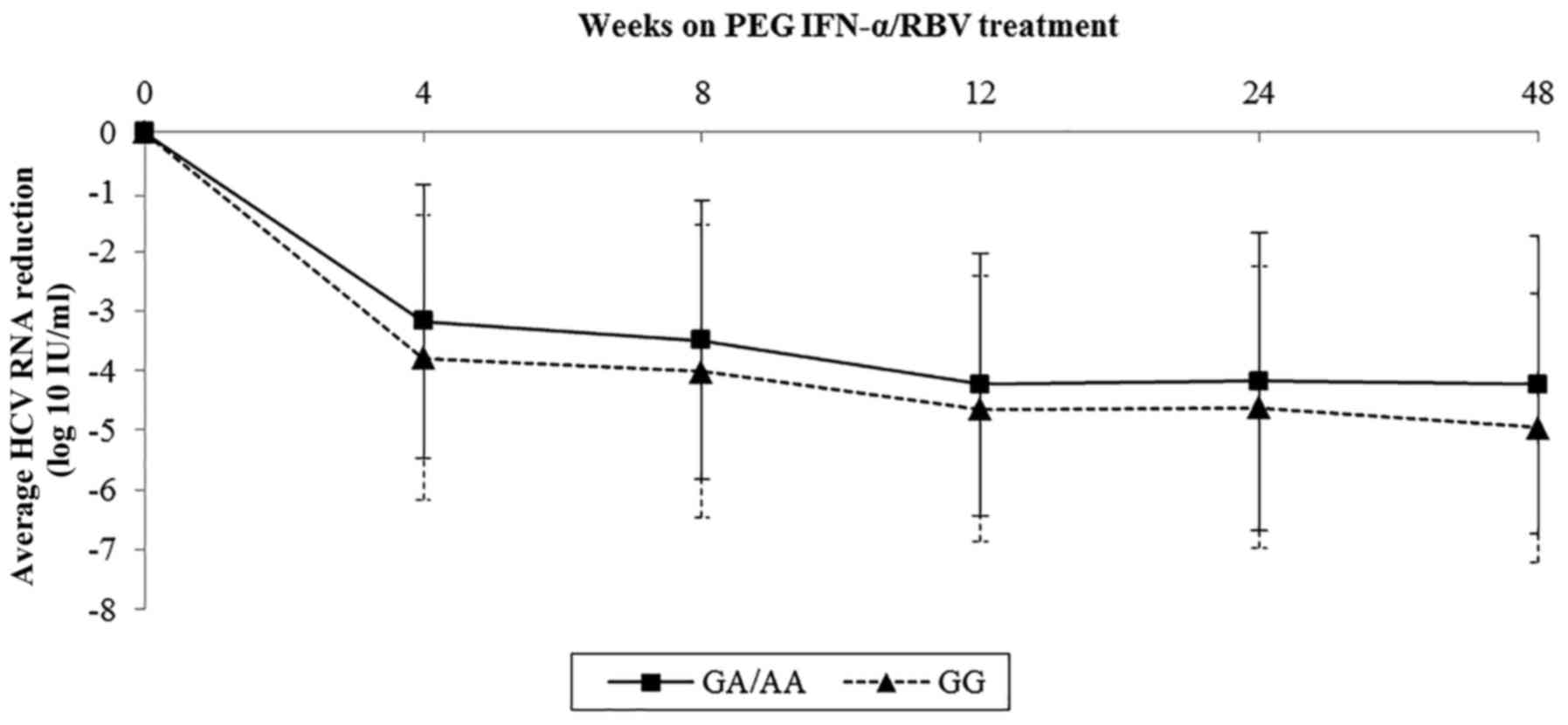
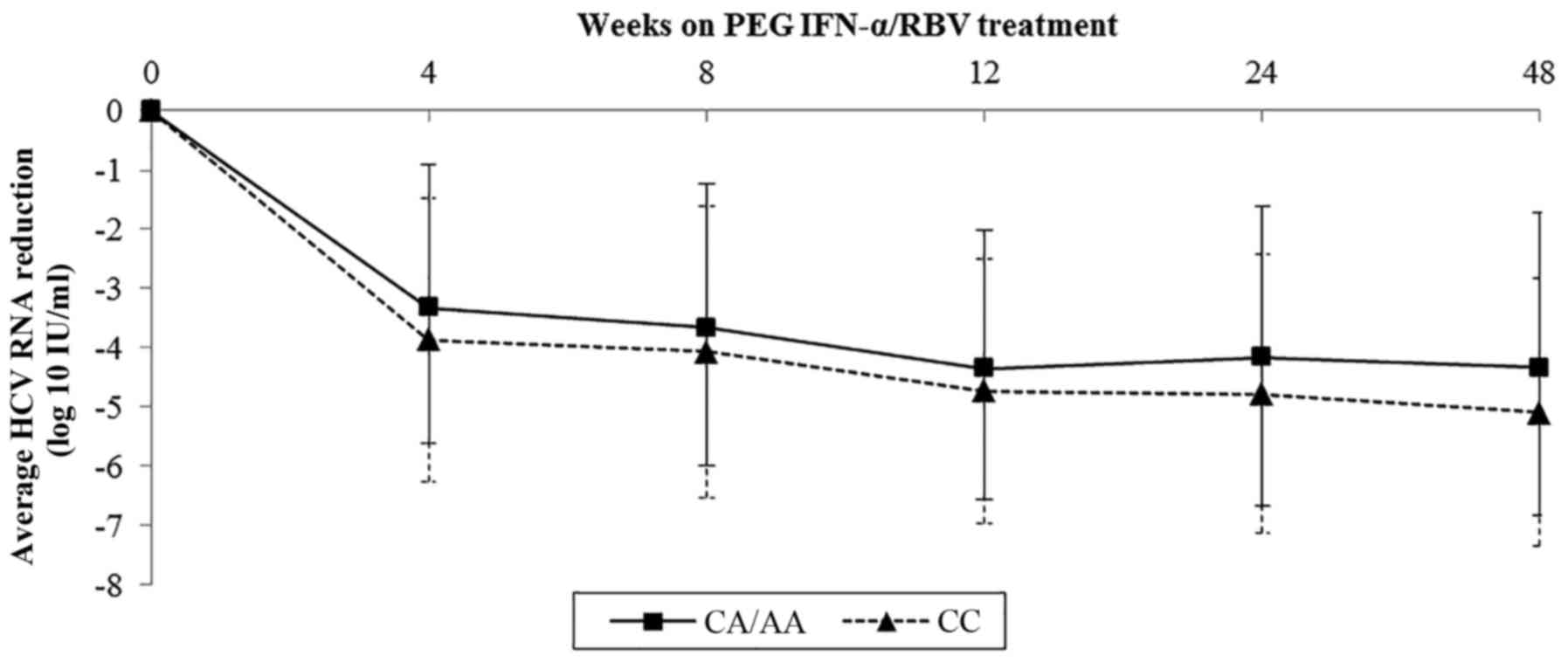
Sample data on viral kinetics of rs1800454 and
rs2071543 were collected and further analyzed by line charts.
Baseline viral load of rs1800454 was higher in patients with the GG
genotype than in those carrying the A allele (mean log10
HCV RNA: 6.03 for GG, 5.70 for GC/CC, P=0.015) (Fig. 3). However, the drop in viral load
was significantly faster in GG patients than in GC/CC patients
during the whole therapy, which was in accordance with the decline
of viral load in rs2071543 (Fig.
4). The results highlighted that the mutation of rs1800454-G
and rs2071543-C may reduce the chance to achieve an SVR.
Discussion
In the present study, the authors attempted to
investigate the viral (HCV viral load) and host (HLA SNPs,
HCV RNA, platelets, GLU and AFP) factors affecting the treatment
response of pegIFN-α/RBV, and to evaluate whether the SNPs were
associated with SVR, RVR and cEVR in Chinese patients with CHC
genotype 1 infection.
MHC class I and II antigens are the central to the
host immune response, which make them ideal candidate genes to
investigate their association with HCV infection (18). HLA is a crucial genetic
factor that initiates and regulates immune response through
presenting endogenous and exogenous antigen to T lymphocytes
(12). An increased and broadly
multi-specific T-cell response is critical to get a favorable
outcome. CD8+ T-cell response to HCV is important to the
occurrence of successful immune response and spontaneous infection
clearance. HLA class II presents viral peptides to CD8+
T-cells to permit detection of infected cells (19). TAP, LMP and
tapasin located in the human MHC class II DNA-binding
loci are playing a crucial role in the HLA class
I-restricted endogenous antigen presenting system (20). A previous study identified some
genomic variants of TAP and LMP which were associated
with chronic hepatitis B and hepatitis C (21). A previous study of the authors
also showed that TAP, LMP and tapasin affected
HCV susceptibility and spontaneous clearance. However, the
relationship between variants of these genes and treatment response
of HCV infection has not been fully studied. Therefore, the present
study was performed to elucidate whether these antigen-presenting
gene polymorphisms could influence the response to pegIFN-α/RBV
treatment in CHC patients.
