Introduction
As the most common malignant brain tumor, glioma
accounts for >80% of cancers in the brain (1,2).
Despite improvements in surgical resection techniques combined with
radiotherapy and chemotherapy, the prognosis of patients with
advanced glioma remains poor (3,4).
Deregulation of oncogenes or tumor suppressors has been implicated
in the development and malignant progression of glioma, and some
have been suggested as potential targets for glioma treatment
(5–7).
MicroRNAs (miRs), a class of non-coding RNAs 18–25
nucleotides in length, act as key regulators of gene expression,
mainly through binding to the complementary regions of their target
mRNAs, resulting in mRNA degradation or protein translation
inhibition (8–10). Through negative regulation of
their target's expression, miRs have been implicated in a variety
of physiological and pathological biological processes, including
cell proliferation, survival, apoptosis, autophagy, cell cycle,
migration and invasion (9,11).
Certain miRs were recently demonstrated to play promoting or
suppressive roles in glioma (12–15). For example, miR-146b acts as a
tumor suppressor in glioma by targeting TRAF6, and its
downregulation is significantly associated with poor prognosis of
glioma patients (16). miR-133b
contributes to arsenic-induced apoptosis in glioma cells by
targeting the human ether-à-go-go-related gene channel (17). miR-503 inhibits glioma cell
proliferation and invasion by inhibiting the expression of L1CAM
(18).
miR-98 is an important member of the let-7/miR-98
family, which has been reported to play oncogenic or tumor
suppressive roles in different human cancers (19,20). For example, Li et al
reported that miR-98 exerted suppressive effects on melanoma
metastasis through a negative feedback loop with interleukin (IL)-6
(21). Du et al
demonstrated that miR-98 plays a suppressive role in oral squamous
cell carcinoma growth and metastasis by directly targeting
insulin-like growth factor 1 receptor (22). The suppressive role of miR-98 in
glioma has been gradually uncovered (23). Chen et al reported that
overexpression of Raf kinase inhibitor protein (RKIP) suppressed
the invasion of glioma cells through upregulation of miR-98
(23). Fan et al
demonstrated that miR-98 overexpression inhibited glioma cell
invasion via targeting inhibitor of nuclear factor kappa-B kinase
subunit ε (24). However, the
regulatory mechanism of miR-98 expression in glioma remains
unclear.
Therefore, the present study aimed to investigate
the molecular mechanism underlying miR-98 expression in glioma and
the regulatory mechanism underlying the role of miR-98 in glioma
progression.
Materials and methods
Tissue collection
The present study was approved by the Ethics
Committee of Xiangya Hospital, Central South University, (Changsha,
China). Glioma tissues (n=84) and normal brain tissues (n=21) were
collected from our hospital between May 2010 and January 2012. The
patients included 52 men and 32 women, aged 23–68 years; 31
patients had WHO grade I-II, while 53 had WHO grade III-IV disease.
All patients provided written informed consent. All tissue samples
were immediately snap-frozen in liquid nitrogen and stored at −80°C
until use.
Cell culture and treatment
Normal human astrocytes were purchased from the IBS
Cell Bank of Fudan University (Shanghai, China) and cultured in
astrocyte media (Science Cell, Carlsbad, CA, USA) with 10% fetal
bovine serum (FBS) at 37°C in a humidified incubator containing 5%
CO2. Human glioma cell lines, including U87, U251, U373
and SHG44, were purchased from the Cell Bank of Central South
University. Cells were cultured in Dulbecco's modified Eagle's
medium (DMEM) with 10% FBS (both from Thermo Fisher Scientific,
Waltham, MA, USA) at 37°C in a humidified incubator containing 5%
CO2. 5-Aza-20-deoxycytidine (5-Aza) was purchased from
Sigma-Aldrich; Merck KGaA (St. Louis, MO, USA), and dissolved in
phosphate-buffered saline (PBS) at indicated concentrations. Glioma
cells were treated with 1 mM 5-Aza for 48 h, followed by assessment
of miR-98 expression.
Cell transfection
Lipofectamine 2000 (Thermo Fisher Scientific) was
used to perform cell transfection according to the manufacturer's
instructions. U87 cells were transfected with scramble miR
(miR-NC), miR-98 mimics, negative control (NC) inhibitor, miR-98
inhibitor, or co-transfected with miR-98 mimics and
pc-DNA3.1-Sal-like protein 4 (SALL4) plasmid, or miR-98 mimics and
blank pc-DNA3.1 vector. The cells were then cultured for 48 h
before the following assays.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA from tissues and cell lines was extracted
using TRIzol reagent, then converted to cDNA using the Reverse
Transcription kit (both from Thermo Fisher Scientific), according
to the manufacturer's instructions. qCR was then performed by using
the qPCR detection kit on ABI 7300 Plus thermocycler (both from
Thermo Fisher Scientific). For miR expression detection, U6 was
used as an internal reference. For mRNA detection, glyceraldehyde
3-phosphate dehydrogenase (GAPDH) was used as internal control. The
primer sequences for SALL4 were as follows: Forward,
5′-AGCACATCAACTCGGAGGAG-3′ and reverse,
5′-CATTCCCTGGGTGGTTCACTG-3′. The primer sequences for GAPDH were as
follows: Forward, 5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse,
5′-GGCTGTTGTCATACTTCTCATGG-3′. The PCR steps were 95°C for 5 min,
and 40 cycles of 95°C for 30 sec and at 60°C for 30 sec. The
relative expression was analyzed by the 2−ΔΔCq method
(25).
Western blot assay
Cells were lysed with ice-cold lysis buffer. Protein
was separated with 12% sodium dodecyl sulfate-polyacrylamide gel
electrophoresis and then transferred onto a polyvinylidene
difluoride membrane (Thermo Fisher Scientific), which was incubated
with PBS containing 5% non-fat milk (Yili, Beijing, China) for 3 h
at room temperature. After washing with PBS 3 times, the membrane
was incubated with rabbit anti-human SALL4 antibody (1:50, ab29112)
and rabbit anti-human GAPDH antibody (1:100, ab9485) (both from
Abcam, Cambridge, MA, USA) at 4°C overnight. After washing with PBS
3 times, the membrane was incubated with goat anti-rabbit secondary
antibody (1:5,000, ab6721; Abcam) at room temperature for 40 min.
The Chemiluminescent Substrate kit (Thermo Fisher Scientific) was
used to detect signals, according to the manufacturer's
instructions. The relative protein expression was analyzed by
Image-Pro Plus software 6.0, and presented as the density ratio vs.
GAPDH.
Detection of cell proliferation
U87 cells were plated at a density of 10,000
cells/well in 96-well plates. After being cultured for 0, 24, 48
and 72 h, the cells were incubated with
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT;
Sigma-Aldrich; Merck KGaA) at a final concentration of 0.5 mg/ml
for 4 h at 37°C. Following removal of the medium, 150 mM dimethyl
sulfoxide solution (Sigma-Aldrich; Merck KGaA) was added. The
absorbance was read at 570 nm using a BioTek™ ELX-800™ Absorbance
Microplate reader (BioTek Instruments, Inc., Winooski, VT,
USA).
