Introduction
Mesenchymal stem cells (MSCs) are multipotent cells
capable of differentiating into myogenic, osteoblastic,
chondrogenic or adipogenic lineages. MSCs can be isolated from a
number of tissues, such as trabecular bone, the periosteum,
synovium, adipose tissue, skeletal muscle and deciduous teeth, as
well as from placenta and umbilical cord blood (1–7).
Due to their wide availability and the characteristic of
multipotent differentiation, MSCs have been widely used in bone
tissue engineering in the clinical setting (2–5,8).
Bone morphogenetic proteins (BMPs), which belong to
the transforming growth factor-β superfamily, are important in stem
cell biology and regulate cell proliferation and differentiation
during development (9,10). Additionally, the roles of BMPs in
bone and skeletal development are well recognized (6,11).
More than 20 BMPs have been identified, and BMP2, BMP4, BMP6 and
BMP7 are of more osteoinductive ability (11). Recombinant human BMP2 and BMP7
proteins have been used in the clinical setting (12–15). However, it remains unclear as to
whether BMP2 and BMP7 are the most potent BMPs in inducing
osteogenic differentiation and bone formation. It has previously
been demonstrated that BMP9 is more potent in promoting the
osteogenic differentiation of MSCs both in vitro and in
vivo (16). Many pivotal
transcription factors, such as inhibitor of differentiation (Id)1,
Id2, Id3, distal-less homeobox 5 (DLX5) and runt-related
transcription factor (RUNX)2, are pivotal mediators of the
BMP9-induced differentiation of MSCs (16). A variety of signaling pathways,
such as the canonical BMP/Smad signaling pathway and non-canonical
mitogen-activated protein kinase (MAPKs), have been identified to
meditate BMP9-induced osteogenic signaling (17–19). We have also previously found that
BMP type I receptors, ALK1 and ALK2, are involved in BMP9-induced
osteogenesis (20). Additionally,
microRNAs (miRNAs or miRs) are important regulators of BMP9-induced
osteogenesis (21–23). Despite these meaningful findings,
BMP9 remains the least studied BMPs and the molecular mechanisms
involved in BMP9 osteoinductive activity remain to be fully
elucidated.
The RUNX proteins, which include RUNX1, RUNX2 and
RUNX3, play an important role in cell growth and differentiation
during embryonic development (24). The RUNX1 null phenotype leads to
the early embryonic lethality of homozygous mice, demonstrating
that RUNX1 is required for definitive hematopoiesis (25). RUNX2 acts as a key regulator
during osteogenesis (26,27). RUNX2 knockout leads to the
impaired hepertrophy of chondrocytes followed by the complete
absence of mineralized bone (26,27). Previous studies on RUNX3 have
mainly focused on the function of gastric tumor suppressor and
central nervous system development (28,29). RUNX3 also plays an important role
in chondrocyte differentiation and bone formation (30–32). The expression of RUNX3 increases
at the stage of prehypertrophic chondrocytes and is maintained in
hypertrophic chondrocytes (32).
However, it decreases in terminal hypertrophic chondrocytes, and
the expression pattern of RUNX3 overlaps with RUNX2 (32). RUNX genes and proteins share
structural and organizational features and all RUNX proteins are
co-expressed in many skeletal elements (33–35). We have validated in our previous
studies that RUNX2 is involved in the BMP9-induced osteogenic
differentiation of MSCs (16,36). In the present study, we examined
the effect of RUNX3 on the BMP9-induced osteogenic differentiation
of MSCs. Our results demonstrated that RUNX3 may play regulatory
roles in the BMP9-induced osteogenic differentiation of MSCs by
affecting the Smad signaling cascade.
Materials and methods
Cell lines, cell culture and
chemicals
293 (for amplification of adenoviruses), HCT116,
C3H10T1/2, C2C12 cells were obtained from ATCC (Manassas, VA, USA)
and maintained in complete Dulbecco's modified Eagle's medium
(DMEM) supplemented with 10% fetal bovine serum and 100 U/ml
streptomycin/penicillin at 37°C in a humidified atmosphere of 5%
CO2. Unless otherwise indicated, all chemicals were
purchased from Sigma-Aldrich (St. Louis, MO, USA).
Construction of recombinant
adenovirus
Recombinant adenoviruses expressing exogenous BMP9
(Ad-BMP9) and RUNX3 (Ad-RUNX3), and recombinant adenovirus
expressing small interfering RNA (siRNA) targeting RUNX3
(Ad-siRUNX3) was generated using hte AdEasy system as previously
described (37). Adenoviruses
only expressing GFP (Ad-GFP) and RFP (Ad-RFP) were used as
controls. Ad-RUNX3 or Ad-BMP9 were used to infect the MSCs with a
multiplicity of infection (MOI) of 5. An MOI of 5 indicates 5
active viral particles per cell. To obtain the indicated MOI, we
first counted the quantity of the active viral particles, then
counted the cell number and finally calculated the volume of
viruses according to the indicated MOI.
Preparation of BMP9 conditioned
medium
BMP9 conditioned medium (BMP9-CM) was prepared as
follows: briefly, HCT116 cells were seeded in 100 mm dish and
infected with an optimal titer of Ad-BMP9. The culture medium was
changed into serum-free DMEM at 8 h following infection. BMP9
conditioned medium (BMP9-CM) was harvested at 24 and 48 h after
infection and used immediately.
Total RNA isolation, semi-quantitative
PCR and reverse transcription-quantitative PCR (RT-qPCR)
Total RNA was isolated from the cells using TRIzol
reagent (Invitrogen, Carlsbad, CA, USA) and used to generate cDNA
hexamer (Takara Bio Inc., Otsu, Japan) and MMLV transcription
reverse transcriptase (Promega, Sunnyvale, CA, USA). RT-PCR was
carried out as previously described (17–19) and PCR primers (Table I) were designed using the Primer3
program. A touchdown cycling program for RT-PCR was carried out as
follows: 94°C for 5 min for 1 cycle, 94°C for 30 sec, 68°C for 30
sec, and 72°C for 12 cycles with decrease in 1°C/cycle and then at
94°C for 30 sec, 55°C for 30 sec, and 72°C for 30 sec for 18–27
cycles depending on the abundance of a given gene. qPCR based on
SYBR Premix Ex Taq (Takara Bio Inc.) was carried out to amplify the
interesting genes with Bio-Rad iQ5 instrument (Bio-Rad, Hercules,
CA, USA). Gene expression results were analyzed using the ΔΔCt
method. A RT-qPCR cycling program was carried out as follows: 95°C
for 3 min, 95°C for 3 sec, 55°C for 30 sec (36 cycles), 95°C for 10
sec, 65°C for 10 sec and 95°C for 50 sec. Triplicate reactions were
carried out for each sample.