The results demonstrated that two tagging SNPs of
tapasin (rs1059288 T>C and rs2282851 C>T) had no
relationship with the treatment response of HCV infection, which
contradicted the results from the study in a European Caucasian
population (22). Another study
found that mutation of tapasin rs9277972 A>T increased
the risk of HCV chronicity, which was not observed in the present
study (23). In line with other
studies, TAP2 rs1800454 was shown to be significantly
associated with increased risk for progression of HCV infection
(19,23). TAP2 rs1800454 G>A makes
a missense mutation and may be an independent risk factor for
failing to achieve SVR, RVR and cEVR. However, another two studies
in European and Japan population respectively failed to delineate a
strong association between TAP gene polymorphism and
response to interferon treatment (19,24). Mutation may alter the activity of
the encoding protein and have an impact on antigen presentation
process. Studies around the world have suggested that LMP7
gene polymorphism had an influence on the outcomes of HCV
infection, which was consistent with the finding of the authors'
previous study (20,25,26). The present study indicated that
LMP7 rs2071543 C>A also renders a missense mutation,
influencing the treatment response in CHC patients (19,27). Of note, few studies have explored
the role of TAP2, LMP7 gene in the treatment response
of CHC patients, with inconsistent study designs, as well as
participants of different physical conditions and genetic
background, which led to a marked knowledge gap in this field
(19,24,27).
In the analysis of rs1800454 and rs2071543 with
those factors in model (baseline HCV RNA level, baseline platelets
level, baseline GLU level and baseline AFP level), the interaction
effect is not significant (data not shown), which illustrated that
all factors above may be independent factors of hepatitis C
treatment response. In addition, when including six factors of
rs1800454, rs2071543, baseline HCV RNA level, baseline platelets
level, baseline GLU level and baseline AFP level, the AUC was
improved to 0.704. The similar rise in AUC could be found in an
Egyptian research where an AUC of 0.68 including serum AFP and
viral load was calculated (28).
The results of the study have a potential implication that making
full use of collected routine data can take effect in the
determination of prognosis and the adjustment for treatment
procedures.
There are several potential limitations that need to
be considered and discussed. In the present study, only six SNPs of
three genes were selected in the HLA class II region, whereas there
are many genes involved in the MHC region. Some small molecular
compounds known as DAAs, have been developed in recent years. The
preliminary results have shown promising clinical application,
however, this study only predicted the response to pegIFN-α/RBV
therapy. PegIFN-α/RBV combination therapy is still the predominant
treatment for hepatitis C in light of the economic burden in
developing countries, such as China. However, the DAA will be a
major direction for the future research. Finally, there are other
well-known predictive factors that the authors did not adjust for
in this study, such as HCV core amino acid 70 and IL28B, which may
influence the results. Therefore, the authors' future research will
focus on the following aspects. First of all, the types of hospital
patients will be considered to test the validity and
generalizability of results of the present study. Secondly, the
authors intend to expand the sample size and include more known
predictive factors, for example HCV core amino acid 70 and IL28B.
Thirdly, the biological information of these genes will be utilized
for further functional studies, exploring the mechanism of the
association between these genes and hepatitis C virus. The benefits
of conducting such research would be barely doubted given good
representativeness, since all patients were remunerated blood
donation population and from the same district. Furthermore, the
patients were exposed during the same period and their infection
outcomes were steady after decades. More importantly, earlier study
of the authors suggested that genetic variants in HLA were
relevant to HCV infection susceptibility and viral clearance, the
present study thoroughly discussed and determined the association
between LMP7, TAP2 gene and treatment response in CHC
patients.
In conclusion, the present study demonstrated that
LMP7 and TAP2 loci were candidate regions that had
some novel SNPs for treatment response to pegIFN-α/RBV in the
Chinese CHC patients.
Acknowledgments
The present study was supported in part by the
National Natural Science Foundation of China (grant nos. 81473029,
81502853 and 81473028), the Science and Technology Development Fund
Key Project of Nanjing Medical University (grant no.
2016NJMUZD012), the Natural Science Foundation of Jiangsu Province
(grant no. BK20151026), Priority Academic Program Development of
Jiangsu Higher Education Institutions (PAPD).
