Introduction
Endometriosis is an estrogen-driven inflammatory
condition, defined by a misplacement of endometrium outside of the
uterine cavity, most commonly in the pelvic cavity and is one of
the common causes of infertility (1,2).
It affects 6 to 10% of women of reproductive age (3), but with varying symptoms including
severe dysmenorrhea (4,5), chronic pelvic pain, dysfunctional
uterine bleeding (5), as well as
urinary tract and gastrointestinal symptoms (6). Notably, endometriosis possesses many
features of a benign neoplastic process with the potential for
malignant transformation (7).
Endometriosis is a major problem of women's health, which affects
dramatically the quality of life. It has been accepted that
multiple factors contribute to the development of this condition,
including genetic and environmental ones. However, the exact
molecular and pathophysiological pathways leading to endometriosis
are still unclear, as at present only various hypotheses have been
suggested (8–10). Thus, it may be assumed that all
cases of endometriosis are not able to be explained by one theory
only. Genetic factors contribute to the heritability of
endometriosis (11–14) and the overall heritability has
been estimated at approximately 50%, as shown from twin studies
(15,16). Notably, the impact of epigenetics
in endometriosis has been under investigation in recent years and
significant progress regarding DNA methylation and histone
post-translational modifications has been achieved (17,18). The epigenetic disruption of gene
expression plays an important role in the development of
endometriosis through interaction with environmental changes.
Candidate gene association studies, genome-wide
association studies and various meta-analyses have led to the
identification of many endometriosis-risk loci that may be
initially involved in the pathogenesis of endometriosis (19–24). These gene loci have been
categorized according to the function of their gene products, i.e.,
growth factors, matrix remodeling, cell cycle regulation and
signaling, oncogenes, hormone receptors and metabolism, adhesion
molecules, transcription regulation, cytokines, inflammation,
immune and oxidative stress (reviewed in ref. 25). However, a portion of these data
has been rather disappointing due to the absence of replication in
independent populations (26). At
present, 19 single nucleotide polymorphisms (SNPs) associated with
endometriosis have been identified, which can explain approximately
5.19% of the disease variance (27). The list of novel
endometriosis-associated loci is enriching considering that recent
studies are focused mainly on the severe stages of the disease
(stage III/IV endometriosis), thus suggesting the greater genetic
burden of moderate to severe endometriosis cases compared to that
of minimal or mild disease (stage I/II) (22,28).
Previous findings have shown that various cytokines
may be used as potential biomarkers for the diagnosis of
endometriosis, given that they were detectable in the serum and
peritoneal fluid (29). In this
framework, IL-16 has been found in amniotic fluid with its levels
declining over gestation, while IL-16 transcripts were detected in
whole tissue extracts of fetal gut, skin and placenta (30,31). Interleukin-16 (IL-16), also known
as a lymphocyte chemoattractant factor, is a polypeptide
proinflammatory cytokine that plays a pivotal (decisive) role in
most immune and inflammatory responses as well as in the
pathogenesis of endometriosis (32). It is produced by a variety of cell
types previously found in association with complex disorders and it
is now clear that this cytokine plays a critical role in the
regulation of cellular functions. The precise mechanism by which
IL-16 functions as an inflammatory mediator is still under
investigation and not fully clarified.
Accumulated data suggest that IL-16 activates T
lymphocytes, thus resulting in the secretion of several
proinflammatory cytokines (33).
It is produced mainly by CD8 lymphocytes as a 67-kDa precursor
protein (34). Human IL-16 is
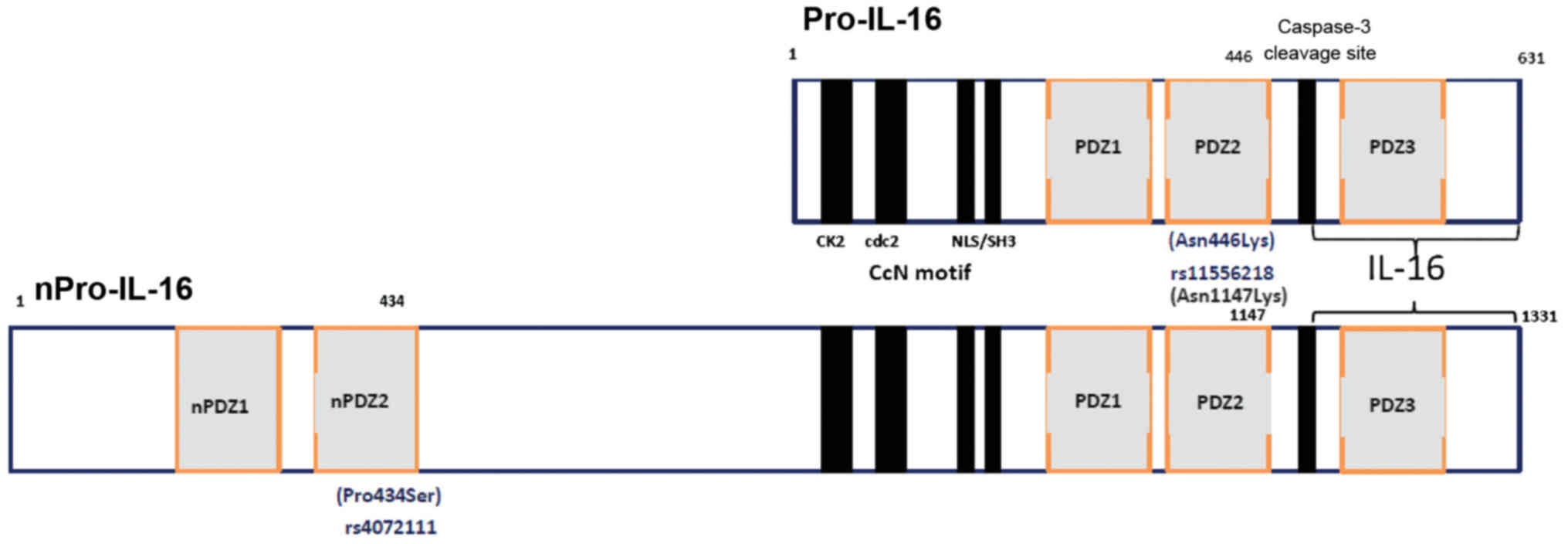
normally produced as a 631-amino acid precursor protein, Pro-IL-16,
which is then cleaved at the subsequent step by the enzyme
caspase-3 to release the biologically active C-terminal domain,
consisting of 121 amino acids (35–37). Two functional polymorphisms in
this gene (rs4072111 C/T and rs11556218 T/G) have been reported to
be associated with various cardiovascular (38–40), neurodegenerative (41), infectious (42), inflammatory or autoimmune diseases
(43–46), as well as with various types of
cancer (47–50). Of the two, rs11556218 is a
missense exon-SNP, located in the exon 6 region, leading to an
amino acid change (Asn446Lys) on position 446 of the shorter
isomorph 2 (631 aminoacids) of Pro-IL-16 (Fig. 1), which may alter protein
structure and function. The rs4072111 is another missense SNP
(Pro434Ser) appearing on position 434 of the second PDZ domain of
the longer isomorph 1, i.e., the neuronal nPro-IL-16 (1,331 amino
acids) (Fig. 1). PDZ domains were
originally identified as repeated sequences conserved in two
proteins, postsynaptic density protein PSD95, Drosophila
discs large tumor suppressor protein DLG (51). PDZ domains are now known to be
present in many proteins (52).
