Introduction
Malignant melanoma is the most dangerous type of
skin cancer that develops from the melanocyte pigment cells
(1). The worldwide incidence of
this disease is increasing yearly (2) and ~68,480 associated mortalities
occur in the United States each year. This disease has the
proclivity to metastasize, therefore, it usually progresses into an
aggressive invasive disease in ~20% of patients (3). Metastatic melanoma has a poor
prognosis, with a median survival time of 6–9 months and a 5-year
survival rate of 5–10% (4).
However, the mechanisms of its invasion and metastasis have not
been clearly understood. Therefore, it is necessary to investigate
melanoma biology to facilitate the development of therapeutic
strategies in melanoma.
Non-coding RNAs (ncRNAs) are emerging as novel
regulators in the cancer paradigm (5). Recent studies show a particular
focus on long ncRNAs (lncRNAs), which are >200 nucleotides in
length without protein-coding capacity (6). lncRNAs have been demonstrated to
serve an important role in tumorigenesis (7,8).
Particularly, several lncRNAs, including long intergenic
non-protein coding RNA 673, BRAF-activated non-protein coding RNA
and HOX transcript antisense RNA, have been reported to be involved
in the tumorgenesis, invasion and metastasis of melanoma via
complicated regulatory mechanisms (5,9,10).
The lncRNA Pvt1 oncogene (non-protein coding) (PVT1) is an oncogene
that is located at 8q24.21, a well-known cancer risk region
(11). PVT1 has been found to be
overexpressed in numerous human cancer types, including ovarian,
breast and non-small cell lung cancer (12–14). However, to the best of our
knowledge, the possible roles and mechanisms of PVT1 in regulating
cell proliferation and metastasis in melanoma has not been
reported.
The present study investigated the expression of
PVT1 in melanoma and investigated the potential effects of PVT1
expression on the proliferation and metastasis of melanoma cells.
The study aimed to elucidate the underlying mechanism of melanoma
and to provide novel insights into the treatment of this
disease.
Materials and methods
Patients and samples
Between January 2014 and February 2016, 37 patients
who were diagnosed with melanoma in the Shandong Provincial
Hospital Affiliated to Shandong University (Jinan, Shandong, China)
were enrolled in the present study. The melanoma diagnosis was
pathologically confirmed. Tumor tissues and their adjacent (3–5 cm
away) normal skin were obtained from clinically ongoing surgical
specimens. All patients provided written informed consent prior to
the study. All procedures in this study were approved by the Human
Ethics Committee of the Shandong Provincial Hospital Affiliated to
Shandong University.
Cell culture and small interfering RNA
(siRNA) transfection
The human melanoma A-375 and sk-mel-5 cell lines and
normal epiderma melanocytes PIG1 cell line were purchased from the
American Type Culture Collection (ATCC; Manassas, VA, USA) to use
in this study. The A-375 cells and PIG1 cells (control) were
cultured in RPMI-1640 medium (Gibco; Thermo Fisher Scientific,
Inc., Waltham, MA, USA) and the sk-mel-5 cells were cultured in
Dulbecco’s modified Eagle’s medium (Gibco; Thermo Fisher
Scientific, Inc.). The media were supplemented with 10% fetal
bovine serum (FBS) (Gibco; Thermo Fisher Scientific, Inc.), 100
U/ml penicillin and 100 μg/ml streptomycin (both Invitrogen;
Thermo Fisher Scientific, Inc.) in an atmosphere of 5%
CO2 at 37°C. Culture medium was changed every 2
days.
For cell transfection, a total of 100 nM silenced
vector of PVT1 (Sangon Biotech Co., Ltd., Shanghai, China) was
transfected into the cells using Lipofectamine 2000®
(Invitrogen; Thermo Fisher Scientific, Inc.). The target sequences
for PVT1 were as follows: Sense, 5′-GCUUGGAGGCUGAGGAGUUTT-3′ and
antisense, 5′-AACUCCUCAGCCUCCAAGCTT-3′. siRNA vector without
silenced PVT1 sequence was transfected into the cells as a
control.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay
Cell proliferation was detected using an MTT assay
as previously described (15).
Briefly, after 24 h of cell transfection, cells were plated into
96-well plates at a density of 5×103 cells/well. After
cultivation for 24 h, the supernatant was discarded, and 20
μl MTT was added for another 4 h incubation in RPMI-1640
supplemented with 10% FBS. Next, 150 μl dimethyl sulfoxide
was mixed with the cells for 10 min. Absorbance of cells in each
well was observed at 570 nm under an absorption spectrophotometer
(Olympus, Tokyo, Japan) for the cell number calculation.
Clonogenic assay
A clonogenic assay was performed with a modification
of a previous method (16). In
brief, following siRNA transfection for 48 h, cells were plated
into the 60-mm tissue culture dishes in triplicate at a density of
100 cells/dish. The cells were then grown in RPMI-1640 medium
containing 10% FBS at 37°C for 14 days. Following this, the cells
were fixed and stained with Diff-Quick (Medion Diagnostics,
Dudingen, Switzerland) according to manufacturer’s protocol,
followed by air drying. Finally, colonies were counted under an
inverted microscope (IX83; Olympus), and the cell number of each
colony was at least 30 cells.
Cell cycle assay
Subsequent to 48 h of siRNA transfection, the cells
were seeded into 6-well plates and cultured for 48 h to reach
75–80% culture confluency. Thereafter the cells were collected and
fixed in 70% ethanol, washed with phosphate-buffered saline (PBS),
and resuspended in 200 μl PBS containing 0.5 mg/ml RNase,
0.05% Triton X-100 and 10 μg/ml propidium iodide (Merck
KGaA, Darmstadt, Germany). Following incubation for 1 h at 37°C in
the dark, the cells were analyzed immediately using a flow
cytometer (BD Biosciences, San Jose, CA, USA) for cell cycle
analysis. The experiment was performed in triplicates.