 | Table ISequences of primers for RT-PCR and
RT-qPCR. |
Table I
Sequences of primers for RT-PCR and
RT-qPCR.
| Gene name | Forward primer | Reverse primer |
|---|
| GAPDH |
GGCTGCCCAGAACATCAT |
CGGACACATTGGGGGTAG |
| Id1 |
ACGACATGAACGGCTGCT |
CAGCTGCAGGTCCCTGAT |
| Id2 |
CAGCATCCCCCAGAACAA |
TCTGGTGATGCAGGCTGA |
| Id3 |
CTACGAGGCGGTGTGCTG |
GCGCGAGTAGCAGTGGTT |
| DLX5 |
CTCAGCCACCACCCTCAT |
TGGCAGGTGGGAATTGAT |
| RUNX2 |
GGTGAAACTCTTGCCTCGTC |
AGTCCCAACTTCCTGTGCT |
| RUNX3 |
CACCGGCAGAAGATAGAAG |
GTCTGAGGAGCCTTGGATTG |
Western blot analysis
For western blot analysis, the cells were lysed with
RIPA buffer and cleared total lysate was denatured by boiling and
loaded onto an 8–15% gradient sodium dodecyl sulfate-polyacrylamide
gel electrophoresis (SDS-PAGE) gel. Following electrophoretic
separation, the proteins were transferred onto an Immobilon-P
membrane and blocked by super-block blocking buffer and probed by
primary antibodies, followed by incubation with secondary antibody.
The proteins of interest were detected using a SuperSignal West
Pico Chemiluminescent Substrate kit. The following primary
antibodies were obtained from Santa Cruz Biotechnology, Inc. (Santa
Cruz, CA, USA) as follows, anti-β-actin (sc-47778), anti-RUNX3
(sc-30197), anti-RUNX2 (sc-12488), anti-DLX5 (sc-18151) and
anti-Smad1/5/8 (sc-6031). The following primary antibodies were
purchased from Cell Signaling Technology (Danvers, MA, USA) as
follows, anti-phospho-p38 (#4511), anti-p38 (#9212),
anti-phospho-ERK1/2 (#4370), anti-ERK1/2 (#4695). The dilution for
all primary antibodies was 1:1,000. The following secondary
antibodies were purchased from Zhongshan Golden Bridge (Guangzhou,
China) as follows, peroxidase-conjugated rabbit anti-goat IgG
(ZB-2306), peroxidase-conjugated goat anti-mouse IgG (ZB-2305) and
peroxidase-conjugated goat anti-rabbit IgG (ZB-2301). The dilution
for all secondary antibodies was 1:5,000.
Measurement of alkaline phosphatase
(ALP)
ALP activity was assessed by a modified Great Escape
SEAP Chemiluminescence assay (BD Clontech, Mountain View, CA, USA)
as previously described (17–19). For the chemilluminescence assays,
each assay condition was performed in triplicate, and the results
were repeated in at least 3 independent experiments. ALP activity
was normalized to the total cellular protein level.
Measurement of matrix mineralization
Mineralized matrix nodules were stained for calcium
precipitation by means of Alizarin Red S staining, as previously
described (19). The cells were
fixed with 0.05% (v/v) glutaraldehyde at room temperature for 10
min. After being washed with distilled water, the fixed cells were
incubated with 0.4% Alizarin Red S for 5 min, followed by extensive
washing with distilled water. The staining of calcium mineral
deposits was recorded under bright field microscopy. The results
were repeated in at least 3 independent experiments.
Statistical analysis
Data were analyzed using One-way analysis of
variance (ANOVA) followed by the Tukey's test. Data are presented
as the means ± SEM and statistical significance was indicated by
p-value <0.05.
Results
BMP9 upregulates the expression of
endogenous RUNX3 in MSCs
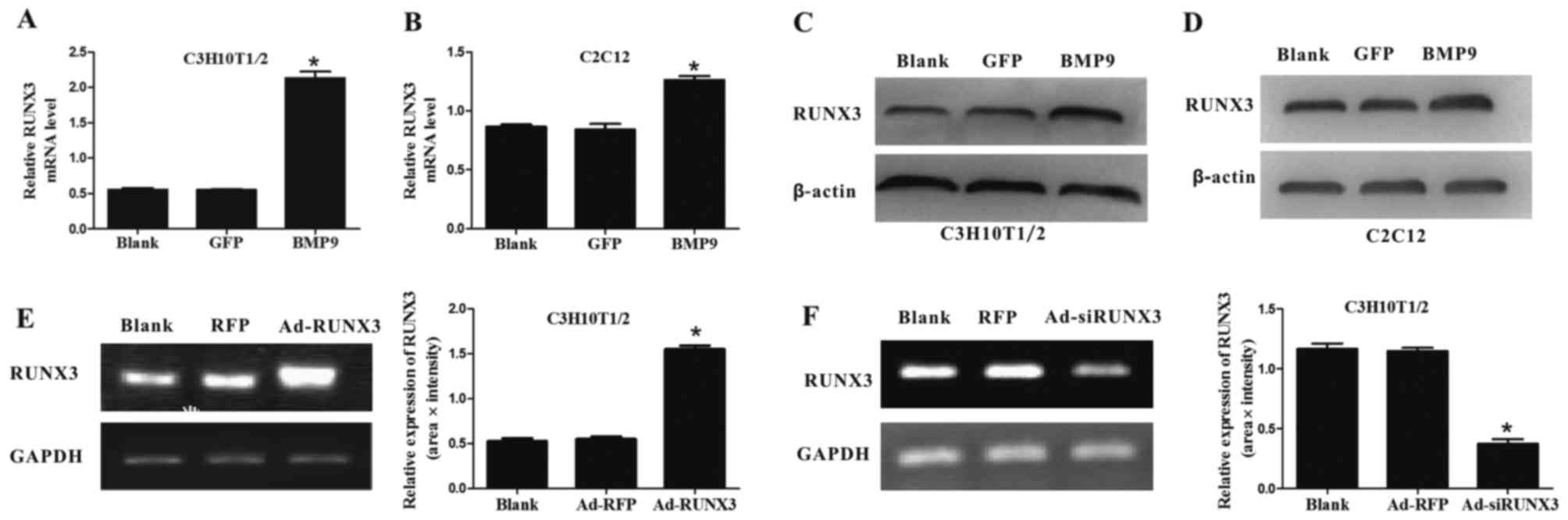
First of all, we sought to determine whether BMP9
can affect the expression of endogenous RUNX3. We found that BMP9
effectively increased the mRNA and protein expression level of
RUNX3 in C3H10T1/2 cells (Fig. 1A and
C) and C2C12 cells (Fig. 1B and
D). Subsequently, we successfully constructed recombinant
adenovirus expressing RUNX3 (Ad-RUNX3) (Fig. 1E) and adenovirus expressing siRNA
against RUNX3 (Ad-siRUNX3) (Fig.
