Introduction
Copper metabolism Murr1 domain containing 1 (COMMD1)
is a member of the COMMD family, which has ten members. This
protein family is distinguished by a unique C-terminal motif called
the COMM domain (1,2). COMMD1 was previously reported to
regulate copper homeostasis by binding to ATPase copper
transporting alpha/beta (ATP7A/B), which are associated with the
copper storage disorders Menkes disease and Wilson's disease
(3,4). COMMD1 regulates the folding,
stability, ubiquitination and protein degradation of its
interaction partner proteins, including ATP7A, ATP7B and nuclear
factor (NF)-κB (5–8). COMMD1 has been reported as a human
immunodeficiency virus (HIV)-1 host factor and inhibits HIV-1
replication by blocking the degradation of the inhibitor of NF-κB
(IκB-α) (9). A previous study by
our group reported that IκB-α expression is increased by HIV-1
latent infection through the induction of COMMD1 expression.
Induction of COMMD1 in HIV-1 latent-infected cells also maintains
latent HIV-1 infection. The expression of the COMMD1 protein and
mRNA in HIV-1 latent-infected myeloid cells is stronger than in
parental cells (10). However, it
remains elusive how COMMD1 transcription and expression are
regulated. To the best of our knowledge, no previous studies have
assessed COMMD1 transcriptional regulation.
The transcription factors specificity protein 1
(Sp1) and Sp3 belong to the Sp family, which has four members. Sp
factors bind to GC-rich DNA sequences in human gene promoters. The
Sp family is ubiquitously expressed and regulates a number of
housekeeping and tissues-specific genes (11,12). Sp1 and Sp3 have 90% DNA sequence
homology in the zinc finger-binding domain and exhibit similar
specificities and affinities for DNA binding (13). However, although there are
structural similarities between them, a functional comparison
identified Sp1 as a transcriptional activator, with Sp3 being
either a gene expression activator or repressor depending on the
promoter structure or cell type (13,14).
The present study hypothesized that basal COMMD1
transcription is regulated by Sp1 because COMMD1 expression is
ubiquitous (1), and due to
prospective consensus Sp binding sites identified in the human
COMMD1 promoter region (Fig. 1A).
In the present study, the −1,192/+83 bp region of the COMMD1
promoter was cloned and the effects of Sp1 and Sp3 on the promoter
activity and COMMD1 mRNA and protein expression were assessed with
a luciferase assay using COMMD1 promoter constructs. It was
demonstrated that Sp1 upregulates COMMD1 promoter activity as a
transcriptional activator, while Sp3 suppresses COMMD1 promoter
activity as a repressor. Sp1 regulates COMMD1 promoter activity via
−11/−1 bp of the Sp1-binding site and is required for the basal
expression of COMMD1 in human cell lines.
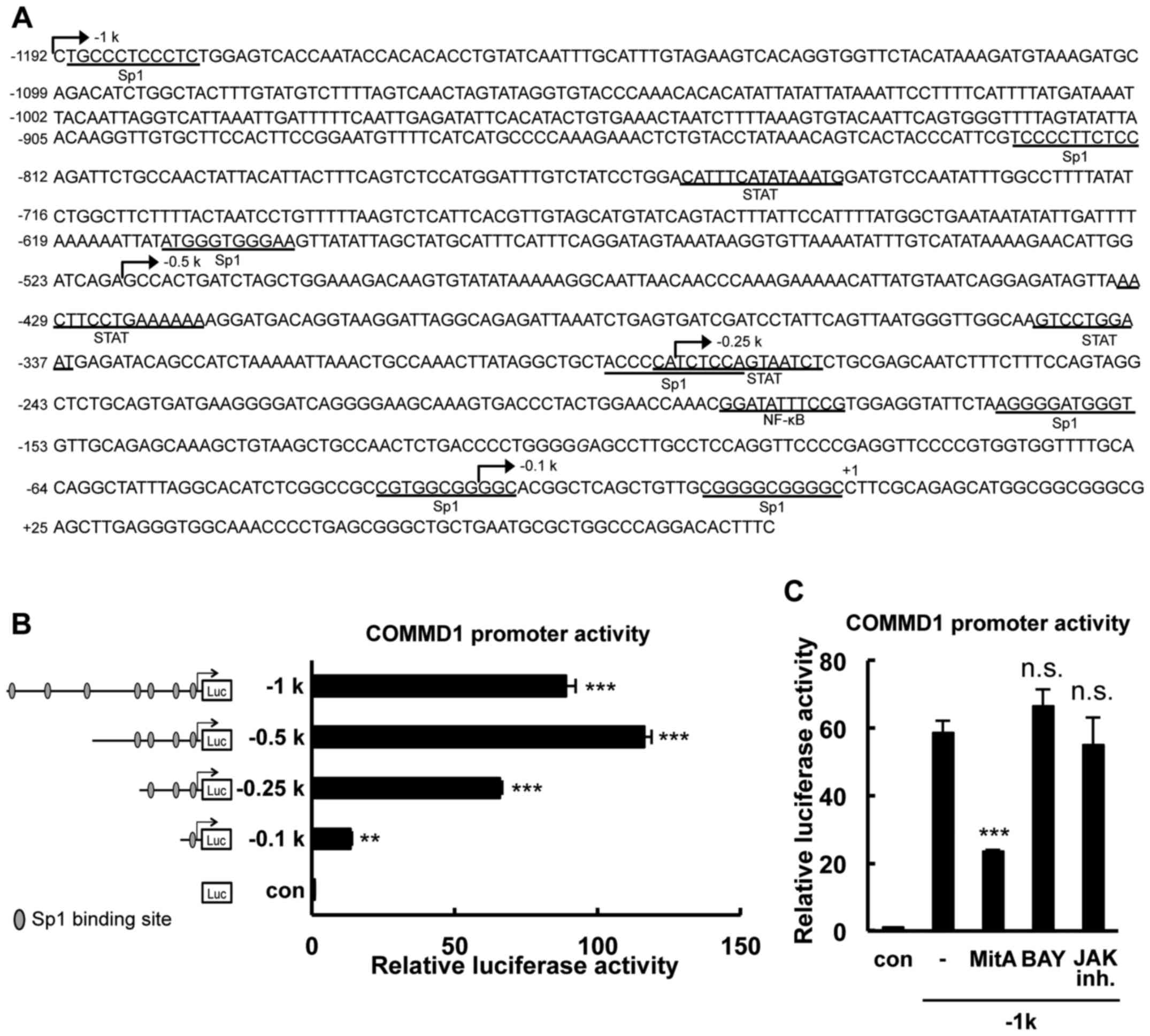 | Figure 1Identification of the minimal promoter
region required for basal COMMD1 promoter activity. (A) Nucleotide
sequence of the 5′flanking region of the human COMMD1 gene. The
site indicated by (+1) denotes the start site of transcription. The
predicted binding sites for Sp1, STAT and NF-κB are marked on the
sequence. The lengths of the different promoter constructs are
indicated by arrows. (B) Deletion analysis of the COMMD1 promoter.