Detection of cell migration
U87 cells were cultured to full confluence. Wounds
~1 mm wide were created using a plastic scriber. Cells were washed
and then cultured in DMEM containing 10% FBS for 48 h.
Subsequently, the cells were observed and photographed under a
microscope.
Detection of cell invasion
Transwell assay was conducted to detect cell
invasion capacity using Transwell chambers (BD Biosciences,
Franklin Lakes, NJ, USA). U87 cell suspension (106
cells/ml) was prepared in DMEM without added FBS, 300 µl of
the cell suspension was added into the upper chamber, and 300
µl of DMEM with 10% FBS was added into the lower chamber.
After incubation at 37°C for 24 h, the U87 cells that did not
migrate through the pores were carefully wiped off using a
cotton-tipped swab. The filters were fixed in 90% alcohol and then
stained by crystal violet (Sigma-Aldrich; Merck KGaA). The invading
cells were counted using an inverted microscope (Olympus, Tokyo,
Japan).
Dual luciferase reporter assay
TargetScan (www.targetscan.org) was used to predict the putative
target of miR-98, according to the manufaturer's instruction. The
wild-type (WT) or mutant type (MT) of SALL4 3′ untranslated region
(3′UTR) was constructed and inserted into the psiCHECK2 luciferase
reporter vector. U87 cells were co-transfected with WT-SALL4-3′UTR
or MT-SALL4-3′UTR reporter plasmid plus miR-98 mimics or miR-NC
using Lipofectamine 2000. Following co-transfection for 48 h, the
luciferase activity was determined using the Dual-Luciferase
Reporter assay system (Promega, Madison, WI, USA), according to the
manufacturer's instructions. Renilla luciferase activity was
normalized to Firefly luciferase activity.
Statistical analysis
Data are expressed as the mean ± standard deviation.
Statistical analysis was performed using SPSS 20 software (SPSS,
Armonk, NY, USA). The differences between two groups were analyzed
using the Student's t-test. The association of gene expression or
methylation status with clinical characteristics in glioma was
analyzed using the Chi-squared test. P<0.05 was considered to
indicate statistically significant differences.
Results
miR-98 is frequently downregulated in
glioma tissues and cell lines
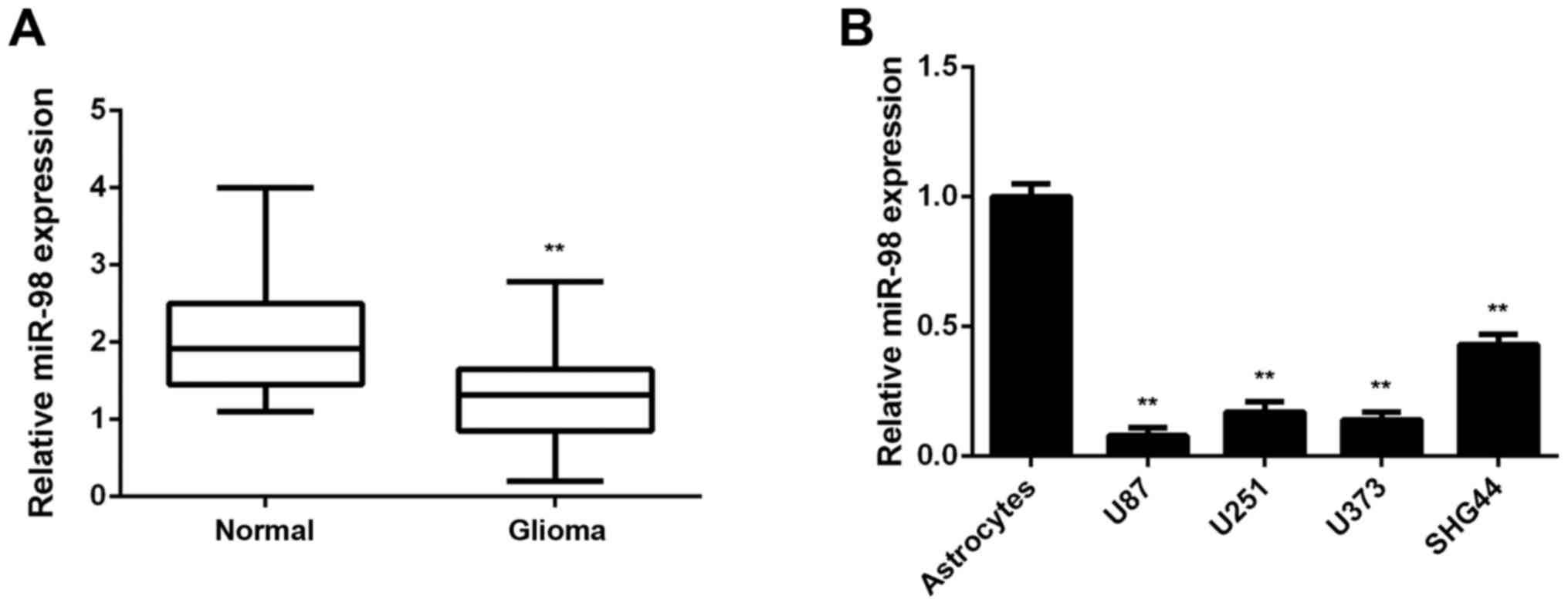
In the present study, the miR-98 levels in glioma
tissues were first examined; normal brain tissues were used as
control. RT-qPCR data indicated that miR-98 was frequently
downregulated in glioma tissues compared with normal brain tissues
(Fig. 1A). To confirm these
findings, the miR-98 levels in glioma cell lines were then
examined; normal human astrocytes were used as control. The
expression of miR-98 was also reduced in glioma cell lines compared
with normal astrocytes (Fig. 1B).
Accordingly, the expression of miR-98 was found to be downregulated
in glioma.