Migration assay
For cell migration assay, a Transwell insert was
used. Subsequent to 48 h of siRNA transfection, the cells were
incubated for 24 h in serum-free RPMI-1640 medium containing 0.01%
bovine serum albumin (BSA) (Merck KGaA). The upper layer of the
Transwell was enveloped with serum-free RPMI-1640 medium, then
air-dried at 4°C. After suctioning out the medium, 50 μl
fresh serum-free medium that contained 10 g/l BSA was added. After
being cultured at 37°C for 30 min, the Transwell was put into the
24-well plates and cultured with RPMI-1640 medium mixed with 10%
FBS. Cells in the Transwell insert were suspended with serum-free
RPMI-1640 medium for 48 h. The Transwell insert was then washed
with PBS to remove the upper cells on the microporous membrane and
fixed in ice-cold alcohol. Finally, the Transwell insert was
stained with 0.1% crystal violet at room temperature for 30 min,
and decolorized with 33% acetic acid to exclude the false positive
staining.
Invasion assay
For the assessment of cell invasion, a scratch assay
was used. Briefly, cells in each group were cultured in 6-well
plates at a density of 5×105/well. A wound was made
across the well with a 200-μl pipette tip. Images of the
wound were captured immediately and then the cellular invasion of
cells across the wound was documented using images captured under
an inverted microscope (×10 magnification). The width of the gap
was detected in 5 different fields and the average width was
calculated.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA from cancer tissues and cells was
extracted using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) and then treated with RNase-free DNase I
(Promega, Madison, WI, USA). The concentration and purity of the
isolated RNA were detected using SMA 400 UV-VIS (Merinton
Instrument, Ltd., Shanghai, China). Next, 0.5 μg/ml purified
RNA that was mixed with nuclease-free water was used for cDNA
synthesis with the PrimerScript 1st Strand cDNA Synthesis kit
(Invitrogen; Thermo Fisher Scientific, Inc.). The expression of the
targets in the tissues or cells was measured in an Eppendorf
Mastercycler (Brinkmann Instruments, Inc., Westbury, NY, USA) using
the SYBR ExScript qRT-PCR kit (Takara Biotechnology Co., Ltd.,
Dalian, China) at a final volume of 20 μl using the standard
protocol, which was as follows: 95°C for 3 min, 40 cycles of 95°C
for 10 sec and 60°C for 30 sec. Each reaction was performed in
triplicate, and the 2−ΔΔCq method (17) was used to determine the relative
gene expression levels. The melting curve of the amplification
products was analyzed at the end of each PCR to confirm that only
one product was amplified and detected. Glyceraldehyde 3-phosphate
dehydrogenase (GAPDH) was chosen as the internal control for mRNA
or lncRNAs. Primers used for targets amplification are shown in
Table I.
 | Table IPrimers used for target
amplification. |
Table I
Primers used for target
amplification.
| Name | Forward primers
(5′-3′) | Reverse primers
(5′-3′) |
|---|
| PVT1 |
TGAGAACTGTCCTTACGTGACC |
AGAGCACCAAGACTGGCTCT |
| Cyclin D1 |
CAAATGGAGCTGCTCCTGGTG |
CTTCGATCTGCTCCTGGCAGG |
| Cyclin E1 |
GCCAGCCTTGGGACAATAATG |
GCCAGCCTTGGGACAATAATG |
| Cyclin A1 |
CAAGGTCCTGATGCTTGTCA |
CCCATGGTCAGAGAGCACTT |
| E-cadherin |
TGTAGTTACGTATTTATTTTTAGTGGCGTC C |
GAATACGATCGAATCGAACCG |
| N-cadherin |
AACTCCAGGGGACCTTTTC |
CAAATGAAACCGGGCTATC |
| Vimentin |
TCCAAGTTGCTGACCTCTC |
TCAACGGCAAAGTTCTCTTC |
| GAPDH |
TTCGACAGTCAGCCGCATCTT |
ATCCGTTGACTCCGACCTTCA |
Western blotting
The cells were cultured for 48 h after transfection,
then they were lysed with RIPA lysis and extraction buffer (Sangon
Biotech Co., Ltd.). The supernatant was collected for the
measurement of protein concentrations using a bicinchoninic acid
protein assay kit (Pierce; Thermo Fisher Scientific.). Next, 20
μg protein was subjected to a 10% sodium dodecyl
sulfate-polyacrylamide gel electrophoresis and then transferred to
polyvinylidene fluoride membrane (EMD Millipore, Billerica, MA,
USA). Subsequent to being blocked with Tris-buffered saline
Tween-20 (TBST) containing 5% skimmed milk at room temperature for
1 h, the membrane was incubated with rabbit anti-human primary
antibodies against cyclin D1 (cat. no. 2978), cyclin E1 (cat. no.
20808) and cyclin A1 (cat. no. PA5-16519), E-cadherin (cat. no.
3195), N-cadherin (cat. no. 13116), vimentin (cat. no. 5741), and
GAPDH (cat. no. 5174) (all 1:1,000 dilution) at 4°C overnight.
Following this, the membrane was washed with 1X TBST 3 times and
then incubated with horseradish peroxidase-labeled goat anti-rabbit
secondary antibody (1:1,000 dilution; cat. no. 14708) at room
temperature for 1 h. All antibodies were purchased from Santa Cruz
Biotechnology, Inc. (Dallas, TX, USA), except for cyclin A1 that
was purchased from Thermo Fisher Scientific, Inc. Detection was
conducted using the development of X-ray after the addition of a
chromogenic substrate with an enhanced chemiluminescence method
(EMD Millipore). GAPDH served as the internal control.
Qualification of protein bands were determined using the Adobe
Photoshop CS5 (Adobe, San Jose, CA, USA).
Chromatin immunoprecipitation (ChIP)
assay
The ChIP assay was conducted with an EZ ChIP
Chromatin Immunoprecipitation kit (EMD Millipore) according to the
manufacturer’s protocols. Cross-linked chromatin was sonicated into
200-1,000-bp fragments. The chromatin was immunoprecipitated using
anti-c-Myc antibody (cat. no. 611451; 1:200 dilution; BD
Biosciences). Normal mouse immunoglobulin G (IgG; cat. no. 554002;
1:200 dilution; BD Biosciences) was chosen as the negative control.
RT-qPCR was conducted using SYBR-Green Mix (Roche Diagnostics,
Indianapolis, IN, USA) as aforementioned.
RNA immunoprecipitation (RIP) assay
The RIP assay was performed using the Magna RIP
RNA-Binding Protein Immunoprecipitation kit (EMD Millipore)
according to the manufacturer’s protocols. Anti-enhancer of zeste
homolog 2 (EZH2) antibody and IgG (control) were used for RIP (Cell
Signaling Technology, Inc., Danvers, MA, USA). Co-precipitated RNAs
were detected by qRT-PCR analysis. Total RNAs (input controls) and
isotype controls were detected simultaneously to demonstrate that
the detected signals were the results of RNAs specifically binding
to E-box.