Additionally, they function as motifs for protein-protein
interaction (PPI) (53,54). Proteins with PDZ motifs have been
associated with neoplasia and alterations in cell proliferation as
originally described (51). While
the majority of PDZ-containing proteins appear to participate in
PPIs in the cytoplasm at the sites of cell-cell contact, a number
of PDZ domain-containing proteins have been identified to localize
in the nucleus (52–54).
Encouraged by the IL-16 association with
endometriosis detected by Azimzadeh et al (55) recently, we conducted the current
study to investigate whether rs4072111 and rs11556218 SNPs of the
IL-16 gene were associated with the risk for endometriosis
and/or with progression to the severe stages (III–IV) of this
condition in the Greek population. Furthermore, we attempted to
detect any ethnic-specific differences regarding the genetic
association of these SNPs with endometriosis, considering that
population differences for endometriosis have been reported
previously in terms of genetic susceptibility and disease
manifestations (56–58).
Patients and methods
Patient population and study design
In this case control association study, 305 women
were enrolled (159 endometriosis patients and 146 controls)
followed in the Department of Obstetrics and Gynecology of
Venizeleion General Hospital of Heraklion (Heraklion, Crete). All
the women had undergone surgery in the aforementioned tertiary care
centre. The average age of the Greek endometriosis and control
cohorts was 32.25±7.1 and 29.49±6.7 years, respectively. The women
with endometriosis were diagnosed surgically (laparotomy or
laparoscopy), and the disease was confirmed histologically from
biopsies. Staging of the disease was performed according to the
revised American Fertility Society classification (59). All the members of the control
group had given birth to 2–5 (2.3±0.6) children and had no previous
medical record of chronic pelvic pain, dysmenorrhea, or
dyspareunia. According to the revised American Fertility Society
Classification (1985), 81 (50.94%) stage I–II patients and 78
(49.06%) patients had moderate to severe endometriosis (stage
III–IV). All the subjects were of self-reported Greek origin.
Written informed consent was obtained from both patients and
controls. The study was performed in the Section of Molecular
Pathology and Human Genetics of the Medical School of Crete, after
obtaining the approval of the Research Committee of the Venizeleion
General Hospital of Heraklion and was carried out in compliance
with the declaration of Helsinki.
Genotyping
Whole blood was collected preoperatively in
ethylenediaminetetraacetic acid (EDTA)-containing tubes. Genomic
DNA was isolated from peripheral blood leukocytes by using the
commercial kit Invitrogen (PureLink® Genomic DNA Mini
kit; Invitrogen Life Technologies, Carlsbad, CA, USA) according to
the manufacturer's instructions. The extracted DNA was stored at
−20°C until analyzed. Genotyping of two common SNPs in the
IL-16 gene, rs4072111 (Pro434Ser) and rs11556218 (Asn446Lys)
was performed by following the restriction fragment length
polymorphism (RFLP) approach, by using BsmAI and
NdeI, respectively, as described elsewhere (47,55). Amplification of the genomic
fragments harboring the polymorphic sites was carried out using the
GoTaq polymerase provided by Promega Corporation (Madison, WI,
USA). Briefly, the upstream primers 5′-CACTGTGATCCCGGTCCAGTC-3′ and
5′-GCTCAGGTTCACAGAGTGTTTCCATA-3′ as well as the downstream primers
5′-TTCAGGTACAAACCCAGCCAGC-3′ and 5′-TGTGACAATCACAGCTTGCCTG-3′,
based on the human IL-16 gene sequence (accession no.
NC_000015.10) were used to generate the regions of the rs4072111
(164 bp) (C>T) and rs11556218 (171 bp) (T>G) SNPs of the
IL-16 gene, respectively.
Polymerase chain reactions (PCR) were carried out in
the final volume of 25 μl containing: 10X PCR buffer and 2
mM MgCl2 (both from Roche Diagnostics GmbH, Mannheim,
Germany), 0.4 mM of each dNTP (Fermentas, St. Leon-Rot, Germany), 5
pmol of each primer, 50 ng template DNA, 1 unit Taq DNA polymerase
(Roche Diagnostics GmbH) and sterile distilled water up to 25
μl. A hot start was used with initial heating at 94°C for 5
min and then 30 cycles of denaturing (at 94°C for 30 sec),
annealing for 30 sec (60°C for rs11556218 and 60°C for rs4072111)
and chain extension (at 72°C for 30 sec), followed with a final
extension step at 72°C for 5 min. Both undigested and digested PCR
products were visualized in 2.5% agarose gel stained with ethidium
bromide in reference to a molecular weight marker (100 bp DNA
ladder; Invitrogen Life Technologies). The genotyping of the
samples for rs4072111 was performed by digesting the 164-bp PCR
product with BsmAI restriction enzyme (New England BioLabs,
Inc., Beverly, MA, USA), which digested the DNA that was amplified
from the 'T' allele, thus generating two fragments of 140 and
24-bp. The presence of allele 'C' resulted in the absence of the
BsmAI restriction site (47). In addition, the genotyping of the
samples for rs11556218 was performed by digesting the 171-bp PCR
product with NdeI restriction enzyme (New England BioLabs,
Inc.), which digested the DNA that was amplified from the 'G'
allele, thus generating two fragments of 147 and 24-bp. The
presence of allele 'T' resulted in the absence of the NdeI
restriction site (47). Genotypes
were scored blindly, and analysis of all the ambiguous samples was
repeated. Furthermore, to ensure accuracy of the results, 10% of
the samples were amplified twice.