References
|
1
|
Chen SR, Wang AQ, Lin LG, Qiu HC, Wang YT
and Wang Y: In vitro study on anti-hepatitis C virus activity of
Spatholobus suberectus Dunn. Molecules. 21:E13672016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cui Y and Jia J: Update on epidemiology of
hepatitis B and C in China. J Gastroenterol Hepatol. 28(Suppl 1):
7–10. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cai L, Gao C, Tang S, Wang J, Xue X, Yue
M, Deng X, Su J, Peng Z, Lu Y, et al: Sex-specific association of
estrogen receptor 2 polymorphisms with hepatitis C virus infection
outcomes in a high-risk Chinese Han population. Infect Genet Evol.
28:118–124. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bandiera S, Billie Bian C, Hoshida Y,
Baumert TF and Zeisel MB: Chronic hepatitis C virus infection and
pathogenesis of hepatocellular carcinoma. Curr Opin Virol.
20:99–105. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Matsuura K and Tanaka Y: Natural history
of hepatitis C virus infection. Nihon Rinsho. 73:195–200. 2015.In
Japanese. PubMed/NCBI
|
|
6
|
Falade-Nwulia O, Suarez-Cuervo C, Nelson
DR, Fried MW, Segal JB and Sulkowski MS: Oral direct-acting agent
therapy for hepatitis C virus infection: A systematic review. Ann
Intern Med. 166:637–648. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fateh A, Aghasadeghi MR, Keyvani H,
Mollaie HR, Yari S, Hadizade Tasbiti AR, Ghazanfari M and Monavari
SH: High resolution melting curve assay for detecting rs12979860
IL28B polymorphisms involved in response of Iranian patients to
chronic hepatitis C treatment. Asian Pac J Cancer Prev.
16:1873–1880. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ragheb MM, Nemr NA, Kishk RM, Mandour MF,
Abdou MM, Matsuura K, Watanabe T and Tanaka Y: Strong prediction of
virological response to combination therapy by IL28B gene variants
rs12979860 and rs8099917 in chronic hepatitis C genotype 4. Liver
Int. 34:890–895. 2014. View Article : Google Scholar
|
|
9
|
Aziz H, Raza A, Ali K, Khattak JZ, Irfan J
and Gill ML: Polymorphism of the IL28B gene (rs8099917, rs12979860)
and virological response of Pakistani hepatitis C virus genotype 3
patients to pegylated interferon therapy. Int J Infect Dis.
30:91–97. 2015. View Article : Google Scholar
|
|
10
|
Garcia RF, Moreira S, de Araújo Ramos AL,
Ferreira LE, de Mattos AA, Tovo CV, Nader LA, Ramos JA, Rondinelli
E, de Jesus Dominici A, et al: Interleukin 28B-related
polymorphisms: A pathway for understanding hepatitis C virus
infection? World J Gastroenterol. 19:7399–7404. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chowdhry M, Makroo RN, Singh M, Agrawal S,
Kumar M and Thakur Y: Human leucocyte antigen class I and II
alleles associated with anti-hepatitis C virus-positive patients of
North India. Indian J Med Microbiol. 34:299–302. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
El-Bendary M, Neamatallah M, Esmat G,
Kamel E, Elalfy H, Besheer T, Eldeib D, Eladl AH, El-Setouhy M,
El-Gilany AH, et al: Associations of human leucocyte antigen class
II-DQB1 alleles with hepatitis C virus infection in Egyptian
population: A multicentre family-based study. J Viral Hepat.
23:961–970. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Huang J, Huang K, Xu R, Wang M, Liao Q,
Xiong H, Li C, Tang X, Shan Z, Zhang M, et al: The associations of
HLA-A*02:01 and DRB1*11:01 with hepatitis C virus spontaneous
clearance are independent of IL28B in the chinese population. Sci
Rep. 6:314852016. View Article : Google Scholar
|
|
14
|
Waldron PR, Belitskaya-Lévy I, Chary A,
Won J, Winters M, Monto A, Ryan J, Lazzeroni LC and Holodniy M:
Genetic variation in the IL-6 and HLA-DQB1 genes is associated with
spontaneous clearance of hepatitis C virus infection. J Immunol
Res. 2016:65304362016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sedighimehr P, Irani S, Sakhaee F, Vaziri
F, Aghasadeghi M, Sadat SM, Jamnani FR, Fateh A and Siadat SD:
IL28B rs12980275 and HLA rs4273729 genotypes as a powerful
predictor factor for rapid, early, and sustained virologic response
in patients with chronic hepatitis C. Arch Virol. 162:181–189.