Hypermethylation causes miR-98
downregulation in glioma
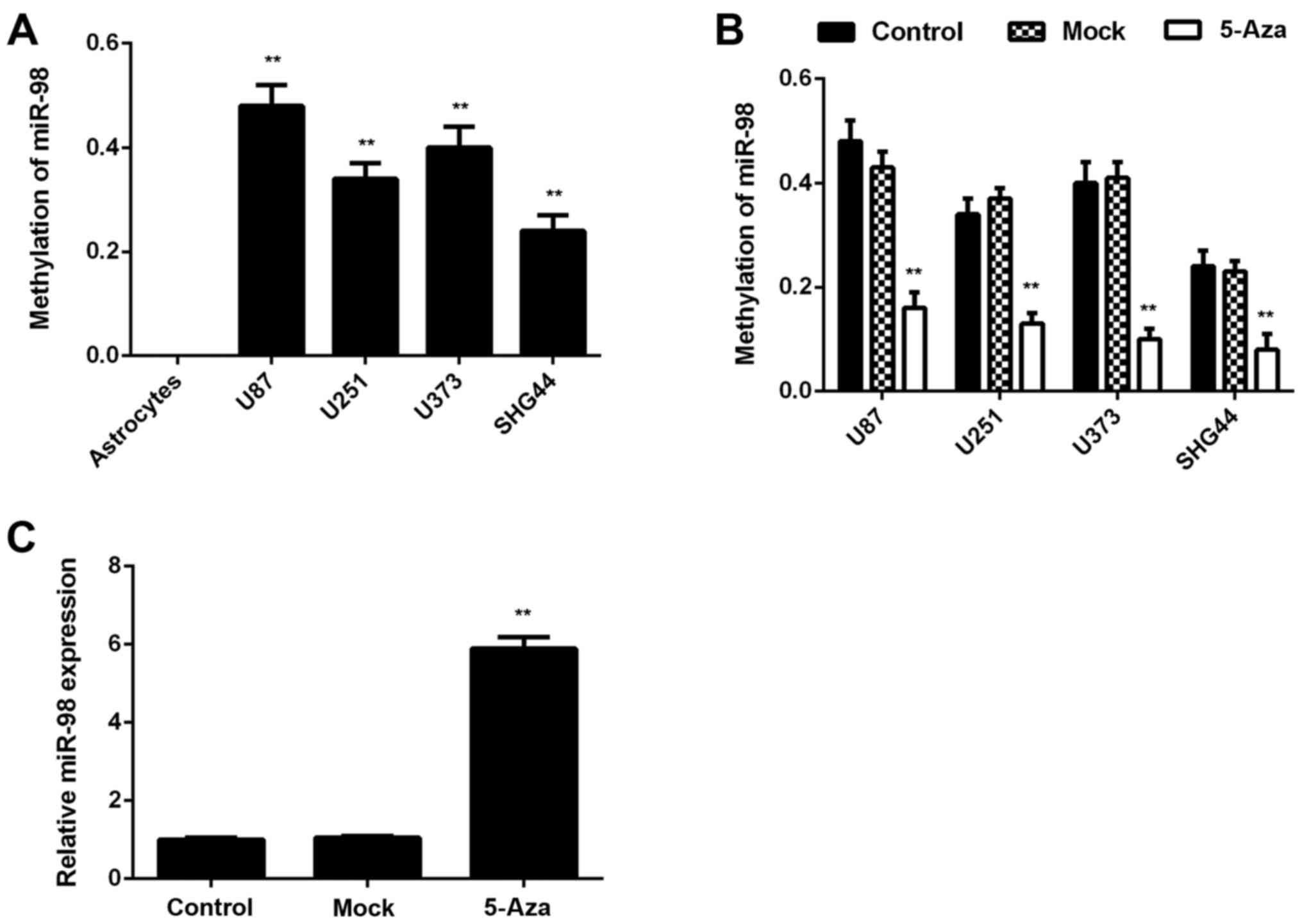
The mechanism underlying miR-98 downregulation in
glioma was further investigated. Methylation-specific PCR was
performed to analyze the methylation status of miR-98 in glioma and
normal brain tissues. It was observed that the methylation status
of miR-98 was significantly higher in glioma tissues compared with
that in normal brain tissues (Table
I). The methylation status of miR-98 in glioma cell lines and
normal human astrocytes was then examined. All glioma cell lines
were positive for methylation, whereas no methylation was
detectable in normal human astrocytes (Fig. 2A). Therefore, that the
downregulation of miR-98 expression in glioma is likely due to the
high methylation. To further confirm these findings, glioma cell
lines were treated with 5-Aza, a DNA methyltransferase inhibitor,
for 48 h. Following treatment, the methylation of miR-98 was
significantly reduced and the expression levels of miR-98 were
significantly upregulated in these glioma cell lines, which further
indicates that the hypermethylation status leads to miR-98
downregulation in glioma (Fig. 2B and
C).
 | Table IMethylation status of miR-98 in glioma
tissues and normal brain tissues. |
Table I
Methylation status of miR-98 in glioma
tissues and normal brain tissues.
| Tissues | No. | Methylated | Unmethylated | P-value |
|---|
| Normal brain | 21 | 1 | 20 | <0.01 |
| Glioma | 84 | 58 | 26 | |
miR-98 downregulation and methylation are
associated with a progressive phenotype and shorter survival in
glioma
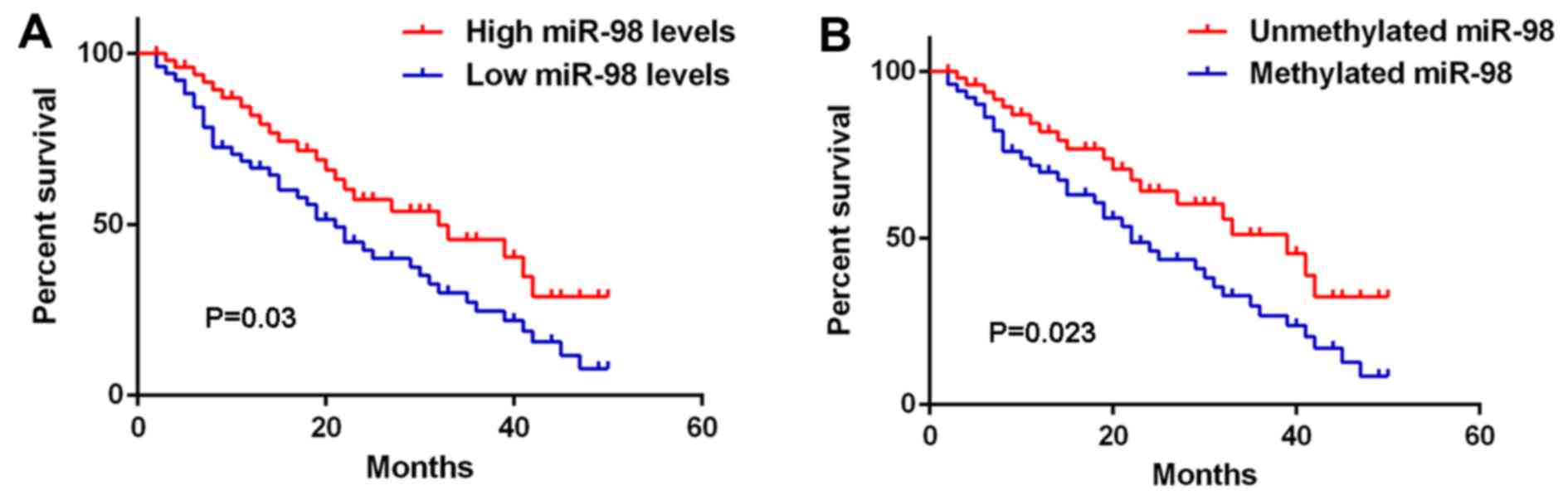
The clinical significance of miR-98 expression in
glioma was further investigated. As shown in Table II, low miR-98 levels were
significantly associated with high pathological grade and low
Karnofsky performance scale (KPS) index in glioma, but were not
associated with age, sex, or type of surgery (gross total resection
or partial resection), suggesting that reduced miR-98 expression
contributes to glioma progression. Interestingly, high methylation
of miR-98 was also found to be associated with high pathological
grade and low KPS index in glioma (Table III). Moreover, glioma patients
with low expression of miR-98 exhibited shorter overall survival
compared with those with high miR-98 levels (Fig. 3A). Consistently, glioma patients
with high miR-98 methylation had a poorer prognosis compared with
those with low miR-98 methylation (Fig. 3B). Therefore, both miR-98
downregulation and methylation in glioma are associated with a
progressive phenotype and shorter survival.