293T cells were transfected with COMMD1 promoter deletion
constructs and lysates were harvested 48 h later for the luciferase
reporter assay. Reporter activity is expressed as fold activation
over the empty vector. (C) 293T cells were transfected with the
COMMD1 promoter (−1 k) and treated with MitA, BAY or JAK inh. for
24 h. Lysates were harvested 48 h later for the luciferase reporter
assay. Values are expressed as the mean ± standard error from three
independent experiments. In (A) and (B), P-values were determined
by analysis of variance followed by with Dunnett's test.
**P<0.01 and ***P<0.001, respectively,
vs. the con or blank group. ns, not significant; SP, specificity
protein; STAT, signal transducer and activator of transcription;
NF, nuclear factor; Luc, luciferase; JAK inh., Janus kinase
inhibitor Pan-JAK; con, control; COMMD1, copper metabolism Murr1
domain containing 1; MitA, mithramycin A; BAY, BAY 11-7085. |
Materials and methods
Reagents, antibodies and plasmids
Mithramycin A was purchased from Sigma-Aldrich
(Merck KGaA, Darmstadt, Germany). BAY 11-7085 was obtained from
Tokyo Chemical Industry (Tokyo, Japan). Pan-JAK, a Janus kinase
(JAK) inhibitor, was from Santa Cruz Biotechnology, Inc. (Dallas,
TX, USA). An antibody to COMMD1 (cat. no. ab58322) was from Abcam
Japan (Tokyo, Japan). Anti-heat shock cognate 71 kDa protein
(Hsc70; cat. no. SPA-815) was from Enzo Life Sciences (Farmingdale,
NY, USA). Anti-Sp1 (cat. no. sc-59) and Sp3 (cat. no. sc-644X) for
the chromatin immunoprecipitation (ChIP) assay were from Santa Cruz
Biotechnology, Inc. Anti-histone deacetylase 1 (HDAC1; cat. no.
607401) was from BioLegend (San Diego, CA, USA). Anti-Sp1 was from
Active Motif (Carlsbad, CA, USA). Horseradish peroxidase
(HRP)-conjugated anti-mouse (cat. no. 7076) and anti-rabbit (cat.
no. 7074) antibodies were from cell signaling technology (Danvers,
MA). The pGL4.11 plasmid was from Promega Corp. (Madison, WI, USA).
The pERV2/Sp1 construct was kindly provided by Dr. G. Suske
(Institute for Molecular Biology and Tumour Research,
Philipps-University of Marburg, Marburg, Germany). pN3-Sp3FL
(plasmid no. 24541) was obtained from Addgene Inc. (Cambridge, MA,
USA).
Cloning of the COMMD1 genomic 5′region
and construction of luciferase reporter gene plasmids
The region 1.2 kb upstream of the COMMD1 gene and
part of its 5′ untranslated region (UTR) were amplified with
primers and Takara LA Taq polymerase with GC buffer (Takara Bio,
Inc., Otsu, Japan) using genomic DNA prepared from U937 cells as a
template. The polymerase chain reaction (PCR) product was cloned
into the pCR2.1 TOPO vector using the TA cloning kit (cat. no.
K4520-01; Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA,
USA). The COMMD1 promoter-luciferase reporter plasmid was
constructed by subcloning the COMMD1 promoter fragment (−1,192/+83)
into the XhoI-KpnI restriction site of the pGL4.11 vector (Promega
Corp.). Deletion mutants of the COMMD1 promoter were constructed by
a PCR-based approach using the −1,192/+83 construct as a template
and inserting the amplified fragments into the pGL4.11 vector.
Point mutant constructs were prepared using the Quick change II XL
site-directed mutagenesis kit (cat no. 200521; Stratagene, La
Jolla, CA, USA) according to the manufacturer's instructions. The
sequences of primers used for mutagenesis are listed in Table I. All mutant plasmids were
generated using the −283/+83 construct in the pGL4.11 vector as a
template. Restriction sites were introduced into mutations for
selection, and the incorporation of the mutation was verified by
restriction digestion and sequencing. All plasmids used in the
present study were sequenced with an ABI 3130 Genetic analyzer
(Applied Biosystems; Thermo Fisher Scientific, Inc.).
 | Table IPrimers for COMMD1 promoter cloning
and mutagenesis of specificity protein 1-binding sites. |
Table I
Primers for COMMD1 promoter cloning
and mutagenesis of specificity protein 1-binding sites.
| Primer name | Sequence (5′–3′) |
|---|
| 5′ COMMD1prom
(−1192_KpnI) |
GGTACCCTGCCCTCCCTCTGGAGTCACCAATAC |
| 5′ COMMD1prom
(−519_KpnI) |
GGTACCGAGCCACTGATCTAGCTGGAAAGAC |
| 5′ COMMD1prom
(−283_KpnI) |
GGTACCTCTCCAGTAATCTCTGCGAGCAATC |
| 5′ COMMD1prom
(−29_KpnI) |
GGTACCGGCACGGCTCAGCTGTTGCGGGGC |
| 3′ COMMD1prom
(+83_XhoI) |
CTCGAGGAAAGTGTCCTGGGCCAGCGCATTC |
| COMMD1prom mt1
Forward |
CCGTGGAGGTATTCTAAGGTCATTGGTGTTGCAGAGCAAAGCT |
| COMMD1prom mt1
Reverse |
AGCTTTGCTCTGCAACACCAATGACCTTAGAATACCTCCACGG |
| COMMD1prom mt2
Forward |
GCACATCTCGGCCGCCGTATACCGGCACGGCTCAGCTG |
| COMMD1prom mt2
Reverse |
CAGCTGAGCCGTGCCGGTATACGGCGGCCGAGATGTGC |
| COMMD1prom mt3
Forward |
CGGCTCAGCTGTTGCGGCCATCGGCCTTCGCAGAGCATG |
| COMMD1prom mt3
Reverse |
CATGCTCTGCGAAGGCCGATGGCCGCAACAGCTGAGCCG |
| COMMD1prom mt2-2
Forward |
GCACATCTCGGCCGCCGTCCATCGGCACGGCTCAGCTG |
| COMMD1prom mt2-2
Reverse |
CAGCTGAGCCGTGCCGATGGACGGCGGCCGAGATGTGC |
Cell culture and transfection
The human embryonic kidney cell line 293T and the
human cervical carcinoma cell line HeLa were obtained from the
American Type Culture Collection (Manassas, VA, USA) were
maintained in Dulbecco's modified Eagle's medium supplemented with
10% fetal bovine serum (Nichirei Biosciences, Inc., Tokyo, Japan)
at 37°C in a humidified atmosphere of 5% CO2. Human
promyeloid U937 were obtained from the American Type Culture
Collection. Cells were cultured in RPMI-1640 (Invitrogen; Thermo
Fisher Scientific, Inc.) supplemented with 10% fetal bovine serum
and antibiotics at 37°C in a humidified atmosphere containing 5%
CO2. Transient transfection of plasmid DNA was performed
using HilyMax reagent (Dojindo Laboratories, Kumamoto, Japan)
following the manufacturer's recommended protocol as described
previously (15). Sp1
small-interfering RNA (si-RNA) was purchased from Sigma Aldrich
(Merck KGaA). The transfection of Sp1 si-RNA was performed using
TransIT-TKO reagent (Mirus Bio LLC, Madison, WI, USA) as described
previously (16). The sequence of
siRNA targeting Sp1 (si-Sp1) was as follows: Forward,
5′-CUACUACUACCACCAGCAATT-3′ and reverse,
5′-UUGCUGGUGGUAGUAGUAGTT-3′. The negative control siRNA (MISSION
siRNA Universal Negative Control; Sigma-Aldrich, Tokyo, Japan) was
also used (con-si).