Statistical analysis
Data are expressed as the mean ± standard deviation
and were analyzed using GraphPrism 5.0 software (GraphPad Software,
Inc., La Jolla, CA, USA). Statistical analyses were performed using
SPSS 19.0 statistical software (IBM Corp., Armonk, NY, USA). An
independent samples t-test was used for the paired data
significance calculation. Analysis of variance was used to
calculate the significance for >2 groups, with Tukey’s post-hoc
test used to calculate the difference among groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
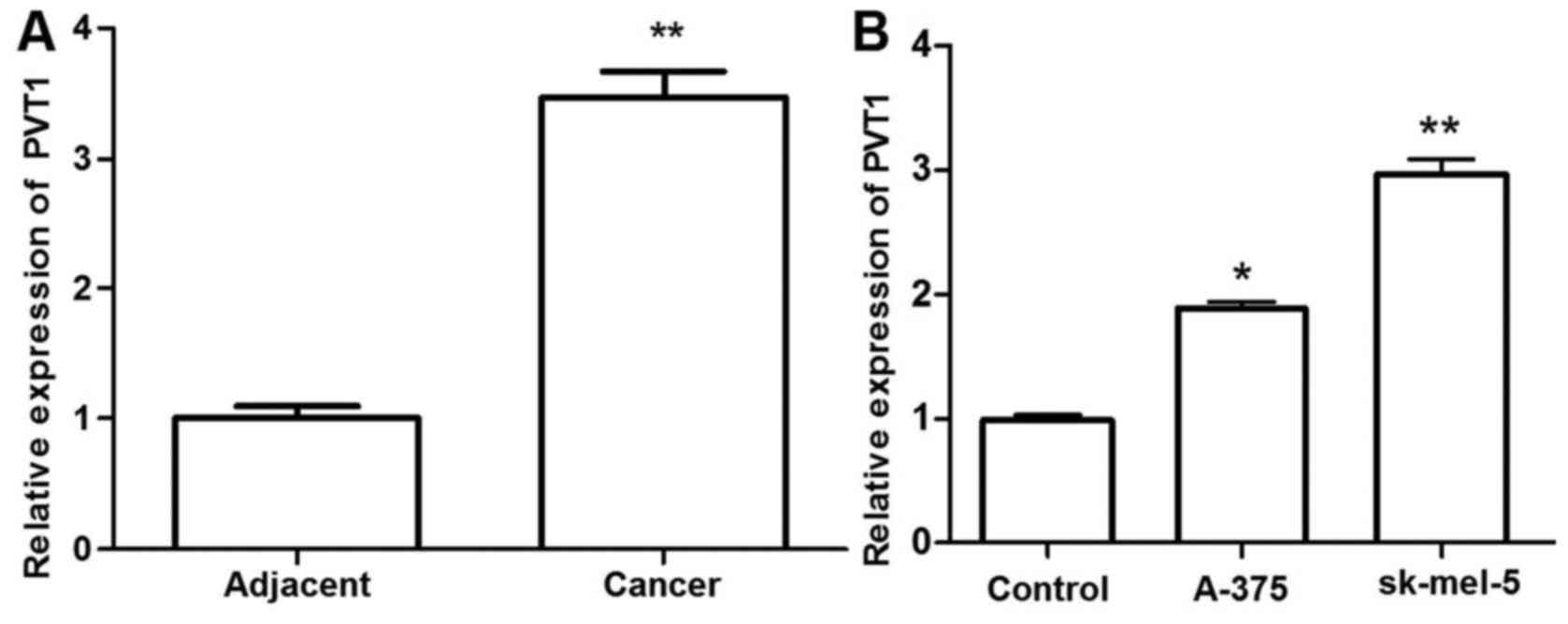
PVT1 is upregulated in melanoma tissue
and cells
To investigate the role of PVT1 in melanoma, the
expression levels of PVT1 in 37 paired melanoma tissues and
adjacent normal skin were determined by RT-qPCR and normalization
to GAPDH. Additionally, the expression levels of PVT1 in 3 melanoma
cell lines were detected. The relative expression level of PVT1 was
significantly higher in melanoma tissues and cells compared with
that in the adjacent normal tissues and control cells (P<0.01).
PVT1 exhibited the highest expression levels in the sk-mel-5 cells
(P<0.01). These results suggested that melanoma progression is
associated with the abnormal expression of PVT1 (Fig. 1).