 | Table IIAssociation between miR-98 expression
and clinicopathological characteristics in glioma. |
Table II
Association between miR-98 expression
and clinicopathological characteristics in glioma.
| Variables | Cases (n=84) | Low miR-98
expression (n=41) | High miR-98
expression (n=43) | P-value |
|---|
| Age, years | | | | 0.512 |
| <55 | 39 | 21 | 18 | |
| ≥55 | 45 | 20 | 25 | |
| Sex | | | | 0.268 |
| Male | 52 | 28 | 24 | |
| Female | 32 | 13 | 19 | |
| WHO grade | | | | 0.025 |
| I–II | 31 | 10 | 21 | |
| III–IV | 53 | 31 | 22 | |
| KPS index | | | | 0.013 |
| >90 | 30 | 9 | 21 | |
| ≤90 | 54 | 32 | 22 | |
 | Table IIIAssociation between methylation
status of miR-98 and clinicopathological characteristics in
glioma. |
Table III
Association between methylation
status of miR-98 and clinicopathological characteristics in
glioma.
| Variables | Cases (n=84) | Methylated
(n=58) | Unmethylated
(n=26) | P-value |
|---|
| Age, years | | | | 0.354 |
| <55 | 39 | 29 | 10 | |
| ≥55 | 45 | 29 | 16 | |
| Sex | | | | 0.339 |
| Male | 52 | 38 | 14 | |
| Female | 32 | 20 | 12 | |
| WHO grade | | | | 0.013 |
| I–II | 31 | 16 | 15 | |
| III–IV | 53 | 42 | 11 | |
| KPS index | | | | 0.001 |
| >90 | 30 | 14 | 16 | |
| ≤90 | 54 | 44 | 10 | |
Restoration of miR-98 expression inhibits
U87 cell migration and invasion
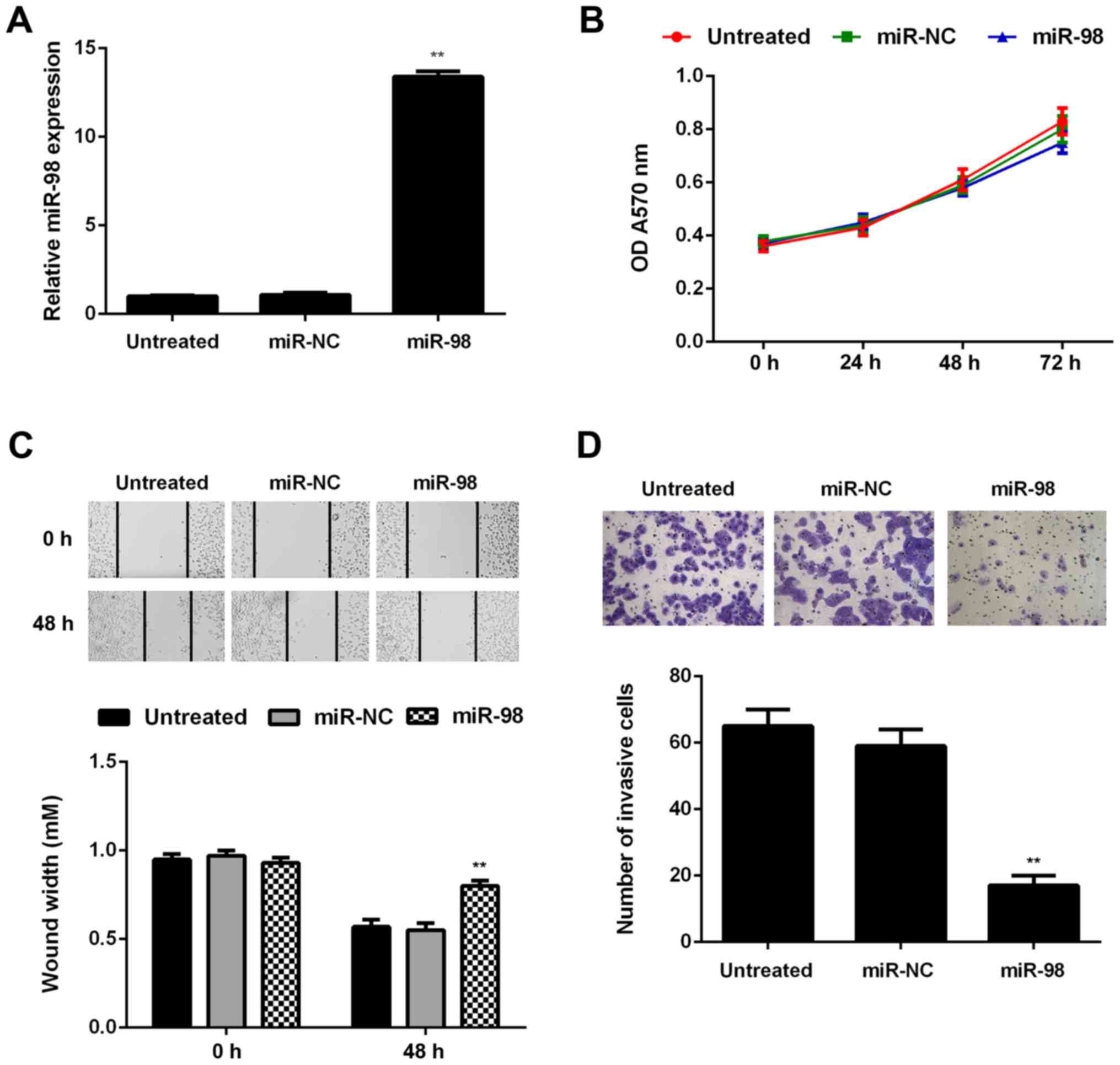
To further investigate the regulatory role of miR-98
in glioma, U87 cells were transfected with miR-98 mimic or miR-NC.
Untreated cells were used as the control group. Following
transfection, the miR-98 levels were significantly higher in the
miR-98 group compared with those in the untreated group (Fig. 4A). The proliferation, migration
and invasion of U87 cells in each group were then assessed using
the MTT, wound healing and Transwell assays, respectively. Although
it did not affect on cell proliferation, restoration of miR-98
expression significantly decreased U87 cell migration and invasion
(Fig. 4B–D).