Reporter gene assays
293T cells seeded onto 24-well plates were
transfected with 0.1 µg of the wild-type or mutant COMMD1
promoter in pGL4.11, together with a control Renilla
luciferase plasmid (Promega Corp.). The amount of co-transfected
Sp1 or Sp3 expression vector was as indicated in the figures. Sp1
inhibitor mithramycin A was added 24 h prior to harvesting the
cells. At 48 h after transfection, cells were assayed for
luciferase activity using a Dual-Luciferase Reporter Assay system
(Promega Corp.) as described previously (16).
Western blot analysis
To analyze COMMD1 and Hsc70 protein expression,
immunoblotting was performed as described previously (17). 293T cells were transfected with
Sp1 and incubated for 72 h. The protein lysate was recovered by
radioimmunoprecipitation assay buffer as we described previously
(18). For western blot analysis
of Sp1 and HDAC1, nuclear extracts were prepared from cells
according to a previously published protocol (17). The concentration of each sample
was determined using a bicinchoninic acid assay kit (cat no. 23225;
Thermo Fisher Scientific, Inc.). Proteins (10 µg) were
fractionated by 10% SDS-PAGE for COMMD1, Hsc70 and HDAC1 or 7.5%
SDS-PAGE for Sp1. Proteins were transferred onto a polyvinylidene
difluoride membranes. After blocking, the membrane was probed with
the appropriate anti-COMMD1 (cat. no. ab58322; 1:1,000), Hsc70
(cat. no. SPA-815, 1:1,000), Sp1 (cat. no. sc-59, 1:1,000) and
HDAC1 (cat. no. 607401, 1:1,000) antibodies for 2 h at room
temperature. Secondary IgG-HRP antibodies [anti-rabbit-HRP (cat.
no. 7074) or mouse-HRP (cat. no. 7076); both 1:2,000] were added
and incubated for 1 h at room temperature. The blot was visualized
with Chemi-Lumi One Super (Nacalai Tesque, Kyoto, Japan). Band
intensity was quantified with Image Gauge software (ver. 4.2; Fuji
Film, Tokyo, Japan).
Reverse transcription-quantitative
(RT-q)PCR
Total RNA was isolated from cells with RNAiso Plus
(Takara Bio, Inc.) and cDNA was synthesized using the PrimeScript
RT regent kit (Takara Bio, Inc.) according to the manufacturer's
protocol. RT reaction was performed using the Takara PCR Thermal
Cycler Dice (Takara Bio, Inc.) with the following conditions: 37°C
for 15 min, 85°C for 5 sec. Real-time PCR amplification of COMMD1,
Sp1 and internal control 18S ribosomal (r)RNA were performed using
the Fast SYBR-Green Master Mix (Applied Biosystems; Thermo Fisher
Scientific, Inc.) following the manufacturer's instructions and as
described previously (10). PCR
amplifications were performed using the StepOne real-time PCR
system with the following amplification conditions: 95°C for 3 min,
40 cycles at 95°C for 10 sec, at 55°C for 30 sec. The Cq
values for each gene amplification were normalized by subtracting
the Cq value calculated for 18S rRNA. Normalized gene
expression values were presented as the relative quantity of each
gene-specific mRNA according to 2−ΔΔCq method (19). The oligonucleotide primers used
for real-time qPCR amplifications are listed in Table II.
 | Table IIPrimers used for the reverse
transcription-quantitative polymerase chain reaction analysis. |
Table II
Primers used for the reverse
transcription-quantitative polymerase chain reaction analysis.
| Gene | Forward
(5′–3′) | Reverse
(5′–3′) |
|---|
| COMMD1 |
CTGGAGGCATTCTTGACTGCTC |
GCTCTCACGGATTTTTGTCTTGTG |
| Sp1 |
TCACTGTGAATGCTGCTCAACTCTC |
AGACCAAGCTGAGCTCCATGATCAC |
| Sp3 |
CTGTCCCAATGTAAAGAAGGTG |
AGAATGCCAACGCAGATGAG |
| rRNA 18s |
CGGCTACCACATCCAAGGAA |
GCTGGAATTACCGCGGCT |
ChIP assay
To examine the binding of Sp1 to the COMMD1
promoter, the nuclear extract of 293T cells was used for the
chromatin immunoprecipitation (ChIP) assay according to a
previously described method (16). Cells were cross-linked using
formaldehyde (1% final concentration) added directly to the cell
culture media at 37°C for 15 min, and the reaction was stopped by
adding glycine (0.125 M final concentration). Cells were rinsed
with cold PBS and resuspended in cell lysis buffer. This mixture
was incubated on ice for 10 min and then homogenized. The nuclei
were resuspended in nucleus lysis buffer and incubated on ice for
10 min. the samples were sonicated on ice with the ultrasonic
homogenizer VP-050 (TAITEC, Saitama, Japan). The chromatin solution
was precleaned using Staphlococcus aureus protein A-positive
cells (Pansorbin, #507862; Merck KGaA). Two micrograms of anti-Sp1
or anti-Sp3 antibody was incubated with pre-cleared chromatin. PCR
was performed using the Fast SYBR green master mix as above. The
primer set for measuring Sp1 and Sp3 binding activities was as
follows: −0.1 kb sense, 5′-GGTACCTCTCCAGTAATCTCTGCGAGCAATC-3232 and
antisense, 5′-CTCGAGGAAAGTGTCCTGGGCCAGCGCATTC-3′.
Statistical analysis
Values are expressed as the mean ± standard error.
Data were analyzed by a one-way analysis of variance with the
Tukey-Kramer multiple-comparisons test or Dunnett's test, or by
Student's t-test. JMP software (ver. 8.0.2; SAS Institute, Cary,
NC, USA) was used for all statistical analyses. P<0.05 was
considered to indicate a statistically significant difference.
Results
Cloning of the 5′-flanking region of
COMMD1 and analysis of putative transcription factor binding
sites
To characterize the COMMD1 transcriptional system, a
genomic DNA fragment containing the 5′-flanking region of the human
COMMD1 gene (GenBank accession no. NC_000002.12) was cloned. The
cloned fragment spans 1.2-kb upstream of exon 1, down the length of
the first exon to ~72 bp of the 5′UTR. An analysis of the COMMD1
5′-flanking region using the JASPAR database (http://jaspar.genereg.net) revealed a GC-rich region
that has 7 putative binding sites for Sp1 within −1,192 bp upstream
of the transcriptional starting site, which was designated as +1
(Fig. 1A). To identify the
probable minimal promoter region required for the basal
transcriptional activity of the COMMD1 gene, deletion constructs
that contained 7, 4, 3 and 1 Sp1 binding sites were generated
(Fig. 1B). These constructs were
transfected into 293T cells that express COMMD1. The −519/+83 bp
construct had the highest activity among all constructs. The
activities of the −1,192/+83, −519/+83, −283/+83 and −29/+83 bp
constructs containing 7, 4, 3 and 1 Sp1 binding sites,
respectively, were 88-, 116-, 65- and 13-fold higher than the basal
activity, respectively. Putative binding sites for NF-κB and signal
transducer and activator of transcription (STAT) transcription
factors were detected in this region in an in silico
analysis. However, blockage of the NF-κB, STAT and JAK activation
pathways did not affect COMMD1 promoter activity (Fig. 1C). Thus, these results indicated
that the cis-regulatory elements required for COMMD1
transcriptional activity are mainly located in the core promoter
region of −283 to −1 bp.