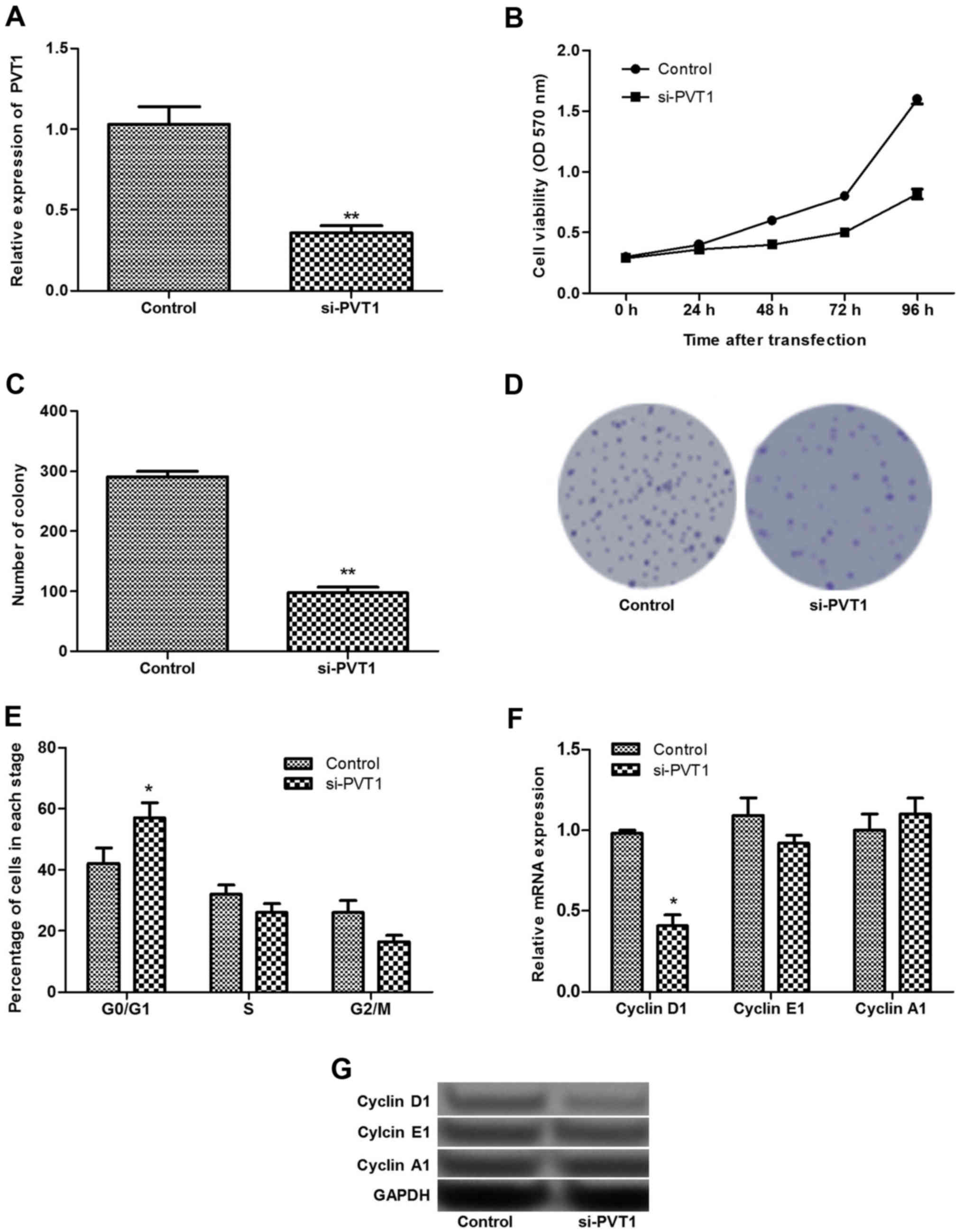
Silenced PVT1 inhibits cell proliferation
and arrests the cell cycle at the G0/G1 stage
As PVT1 exhibited the highest expression levels in
sk-mel-5 cells, these cells were selected for cell transfection and
the subsequent experiments. As shown in Fig. 2A, after sk-mel-5 cells were
transfected with siRNA-PVT1, the expression of PVT1 decreased
significantly (P<0.01). MTT and clonogenic assays found that
silenced PVT1 could inhibit cell proliferation significantly
(P<0.01; Fig. 2B–D).
Additionally, the cell cycle assay revealed that
silenced PVT1 significantly increased the percentage of cells at
the G0/G1 stage (P<0.05), suggesting that the cells were
arrested in the G0/G1 stage (Fig.
2E). To further verify these findings, the expression levels of
cell cycle-related proteins (cyclin D1, E1 and A1) were detected.
The results showed that the relative expression level of cyclin D1
decreased significantly in siRNA-PVT1 group compared with that in
the control group (P<0.05), while the expression of cyclin A1
and E1 did not change significantly between the two groups
(Fig. 2F and G).
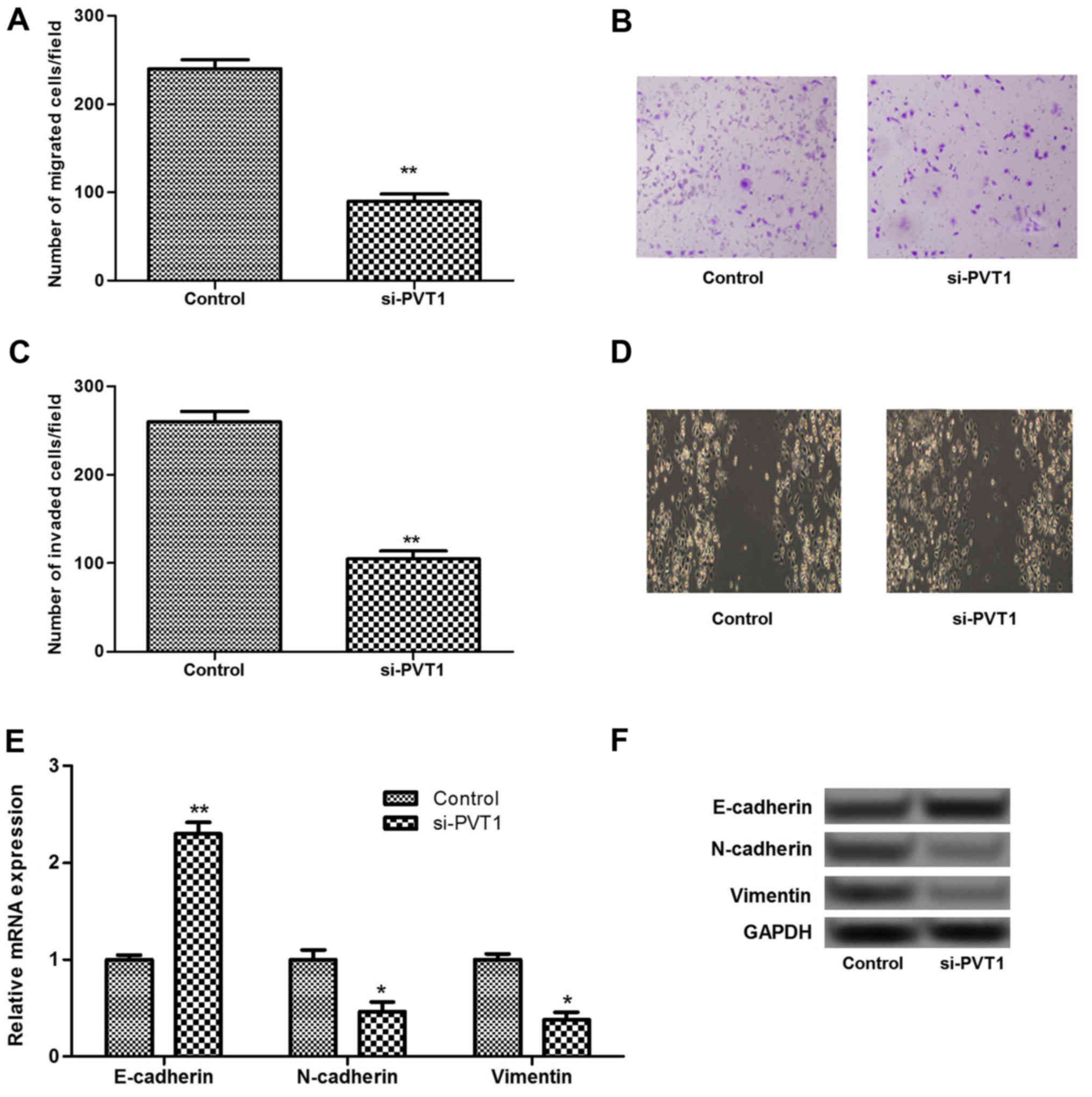
Silenced PVT1 suppresses cell migration
and invasion by inhibiting epithelial-mesenchymal transition
(EMT)
To investigate the role of PVT1 in melanoma cell
migration and invasion, Transwell and scratch assays were
performed. The results showed that silenced PVT1 significantly
inhibited the migration and invasion of sk-mel-5 cells compared
with that in the control (P<0.01; Fig. 3A–D). It has been reported that EMT
plays an important role in the progression of primary tumors toward
metastasis (18). Therefore, the
expression levels of EMT associated biomarkers, including
E-cadherin, N-cadherin and vimentin, were investigated in the
present study. As shown in Fig. 3E
and F, the expression of E-cadherin increased significantly in
the PVT1-silenced group compared with that in the control group
(P<0.01). By contrast, the expressions of N-cadherin and
vimentin decreased significantly in the PVT1-silenced group
(P<0.05). These results suggested that silenced PVT1 may inhibit
cell migration and invasion by inhibiting EMT.