Construction of IL-16 domain
three-dimensional (3D) model
SNPs bioinformatics analysis was performed using
NCBI dbSNP (for nucleotide sequence analysis), UNIPROT (for protein
sequence analysis) and PDB (for protein structure analysis)
databases. PYMOL (DeLano Scientific, San Carlos, CA, USA) was used
for 3D structural positioning, mutation analysis and visualization.
The crystal structures of IL-16 PDZ and PDZ12 domains (PDB codes,
1X6D2QT5 and 2KA9) were used as the initial models. The 3D
structure modeling was performed using SWISS-MODEL (60).
Statistical analysis
All cases and controls used in the analysis were
unrelated. The GraphPad Prism statistical program (GraphPad
Software, San Diego, CA, USA) was used in the framework of the
analyses performed. A two-tailed P-value <0.05 was defined as
statistically significant. Odds ratios (OR) and 95% confidence
intervals (CI) were calculated. The χ2 test, with one or
two degrees of freedom or Fisher's exact test was used to examine
differences of genotype and allele frequencies between patients and
controls, where all SNPs had a call rate of >98%. The possible
deviation from Hardy-Weinberg equilibrium (HWE) was performed by
using the program 'Calculate' (copyright TRG, SR, INMD, 2008). The
distribution of genotypes in case group for both SNPs examined was
found to be under HWE (P>0.01).
Results
rs11556218 IL-16 T/G SNP
In the case of the rs11556218 SNP, a statistical
significant difference was found in the frequencies of GG and GT
genotypes in patients and controls (P<0.0001, OR=7.01; 95% CI,
3.65–13.46; and P<0.0001, OR=4.17; 95% CI, 2.30–7.56,
respectively) (Table I).
Additionally, we observed a significant difference in the
distribution of the 'G' allele between the endometriosis and
control groups (P<0.0001, OR=3.02; 95% CI, 2.17–4.20), being
associated with an increased susceptibility for endometriosis.
 | Table IGenotypes and alleles frequency of
the IL-16 rs11556218 SNP analyzed in 159 women with
endometriosis and 146 healthy controls. |
Table I
Genotypes and alleles frequency of
the IL-16 rs11556218 SNP analyzed in 159 women with
endometriosis and 146 healthy controls.
| rs11556218 | Patients | Controls | P-value | OR (95% CI) |
|---|
| Genotypes | n=159 | n=146 | | |
| G/G | 64 (40.25%) | 27 (18.49%) |
<0.0001 | 7.01
(3.65–13.46) |
| G/T | 72 (45.28%) | 51 (34.93%) |
<0.0001 | 4.17
(2.30–7.56) |
| T/T | 23 (14.46%) | 68 (46.58%) | | 1.00
(Reference) |
| Alleles | n=318 | n=292 | | |
| G | 200 (62.89%) | 105 (35.96%) |
<0.0001 | 3.02
(2.17–4.20) |
| T | 118 (37.11%) | 187 (64.04%) | | 1.00
(Reference) |
Of note, when patients were analyzed according to
the severity of the disease, a significant association was detected
regarding the GG and GT genotypes of this SNP in patients with
stage III/IV of the disease and controls (P=0.0004, OR=3.92; 95%
CI, 1.87–8.22; and P=0.0178, OR=2.37; 95% CI, 1.20–4.69,
respectively) (Table II). The
'G' allele was also associated with endometriosis in this analysis
(P<0.0001, OR=2.30; 95% CI, 1.55–3.43).
 | Table IIGenotypes and alleles frequency of
the IL-16 rs11556218 SNP analyzed in 78 women with
endometriosis (stage III and IV) and 146 healthy controls. |
Table II
Genotypes and alleles frequency of
the IL-16 rs11556218 SNP analyzed in 78 women with
endometriosis (stage III and IV) and 146 healthy controls.
| rs11556218 | Patients | Controls | P-value | OR (95% CI) |
|---|
| Genotypes | n=78 | n=146 | | |
| G/G | 28 (35.88%) | 27 (18.49%) | 0.0004 | 3.92
(1.87–8.22) |
| G/T | 32 (41.04%) | 51 (34.93%) | 0.0178 | 2.37
(1.20–4.69) |
| T/T | 18 (23.08%) | 68 (46.58%) | | 1.00
(Reference) |
| Alleles | n=156 | n=292 | | |
| G | 88 (56.41%) | 105 (35.96%) |
<0.0001 | 2.30
(1.55–3.43) |
| T | 68 (43.59%) | 187 (64.04%) | | 1.00
(Reference) |
The rs4072111 IL-16 SNP
We further evaluated the effect of the rs4072111 SNP
in the development of endometriosis. Based on the genotyping as
well as the allelic data obtained, no association with
endometriosis was detected either for CC genotype or for allele 'C'
(Table III) (P=0.46, OR=3.51;
95% CI, 0.14–87.07; and P=0.15, OR=1.54; 95% CI, 0.87–2.74,
respectively). Furthermore, when patients were analyzed according
to the severity of the disease (stage III/IV), no association was
detected either (data not shown). The genotyping success for all
the SNPs analyzed was >98%.
 | Table IIIGenotypes and alleles frequency of
the IL-16 rs4072111 SNP analyzed in 159 women with
endometriosis and 146 healthy controls. |
Table III
Genotypes and alleles frequency of
the IL-16 rs4072111 SNP analyzed in 159 women with
endometriosis and 146 healthy controls.