1F). Our results indicate that BMP9 increases the expression of
RUNX3 in MSCs.
Effect of RUNX3 on the BMP9-induced
increase in the level of the early osteogenic marker, ALP
BMP9 increased RUNX3 expression in MSCs, and this
prompted us to evaluate the effect of RUNX3 on the BMP9-induced
osteogenic differentiation of MSCs. We infected the C3H10T1/2 and
C2C12 cells with Ad-RFP or Ad-RUNX3, followed by treatment with
BMP9-CM. The results revealed that the BMP9-induced ALP activity
was further increased following transfection of the C3H10T1/2 cells
(Fig. 2A and B) and C2C12 cells
(Fig. 2E and F) with Ad-RUNX3. To
complement these results, we also infected the C3H10T1/2 and C2C12
cells with siRNA against RUNX3 (Ad-siRUNX3). Still, an increased
level of ALP activity was observed in the C3H10T1/2 cells (Fig. 2C and D) and C2C12 cells (Fig. 2G and H). Collectively, both the
overexpression and/or knockdown of RUNX3 enhanced the BMP9-induced
early osteogenenic differentiation of MSCs.
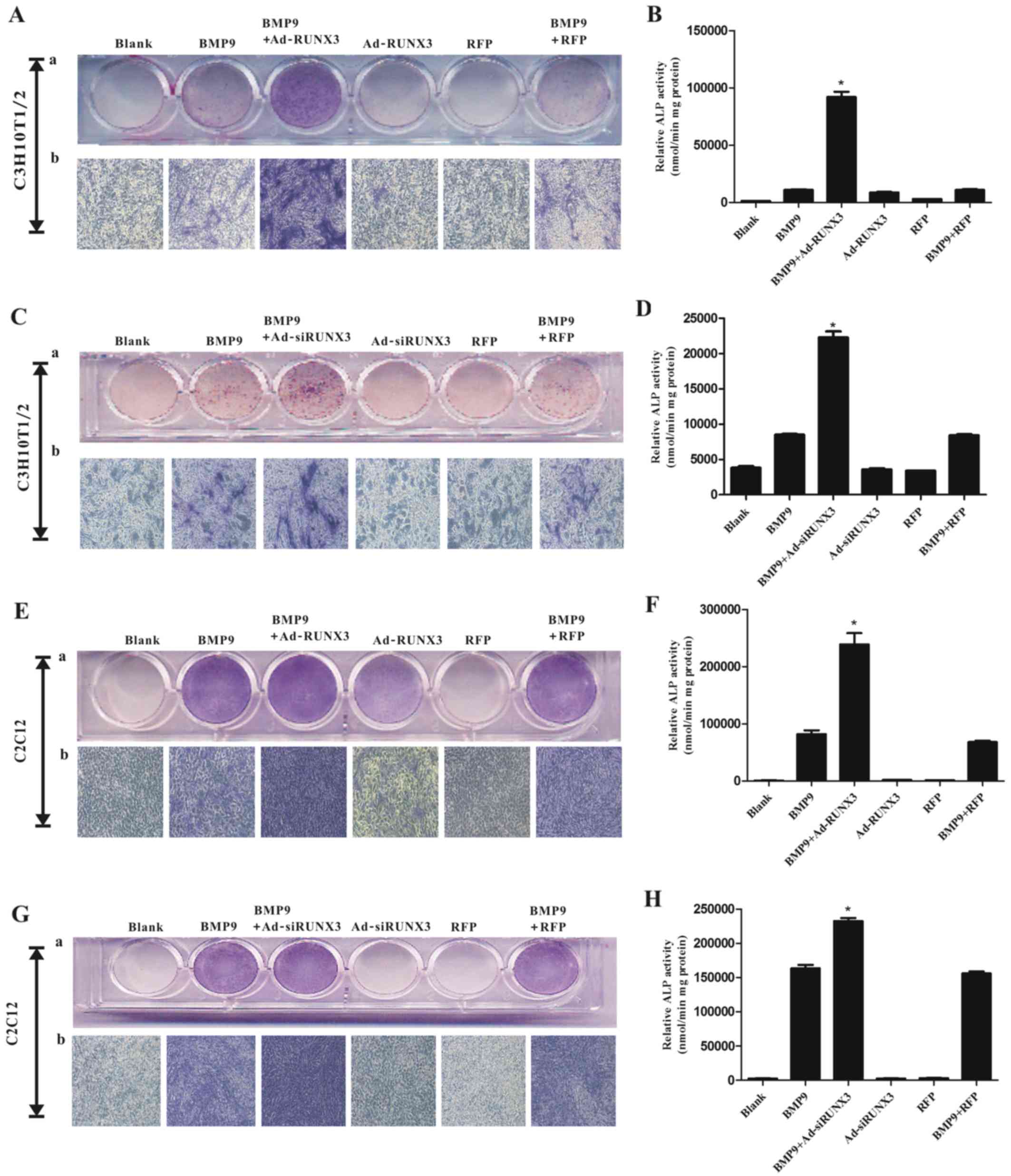 | Figure 2Effect of runt-related transcription
factor 3 (RUNX3) on bone morphogenetic protein 9 (BMP9)-induced
alkaline phosphatase (ALP) activity in mesenchymal stem cells
(MSCs). (A) Adenovirus (Ad)-RUNX3 enhanced BMP9-induced ALP
activity in C3H10T1/2 cells, asdetected by staining assay,
magnification, ×100. (B) Ad-RUNX3 enhanced BMP9-induced ALP
activity in C3H10T1/2 cells, as detected by quantitive assay. (C)
Adenovirus expressing small interfering RNA (siRNA) targeted
against RUNX3 (Ad-siRUNX3) enhanced BMP9-induced ALP activity in
C3H10T1/2 cells, as detected by staining assay. (D) Ad-siRUNX3
enhanced BMP9-induced ALP activity in C3H10T1/2 cells, as detected
by quantitive assay. (E) Ad-RUNX3 enhanced BMP9-induced ALP
activity in C2C12 cells, as detected by staining assay. (F)
Ad-RUNX3 enhanced BMP9-induced ALP activity in C2C12 cells, as
detected by quantitive assay. (G) Ad-siRUNX3 enhanced BMP9-induced
ALP activity in C2C12 cells, as detected by staining assay. (H)
Ad-siRUNX3 enhanced BMP9-induced ALP activity in C2C12 cells, as
detected by quantitive assay. Magnification, ×100.