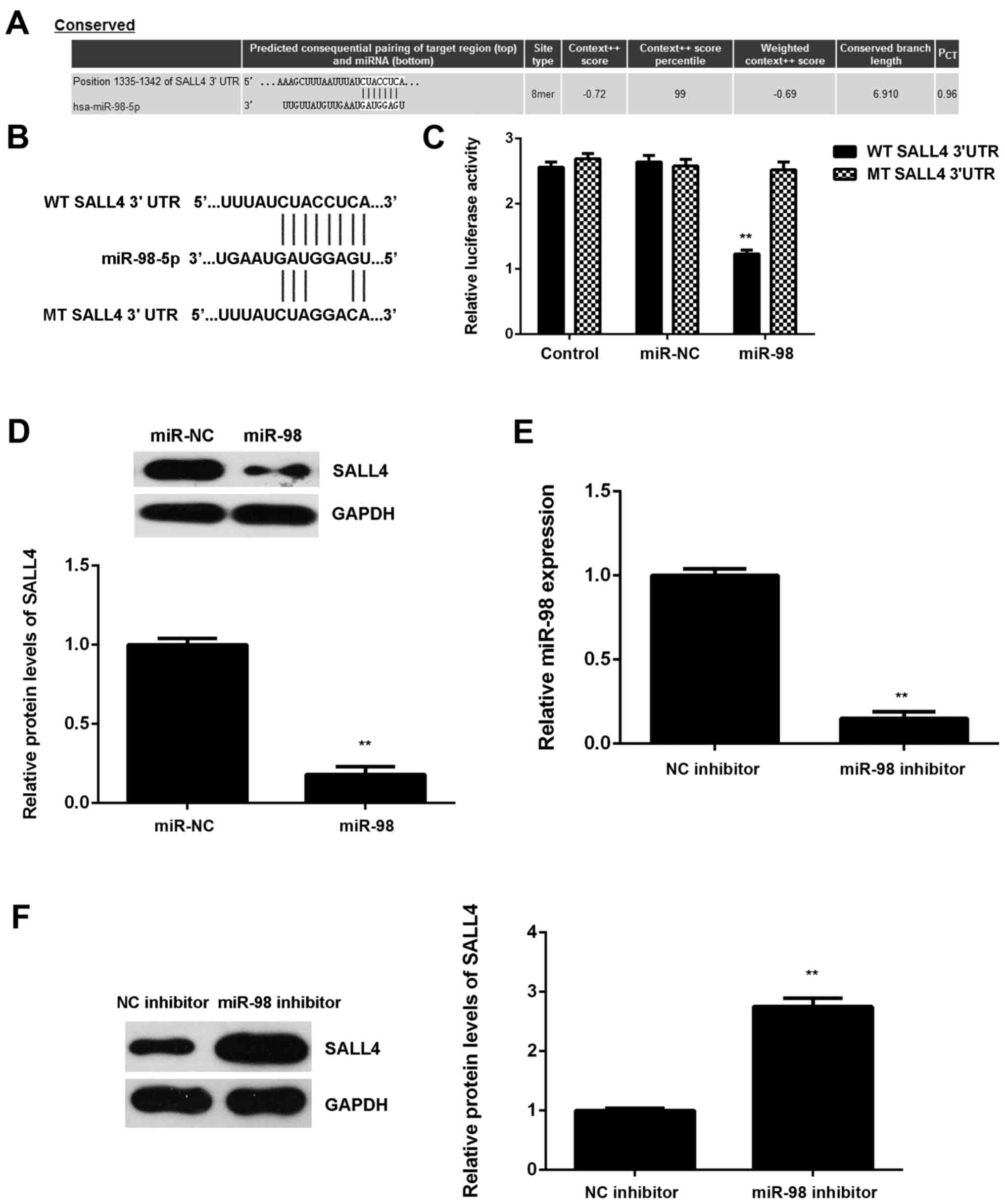
SALL4 is a target gene of miR-98 in U87
cells
Subsequently, the potential targets of miR-98 in
glioma were examined. SALL4 was predicted to be a potential target
of miR-98 (Fig. 5A). To confirm
this association, luciferase reporter plasmids containing WT or MT
of SALL4 3′UTR were generated (Fig.
5B). The luciferase reporter gene assay revealed that
luciferase activity was markedly downregulated in U87 cells
co-transfected with the WT-SALL4-3′UTR luciferase reporter plasmid
and miR-98 mimic compared with the control group, which was
eliminated by transfection with MT-SALL4-3′UTR luciferase reporter
plasmid (Fig. 5C). These findings
confirm that SALL4 is a direct target gene of miR-98 in U87
cells.
The regulatory effect of miR-98 on the protein
expression of SALL4 in U87 cells was then studied. The protein
levels of SALL4 were significantly lower in the miR-98 group
compared with those in the miR-NC group, indicating that
overexpression of miR-98 inhibited SALL4 protein expression in U87
cells (Fig. 5D). To further
confirm these data, U87 cells were then transfected with miR-98
inhibitor. Transfection with NC inhibitor was used as the control
group. Following transfection, the miR-98 levels were significantly
lower in the miR-98 inhibitor group compared with the NC inhibitor
group (Fig. 5E). Western blot
data further demonstrated that downregulation of miR-98 caused a
significant increase in the SALL4 protein expression in U87 cells
(Fig. 5F). Based on these
findings, miR-98 appears to directly bind to the 3′UTR of SALL4
mRNA and downregulate its protein expression in U87 cells.