| rs4072111 | Patients | Controls | P-value | OR (95% CI) |
|---|
| Genotypes | n=159 | n=146 | | |
| C/C | 137 (86.16%) | 117 (80.14%) | 0.46 | 3.51
(0.14–87.07) |
| C/T | 22 (13.84%) | 28 (19.18%) | 1 | 2.37
(0.09–61.01) |
| T/T | 0 (0%) | 1 (0.68%) | | |
| Alleles | n=318 | n=292 | | |
| C | 296 (93.08%) | 262 (89.72%) | 0.15 | 1.54
(0.87–2.74) |
| T | 22 (6.92%) | 30 (10.28%) | | |
Developing 3D models of IL-16
protein
We located the rs4072111 and rs11556218 SNPs on the
aminoacid sequences of the isomorphs of Pro-IL-16 (Fig. 1). We then attempted to approach
the functional role of both SNPs under investigation, by
constructing 3D models of the respective PDZ domains of nPro-IL-16
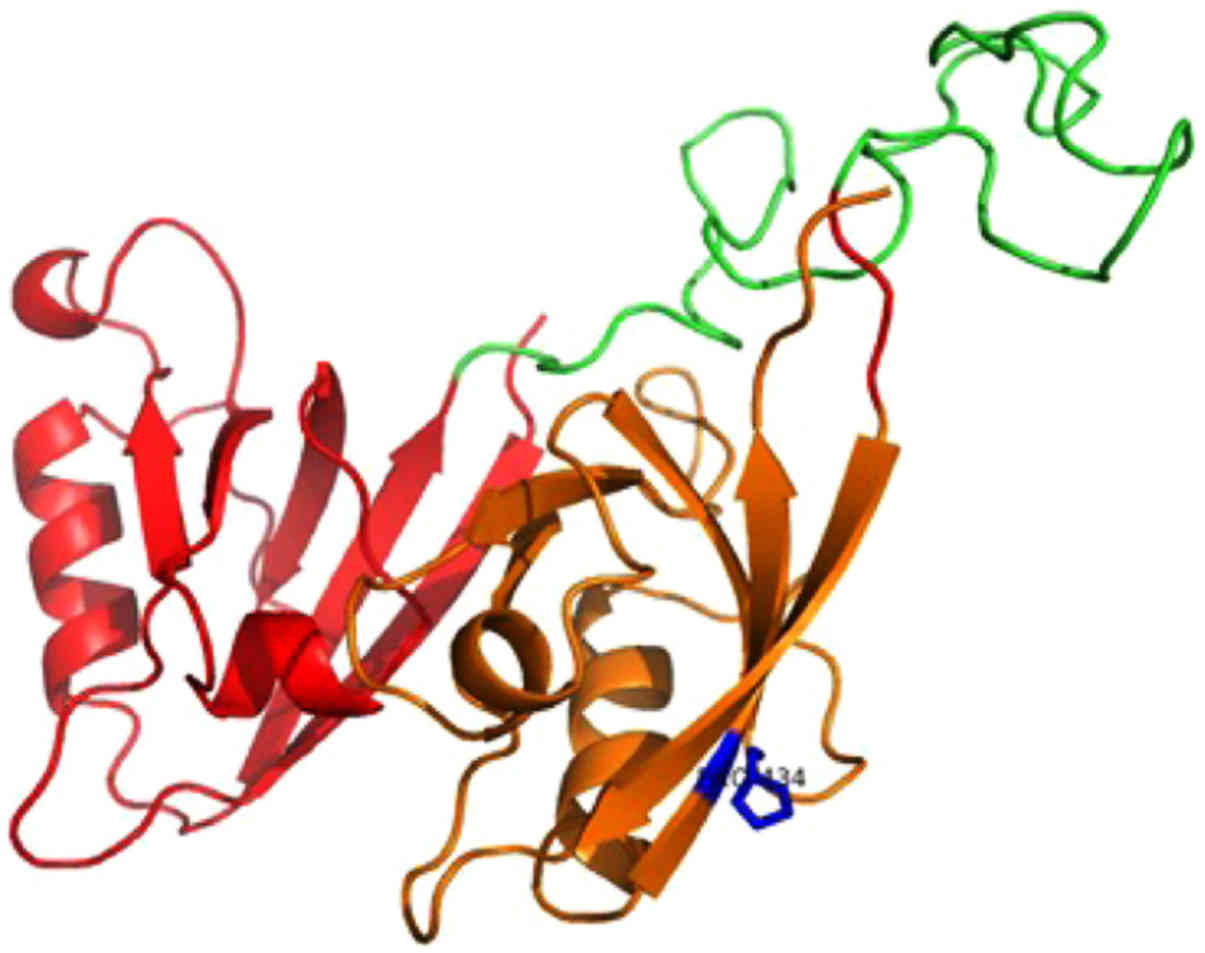
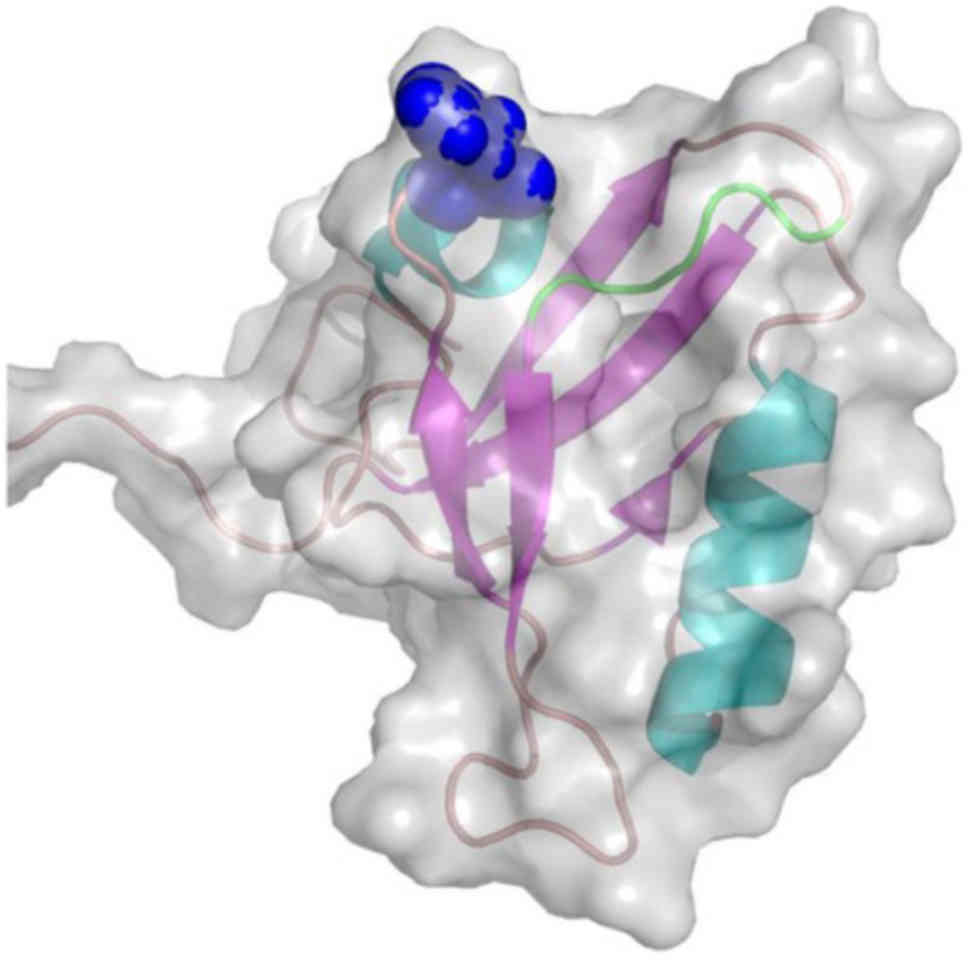
isomorph 1 (Fig. 2) and Pro-IL-16
isomorph 2 (Fig. 3) proteins. The
first model consists of the N-terminal two PDZ domains (residues
211-443) of nPro-IL-16. The location of the mutant residue is
predicted on the last β-strand of the second PDZ domain (Fig. 2). The second model is based on the
NMR structure of the second PDZ domain (PDB code, 1X6D) (61) of the N-terminal Pro-IL-16 isomorph
2. The rs11556218SNP-derived mutant Asn446Lys is located on the
β3/α1 loop, at the entrance of the recognition cavity of the PDZ
domain and in close proximity to the GLGF motif, known as the
'carboxylate binding loop' (Fig.