*P<0.05 vs. BMP9 group. |
Effect of RUNX3 on the BMP9-induced
increase of the late osteogenic marker, matrix mineralization
Although ALP is an early osteogenic marker, it is
hardly an accurate late osteogenic marker. Thus, we sought to
examine the effect of RUNX3 on the BMP9-induced late osteogenic
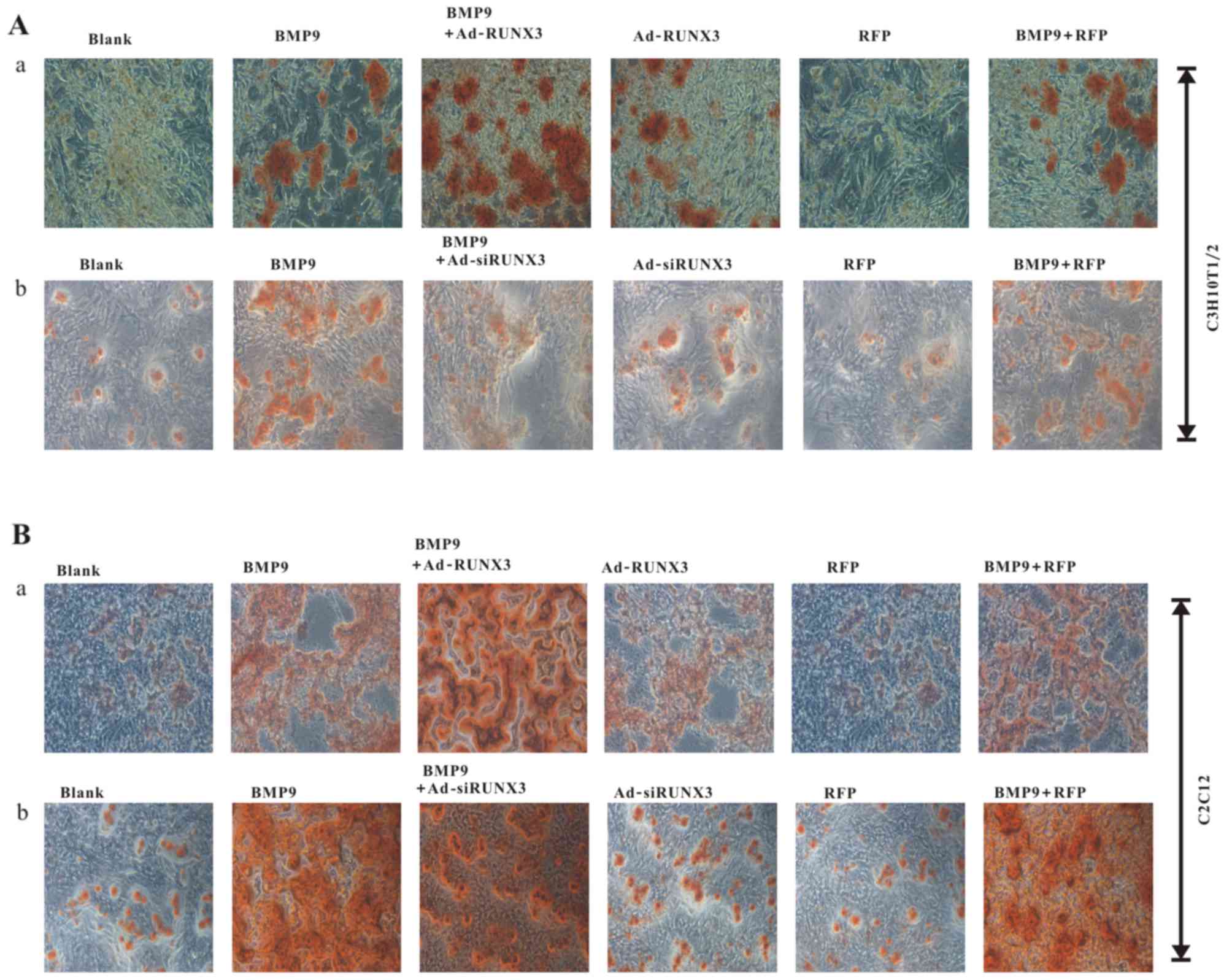
marker, matrix mineralization. We found that transfection with
Ad-RUNX3 increased BMP9-induced matrix mineralization in C3H10T1/2
cells (Fig. 3A–a) and C2C12 cells
(Fig. 3B–a). On the contrary
however, transfection with Ad-siRUNX3 decreased BMP9-induced matrix
mineralization, as shown in the C3H10T1/2 cells (Fig. 3A–b) and C2C12 cells (Fig. 3B–b). These results suggest that
RUNX3 affects the BMP9-induced osteogenic differentiation of
MSCs.
Effect of RUNX3 on the BMP9-induced
expression of pivotal osteogenic transcription factors
It has previously been demonstrated that a number of
pivotal osteogenic transcription factors, such as Id1, Id2, Id3,
DLX5 and RUNX2 are involved in the BMP9-induced osteogenic
differentiation of MSCs (39).