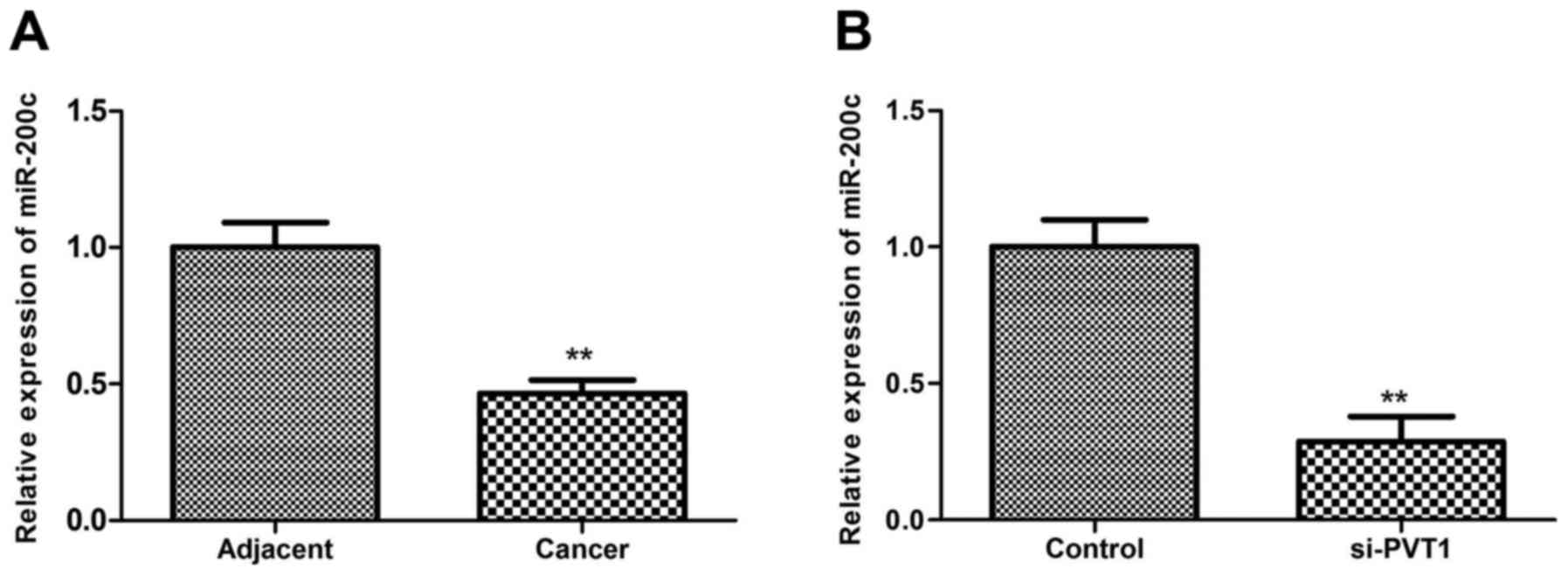
Effects of PVT1 suppression on miR-200c
expression
MicroRNA (miRNA/miR)-200 family members have been
found to play a key role in tumor development (19). miR-200c, a member of the miR-200
family, has been reported to be down-regulated in melanoma
(20). The present study
investigated the association between miR-200c and PVT1 in melanoma.
In accordance with the aforementioned previous study, the present
study also found that miR-200c was downregulated in melanoma tissue
compared with the adjacent tissue (P<0.01; Fig. 4A). Additionally, the results
showed that miR-200c expression decreased significantly when
sk-mel-5 cells were transfected with siRNA-PVT1 (P<0.01), which
suggested that silenced PVT1 may reduce the miR-200c expression
(Fig. 4B).