3).
Discussion
Recent advances in genetics and relevant technology
during the current post-genomic era resulted in the identification
and a better understanding of genetic risk factors associated with
endometriosis. In the present study, we investigated the role of
two SNPs of the IL-16 gene with regard to risk of
endometriosis susceptibility in Greek women. To the best of our
knowledge, this is the first study to screen for IL-16 gene
polymorphisms in patients with endometriosis in a European
population. The IL-16 gene is located on chromosome 15
(15q26.1) (62).
Although GWAS have detected numerous endometriosis
susceptibility genes, it is clear that there are many differences
in genetic associations with endometriosis across different world
populations and, therefore, it is important to study the genetic
basis of the disease in multiple populations (63,64). This would be particularly
important considering that some major endometriosis risk factors
such as WNT4, VEZT and FSHB were shown by a
recent study of our group, focused on the same (Greek) population
analyzed in the present study, to have a specific geographic
distribution (58). In this
framework, genetic association studies involving rs4072111 and
rs11556218 SNPs of the IL-16 gene in the Greek population
showed that rs11556218 only is strongly associated with an
increased susceptibility for the development of endometriosis at
both the genotypic and allelic level.
The results of a recent study conducted in Iran
showed that genotype and allelic distribution in the two
IL-16 exonic polymorphisms rs4072111 and rs11556218 was
significantly different between endometriosis patients and healthy
individuals (65). Of note,
allele 'G' of rs11556218 was found to be protective for
endometriosis and did not increase the risk for the disease, as
found in the Greek population in the present study. No significant
differences were detected in the genotype and allele frequencies of
the rs11556218 SNP between patients with endometriosis and controls
either in a Chinese (66) or a
Korean population (67). Of note,
the allele frequencies that we obtained for the Greek control
population for rs11556218 SNP vary significantly in comparison with
Iranian (55) and Chinese
(18,40,47,66), as published in the literature.
These observations underline the importance of assessing genetic
variants in different ethnic and/or racial populations in any
attempt to approach the genetic basis of endometriosis and the
specific effects of various alleles in different populations.
Genetic variation in the DNA sequence of the
IL-16 gene may lead to altered cytokine production and/or
activity, and this variation may modulate an individual's
susceptibility to endometriosis. Notably, in patients with
colorectal cancer or gastric cancer, IL-16 serum levels were
significantly higher than those in the healthy controls, although
no significant association between IL-16 polymorphisms and
serum levels of IL-16 was observed (47). Several cytokines have been shown
to appear differences in women with endometriosis in comparison
with controls. Thus, among members of the interleukin family
evaluated, IL-16 exhibited elevated levels (68). Nevertheless, few studies have
directly examined the mechanisms by which IL-16 is involved in the
development and progression of endometriosis. IL-6 was found to be
elevated in the peritoneal fluid and serum of women with
endometriosis (69,70). However, in another study, it was
found that although the concentrations of IL-16 in peritoneal fluid
and sera were both lower in women with endometriosis, the observed
differences did not reach a level of statistical significance
(71). Apparently, these findings
should be validated in larger studies, in order to clarify the role
of this molecule in endometriosis.
Although the molecular mechanisms by which
IL-16 gene polymorphisms are associated with endometriosis
remain unclear, additional functional studies, in combination with
genetic studies involving subjects from various ethnicities, may
provide valuable information concerning this issue. GWAS have
detected many regions harboring interesting disease-candidate genes
but the risk alleles may not always act in obvious ways. As a
consequence, it is necessary to accumulate evidence of their
functional significance by performing gene expression studies,
epigenetic analyses or further functional studies. In our attempt
to approach the functional role of the SNPs studied, we constructed
3D models of the respective PDZ domains of nPro-IL-16 isomorph 1
and Pro-IL-16 isomorph 2 proteins. It is apparent that the
rs11556218 SNP polymorphism leading to the Asn446Lys mutation on
the PDZ2 domain of Pro-IL-16 affected the function of the protein.
The introduction of the positive charge on the side chain of the
lysine no. 446 amino acid residue, located on the rim of the GLGF
carboxylate binding loop, may deregulate protein-protein
recognition.
A recent study by Xiao et al (72) presented a disease network of
endometriosis that integrated human PPIs and known disease-causing
genes. Considering that most human diseases reflect the phenotype
of the co-operative function of many causative gene alleles
(73), gene networks confer
information for the underlying disease mechanisms. The construction
of this network was based on endometriosis-causing genes that were
identified from disease-gene databases and subsequent calculations
and approaches using bioinformatics. However, IL-16 was not
included in this network.
The pathogenesis of endometriosis is highly complex
given that it involves genetic, epigenetic and environmental
factors, with all of them interacting with each other in order to
yield the disease phenotype. Thus, conflicting studies appear
frequently and the interpretation of the data collected remains a
challenge. A major advantage of our study was the homogeneous
patient cohort and control group selected, thus minimizing the
possibility that our results are biased by sampling. The major
weakness of the present study was the small sample size, leading to
a low statistical power, probably non-efficient to detect a weak
genetic effect. Furthermore, it should be mentioned that
laparoscopy is an expensive and invasive procedure, and women with
no symptoms of endometriosis have had low adherence to accepting
this diagnostic procedure. The failure to confirm previous findings
from another study for rs4072111 is largely attributed to
population differences or, probably, to interactions with genetic
and/or non-genetic factors (74).
Together, the results from the present study demonstrate the
difficulty in identifying common, generalizable risk alleles in a
complex disease, such as endometriosis.