Thus, in order to evaluate the effect of RUNX3 on the BMP9-induced
expression of pivotal osteogenic transcription factors, we infected
the C3H10T1/2 cells with Ad-RUNX3, Ad-siRUNX3 or Ad-RFP and then
exposed the cells to BMP9-CM. The gene expression levels of Id1,
Id2, Id3, DLX5 and RUNX2 were determined by RT-wPCR and the protein
levels of DLX5 and RUNX2 were determined by western blot analysis.
We found that the expression levels of Id3, DLX5 and RUNX2 were
significantly increased by infection with Ad-RUNX3 and decreased by
infection with Ad-siRUNX3 (Fig.
4C–E). However, infection with Ad-RUNX3 and Ad-siRUNX3 both
increased the BMP9-induced expression of Id1, Id2 (Fig. 4A and B). Subsequently, we further
examined the protein expression levels of DLX5 and RUNX2 by western
blot analysis, and found that Ad-RUNX3 increased the BMP9-induced
expression of DLX5 and RUNX2, while Ad-siRUNX3 decreased the
expression of DLX5 and RUNX2 at the protein level (Fig. 4F). Collectively, these results
suggest that RUNX3 is involved in regulating the BMP9-induced
osteogenic differentiation of MSCs.
Effect of RUNX3 on BMP9-induced classical
Smad-dependent signaling and Smad-independent MAPK signaling
We then sought to examine the possible mechanisms
behind the effect of RUNX3 on the BMP9-induced osteogenic
differentiation of MSCs. Previous studies have reported that the
Smad-dependent signaling pathway and Smad-independent MAPK pathways
are crucial in regulating BMP9 osteoinductive signaling (17–19). Thus, we wished to confirm whether
RUNX3 can affect BMP9-activated Smad signaling and MAPK signaling
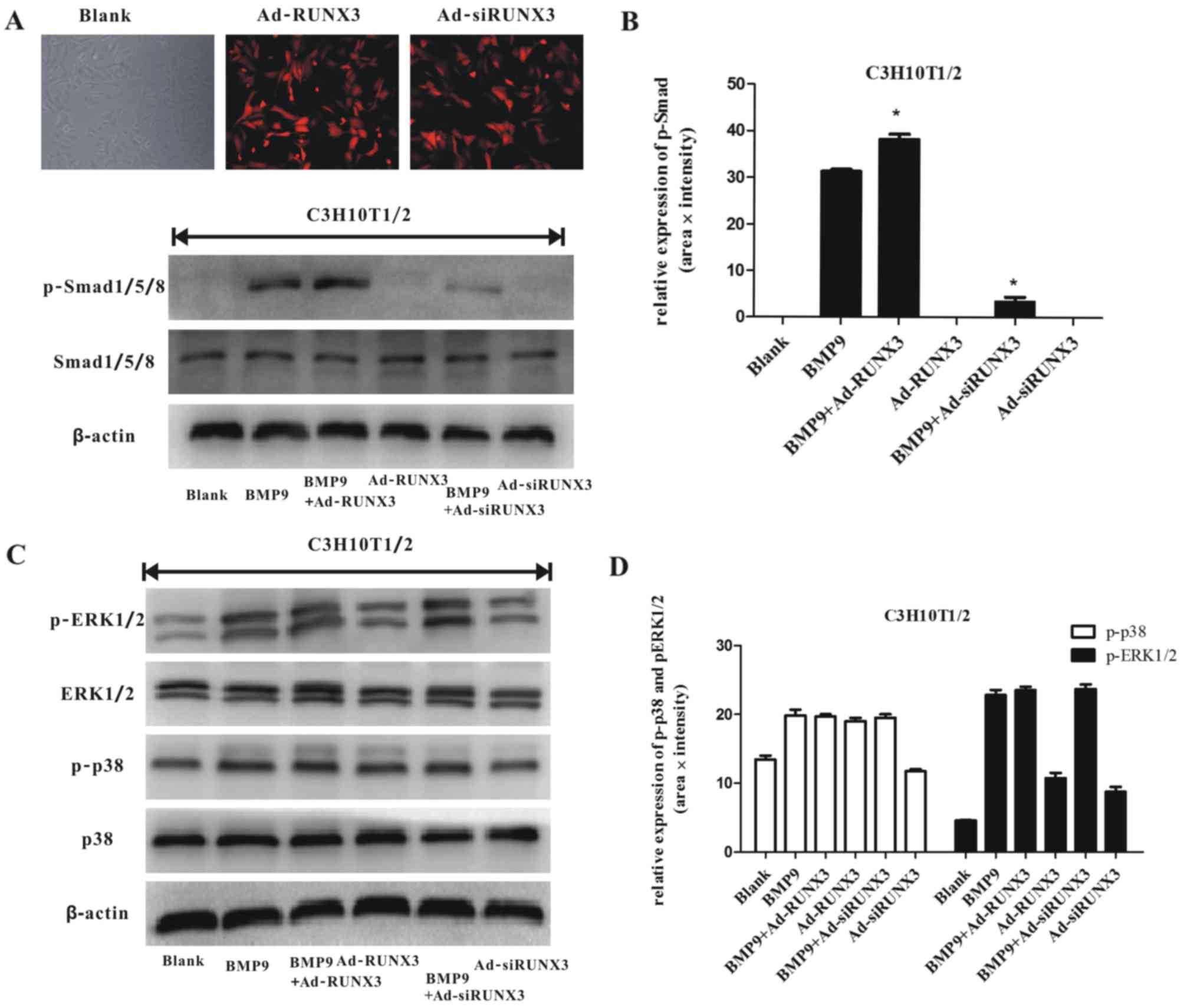
in MSCs. We infected the C3H10T1/2 cells with Ad-RUNX3 or/and
Ad-siRUNX3 (Fig. 5A). We found
that Ad-RUNX3 increased the BMP9-induced phosphorylation of
Smad1/5/8, whereas Ad-siRUNX3 decreased the BMP9-induced
phosphorylation of Smad1/5/8 (Fig. 5A
and B). We also found that BMP9 simultaneously stimulated the
phosphorylation of p38 and ERK1/2 in the osteogenic differentiation
process of MSCs (Fig. 5C and D),
consistent with previous results (18). However, RUNX3 did not alter the
BMP9-induced activation of p38 and ERK1/2 (Fig. 5C and D). These results indicate
that RUNX3 regulates the BMP9-induced differentiation of MSCs via
canonical Smad signaling.