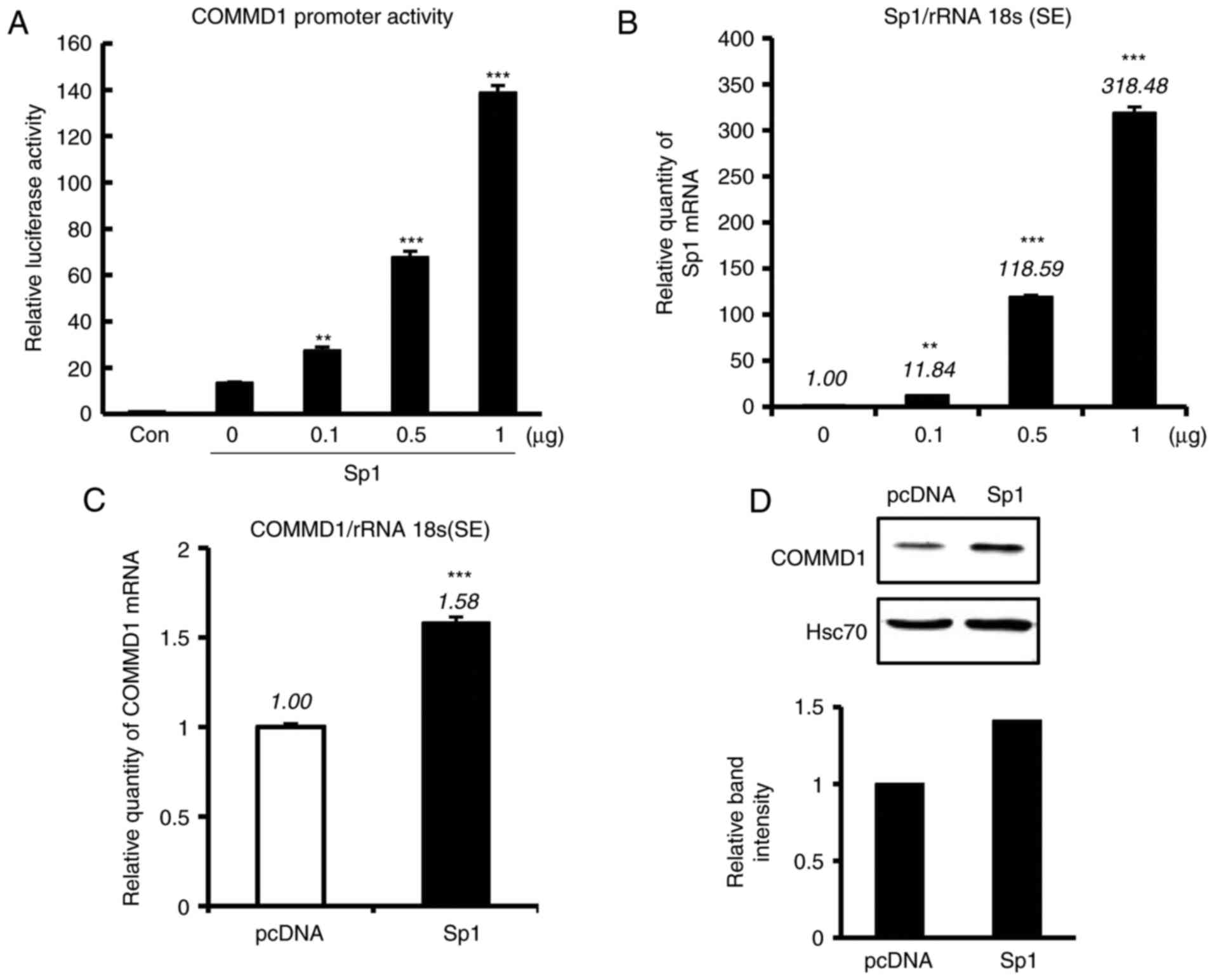
Sp1 activates COMMD1 promoter
activity
To clarify whether these Sp1 sites are involved in
COMMD1 transcription, the −0.25-kb construct and increasing amounts
of the Sp1-expressing plasmid were co-transfected into 293T cells.
An increase in COMMD1 reporter activity was identified in 293T
cells that was dependent on the amount of Sp1 expression vector
(Fig. 2A), which was in parallel
with the amount of Sp1 mRNA expression (Fig. 2B). The overexpression of Sp1 also
increased COMMD1 mRNA and protein levels (Fig. 2C and D). These results suggested
that Sp1 enhances the promoter activity of COMMD1.
Inhibition of Sp1 downregulates
endogenous COMMD1 expression
The present study analyzed the effects of
mithramycin A, a drug known to modify the GC-rich region of DNA and
inhibit Sp1 binding (20). As
presented in Fig. 3A, treatment
with mithramycin A downregulated the promoter activity of COMMD1.
The mRNA and protein expression of COMMD1 was also decreased with
mithramycin A treatment (Fig. 3B and
C). As mithramycin A has broad effects and induces cell death,
the present study also applied Sp1-specific si-RNA, si-Sp1, which
was transfected into HeLa cells (Fig.
3D). Mild Sp1 knockdown suppressed the mRNA and protein
expression of COMMD1 (Fig. 3E and
F). Collectively, these results suggest a role for Sp1 in
regulating the COMMD1 promoter.