PVT1 expression is associated with EZH2
and regulates cell metastasis
A previous study found that EZH2 often functions as
an inhibitor in cancer suppressor genes, and that lncRNAs could
modulate tumor cells growth and metastasis via binding to EZH2
(21). Therefore, the present
study analyzed the interactions between PVT1 and EZH2 using an RIP
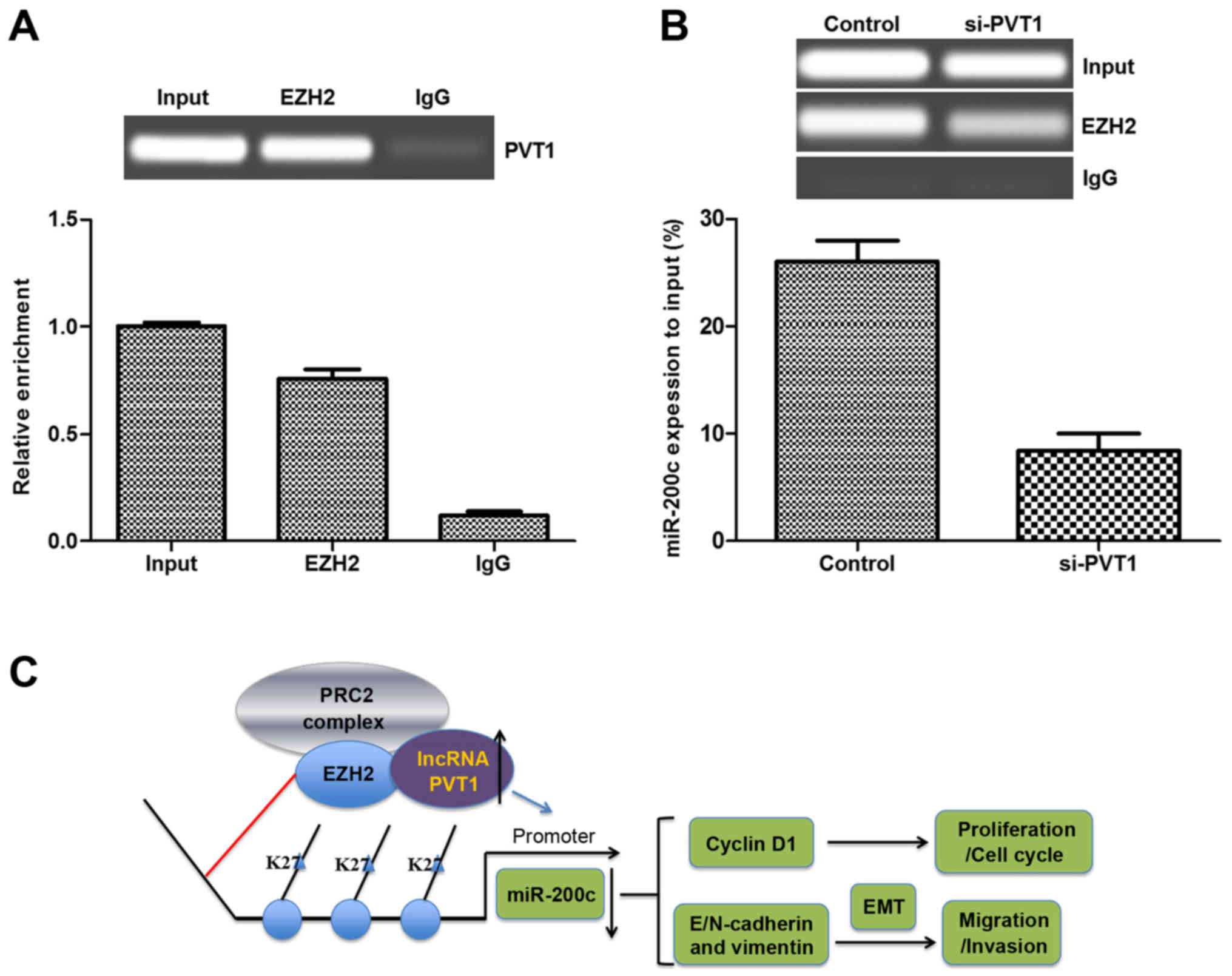
assay in melanoma cells. As shown in Fig. 5A, endogenous PVT1 was highly
enriched by EZH2 RIP compared with the negative control (IgG RIP),
implying that PVT1 may bind to EZH2. Additionally, a ChIP assay was
performed to determine if silenced PVT1 affected the expression of
miR-200c. As shown in Fig. 5B,
the expression of miR-200c decreased significantly in the
PVT1-silenced group compared with that in the control in the
sk-mel-5 cells, which suggested that silenced PVT1 may alter the
expression of miR-200c by targeting the promoter of miR-200c
(Fig. 5C). Taken together, these
results suggested that PVT1 silencing may inhibit the expression of
miR-200c to inhibit the metastasis of melanoma via binding to
EZH2.
Discussion
The present study examined the expression of PVT1 in
melanoma tissues and cell lines. The results showed that PVT1 was
overexpressed in melanoma tissues and cell lines compared with that
in controls. Silenced PVT1 significantly inhibited cell
proliferation, migration and invasion, and arrested the cell cycle
at the G0/G1 stage. Furthermore, silenced PVT1 affected the
expression of cell cycle-related proteins and EMT-associated
biomarkers. Silenced PVT1 also significantly reduced the expression
of miR-200c by binding to EZH2.
An increasing number of studies have determined a
role for lncRNAs in cancer (7,22).
In particular, an increasing amount of evidence has shown that PVT1
plays an important role in the progression of cancer, since it
resides in the well-known cancer risk region (11,23). For instance, Takahashi et
al (24) reported that
overexpression of PVT1 is involved in the prognosis of colorectal
cancer by regulating cell apoptosis. Wang et al (25) found that upregulated PVT1 promotes
stem cell proliferation in hepatocellular carcinoma. In accordance
with these findings, the present study found that PVT1 was
overexpressed in melanoma tissues and cell lines compared with that
in controls, which suggested the potential role of PVT1 in
melanoma.
To demonstrate the role of PVT1 in melanoma,
siRNA-mediated gene silencing was used to suppress the expression
of PVT1 in melanoma cells, and then the effect of silenced PVT1 on
cell proliferation was investigated. The results showed that
silenced PVT1 significantly inhibited cell proliferation, which was
in agreement with other studies. Kong et al (26) found that PVT1-knockdown
significantly inhibits gastric cancer cell proliferation in
vitro and in vivo. Yang et al (14) also found that knockdown of PVT1
inhibited cell proliferation in non-small cell lung cancer. As cell
proliferation is closely associated with the cell cycle,
accordingly, the effects of PVT1 expression on the melanoma cell
cycle and cell cycle-associated protein expression were analyzed.
It was found that the silencing of PVT1 arrested the melanoma cell
cycle at the G0/G1 stage, and that the expression of cyclin D1
decreased significantly. Cyclin D1 serves a vital role in promoting
cancer cell proliferation (27),
and has been considered as a pivotal element of malignant
transformation in human cancer (28). A previous study also found that
cyclin D1 was downregulated by silenced PVT1 in thyroid cancer
(29). Taken together, these
results indicated that downregulated cyclin D1 may be associated
with the suppression of melanoma cell proliferation via the arrest
of the cell cycle at the G0/G1 stage.