Discussion
BMP9 [also known as growth differentiation factor-2
(GDF-2)], was originally isolated from fetal mouse liver and has
been shown to stimulate hepatocyte proliferation (38). Other roles of BMP9 include
regulating glucose and lipid metabolism in the liver (39), inducing the cholinergic phenotype
of embryonic basal forebrain cholinergic neurons (40) and maintaining homeostasis of iron
metabolism (41). Previously,
BMP9 was identified as one of the most potent osteogenic BMPs both
in vitro and in vivo (16), and yet it remains one of the least
characterized BMPs. RUNX family members are similar in mRNA and
protein structure and are co-distributed in skeletal elements
(33–35). It also has been reported that
RUNX2 is essential for osteogenesis and is involved in BMP9-induced
osteogenesis (16,26,27,36). Although RUNX3 is an important
regulator during chondrocyte differentiation and bone formation
(42–44), the detailed role of RUNX3 in
BMP9-induced osteogenesis is largely unknown. In the present study,
we found RUNX3 regulated BMP9-induced osteogenesis via Smad
signaling.
BMP9 was found to increase endogenous RUNX3
expression in MSCs in the present study. Notably, the
overexpression of RUNX3 and the knockdown of RUNX3 both led to a
potent promotion of ALP levels. RUNX1, RUNX2 and RUNX3 are all
expressed in mesenchymal condensation (34). It has been previously demonstrated
that after achieving the knockdown of one of the three RUNX
members, the remaining RUNX proteins may compensate for the lack of
the targeted one (32).
Therefore, we speculate that this redundant function among the RUNX
family may lead to the increased ALP activity following the
knockdown of RUNX3. BMP9-induced late osteogenic marker matrix
mineralization was enhanced by the overexpression of RUNX3, whereas
it was inhibited by the knockdown of RUNX3. We noticed that the
results of the ALP content and matrix mineralization were not
consistent. We therefore hypothesized that this phenomenon may due
to the following reasons: i) the sustained expression of RUNX3 and
the knockdown of RUNX3 cannot reproduce the spatial and temporal
expression pattern of RUNX3 during osteogenic differentiation; ii)
the redundant function among the RUNX family may exert a
differential influence on ALP and matrix mineralization.
Many pivotal transcription factors, such as Id, DLX5
and RUNX2 are involved in the BMP9-induced osteogenesis of MSCs
(11). DLX5 and RUNX2 are also
key transcription factors associated with osteogenesis (26,27,45). In this study, however, we find
that the transcriptional levels of Id1 and Id2 were both
upregulated by the overexpression and/or knockdown of RUNX3. We
speculate that the redundant function among the RUNX family may
have lead to this phenomenon. The results revealed that RUNX3
regulated important transcription factors involved in the
BMP9-induced osteogenesis of MSCs.
We validated that Smad1/5/8, ERK1/2, p38 and JNKs
are all involved in BMP9 osteoiductive signaling (17–19), implying that Smad-dependent and
Smad-independent MAPK pathways are most important in regulating the
BMP9-induced osteogenic differentiation of MSCs. In the present
study, we found that the phosphorylation of Smad1/5/8 was increased
by the overexpression of RUNX3 and was inhibited by the knockdown
of RUNX3. This result suggests that RUNX3 may affect BMP9-induced
Smad signaling. One possible explanation of the RUNX3 effect on
Smad signaling is that RUNX3 may facilitate Smad1/5/8 to bind to
BMP type I receptor which can directly phosphorylate Smad1/5/8.
Additionally, nuclear export factor RanBP3 can regulate the
phosphorylation of Smad1/5/8 by controlling the nucleocytoplasmic
shuttling of Smad1, Smad5 and Smad8 (Smad1/5/8) (46,47). Nuclear phosphatases, such as PPM1A
can also affect the phosphorylation of Smad1/5/8 by
dephosphorylating Smad1/5/8 (48,49). Therefore another explanation of
the RUNX3 effect on Smad signaling is that RUNX3 may interact with
RanBP3 and/or PPM1A to affect Smad1/5/8 phosphorylation. BMP9 can
simultaneously stimulate the phosphorylation/activation of p38 and
ERK1/2 (18). However, RUNX3 did
not affect the BMP9-induced phosphorylation of p38 and ERK1/2.
Taken together, our results strongly indicate that RUNX3 is likely
to play regulatory roles in the BMP9-induced osteogenic
differentiation of MSCs mainly by affecting the Smad signaling
cascade.
Previous studies have demonstrated that RUNX3 is an
important regulator of chondrocyte differentiation (30–32,50,51). The loss of osteoblastic RUNX3
produces severe congenital osteopenia (33). To date, little is known about
RUNX3 during osteogenesis. In this study, we explored the detailed
roles of RUNX3 during the BMP9-induced osteogenic differentiation
of MSCs. We found that RUNX3 played an important role during
BMP9-induced osteogenesis. These finding may contribute not only to
enrich our understanding of the osteoinductive activity of BMP9,
but may also provide a solid basis for the use of BMP9 in bone
tissue engineering. Future studies are warranted to identity the
direct target gene of RUNX3 during BMP9-induced osteogenesis and
explore the precise mechanisms responsible for the effects of RUNX3
on the phosphorylation of Smad1/5/8.
Acknowledgments
This study was supported by grants from the National
Natural Science Foundation of China (no. 81272006) and the Natural
Science Foundation Project of Chongqing Science and Technology
Commission (no. cstc2013jcyjA10061)
References
|
1
|
Deschaseaux F, Pontikoglou C and Sensébé
L: Bone regeneration: The stem/progenitor cells point of view. J
Cell Mol Med. 14:103–115. 2010. View Article : Google Scholar
|
|
2
|
Shenaq DS, Rastegar F, Petkovic D, Zhang
BQ, He BC, Chen L, Zuo GW, Luo Q, Shi Q, Wagner ER, et al:
Mesenchymal progenitor cells and their orthopedic applications:
Forging a path towards clinical trials. Stem Cells Int.
2010:5190282010. View Article : Google Scholar
|
|
3
|
Thakker R and Yang P: Mesenchymal stem
cell therapy for cardiac repair. Curr Treat Options Cardiovasc Med.
16:3232014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Myers TJ, Granero-Molto F, Longobardi L,
Li T, Yan Y and Spagnoli A: Mesenchymal stem cells at the
intersection of cell and gene therapy. Expert Opin Biol Ther.