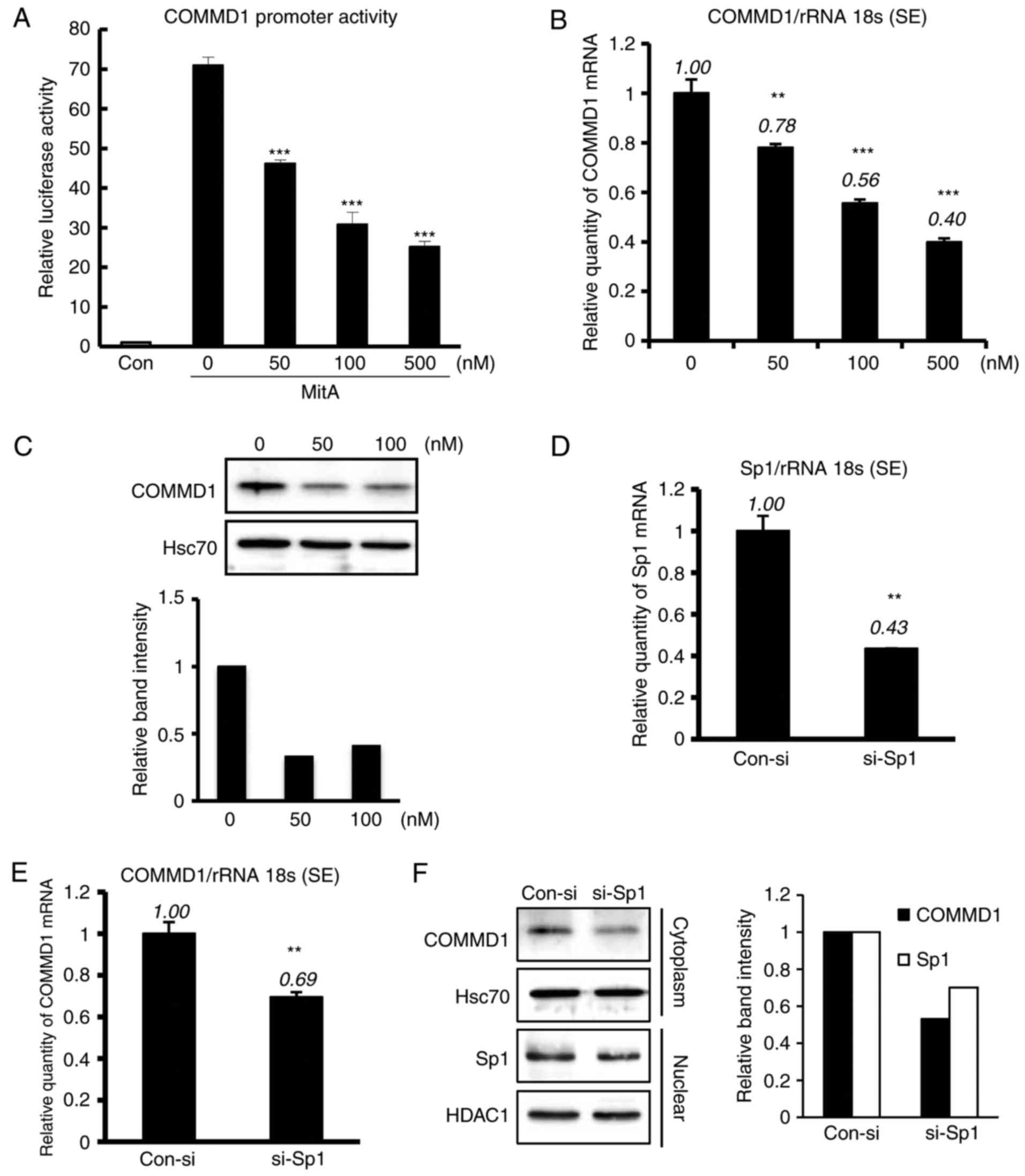 | Figure 3MitA and si-Sp1 suppress the
expression and promoter activity of COMMD1. (A) 293T cells were
transfected with the COMMD1 promoter (−0.25 kb) and treated with
MitA for 24 h at the indicated concentration. (B and C) COMMD1 mRNA
and protein expression in 293T cells treated with MitA for 24 and
72 h, respectively. (D) Sp1 mRNA expression in HeLa cells
transfected with si-Sp1 or con-si. (E and F) COMMD1 mRNA and
protein expression, respectively, in HeLa cells transfected with
si-Sp1 or con-si. Band intensities were quantified and the relative
ratios of the proteins indicated are presented in a bar graph.
Values are expressed as the mean ± standard error from three
independent experiments. In (A) and (B), P-values were determined
by analysis of variance followed by the Tukey-Kramer test and
Dunnett's test, respectively. In E and F, P-values were assessed
using Student's t-test. **P<0.01 and
***P<0.001, respectively, vs. pcDNA or the control.
si-SP1, small interfering RNA targeting specificity protein 1;
COMMD1, copper metabolism Murr1 domain containing 1; MitA,
mithramycin A; rRNA 18s, 18 s ribosomal RNA; con, control; HSC70,
heat shock cognate 71 kDa protein; HDAC1, histone deacetylase
1. |
Sp3 suppresses the activity of the
promoter of COMMD1, but not its expression
Sp1 and Sp3 have similar structures and bind to the
same Sp1-binding sites. However, their DNA-binding properties and
regulatory functions differ depending on the promoter region or
cellular background (14). The
present study investigated whether Sp3 regulates COMMD1 promoter
activity (Fig. 4). 293T cells
were co-transfected with the −0.25-kb construct and increasing
amounts of the Sp3 expression plasmid, and decrease in reporter
activity was observed (Fig. 4A).
In addition, to assess the association between Sp1 and Sp3 in the
COMMD1 promoter region, the −0.25-kb construct was co-transfected
with expression vector of Sp1 and Sp3 into 293T cells. Of note,
COMMD1 promoter activity was upregulated by Sp1 and was suppressed
by Sp3 (Fig. 4B). However, when
Sp3 was transfected into 293T cells (Fig. 4D), the expression of COMMD1 mRNA
and protein was not suppressed (Fig.
4C and E). It was then investigated whether Sp3 decreases
COMMD1 expression in Sp1-silenced cells. Sp1 knockdown decreased
the expression of COMMD1 mRNA, and this decrease was enhanced by
Sp3 overexpression (Fig. 4F).
Thus, taken together, the results indicated that Sp3 regulates the
transcription of COMMD1 in the absence of Sp1.
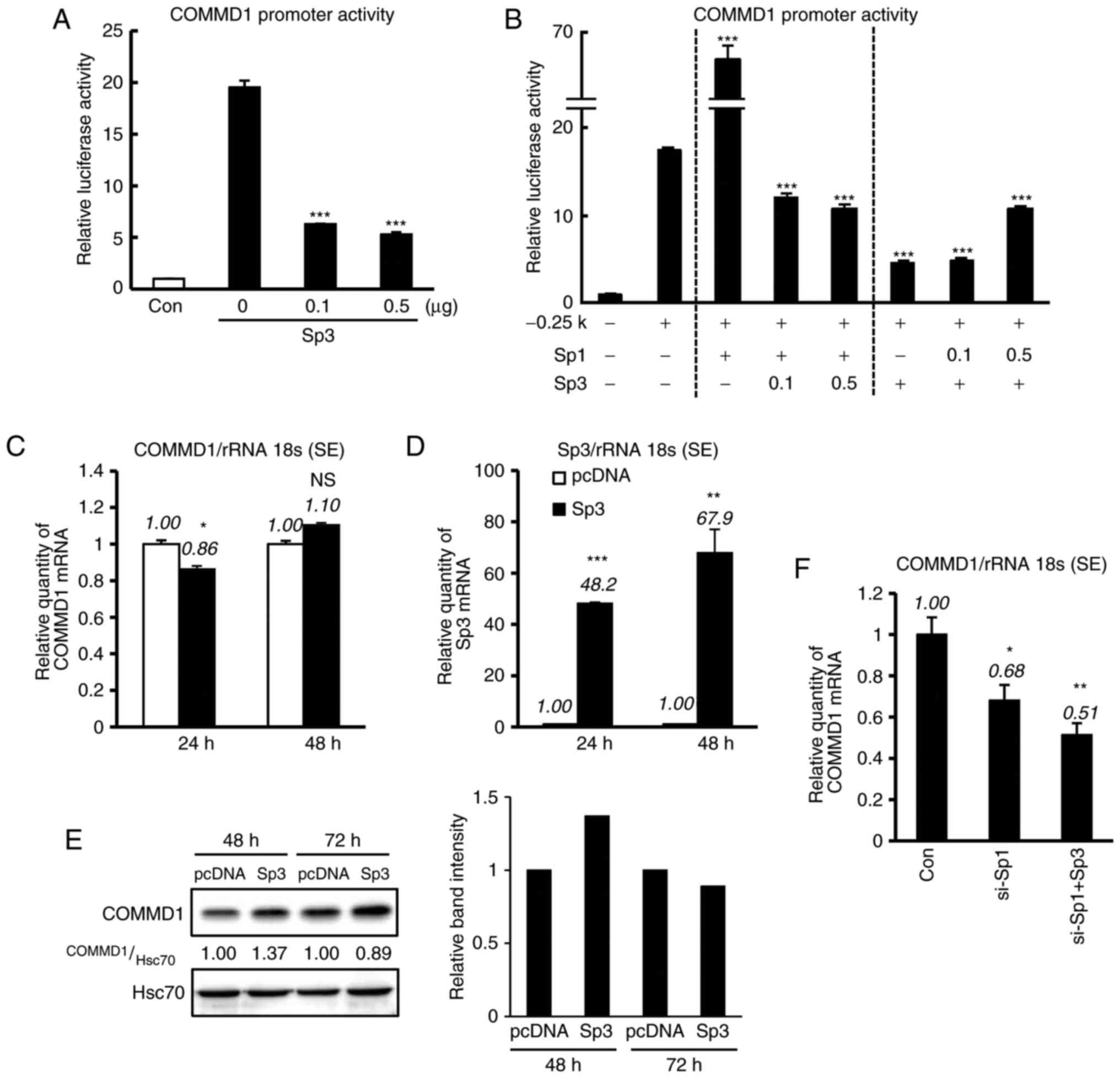 | Figure 4Sp3 downregulates COMMD1 promoter
activity, but not its expression. (A) The COMMD1 promoter (−0.25
kb) was co-transfected with the indicated amount of Sp3 into 293T
cells. Empty vectors were added to ensure a constant input of DNA.