As aforementioned, melanoma has a proclivity to
metastasize and invade (3). EMT
is an biological process that gives rise to the dissemination of
single carcinoma cells from the sites of the primary tumors,
contributing to the progression of primary tumors toward metastasis
(18). Various carcinoma cell
lines undergo partial or complete EMT (30). Therefore, the present study
investigated the effect of PVT1 expression on melanoma cell
migration and invasion, and the expression of EMT-associated
biomarkers. The data showed that silenced PVT1 significantly
inhibited melanoma cell migration and invasion, increased the
expression of E-cadherin and decreased the expression of N-cadherin
and vimentin significantly. During melanoma development, loss of
functional E-cadherin accompanies the gain of expression of
N-cadherin (31). E-cadherin is a
tumor/invasion suppressor that has been found to be downregulated
in various human carcinomas, including melanoma (32). Disruption of E-cadherin-mediated
homotypic cell adhesion facilitates tumor invasion (33). Additionally, one previous study
also found that overexpression of vimentin in primary melanoma
tissues may cause a high risk of hematogenous metastasis (34). We speculate that silenced PVT1 may
inhibit melanoma cell migration and invasion by regulating EMT.
miR-200c belongs to the miR-200 family, which has
been found to be involved in cancer progression (35). Particularly, the miR-200 family
has been shown to regulate EMT and cell migration in cancer cell
lines (36). Importantly,
miR-200c has been reported to be downregulated in melanoma
(20). In the present study, it
was found that silenced PVT1 reduced the expression of miR-200c,
indicating that silencing PVT1 may suppress the development of
melanoma by downregulating miR-200c expression.
Recently, the histone methyltransferase EZH2 has
attracted considerable attention due to its dysregulation in human
cancer (37). Notably, PVT1 was
reported to promote cell proliferation in gastric cancer by the
recruitment of EZH2 (26). Thus,
the interaction between EZH2 and PVT1 in melanoma was investigated
in the present study and PVT1 was found to be enriched by EZH2,
indicating the interaction between PVT1 and EZH2 in melanoma.
Moreover, Ning et al (38)
found that EZH2-mediated methylation inhibits the expression of
miR-200 and promotes tumor progression. Therefore, a ChIP assay was
performed in the present study to investigate whether silenced PVT1
affected the expression of miR-200c, and the resultant data found
that the expression of miR-200c was decreased significantly by the
silencing of PVT1 in EZH2-treated melanoma cell lines, suggesting
that PVT1 suppression may decrease miR-200c level and that PVT1 may
be required for EZH2 binding to the miR-200c promoter. Taken
together, these results led to the hypothesis that silenced PVT1
may inhibit miR-200c expression via binding to EZH2.
In conclusion, the present study indicates that PVT1
overexpression plays an acceleratory role in the development of
melanoma. Downregulation of PVT1 may inhibit melanoma cell
proliferation by regulating the cell cycle, and may inhibit cell
migration and invasion through the regulation of EMT, which may be
achieved by inhibiting the expression of miR-200c via the binding
of PVT1 to EZH2. Therefore, lncRNA PVT1 may serve as an important
target in the treatment of melanoma.
Acknowledgments
The authors would like to thank Professor Xiulian Xu
and Dr Wai Zhang (Chinese Academy of Medical Sciences and Peking
Union Medical College, Beijing, China) for supporting this study.
The authors would also like to thank Dr Jiang Hong (Key Laboratory
of Cardiovascular Remodeling and Function Research, Shandong,
China) for providing technical assistance.
References
|
1
|
Gray-Schopfer V, Wellbrock C and Marais R:
Melanoma biology and new targeted therapy. Nature. 445:851–857.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2016. CA Cancer J Clin. 66:7–30. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Arozarena I, Sanchez-Laorden B, Packer L,
Hidalgo-Carcedo C, Hayward R, Viros A, Sahai E and Marais R:
Oncogenic BRAF induces melanoma cell invasion by downregulating the
cGMP-specific phosphodiesterase PDE5A. Cancer Cell. 19:45–57. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cummins DL, Cummins JM, Pantle H,
Silverman MA, Leonard AL and Chanmugam A: Cutaneous malignant
melanoma. Mayo Clin Proc. 81:500–507. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tang L, Zhang W, Su B and Yu B: Long
noncoding RNA HOTAIR is associated with motility, invasion, and
metastatic potential of metastatic melanoma. BioMed Res Int.
2013:251098. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Muers M: RNA: genome-wide views of long
non-coding RNAs. Nat Rev Genet. 12:742–743. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shore AN, Herschkowitz JI and Rosen JM:
Noncoding RNAs involved in mammary gland development and
tumorigenesis: there’s a long way to go. J Mammary Gland Biol
Neoplasia. 17:43–58. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Huarte M, Guttman M, Feldser D, Garber M,
Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M,
et al: A large intergenic noncoding RNA induced by p53 mediates
global gene repression in the p53 response. Cell. 142:409–419.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Schmidt K, Joyce CE, Buquicchio F, Brown
A, Ritz J, Distel RJ, Yoon CH and Novina CD: The lncRNA SLNCR1
mediates melanoma invasion through a conserved SRA1-like region.
Cell Reports. 15:2025–2037. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Flockhart RJ, Webster DE, Qu K,
Mascarenhas N, Kovalski J, Kretz M and Khavari PA:
BRAFV600E remodels the melanocyte transcriptome and
induces BANCR to regulate melanoma cell migration. Genome Res.