In conclusion, the present study has shown that the
IL-16 polymorphisms analyzed may be involved in the
development of endometriosis but additional studies in different
populations are needed in an attempt to validate the present
results. Moreover, further investigations to clarify the possible
role of the gene studied in the clinical course of endometriosis
are required to provide functional insight into the role of
IL-16 in endometriosis and elucidate the mechanism by which
the IL-16 gene polymorphisms affect the risk for this
disease.
Acknowledgments
We would like to thank all the practitioners for
providing the data and pathology reports used in the present study.
This study was supported partially by a grant by ELKE (KA 4848,
University of Crete) to I.M.
Notes
[1] Competing
interests
K.Z. has scientific collaborations in the area of
endometriosis with Bayer AG (Leverkusen, Germany), Roche
Diagnostics (Basel, Switzerland) and MDNA. Demetrios A. Spandidos
is the Editor-in-Chief for the journal, but had no personal
involvement in the reviewing process, or any influence in terms of
adjudicating on the final decision, for this article.
References
|
1
|
Giudice LC and Kao LC: Endometriosis.
Lancet. 364:1789–1799. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Halis G and Arici A: Endometriosis and
inflammation in infertility. Ann N Y Acad Sci. 1034:300–315. 2004.
View Article : Google Scholar
|
|
3
|
Burney RO and Giudice LC: Pathogenesis and
pathophysiology of endometriosis. Fertil Steril. 98:511–519. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Bazot M, Lafont C, Rouzier R, Roseau G,
Thomassin-Naggara I and Daraï E: Diagnostic accuracy of physical
examination, trans-vaginal sonography, rectal endoscopic
sonography, and magnetic resonance imaging to diagnose deep
infiltrating endometriosis. Fertil Steril. 92:1825–1833. 2009.
View Article : Google Scholar
|
|
5
|
Koninckx PR, Ussia A, Adamyan L, Wattiez A
and Donnez J: Deep endometriosis: Definition, diagnosis, and
treatment. Fertil Steril. 98:564–571. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Maroun P, Cooper MJ, Reid GD and Keirse
MJ: Relevance of gastrointestinal symptoms in endometriosis. Aust N
Z J Obstet Gynaecol. 49:411–414. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Varma R, Rollason T, Gupta JK and Maher
ER: Endometriosis and the neoplastic process. Reproduction.
127:293–304. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Sampson J: Peritoneal endometriosis due to
menstrual dissemination of endometrial tissue into the peritoneal
cavity. Am J Obstet Gynecol. 14:422–469. 1927. View Article : Google Scholar
|
|
9
|
Batt RE, Smith RA, Buck GM, Severino MF
and Naples JD: Müllerianosis. Prog Clin Biol Res. 323:413–426.
1990.
|
|
10
|
Vercellini P, Viganò P, Somigliana E and
Fedele L: Endometriosis: Pathogenesis and treatment. Nat Rev
Endocrinol. 10:261–275. 2014. View Article : Google Scholar
|
|
11
|
Simpson JL, Elias S, Malinak LR and
Buttram VC Jr: Heritable aspects of endometriosis. I. Genetic
studies. Am J Obstet Gynecol. 137:327–331. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lamb K, Hoffmann RG and Nichols TR: Family
trait analysis: A case-control study of 43 women with endometriosis
and their best friends. Am J Obstet Gynecol. 154:596–601. 1986.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Coxhead D and Thomas EJ: Familial
inheritance of endometriosis in a British population. A case
control study. J Obstet Gynaecol. 13:42–44. 1993. View Article : Google Scholar
|
|
14
|
Stefansson H, Geirsson RT,
Steinthorsdottir V, Jonsson H, Manolescu A, Kong A, Ingadottir G,
Gulcher J and Stefansson K: Genetic factors contribute to the risk
of developing endometriosis. Hum Reprod. 17:555–559. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Treloar SA, O'Connor DT, O'Connor VM and
Martin NG: Genetic influences on endometriosis in an Australian
twin sample. simplesueT@qimr.edu.au. Fertil
Steril. 71:701–710. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Saha R, Pettersson HJ, Svedberg P,
Olovsson M, Bergqvist A, Marions L, Tornvall P and Kuja-Halkola R:
Heritability of endometriosis. Fertil Steril. 104:947–952. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang Z, Zang C, Rosenfeld JA, Schones DE,
Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ and Zhao K:
Combinatorial patterns of histone acetylations and methylations in
the human genome. Nat Genet. 40:897–903. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tan M, Luo H, Lee S, Jin F, Yang JS,
Montellier E, Buchou T, Cheng Z, Rousseaux S, Rajagopal N, et al:
Identification of 67 histone marks and histone lysine crotonylation
as a new type of histone modification. Cell. 146:1016–1028. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Falconer H, D'Hooghe T and Fried G:
Endometriosis and genetic polymorphisms. Obstet Gynecol Surv.
62:616–628. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Nyholt DR, Low SK, Anderson CA, Painter
JN, Uno S, Morris AP, MacGregor S, Gordon SD, Henders AK, Martin
NG, et al: Genome-wide association meta-analysis identifies new
endometriosis risk loci. Nat Genet. 44:1355–1359. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Albertsen HM, Chettier R, Farrington P and
Ward K: Genome-wide association study link novel loci to
endometriosis. PLoS One. 8:e582572013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rahmioglu N, Nyholt DR, Morris AP, Missmer
SA, Montgomery GW and Zondervan KT: Genetic variants underlying
risk of endometriosis: Insights from meta-analysis of eight
genome-wide association and replication datasets. Hum Reprod
Update. 20:702–716. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zondervan KT, Rahmioglu N, Morris AP,
Nyholt DR, Montgomery GW, Becker CM and Missmer SA: Beyond
endometriosis genome-wide association study: From genomics to
phenomics to the patient. Semin Reprod Med. 34:242–254. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Uimari O, Rahmioglu N, Nyholt DR, Vincent
K, Missmer SA, Becker C, Morris AP, Montgomery GW and Zondervan KT:
Genome-wide genetic analyses highlight mitogen-activated protein
kinase (MAPK) signaling in the pathogenesis of endometriosis. Hum
Reprod. 32:780–793. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kobayashi H, Imanaka S, Nakamura H and
Tsuji A: Understanding the role of epigenomic, genomic and genetic
alterations in the development of endometriosis (Review). Mol Med
Rep. 9:1483–1505. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Rahmioglu N, Montgomery GW and Zondervan
KT: Genetics of endometriosis. Wom Health Lond. 11:577–586. 2015.