(B) The COMMD1 promoter (−0.25 kb) was co-transfected with the
indicated amounts of Sp1 and Sp3 into 293T cells. Luciferase
activity in lysates was measured 48 h after transfection. (C and D)
COMMD1 and Sp3 mRNA expression in 293T cells transfected with pcDNA
(white bar) or Sp3 (black bar). (E) COMMD1 protein expression in
293T cells transfected with pcDNA or Sp3. (F) si-Sp1 with or
without Sp3 was co-transfected into HeLa cells for 24 h. Band
intensities were quantified and the relative ratios of the
indicated proteins are displayed in bar graph. Values are expressed
as the mean ± standard error from three independent experiments. In
(A), (B) and (F), P-values were determined by ANOVA followed by the
Tukey-Kramer test. In (C) and (D), P-values were assessed using
Student's t-test. *P<0.05, **P<0.01 and
***P<0.001 vs. pcDNA, −0.25 kb or the control. ns,
not significant; COMMD1, copper metabolism Murr1 domain containing
1; si-SP1, small interfering RNA targeting specificity protein 1;
rRNA 18s, 18 s ribosomal RNA; con, control; HSC70, heat shock
cognate 71 kDa protein. |
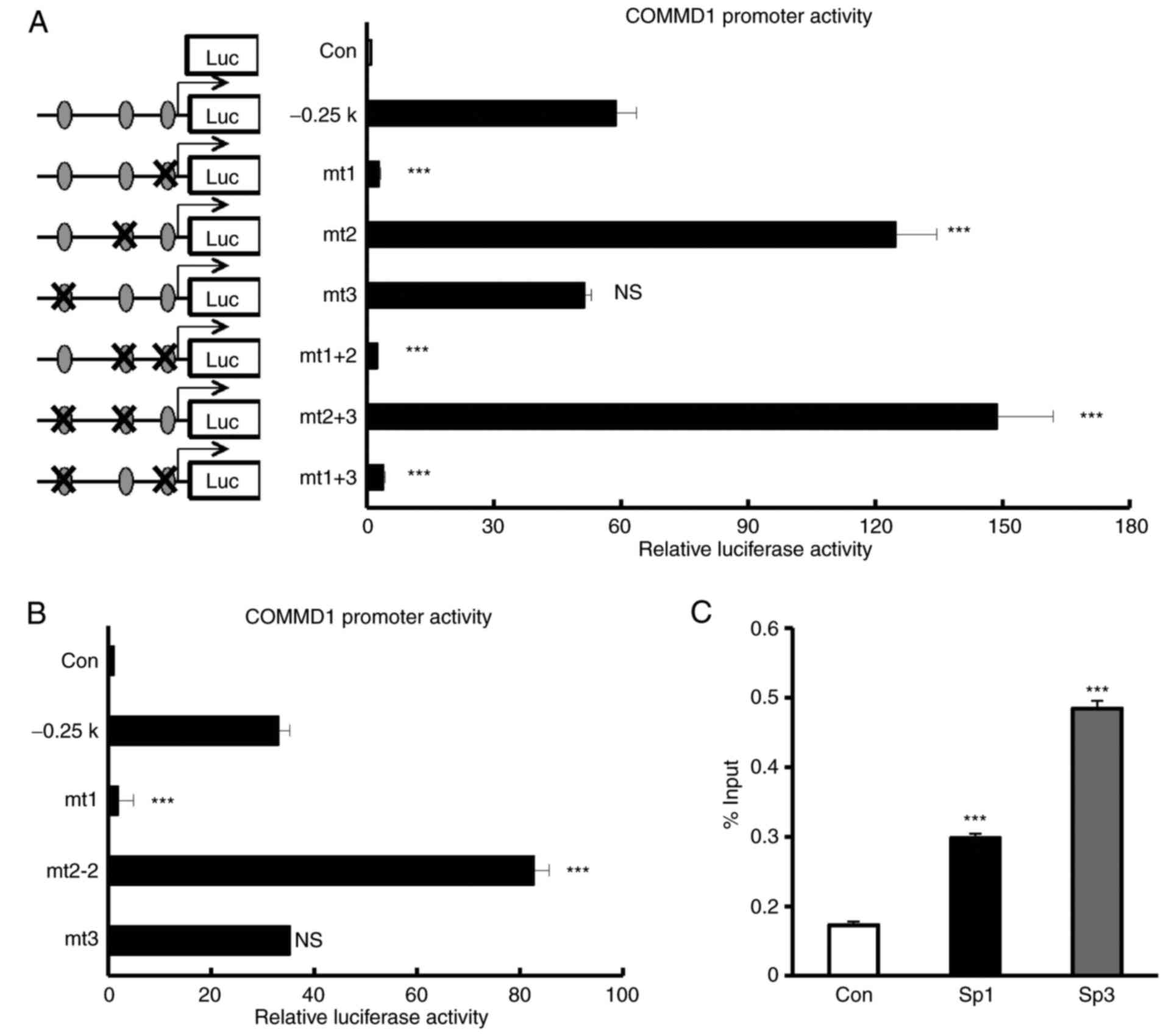
The −11/−1 bp Sp1-binding site is
essential for COMMD1 promoter activity
Next, the present study investigated which Sp1 site
is indispensable for the proximal promoter activity of COMMD1.