2017. View Article : Google Scholar
|
|
16
|
McKiernan SM, Hagan R, Curry M, McDonald
GS, Kelly A, Nolan N, Walsh A, Hegarty J, Lawlor E and Kelleher D:
Distinct MHC class I and II alleles are associated with hepatitis C
viral clearance, originating from a single source. Hepatology.
40:108–114. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang P, Zhang Y, Lu X, Xu Y, Wang J,
Zhang Y, Yu R and Su J: Association of polymorphisms in HLA antigen
presentation-related genes with the outcomes of HCV infection. PLoS
One. 10:e01235132015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Duggal P, Thio CL, Wojcik GL, Goedert JJ,
Mangia A, Latanich R, Kim AY, Lauer GM, Chung RT, Peters MG, et al:
Genome-wide association study of spontaneous resolution of
hepatitis C virus infection: data from multiple cohorts. Ann Intern
Med. 158:235–245. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sugimoto Y, Kuzushita N, Takehara T, Kanto
T, Tatsumi T, Miyagi T, Jinushi M, Ohkawa K, Horimoto M, Kasahara
A, et al: A single nucleotide polymorphism of the low molecular
mass polypeptide 7 gene influences the interferon response in
patients with chronic hepatitis C. J Viral Hepat. 9:377–384. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Cui Q, Zhang Y, Su J, Shi C, Lei N, Ding
K, Li J, Yu R, Wang L and Wang N: The association between the
genetic polymorphisms of LMP2/LMP7 and the outcomes of HCV
infection among drug users. J Biomed Res. 24:374–380. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Huang P, Dong L, Lu X, Zhang Y, Chen H,
Wang J, Zhang Y, Su J and Yu R: Genetic variants in antigen
presentation-related genes influence susceptibility to hepatitis C
virus and viral clearance: A case control study. BMC Infect Dis.
14:7162014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ashraf S, Nitschke K, Warshow UM, Brooks
CR, Kim AY, Lauer GM, Hydes TJ, Cramp ME, Alexander G, Little AM,
et al: Synergism of tapasin and human leukocyte antigens in
resolving hepatitis C virus infection. Hepatology. 58:881–889.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kuzushita N, Hayashi N, Kanto T, Takehara
T, Tatsumi T, Katayama K, Ohkawa K, Ito A, Kasahara A, Moribe T, et
al: Involvement of transporter associated with antigen processing 2
(TAP2) gene polymorphisms in hepatitis C virus infection.
Gastroenterology. 116:1149–1154. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Airoldi A, Zavaglia C, Silini E, Tinelli
C, Martinetti M, Asti M, Rossini A, Vangeli M, Salvaneschi L and
Pinzello G: Lack of a strong association between HLA class II,
tumour necrosis factor and transporter associated with antigen
processing gene polymorphisms and virological response to
alpha-interferon treatment in patients with chronic hepatitis C.
Eur J Immunogenet. 31:259–265. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
El Awady MK, Omran MH, Ibrahim MK,
Moustafa AM, Dawood RM, Bader El Din NG, Elsharkawy A, Abdel Aziz
MS, El Shenawy R, El Abd YS, et al: Low Molecular Mass Polypeptide
7 single nucleotide polymorphism is associated with the progression
of liver fibrosis in patients infected with hepatitis C virus
genotype 4. Clin Lab. 62:381–387. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ibrahim MK, Salama H, Abd El Rahman M,
Dawood RM, Bader El Din NG, Salem HF, Abdelrahim ME, Omran D, Omran
MH, El-Wakeel KH, et al: Three gene signature for predicting the
development of hepatocellular carcinoma in chronically infected
hepatitis C virus patients. J Interferon Cytokine Res. 36:698–705.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ibrahim MK, Salama H, Abd El Rahman M,
Dawood RM, Bader El Din NG, Salem HF, Abdelrahim ME, Omran D, Omran
MH, El-Wakeel KH, et al: Three gene signature for predicting the
development of hepatocellular carcinoma in chronically infected
hepatitis C virus patients. J Interferon Cytokine Res. 36:698–705.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
El Raziky M, Fathalah WF, Zakaria Z,
Eldeen HG, Abul-Fotouh A, Salama A, Awad A, Esmat G and Mabrouk M:
Predictors of virological response in 3,235 chronic HCV Egyptian
patients treated with peginterferon alpha-2a compared with
peginterferon alpha-2b using statistical methods and data mining
techniques. J Interferon Cytokine Res. 36:338–346. 2016. View Article : Google Scholar : PubMed/NCBI
|