View Article : Google Scholar
|
|
27
|
Sapkota Y, Steinthorsdottir V, Morris AP,
Fassbender A, Rahmioglu N, De Vivo I, Buring JE, Zhang F, Edwards
TL, Jones S, et al: iPSYCH-SSI-Broad Group: Meta-analysis
identifies five novel loci associated with endometriosis
highlighting key genes involved in hormone metabolism. Nat Commun.
8:155392017. View Article : Google Scholar
|
|
28
|
Sapkota Y, Attia J, Gordon SD, Henders AK,
Holliday EG, Rahmioglu N, MacGregor S, Martin NG, McEvoy M, Morris
AP, et al: Genetic burden associated with varying degrees of
disease severity in endometriosis. Mol Hum Reprod. 21:594–602.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Drosdzol-Cop A and Skrzypulec-Plinta V:
Selected cytokines and glycodelin A levels in serum and peritoneal
fluid in girls with endometriosis. J Obstet Gynaecol Res.
38:1245–1253. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Athayde N, Romero R, Maymon E, Gomez R,
Pacora P, Araneda H and Yoon BH: A role for the novel cytokine
RANTES in pregnancy and parturition. Am J Obstet Gynecol.
181:989–994. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Thornton CA, Holloway JA, Shute JK,
Holloway JW, Diaper ND and Warner JO: Human mid-gestation amniotic
fluid contains interleukin-16 bioactivity. Immunology. 126:543–551.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mathy NL, Scheuer W, Lanzendörfer M,
Honold K, Ambrosius D, Norley S and Kurth R: Interleukin-16
stimulates the expression and production of pro-inflammatory
cytokines by human monocytes. Immunology. 100:63–69. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Brait VH, Arumugam TV, Drummond GR and
Sobey CG: Importance of T lymphocytes in brain injury,
immunodeficiency, and recovery after cerebral ischemia. J Cereb
Blood Flow Metab. 32:598–611. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Baier M, Bannert N, Werner A, Lang K and
Kurth R: Molecular cloning, sequence, expression, and processing of
the interleukin 16 precursor. Proc Natl Acad Sci USA. 94:5273–5277.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Center DM, Kornfeld H and Cruikshank WW:
Interleukin-16. Int J Biochem Cell Biol. 29:1231–1234. 1997.
View Article : Google Scholar
|
|
36
|
Chupp GL, Wright EA, Wu D,
Vallen-Mashikian M, Cruikshank WW, Center DM, Kornfeld H and Berman
JS: Tissue and T cell distribution of precursor and mature IL-16. J
Immunol. 161:3114–3119. 1998.PubMed/NCBI
|
|
37
|
Zhang Y, Center DM, Wu DM, Cruikshank WW,
Yuan J, Andrews DW and Kornfeld H: Processing and activation of
pro-interleukin-16 by caspase-3. J Biol Chem. 273:1144–1149. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Bowler RP, Bahr TM, Hughes G, Lutz S, Kim
YI, Coldren CD, Reisdorph N and Kechris KJ: Integrative omics
approach identifies interleukin-16 as a biomarker of emphysema.
OMICS. 17:619–626. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Tong Z, Li Q, Zhang J, Wei Y, Miao G and
Yang X: Association between interleukin 6 and interleukin 16 gene
polymorphisms and coronary heart disease risk in a Chinese
population. J Int Med Res. 41:1049–1056. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Liu XL, Du JZ, Zhou YM, Shu QF and Li YG:
Interleukin-16 polymorphism is associated with an increased risk of
ischemic stroke. Mediators Inflamm. 2013:5647502013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Khoshbakht T, Soosanabadi M, Neishaboury
M, Kamali K, Karimlou M, Bazazzadegan N and Khorram Khorshid HR: An
association study on IL16 gene polymorphisms with the risk of
sporadic Alzheimer's disease. Avicenna J Med Biotechnol. 7:128–132.
2015.PubMed/NCBI
|
|
42
|
Romani S, Hosseini SM, Mohebbi SR,
Kazemian S, Derakhshani S, Khanyaghma M, Azimzadeh P, Sharifian A
and Zali MR: Interleukin-16 gene polymorphisms are considerable
host genetic factors for patients' susceptibility to chronic
hepatitis B infection. Hepat Res Treat. 2014:7907532014.
|
|
43
|
Glas J, Török H-P, Unterhuber H, Radlmayr
M and Folwaczny C: The -295T-to-C promoter polymorphism of the
IL-16 gene is associated with Crohn's disease. Clin Immunol.
106:197–200. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Gu XJ, Cui B, Zhao ZF, Chen HY, Li XY,
Wang S, Ning G and Zhao YJ: Association of the interleukin (IL)-16
gene polymorphisms with Graves' disease. Clin Immunol. 127:298–302.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Xue H, Gao L, Wu Y, Fang W, Wang L, Li C,
Li Y, Liang W and Zhang L: The IL-16 gene polymorphisms and the
risk of the systemic lupus erythematosus. Clin Chim Acta.
403:223–225. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Luo SX, Li S, Zhang XH, Zhang JJ, Long GH,
Dong GF, Su W, Deng Y, Liu Y, Zhao JM and Qin X: Genetic
polymorphisms of interleukin-16 and risk of knee osteoarthritis.
PLoS One. 10:e01234422015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Gao LB, Rao L, Wang YY, Liang WB, Li C,
Xue H, Zhou B, Sun H, Li Y, Lv ML, et al: The association of
interleukin-16 polymorphisms with IL-16 serum levels and risk of
colorectal and gastric cancer. Carcinogenesis. 30:295–299. 2009.
View Article : Google Scholar
|
|
48
|
Li S, Deng Y, Chen ZP, Huang S, Liao XC,
Lin LW, Li H, Peng T, Qin X and Zhao JM: Genetic polymorphism of
interleukin-16 influences susceptibility to HBV-related
hepatocellular carcinoma in a Chinese population. Infect Genet
Evol. 11:2083–2088. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Batai K, Shah E, Murphy AB, Newsome J,
Ruden M, Ahaghotu C and Kittles RA: Fine-mapping of IL16 gene and
prostate cancer risk in African Americans. Cancer Epidemiol
Biomarkers Prev. 21:2059–2068. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Mo CJ, Peng QL, He Y, Wang J, Xie L, Li
TJ, Li S and Qin X: Positive association between IL-16 rs11556218
T/G polymorphism and cancer risk: A meta-analysis. Asian Pac J
Cancer Prev. 15:4697–4703. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Poulat F, de Santa Barbara P, Desclozeaux
M, Soullier S, Moniot B, Bonneaud N, Boizet B and Berta P: The
human testis determining factor SRY binds a nuclear factor
containing PDZ protein interaction domains. J Biol Chem.