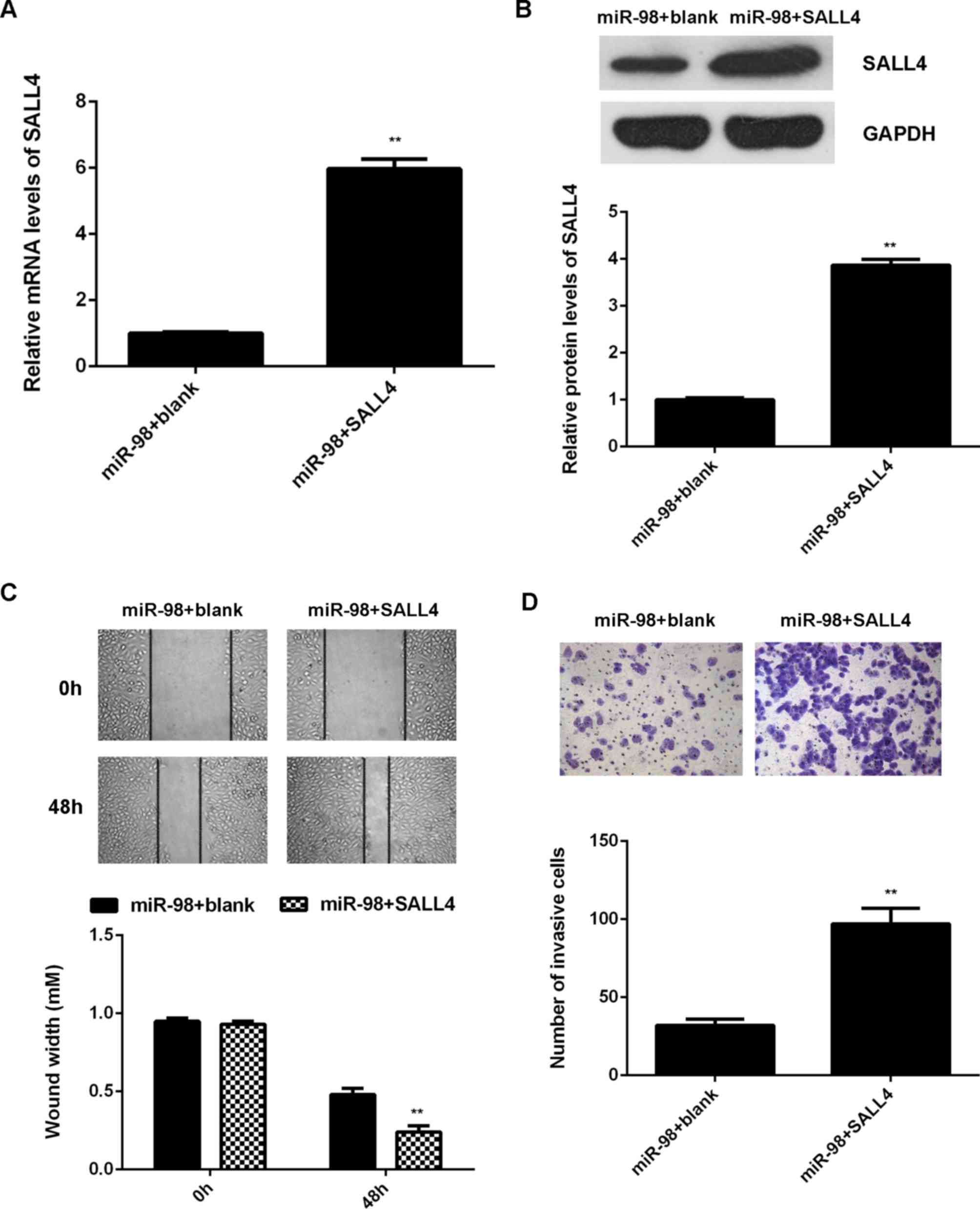
Restoration of SALL4 expression
attenuates the miR-145-mediated inhibition of U87 cell migration
and invasion
Subsequently, we investigated whether SALL4 was
involved in the miR-98-mediated malignant phenotypes of U87 cells.
miR-98-overexpressing U87 cells were transfected with
pcDNA3.1-SALL4 plasmid. miR-98-overexpressing U87 cells transfected
with blank pcDNA3.1 vector were used as the control group. After
transfection, the mRNA and protein levels of SALL4 were found to be
significantly higher in the miR-98 + SALL4 group compared with the
miR-98 + blank group (Fig. 6A and
B). As overexpression of miR-98 promoted U87 cell migration and
invasion, these two phenotypes were examined by conducting wound
healing and Transwell assays. The migration and invasion of U87
cells were significantly upregulated in the miR-98 + SALL4 group
compared with those in the miR-98 + blank group (Fig. 6C and D). According to the
abovementioned data, it was demonstrated that restoration of SALL4
expression attenuates the miR-145-mediated inhibition of migration
and invasion of glioma cells. Therefore, the inhibitory effects of
miR-98 on glioma are at least partly mediated through direct
targeting of SALL4.
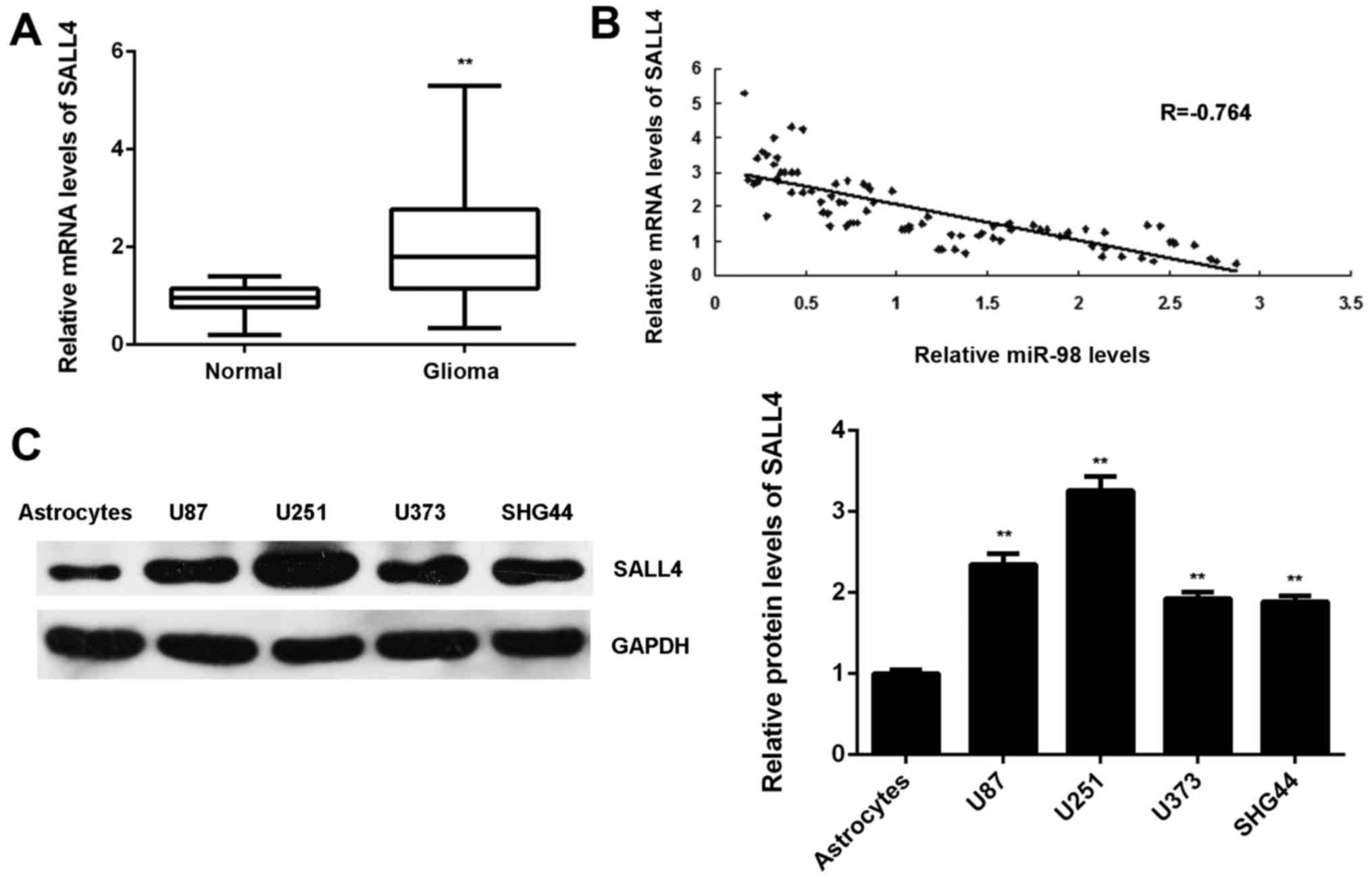
SALL4 is upregulated in glioma, with an
inverse correlation to miR-98 levels
Finally, the expression levels of SALL4 in glioma
was examined. The mRNA levels of SALL4 were significantly higher in
glioma tissues compared with those in normal brain tissues
(Fig. 7A). Moreover, our findings
demonstrated that the increased expression of SALL4 was associated
with malignant progression of glioma (Table IV). Interestingly, an inverse
correlation between the SALL4 and miR-98 levels in glioma tissues
was observed (Fig. 7B). These
findings suggest that the decreased miR-98 expression contributes
to the increased expression of SALL4 in glioma. In addition, the
protein levels of SALL4 were also higher in glioma cell lines
compared with those in normal astrocytes (Fig. 7C).