10:1663–1679. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
García-Gómez I, Elvira G, Zapata AG,
Lamana ML, Ramírez M, Castro JG, Arranz MG, Vicente A, Bueren J and
García-Olmo D: Mesenchymal stem cells: Biological properties and
clinical applications. Expert Opin Biol Ther. 10:1453–1468. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Deng ZL, Sharff KA, Tang N, Song WX, Luo
J, Luo X, Chen J, Bennett E, Reid R, Manning D, et al: Regulation
of osteogenic differentiation during skeletal development. Front
Biosci. 13:2001–2021. 2008. View
Article : Google Scholar
|
|
7
|
Arthur A, Zannettino A and Gronthos S: The
therapeutic applications of multipotential mesenchymal/stromal stem
cells in skeletal tissue repair. J Cell Physiol. 218:237–245. 2009.
View Article : Google Scholar
|
|
8
|
Stappenbeck TS and Miyoshi H: The role of
stromal stem cells in tissue regeneration and wound repair.
Science. 324:1666–1669. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Botchkarev VA: Bone morphogenetic proteins
and their antagonists in skin and hair follicle biologyn. J Invest
Dermatol. 120:37–47. 2003. View Article : Google Scholar
|
|
10
|
Zhao GQ: Consequences of knocking out BMP
signaling in the mouse. Genesis. 35:43–56. 2003. View Article : Google Scholar
|
|
11
|
Luu HH, Song WX, Luo X, Manning D, Luo J,
Deng ZL, Sharff KA, Montag AG, Haydon RC and He TC: Distinct roles
of bone morphogenetic proteins in osteogenic differentiation of
mesenchymal stem cells. J Orthop Res. 25:665–677. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Varady P, Li JZ, Alden TD, Kallmes DF,
Williams MB and Helm GA: CT and radionuclide study of BMP-2 gene
therapy-induced bone formation. Acad Radiol. 9:632–637. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Krebsbach PH, Gu K, Franceschi RT and
Rutherford RB: Gene therapy-directed osteogenesis: BMP-7-transduced
human fibroblasts form bone in vivo. Hum Gene Ther. 11:1201–1210.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Boraiah S, Paul O, Hawkes D, Wickham M and
Lorich DG: Complications of recombinant human BMP-2 for treating
complex tibial plateau fractures: A preliminary report. Clin Orthop
Relat Res. 467:3257–3262. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Rutherford RB, Nussenbaum B and Krebsbach
PH: Bone morphogenetic protein 7 ex vivo gene therapy. Drug News
Perspect. 16:5–10. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kang Q, Sun MH, Cheng H, Peng Y, Montag
AG, Deyrup AT, Jiang W, Luu HH, Luo J, Szatkowski JP, et al:
Characterization of the distinct orthotopic bone-forming activity
of 14 BMPs using recombinant adenovirus-mediated gene delivery.
Gene Ther. 11:1312–1320. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao YF, Xu J, Wang WJ, Wang J, He JW, Li
L, Dong Q, Xiao Y, Duan XL, Yang X, et al: Activation of JNKs is
essential for BMP9-induced osteogenic differentiation of
mesenchymal stem cells. BMB Rep. 46:422–427. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhao Y, Song T, Wang W, Wang J, He J, Wu
N, Tang M, He B and Luo J: P38 and ERK1/2 MAPKs act in opposition
to regulate BMP9-induced osteogenic differentiation of mesenchymal
progenitor cells. PLoS One. 7:e433832012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xu DJ, Zhao YZ, Wang J, He JW, Weng YG and
Luo JY: Smads, p38 and ERK1/2 are involved in BMP9-induced
osteogenic differentiation of C3H10T1/2 mesenchymal stem cells. BMB
Rep. 45:247–252. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Luo J, Tang M, Huang J, He BC, Gao JL,
Chen L, Zuo GW, Zhang W, Luo Q, Shi Q, et al: TGFbeta/BMP type I
receptors ALK1 and ALK2 are essential for BMP9-induced osteogenic
signaling in mesenchymal stem cells. J Biol Chem. 285:29588–29598.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang W, Zhang L, Zhou Y, Ji X, Liu J, Liu
D, Yin P, Peng Y, Hao M, Zhang L, et al: Synergistic effects of
BMP9 and miR-548d-5p on promoting osteogenic differentiation of
mesenchymal stem cells. Biomed Res Int. 2015:3097472015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang R, Weng Y, Li B, Jiang Y, Yan S, He
F, Chen X, Deng F, Wang J and Shi Q: BMP9-induced osteogenic
differentiation is partially inhibited by miR-30a in the
mesenchymal stem cell line C3H10T1/2. J Mol Histol. 46:399–407.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Song Q, Zhong L, Chen C, Tang Z, Liu H,
Zhou Y, Tang M, Zhou L, Zuo G, Luo J, et al: miR-21 synergizes with
BMP9 in osteogenic differentiation by activating the BMP9/Smad
signaling pathway in murine multilineage cells. Int J Mol Med.
36:1497–1506. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Choi JY, Pratap J, Javed A, Zaidi SK, Xing
L, Balint E, Dalamangas S, Boyce B, van Wijnen AJ, Lian JB, et al:
Subnuclear targeting of Runx/Cbfa/AML factors is essential for
tissue-specific differentiation during embryonic development. Proc
Natl Acad Sci USA. 98:8650–8655. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wang Q, Stacy T, Binder M, Marin-Padilla
M, Sharpe AH and Speck NA: Disruption of the Cbfa2 gene causes
necrosis and hemorrhaging in the central nervous system and blocks
definitive hematopoiesis. Proc Natl Acad Sci USA. 93:3444–3449.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Otto F, Thornell AP, Crompton T, Denzel A,
Gilmour KC, Rosewell IR, Stamp GW, Beddington RS, Mundlos S, Olsen
BR, et al: Cbfa1, a candidate gene for cleidocranial dysplasia
syndrome, is essential for osteoblast differentiation and bone
development. Cell. 89:765–771. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Komori T, Yagi H, Nomura S, Yamaguchi A,
Sasaki K, Deguchi K, Shimizu Y, Bronson RT, Gao YH, Inada M, et al:
Targeted disruption of Cbfa1 results in a complete lack of bone
formation owing to maturational arrest of osteoblasts. Cell.