272:7167–7172. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Fanning AS and Anderson JM:
Protein-protein interactions: PDZ domain networks. Curr Biol.
6:1385–1388. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Sherman DL and Brophy PJ: A tripartite
nuclear localization signal in the PDZ-domain protein L-periaxin. J
Biol Chem. 275:4537–4540. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Hsueh YP, Wang TF, Yang FC and Sheng M:
Nuclear translocation and transcription regulation by the
membrane-associated guanylate kinase CASK/LIN-2. Nature.
404:298–302. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Azimzadeh P, Khorram Khorshid HR, Akhondi
MM and Shirazi A: Association of interleukin-16 polymorphisms with
disease progression and susceptibility in endometriosis. Int J
Immunogenet. 43:297–302. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Hsieh YY, Bau DT, Chang CC, Tsai CH, Chen
CP and Tsai FJ: XRCC4 codon 247*A and XRCC4 promoter -1394*T
related genotypes but not XRCC4 intron 3 gene polymorphism are
associated with higher susceptibility for endometriosis. Mol Reprod
Dev. 75:946–951. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Altinkaya SO, Ugur M, Ceylaner G, Ozat M,
Gungor T and Ceylaner S: Vascular endothelial growth factor +405
C/G poly morphism is highly associated with an increased risk of
endometriosis in Turkish women. Arch Gynecol Obstet. 283:267–272.
2011. View Article : Google Scholar
|
|
58
|
Matalliotakis M, Zervou MI, Matalliotaki
C, Rahmioglu N, Koumantakis G, Kalogiannidis I, Prapas I, Zondervan
K, Spandidos DA, Matalliotakis I and Goulielmos GN: The role of
gene polymorphisms in endometriosis. Mol Med Rep. 16:5881–5886.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
The American Fertility Society: Revised
American Fertility Society classification of endometriosis: 1985.
Fertil Steril. 43:351–352. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Biasini M, Bienert S, Waterhouse A, Arnold
K, Studer G, Schmidt T, Kiefer F, Gallo Cassarino T, Bertoni M,
Bordoli L and Schwede T: SWISS-MODEL: Modelling protein tertiary
and quaternary structure using evolutionary information. Nucleic
Acids Res. 42:W252–258. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Sato M, Koshiba S, Inoue M, Kigawa T and
Yokoyama S: RIKEN Structural Genomics/Proteomics Initiative,
deposition. 2005 05 23;
|
|
62
|
Kim HS: Assignment of human interleukin 16
(IL16) to chromosome 15q26.3 by radiation hybrid mapping. Cytogenet
Cell Genet. 84:931999. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Mori M, Yamada R, Kobayashi K, Kawaida R
and Yamamoto K: Ethnic differences in allele frequency of
autoimmune-disease- associated SNPs. J Hum Genet. 50:264–266. 2005.
View Article : Google Scholar
|
|
64
|
Gregersen PK and Olsson LM: Recent
advances in the genetics of autoimmune disease. Annu Rev Immunol.
27:363–391. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Azimzadeh P, Romani S, Mohebbi SR,
Kazemian S, Vahedi M, Almasi S, Fatemi SR and Zali MR:
Interleukin-16 (IL-16) gene polymorphisms in Iranian patients with
colorectal cancer. J Gastrointestin Liver Dis. 20:371–376.
2011.PubMed/NCBI
|
|
66
|
Gan XL, Lin YH, Zhang Y, Yu TH and Hu LN:
Association of an interleukin-16 gene polymorphism with the risk
and pain phenotype of endometriosis. DNA Cell Biol. 29:663–667.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Kim JG, Kim H, Ku S-y, Kim SH, Choi YM and
Kim JH: The association between single nucleotide polymorphisms of
interleukin (IL)-10, IL-10 receptor antagtonist and IL-16 genes and
endometriosis. Fertil Steril. 100(Suppl): S3642013. View Article : Google Scholar
|
|
68
|
Koga K, Osuga Y, Yoshino O, Hirota Y, Yano
T, Tsutsumi O and Taketani Y: Elevated interleukin-16 levels in the
peritoneal fluid of women with endometriosis may be a mechanism for
inflammatory reactions associated with endometriosis. Fertil
Steril. 83:878–882. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Cheong YC, Shelton JB, Laird SM, Richmond
M, Kudesia G, Li TC and Ledger WL: IL-1, IL-6 and TNF-alpha
concentrations in the peritoneal fluid of women with pelvic
adhesions. Hum Reprod. 17:69–75. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Somigliana E, Viganò P, Tirelli AS,
Felicetta I, Torresani E, Vignali M and Di Blasio AM: Use of the
concomitant serum dosage of CA 125, CA 19-9 and interleukin-6 to
detect the presence of endometriosis. Results from a series of
reproductive age women undergoing laparoscopic surgery for benign
gynaecological conditions. Hum Reprod. 19:1871–1876. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Zhang X, Lin J, Deng L, Chen Z and Chen L:
Peritoneal fluid and serum concentration of interleukin-16 in women
with endometriosis. Acta Obstet Gynecol Scand. 84:297–298. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Xiao H, Yang L, Liu J, Jiao Y, Lu L and
Zhao H: Protein-protein interaction analysis to identify biomarker
networks for endometriosis. Exp Ther Med. 14:4647–4654.
2017.PubMed/NCBI
|
|
73
|
Schadt EE: Molecular networks as sensors
and drivers of common human diseases. Nature. 461:218–223. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Ma Y, Tian J, Cen H, Li J, Xu WD, Wang DG,
Pan HF and Ye DQ: Association of c-Jun gene polymorphism with
susceptibility to systemic lupus erythematosus in a Chinese
population. DNA Cell Biol. 31:1274–1278. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Bannert N, Vollhardt K, Asomuddinov B,
Haag M, König H, Norley S and Kurth R: PDZ Domain-mediated
interaction of interleukin-16 precursor proteins with myosin
phosphatase targeting subunits. J Biol Chem. 278:42190–42199. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Berman HM, Westbrook J, Feng Z, Gilliland
G, Bhat TN, Weissig H, Shindyalov IN and Bourne PE: The Protein
Data Bank. Nucleic Acids Res. 28:235–242. 2000. View Article : Google Scholar
|