22:1006–1014. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Colombo T, Farina L, Macino G and Paci P:
PVT1: a rising star among oncogenic long noncoding RNAs. BioMed Res
Int. 2015:3042082015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Guan Y, Kuo WL, Stilwell JL, Takano H,
Lapuk AV, Fridlyand J, Mao JH, Yu M, Miller MA, Santos JL, et al:
Amplification of PVT1 contributes to the pathophysiology of ovarian
and breast cancer. Clin Cancer Res. 13:5745–5755. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Barsotti AM, Beckerman R, Laptenko O,
Huppi K, Caplen NJ and Prives C: p53-dependent induction of PVT1
and miR-1204. J Biol Chem. 287:2509–2519. 2012. View Article : Google Scholar :
|
|
14
|
Yang YR, Zang SZ, Zhong CL, Li YX, Zhao SS
and Feng XJ: Increased expression of the lncRNA PVT1 promotes
tumori-genesis in non-small cell lung cancer. Int J Clin Exp
Pathol. 7:6929–6935. 2014.
|
|
15
|
Zhou Q, Chen J, Feng J and Wang J: Long
noncoding RNA PVT1 modulates thyroid cancer cell proliferation by
recruiting EZH2 and regulating thyroid-stimulating hormone receptor
(TSHR). Tumour Biol. 37:3105–3113. 2016. View Article : Google Scholar
|
|
16
|
Franken NAP, Rodermond HM, Stap J, Haveman
J and van Bree C: Clonogenic assay of cells in vitro. Nat Protoc.
1:2315–2319. 2006. View Article : Google Scholar
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar
|
|
18
|
Biddle A and Mackenzie IC: Cancer stem
cells and EMT in carcinoma. Cancer Metastasis Rev. 31:285–293.
2012. View Article : Google Scholar
|
|
19
|
Park SM, Gaur AB, Lengyel E and Peter ME:
The miR-200 family determines the epithelial phenotype of cancer
cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes
Dev. 22:894–907. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu Y, Brenn T, Brown ER, Doherty V and
Melton DW: Differential expression of microRNAs during melanoma
progression: miR-200c, miR-205 and miR-211 are downregulated in
melanoma and act as tumour suppressors. Br J Cancer. 106:553–561.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chase A and Cross NC: Aberrations of EZH2
in cancer. Clin Cancer Res. 17:2613–2618. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chen H, Xu J, Hong J, Tang R, Zhang X and
Fang JY: Long noncoding RNA profiles identify five distinct
molecular subtypes of colorectal cancer with clinical relevance.
Mol Oncol. 8:1393–1403. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang C, Yu W, Wang Q, Cui H, Wang Y,
Zhang L, Han F and Huang T: Increased expression of the lncRNA PVT1
is associated with poor prognosis in pancreatic cancer patients.
Minerva Med. 106:143–149. 2015.PubMed/NCBI
|
|
24
|
Takahashi Y, Sawada G, Kurashige J, Uchi
R, Matsumura T, Ueo H, Takano Y, Eguchi H, Sudo T, Sugimachi K, et
al: Amplification of PVT-1 is involved in poor prognosis via
apoptosis inhibition in colorectal cancers. Br J Cancer.
110:164–171. 2014. View Article : Google Scholar :
|
|
25
|
Wang F, Yuan JH, Wang SB, Yang F, Yuan SX,
Ye C, Yang N, Zhou WP, Li WL, Li W, et al: Oncofetal long noncoding
RNA PVT1 promotes proliferation and stem cell-like property of
hepatocellular carcinoma cells by stabilizing NOP2. Hepatology.
60:1278–1290. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kong R, Zhang EB, Yin DD, You LH, Xu TP,
Chen WM, Xia R, Wan L, Sun M, Wang ZX, et al: Long noncoding RNA
PVT1 indicates a poor prognosis of gastric cancer and promotes cell
proliferation through epigenetically regulating p15 and p16. Mol
Cancer. 14:822015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Gautschi O, Ratschiller D, Gugger M,
Betticher DC and Heighway J: Cyclin D1 in non-small cell lung
cancer: a key driver of malignant transformation. Lung Cancer.
55:1–14. 2007. View Article : Google Scholar
|
|
28
|
Li Z, Tian T, Lv F, Chang Y, Wang X, Zhang
L, Li X, Li L, Ma W, Wu J, et al: Six1 promotes proliferation of
pancreatic cancer cells via upregulation of cyclin D1 expression.
PLoS One. 8:e592032013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Khoo ML, Beasley NJ, Ezzat S, Freeman JL
and Asa SL: Overexpression of cyclin D1 and underexpression of p27
predict lymph node metastases in papillary thyroid carcinoma. J
Clin Endocrinol Metab. 87:1814–1818. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Thiery JP: Epithelial-mesenchymal
transitions in tumour progression. Nat Rev Cancer. 2:442–454. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li G, Satyamoorthy K and Herlyn M:
N-cadherin-mediated intercellular interactions promote survival and
migration of melanoma cells. Cancer Res. 61:3819–3825.
2001.PubMed/NCBI
|
|
32
|
Hsu MY, Meier FE, Nesbit M, Hsu JY, Van
Belle P, Elder DE and Herlyn M: E-cadherin expression in melanoma
cells restores keratinocyte-mediated growth control and
down-regulates expression of invasion-related adhesion receptors.
Am J Pathol. 156:1515–1525. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Slagle BL, Zhou YZ, Birchmeier W and
Scorsone KA: Deletion of the E-cadherin gene in hepatitis B
virus-positive Chinese hepatocellular carcinomas. Hepatology.
18:757–762. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li M, Zhang B, Sun B, Wang X, Ban X, Sun
T, Liu Z and Zhao X: A novel function for vimentin: the potential
biomarker for predicting melanoma hematogenous metastasis. J Exp
Clin Cancer Res. 29:1092010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
du Rieu MC, Torrisani J, Selves J, Al
Saati T, Souque A, Dufresne M, Tsongalis GJ, Suriawinata AA,
Carrère N, Buscail L, et al: MicroRNA-21 is induced early in
pancreatic ductal adenocarcinoma precursor lesions. Clin Chem.
56:603–612. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Park SM, Gaur AB, Lengyel E and Peter ME:
The miR-200 family determines the epithelial phenotype of cancer
cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes
Dev. 22:894–907. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tao T, Liu D, Liu C, Xu B, Chen S, Yin Y,
Ang L, Huang Y, Zhang X and Chen M: Autoregulatory feedback loop of
EZH2/miR-200c/E2F3 as a driving force for prostate cancer
development. Biochim Biophys Acta. 1839:858–865. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ning X, Shi Z, Liu X, Zhang A, Han L,
Jiang K, Kang C and Zhang Q: DNMT1 and EZH2 mediated methylation
silences the microRNA-200b/a/429 gene and promotes tumor
progression. Cancer Lett. 359:198–205. 2015. View Article : Google Scholar : PubMed/NCBI
|