 | Table IVAssociation between SALL4 expression
and clinicopathological characteristics in glioma. |
Table IV
Association between SALL4 expression
and clinicopathological characteristics in glioma.
| Variables | Cases (n=84) | Low SALL4
expression (n=44) | High SALL4
expression (n=40) | P-value |
|---|
| Age, years | | | | 0.282 |
| <55 | 39 | 23 | 16 | |
| ≥55 | 45 | 21 | 24 | |
| Sex | | | | 0.263 |
| Male | 52 | 30 | 22 | |
| Female | 32 | 14 | 18 | |
| WHO grade | | | | 0.024 |
| I–II | 31 | 11 | 20 | |
| III–IV | 53 | 33 | 20 | |
| KPS index | | | | 0.012 |
| >90 | 30 | 10 | 20 | |
| ≤90 | 54 | 34 | 20 | |
Discussion
Recently, miR-98 was found to play a suppressive
role in glioma (23,24), but the underlying mechanism
remains largely unknown. In the present study, it was demonstrated
that miR-98 was frequently downregulated in glioma, which was
associated with DNA methylation. Both miR-98 downregulation and
methylation were found to be significantly associated with a more
aggressive tumor phenotype in glioma, as well as shorter survival
of glioma patients. Further investigation revealed that miR-98
inhibited the migration and invasion of U87 cells at least partly
via directly targeting SALL4, which was upregulated in glioma, and
its upregulation was associated with glioma progression.
Recently, miR-98 was found to act as a tumor
suppressor in glioma (23,24).
Chen et al reported that the expression of RKIP and miR-98
in glioma tissues was significantly lower compared with that in
normal brain tissues, and overexpression of RKIP may promote the
expression of miR-98, which further inhibits glioma cell invasion,
possibly through targeting HMGA2 (23). Another study revealed that
overexpression of miR-98 inhibited glioma cell migration and
invasion through inhibition of IκB kinase and matrix
metalloproteinase-9 expression, as well as nuclear factor-κB p65
nuclear translocation (24).
However, the regulatory mechanism of miR-98 expression in glioma
and its clinical significance remain unknown. In the present study,
miR-98 was frequently found to be downregulated in glioma tissues
and cell lines. Moreover, DNA methylation may be the main cause of
the reduced miR-98 expression in glioma, as the methylation status
of miR-98 was found to be significantly higher in glioma tissues
compared with that in normal brain tissues, and treatment with DNA
methyltransferase inhibitor significantly upregulated miR-98
expression in glioma cell lines. Furthermore, it was demonstrated
that both miR-98 downregulation and methylation were significantly
associated with higher WHO grade, low KPS and shorter survival in
glioma, suggesting that miR-98 expression or methylation status may
be used as important predictors of prognosis of glioma patients.
Furthermore, it was observed that restoration of miR-98 expression
inhibited the migration and invasion of glioma U87 cells, but did
not affect cell proliferation, which was consistent with the
findings of previous studies (23,24).
SALL4, a zinc finger transcription factor, was then
identified as a novel target gene of miR-98 in glioma cells by
using luciferase reporter gene assay. In fact, SALL4 was previously
reported to be an important marker for stem cells, as it plays a
key role in maintaining the self-renewal capacity of embryonic stem
cells (26). In recent years,
SALL4 was reported to be frequently upregulated in some cancer
types, and to act as an oncogene in esophageal squamous cell
carcinoma, gastric cancer, hepatocellular carcinoma and
intrahepatic cholangiocarcinoma (27–30). Moreover, the promoting role of
SALL4 in glioma has been gradually revealed. Zhang et al
found that the SALL4 expression levels in glioma were significantly
higher compared with those in normal brain tissues, and the
expression of SALL4 was closely correlated with glioma pathological
grade (31), which was consistent
with our data. In addition, they demonstrated that high SALL4
expression was correlated with poor prognosis of glioma patients.
In the present study, the expression of SALL4 was found to be
inversely correlated with miR-98 expression in glioma tissues,
suggesting that downregulation of miR-98 may contribute to SALL4
upregulation.
Moreover, the protein expression of SALL4 was found
to be negatively regulated by miR-98 in U87 cells. Therefore, it
was hypothesized that SALL4 may be involved in the miR-98-mediated
migration and invasion of glioma cells. To confirm this hypothesis,
miR-98-overexpressing U87 cells were transfected with SALL4
expression plasmid, and it was observed that restoration of SALL4
expression significantly attenuated the inhibitory effects of
miR-98 overexpression on U87 cell migration and invasion.
Accordingly, it was demonstrated that miR-98 plays a suppressive
role in the migration and invasion of U87 cells at least partly
through direct targeting of SALL4. In fact, similar findings were
reported in hepatocellular carcinoma. Zhou et al recently
found that miR-98 plays a suppressive role in the proliferation,
migration, invasion and epithelial-to-mesenchymal transition of
hepatocellular carcinoma cells via direct inhibition of SALL4
(32). Therefore, the present
study highlights the significance of miR-98/SALL4 signaling in
human cancers.
To the best of our knowledge, this is the first
study to demonstrate that high methylation induces the
downregulation of miR-98 in glioma, which further promotes glioma
cell migration and invasion via targeting SALL4. Therefore, miR-98
may be a potential therapeutic candidate for glioma.
Notes
[1] Competing
interests
The authors declare that they have no competing
interests.
References
|
1
|
Torre LA, Bray F, Siegel RL, Ferlay J,
Lortet-Tieulent J and Jemal A: Global cancer statistics, 2012. CA
Cancer J Clin. 65:87–108. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2015. CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Alireza M, Amelot A, Chauvet D, Terrier
LM, Lot G and Bekaert O: Poor prognosis and challenging in
treatment of optic nerve malignant gliomas: A literature review and
case report series. World Neurosurg. 97:751.e1–751.e6. 2017.