Reporter constructs carrying a mutation in the Sp1-binding sites
[mutant (mt)1, −11/−1; mt2, −37/−27; mt3, −164/−154] were created
and their activities in 293T cells were analyzed (Fig. 5A and B). The mutation in the
−164/−154 Sp1-binding site (mt3) had no significant effect on
promoter activity. The mutation in the −11/−1 Sp1-binding site
(mt1) completely abolished COMMD1 promoter activity. By contrast,
the mutation in the −37/−27 Sp1-binding site (mt2) led to a
stronger COMMD1 promoter activity than that with the wild-type
promoter. To confirm whether Sp1-binding site (mt2) led to a COMMD1
promoter activity, we created another mutant construct of different
sequence of mt2 (mt2-2). Mt2-2 also led to a stronger COMMD1
promoter activity (Fig. 5B). The
double mutant constructs containing mt1 (mt1+mt2 and mt1+mt3)
abolished the promoter activity. These results indicated that Sp1
regulates COMMD1 through the −11/−1 Sp1-binding site, which is
crucial for the induction of COMMD1 promoter activity. To examine
whether Sp1 or Sp3 binds to the promoter of COMMD1, a ChIP assay
was performed using nuclear extracts from 293T cells and anti-Sp1
or anti-Sp3 antibody. We observed that endogenous Sp1 or Sp3 binds
to the promoter of COMMD1 as determined by ChIP assay (Fig. 5C). Taken together, these results
indicate that Sp1 binds to the −11/−1 bp Sp1-binding site of the
COMMD1 proximal promoter region to activate COMMD1 promoter
activity and is required for the basal expression of COMMD1.
Discussion
COMMD1 was previously reported to interact with the
copper ion channel ATP7A/B and is involved in copper transport in
hepatocytes (2). COMMD1
expression is suggested to correlate with tumor malignancy,
inflammation and anti-viral host defenses. In human cancers,
including breast and prostate cancers, COMMD1 expression is
frequently suppressed, which leads to increased tumor invasion in
these patients (8). COMMD1 mRNA
expression was demonstrated to be reduced in circulating leukocytes
from inflammatory bowel disease (IBD) patients and the decrease in
COMMD1 expression induced constitutive inflammation (21). A recent study indicated that
microRNA-205 suppressed COMMD1 expression in stemness-enriched
cancer cells (22). The present
study was the first to report the cloning and characterization of
the COMMD1 promoter region. The 5′-flanking region of COMMD1 is
GC-rich and contains putative Sp1 consensus sites. COMMD1 is mainly
regulated by the Sp1 transcription factor through the direct
binding of Sp1 to the COMMD1 promoter region.
Sp1 upregulates a variety of genes, including
house-keeping genes and tissue- or cell type-specific genes
(23). An early study
demonstrated that the expression of the human CD14 gene is
upregulated by Sp1 in monocytic cells (24). Interleukin-10 expression is also
regulated by Sp1 and Sp3 in a manner similar to that of other of
house-keeping genes (25). The
present study examined whether COMMD1 expression is regulated by
Sp1. COMMD1 transcription and expression were identified to be
upregulated by Sp1. Sp1 is essential for early embryonic
development in mice study. When we performed mildly knockdown Sp1,
COMMD1 expression decreased (Fig. 3E
and F). Sp1 is important transcription factor for COMMD1
transcription.
Although Sp3 clearly suppressed COMMD1 promoter
activity, Sp3 overexpression did not suppress the mRNA expression
of COMMD1. COMMD1 protein expression was upregulated by Sp3
overexpression; however, Sp3 overexpression did not increase COMMD1
mRNA levels and did not regulate COMMD1 transcription in the
presence of Sp1. Sp1 and Sp3 expression levels are important for
the transcription of certain genes (26,27). While Sp3 decreased COMMD1
expression in Sp1-knockdown cells, the transcriptional regulation
of the COMMD1 gene by Sp1 and Sp3 has remained to be fully
elucidated. However, it is apparent that Sp1 upregulates COMMD1
transcription and expression. By contrast, Sp3 may be involved in
COMMD1 transcription and expression but its effect is dependent on
the presence of Sp1.
A mutagenesis analysis demonstrated that the
mutation in the first Sp1 site (mt1) suppressed promoter activity,
while the mutation in the second site (mt2) enhanced it. These
results suggested that the first site was the activator site of Sp1
binding and the second site was the inhibitory site. Previous
studies reported that Sp1 acts as an activator and repressor in the
same promoter region (28,29).
The present study investigated different mutant sequences in the
second Sp1-binding site (mt2-2) by mutation analysis and identified
that they also enhanced COMMD1 promoter activity. Although each of
the two sites regulated COMMD1 promoter activity in the reporter
assay, the transcription of endogenous COMMD1 by Sp1 may be
regulated in the most closed Sp1-binding site.
The transcription factor Sp1 upregulates the
promoter activity and expression of COMMD1, while Sp3 suppresses
the promoter activity at the steady state. Therefore, the present
study indicated that Sp1 and Sp3 regulate the basal transcription
and expression of COMMD1. However, a previous study by our group
indicated that interferon-α (IFNα) and the Toll-like receptor 7/8
agonist R848 induced COMMD1 mRNA in a promonocyte cell line
(10). In pathogen infection,
particularly with viruses, IFNα produced in response to infection
may regulate COMMD1 transcription. The present in silico
analysis revealed the presence of several putative transcription
factor-binding sites for STAT and interferon regulatory factor in
the COMMD1 promoter region. However, in the present study, NF-κB
and JAK inhibitor treatment did not suppress COMMD1 promoter
activity in 293T cells. In several cancers and IBD, COMMD1
expression has been reported to be suppressed (8,21),
suggesting that inflammation-associated proteins regulate COMMD1
transcription. Further investigation of the transcriptional
regulation of COMMD1 under these conditions or in specific cell
types is required. COMMD1 is a protein associated with multiple
cellular pathways, including copper homeostasis and NF-κB and
hypoxia-inducible factor-1 signaling. In the present study, Sp1 was
identified as a transcriptional regulator of COMMD1. These results
demonstrated the molecular mechanisms of the regulation of COMMD1
gene expression by Sp1 at the steady state.
Acknowledgments
The present study was supported by the Research
Program on HIV/AIDS (grant no. 17fk0410208h002 to S.O. and E.K.) of
the Japan Agency for Medical Research and Development and a
Grant-in-Aid for Research Activity Start-up (grant no. 16H07080 to
E.K.) from the Ministry of Education, Science, Sports and Culture
of Japan. The authors would like to thank Ms. Y. Endo and Ms. Y.
Kanagawa for their secretarial assistance and Ms. I. Suzu and Ms.
S. Fujikawa (Center for AIDS Research, Kumamoto University,
Kumamoto, Japan) for their research assistance.
Notes
[1] Competing
interests
The authors declare that they have no competing
interests.
References
|
1
|
Burstein E, Hoberg JE, Wilkinson AS,
Rumble JM, Csomos RA, Komarck CM, Maine GN, Wilkinson JC, Mayo MW
and Duckett CS: COMMD proteins, a novel family of structural and
functional homologs of MURR1. J Biol Chem. 280:22222–22232. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tao TY, Liu F, Klomp L, Wijmenga C and
Gitlin JD: The copper toxicosis gene product Murr1 directly
interacts with the Wilson disease protein. J Biol Chem.