89:755–764. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Levanon D, Bettoun D, Harris-Cerruti C,
Woolf E, Negreanu V, Eilam R, Bernstein Y, Goldenberg D, Xiao C,
Fliegauf M, et al: The Runx3 transcription factor regulates
development and survival of TrkC dorsal root ganglia neurons. EMBO
J. 21:3454–3463. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Brenner O, Levanon D, Negreanu V, Golubkov
O, Fainaru O, Woolf E and Groner Y: Loss of Runx3 function in
leukocytes is associated with spontaneously developed colitis and
gastric mucosal hyperplasia. Proc Natl Acad Sci USA.
101:16016–16021. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Soung Y, Dong Y, Wang Y, Zuscik MJ,
Schwarz EM, O'Keefe RJ and Drissi H: Runx3/AML2/Cbfa3 regulates
early and late chondrocyte differentiation. J Bone Miner Res.
22:1260–1270. 2007. View Article : Google Scholar
|
|
31
|
Yoshida CA and Komori T: Role of Runx
proteins in chondrogenesis. Crit Rev Eukaryot Gene Expr.
15:243–254. 2005. View Article : Google Scholar
|
|
32
|
Stricker S, Fundele R, Vortkamp A and
Mundlos S: Role of Runx genes in chondrocyte differentiation. Dev
Biol. 245:95–108. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Bauer O, Sharir A, Kimura A, Hantisteanu
S, Takeda S and Groner Y: Loss of osteoblast Runx3 produces severe
congenital osteopenia. Mol Cell Biol. 35:1097–1109. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Levanon D and Groner Y: Structure and
regulated expression of mammalian RUNX genes. Oncogene.
23:4211–4219. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Levanon D, Brenner O, Negreanu V, Bettoun
D, Woolf E, Eilam R, Lotem J, Gat U, Otto F, Speck N, et al:
Spatial and temporal expression pattern of Runx3 (Aml2) and Runx1
(Aml1) indicates non-redundant functions during mouse
embryogenesis. Mech Dev. 109:413–417. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sharff KA, Song WX, Luo X, Tang N, Luo J,
Chen J, Bi Y, He BC, Huang J, Li X, et al: Hey1 basic
helix-loop-helix protein plays an important role in mediating
BMP9-induced osteogenic differentiation of mesenchymal progenitor
cells. J Biol Chem. 284:649–659. 2009. View Article : Google Scholar :
|
|
37
|
He TC, Zhou S, da Costa LT, Yu J, Kinzler
KW and Vogelstein B: A simplified system for generating recombinant
adenoviruses. Proc Natl Acad Sci USA. 95:2509–2514. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Song JJ, Celeste AJ, Kong FM, Jirtle RL,
Rosen V and Thies RS: Bone morphogenetic protein-9 binds to liver
cells and stimulates proliferation. Endocrinology. 136:4293–4297.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Chen C, Grzegorzewski KJ, Barash S, Zhao
Q, Schneider H, Wang Q, Singh M, Pukac L, Bell AC, Duan R, et al:
An integrated functional genomics screening program reveals a role
for BMP-9 in glucose homeostasis. Nat Biotechnol. 21:294–301. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
López-Coviella I, Berse B, Krauss R, Thies
RS and Blusztajn JK: Induction and maintenance of the neuronal
cholinergic phenotype in the central nervous system by BMP-9.
Science. 289:313–316. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Truksa J, Peng H, Lee P and Beutler E:
Bone morphogenetic proteins 2, 4, and 9 stimulate murine hepcidin 1
expression independently of Hfe, transferrin receptor 2 (Tfr2), and
IL-6. Proc Natl Acad Sci USA. 103:10289–10293. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Smith N, Dong Y, Lian JB, Pratap J,
Kingsley PD, van Wijnen AJ, Stein JL, Schwarz EM, O'Keefe RJ, Stein
GS, et al: Overlapping expression of Runx1(Cbfa2) and Runx2 (Cbfa1)
transcription factors supports cooperative induction of skeletal
development. J Cell Physiol. 203:133–143. 2005. View Article : Google Scholar
|
|
43
|
Yoshida CA, Yamamoto H, Fujita T, Furuichi
T, Ito K, Inoue K, Yamana K, Zanma A, Takada K, Ito Y, et al: Runx2
and Runx3 are essential for chondrocyte maturation, and Runx2
regulates limb growth through induction of Indian hedgehog. Genes
Dev. 18:952–963. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kim EJ, Cho SW, Shin JO, Lee MJ, Kim KS
and Jung HS: Ihh and Runx2/Runx3 signaling interact to coordinate
early chondrogenesis: A mouse model. PLoS One. 8:e552962013.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ferrari D, Harrington A, Dealy CN and
Kosher RA: Dlx-5 in limb initiation in the chick embryo. Dev Dyn.
216:10–15. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Chen F, Lin X, Xu P, Zhang Z, Chen Y, Wang
C, Han J, Zhao B, Xiao M and Feng XH: Nuclear export of smads by
ranBP3L regulates bone morphogenetic protein signaling and
mesenchymal stem cell differentiation. Mol Cell Biol. 35:1700–1711.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Dai F, Lin X, Chang C and Feng XH: Nuclear
export of Smad2 and Smad3 by RanBP3 facilitates termination of
TGF-beta signaling. Dev Cell. 16:345–357. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Lin X, Duan X, Liang YY, Su Y, Wrighton
KH, Long J, Hu M, Davis CM, Wang J, Brunicardi FC, et al: PPM1A
functions as a Smad phosphatase to terminate TGFbeta signaling.
Cell. 125:915–928. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Dai F, Shen T, Li Z, Lin X and Feng XH:
PPM1A dephosphorylates RanBP3 to enable efficient nuclear export of
Smad2 and Smad3. EMBO Rep. 12:1175–1181. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wigner NA, Soung Y, Einhorn TA, Drissi H
and Gerstenfeld LC: Functional role of Runx3 in the regulation of
aggrecan expression during cartilage development. J Cell Physiol.
228:2232–2242. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Dalcq J, Pasque V, Ghaye A, Larbuisson A,
Motte P, Martial JA and Muller M: RUNX3, EGR1 and SOX9B form a
regulatory cascade required to modulate BMP-signaling during
cranial cartilage development in zebrafish. PLoS One. 7:e501402012.
View Article : Google Scholar : PubMed/NCBI
|