View Article : Google Scholar
|
|
4
|
Ashby LS, Smith KA and Stea B: Gliadel
wafer implantation combined with standard radiotherapy and
concurrent followed by adjuvant temozolomide for treatment of newly
diagnosed high-grade glioma: A systematic literature review. World
J Surg Oncol. 14:2252016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Guo E and Liu X: Overexpression of SCUBE2
inhibits proliferation, migration, and invasion in glioma cells.
Oncol Res. 25:437–444. 2017. View Article : Google Scholar
|
|
6
|
Liu ZJ, Liu HL, Zhou HC and Wang GC: TIPE2
inhibits hypoxia-Induced Wnt/β-catenin pathway activation and EMT
in glioma cells. Oncol Res. 24:255–261. 2016. View Article : Google Scholar
|
|
7
|
Li Z, Xu C, Gao M, Ding B, Wei X and Ji N:
Reduced expression of Jumonji, AT-rich interactive domain 2
(JARID2) in glioma inhibits tumor growth in vitro and in vivo.
Oncol Res. 25:365–372. 2017. View Article : Google Scholar
|
|
8
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
John B, Enright AJ, Aravin A, Tuschl T,
Sander C and Marks DS: Human MicroRNA targets. PLoS Biol.
2:e3632004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Ambros V: microRNAs: Tiny regulators with
great potential. Cell. 107:823–826. 2001. View Article : Google Scholar
|
|
12
|
Song H, Zhang Y, Liu N, Wan C, Zhang D,
Zhao S, Kong Y and Yuan L: miR-92b regulates glioma cells
proliferation, migration, invasion, and apoptosis via PTEN/Akt
signaling pathway. J Physiol Biochem. 72:201–211. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wei J, Nduom EK, Kong LY, Hashimoto Y, Xu
S, Gabrusiewicz K, Ling X, Huang N, Qiao W, Zhou S, et al: miR-138
exerts anti-glioma efficacy by targeting immune checkpoints. Neuro
Oncol. 18:639–648. 2016. View Article : Google Scholar :
|
|
14
|
Shi L, Fei X, Wang Z and You Y: PI3K
inhibitor combined with miR-125b inhibitor sensitize TMZ-induced
anti-glioma stem cancer effects through inactivation of
Wnt/beta-catenin signaling pathway. In Vitro Cell Dev Biol Anim.
51:1047–1055. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liang ML, Hsieh TH, Ng KH, Tsai YN, Tsai
CF, Chao ME, Liu DJ, Chu SS, Chen W, Liu YR, et al: Downregulation
of miR-137 and miR-6500-3p promotes cell proliferation in pediatric
high-grade gliomas. Oncotarget. 7:19723–19737. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu J, Xu J, Li H, Sun C, Yu L, Li Y, Shi
C, Zhou X, Bian X, Ping Y, et al: miR-146b-5p functions as a tumor
suppressor by targeting TRAF6 and predicts the prognosis of human
gliomas. Oncotarget. 6:29129–29142. 2015.PubMed/NCBI
|
|
17
|
Wang J, Li Y and Jiang C: miR-133b
contributes to arsenic-induced apoptosis in U251 glioma cells by
targeting the hERG channel. J Mol Neurosci. 55:985–994. 2015.
View Article : Google Scholar
|
|
18
|
Liu H, Song Z, Liao D, Zhang T, Liu F,
Zheng W, Luo K and Yang L: miR-503 inhibits cell proliferation and
invasion in glioma by targeting L1CAM. Int J Clin Exp Med.
8:18441–18447. 2015.
|
|
19
|
Wang CY, Zhang JJ, Hua L, Yao KH, Chen JT
and Ren XQ: MicroRNA-98 suppresses cell proliferation, migration
and invasion by targeting collagen triple helix repeat containing 1
in hepatocellular carcinoma. Mol Med Rep. 13:2639–2644. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang G, Zhang X and Shi J: miR-98 inhibits
cell proliferation and invasion of non-small cell carcinoma lung
cancer by targeting PAK1. Int J Clin Exp Med. 8:20135–20145.
2015.
|
|
21
|
Li F, Li XJ, Qiao L, Shi F and Liu W, Li
Y, Dang YP, Gu WJ, Wang XG and Liu W: miR-98 suppresses melanoma
metastasis through a negative feedback loop with its target gene
IL-6. Exp Mol Med. 46:e1162014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Du Y, Li Y, Lv H, Zhou S, Sun Z and Wang
M: miR-98 suppresses tumor cell growth and metastasis by targeting
IGF1R in oral squamous cell carcinoma. Int J Clin Exp Pathol.
8:12252–12259. 2015.
|
|
23
|
Chen Z, Cheng Q, Ma Z, Xi H, Peng R and
Jiang B: Overexpression of RKIP inhibits cell invasion in glioma
cell lines through upregulation of miR-98. BioMed Res Int.
2013:6951792013. View Article : Google Scholar
|
|
24
|
Fan YH, Ye MH, Wu L, Lv SG, Wu MJ, Xiao B,
Liao CC, Ji QK, Chai Y and Zhu XG: Overexpression of miR-98
inhibits cell invasion in glioma cell lines via downregulation of
IKKε. Eur Rev Med Pharmacol Sci. 19:3593–3604. 2015.PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
26
|
Chen X, Vega VB and Ng HH: Transcriptional
regulatory networks in embryonic stem cells. Cold Spring Harb Symp
Quant Biol. 73:203–209. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Forghanifard MM, Ardalan Khales S,
Javdani-Mallak A, Rad A, Farshchian M and Abbaszadegan MR: Stemness
state regulators SALL4 and SOX2 are involved in progression and
invasiveness of esophageal squamous cell carcinoma. Med Oncol.
31:9222014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang L, Xu Z, Xu X, Zhang B, Wu H, Wang
M, Zhang X, Yang T, Cai J, Yan Y, et al: SALL4, a novel marker for
human gastric carcinogenesis and metastasis. Oncogene. 33:5491–500.
2014. View Article : Google Scholar
|
|
29
|
Han SX, Wang JL, Guo XJ, He CC, Ying X, Ma
JL, Zhang YY, Zhao Q and Zhu Q: Serum SALL4 is a novel prognosis
biomarker with tumor recurrence and poor survival of patients in
hepatocellular carcinoma. J Immunol Res. 2014:2623852014.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Deng G, Zhu L, Huang F, Nie W, Huang W, Xu
H, Zheng S, Yi Z and Wan T: SALL4 is a novel therapeutic target in
intrahepatic cholangiocarcinoma. Oncotarget. 6:27416–27426. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang L, Yan Y, Jiang Y, Cui Y, Zou Y,
Qian J, Luo C, Lu Y and Wu X: The expression of SALL4 in patients
with gliomas: High level of SALL4 expression is correlated with
poor outcome. J Neurooncol. 121:261–268. 2015. View Article : Google Scholar
|
|
32
|
Zhou W, Zou B, Liu L, Cui K, Gao J, Yuan S
and Cong N: MicroRNA-98 acts as a tumor suppressor in
hepatocellular carcinoma via targeting SALL4. Oncotarget.
7:74059–74073. 2016. View Article : Google Scholar : PubMed/NCBI
|