278:41593–41596. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
de Bie P, van de Sluis B, Burstein E, van
de Berghe PV, Muller P, Berger R, Gitlin JD, Wijmenga C and Klomp
LW: Distinct Wilson's disease mutations in ATP7B are associated
with enhanced binding to COMMD1 and reduced stability of ATP7B.
Gastroenterology. 133:1316–1326. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vonk WI, de Bie P, Wichers CG, van den
Berghe PV, van der Plaats R, Berger R, Wijmenga C, Klomp LW and van
de Sluis B: The copper-transporting capacity of ATP7A mutants
associated with Menkes disease is ameliorated by COMMD1 as a result
of improved protein expression. Cell Mol Life Sci. 69:149–163.
2012. View Article : Google Scholar :
|
|
5
|
Ke Y, Butt AG, Swart M, Liu YF and
McDonald FJ: COMMD1 downregulates the epithelial sodium channel
through Nedd4-2. Am J Physiol Renal Physiol. 298:F1445–F1456. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Maine GN, Mao X, Komarck CM and Burstein
E: COMMD1 promotes the ubiquitination of NF-kappaB subunits through
a cullin-containing ubiquitin ligase. EMBO J. 26:436–447. 2007.
View Article : Google Scholar
|
|
7
|
Drevillon L, Tanguy G, Hinzpeter A, Arous
N, de Becdelièvre A, Aissat A, Tarze A, Goossens M and Fanen P:
COMMD1-mediated ubiquitination regulates CFTR trafficking. PLoS
One. 6:e183342011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
van de Sluis B, Mao X, Zhai Y, Groot AJ,
Vermeulen JF, van der Wall E, van Diest PJ, Hofker MH, Wijmenga C,
Klomp LW, et al: COMMD1 disrupts HIF-1alpha/beta dimerization and
inhibits human tumor cell invasion. J Clin Invest. 120:2119–2130.
2010. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ganesh L, Burstein E, Guha-Nijogi A,
Louder MK, Mascola JR, Klomp LW, Wijmenga C, Duckett CS and Nabel
GJ: The gene product Murr1 restricts HIV-1 replication in resting
CD4+ lymphocytes. Nature. 426:853–857. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Taura M, Kudo E, Kariya R, Goto H, Matsuda
K, Hattori S, Vaeteewoottacharn K, McDonald F, Suico MA, Shuto T,
et al: COMMD1/Murr1 reinforces HIV-1 latent infection through IκB-α
stabilization. J Virol. 89:2643–2658. 2015. View Article : Google Scholar
|
|
11
|
Beishline K and Azizkhan-Clifford J: Sp1
and the 'hallmarks of cancer'. FEBS J. 282:224–258. 2015.
View Article : Google Scholar
|
|
12
|
Resendes KK and Rosmarin AG: Sp1 control
of gene expression in myeloid cells. Crit Rev Eukaryot Gene Expr.
14:171–181. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li L, He S, Sun J and Davie J: Gene
regulation by Sp1 and Sp3. Biochem Cell Biol. 82:460–471. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Majello B, De Luca P and Lania L: Sp3 is a
bifunctional transcription regulator with modular independent
activation and repression domains. J Biol Chem. 272:4021–4026.
1997. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Taura M, Kariya R, Kudo E, Goto H, Iwawaki
T, Amano M, Suico MA, Kai H, Mitsuya H and Okada S: Comparative
analysis of ER stress response into HIV protease inhibitors:
Lopinavir but not darunavir induces potent ER stress response via
ROS/JNK pathway. Free Radic Biol Med. 65:778–788. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Suico MA, Taura M, Kudo E, Gotoh K, Shuto
T, Okada S and Kai H: The ETS factor myeloid Elf-1-like factor
(MEF)/Elf4 is transcriptionally and functionally activated by
hypoxia. Biol Pharm Bull. 39:641–647. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kudo E, Taura M, Matsuda K, Shimamoto M,
Kariya R, Goto H, Hattori S, Kimura S and Okada S: Inhibition of
HIV-1 replication by a tricyclic coumarin GUT-70 in acutely and
chronically infected cells. Bioorg Med Chem Lett. 23:606–609. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Taura M, Eguma A, Suico MA, Shuto T, Koga
T, Komatsu K, Komune T, Sato T, Saya H, Li JD and Kai H: p53
regulates Toll-like receptor 3 expression and function in human
epithelial cell lines. Mol Cell Biol. 28:6557–6567. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Greenwel P, Inagaki Y, Hu W, Walsh M and
Ramirez F: Sp1 is required for the early response of alpha2(I)
collagen to transforming growth factor-beta1. J Biol Chem.
272:19738–19745. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li H, Chan L, Bartuzi P, Melton SD, Weber
A, Ben-Shlomo S, Varol C, Raetz M, Mao X, Starokadomskyy P, et al:
Copper metabolism domain-containing 1 represses genes that promote
inflammation and protects mice from colitis and colitis-associated
cancer. Gastroenterology. 147:184–195.e3. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yeh DW, Chen YS, Lai CY, Liu YL, Lu CH, Lo
JF, Chen L, Hsu LC, Luo Y, Xiang R and Chuang TH: Downregulation of
COMMD1 by miR-205 promotes a positive feedback loop for amplifying
inflammatory- and stemness-associated properties of cancer cells.
Cell Death Differ. 23:841–852. 2016. View Article : Google Scholar :
|
|
23
|
O'Connor L, Gilmour J and Bonifer C: The
role of the ubiquitously expressed transcription factor Sp1 in
tissue-specific transcriptional regulation and in disease. Yale J
Biol Med. 89:513–525. 2016.PubMed/NCBI
|
|
24
|
Zhang DE, Hetherington CJ, Tan S, Dziennis
SE, Gonzalez DA, Chen HM and Tenen DG: Sp1 is a critical factor for
the monocytic specific expression of human CD14. J Biol Chem.
269:11425–11434. 1994.PubMed/NCBI
|
|
25
|
Tone M, Powell MJ, Tone Y, Thompson SA and
Waldmann H: IL-10 gene expression is controlled by the
transcription factors Sp1 and Sp3. J Immunol. 165:286–291. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Le Goff W, Guerin M, Petit L, Chapman MJ
and Thillet J: Regulation of human CETP gene expression: Role of
SP1 and SP3 transcription factors at promoter sites-690, -629, and
-37. J Lipid Res. 44:1322–1331. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Apt D, Watts RM, Suske G and Bernard HU:
High Sp1/Sp3 ratios in epithelial cells during epithelial
differentiation and cellular transformation correlate with the
activation of the HPV-16 promoter. Virology. 224:281–291. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Encarnacao PC, Ramirez VP, Zhang C and
Aneskievich BJ: Sp sites contribute to basal and inducible
expression of the human TNIP1 (TNFα-inducible protein 3-interacting
protein 1) promoter. Biochem J. 452:519–529. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li R, Hodny Z, Luciakova K, Barath P and
Nelson BD: Sp1 activates and inhibits transcription from separate
elements in the proximal promoter of the human adenine nucleotide
translocase 2 (ANT2) gene. J Biol Chem. 271:18925–18930. 1996.
View Article : Google Scholar : PubMed/NCBI
|