Introduction
Osteoporosis is a systemic skeletal disorder
characterized by a reduction in bone mineral density (BMD) and
disrupted bone architecture, which results in a higher risk of bone
fractures (1,2). It is reported that ~50% of
postmenopausal women suffer from osteoporosis, which is defined as
postmenopausal osteoporosis (PMOP) (3,4).
The basic pathogenesis of PMOP involves an imbalance between bone
resorption by osteoclasts and bone formation by osteoblasts, which
is mainly induced by decreased estrogen. As PMOP is a chronic
disease, it imposes a significant financial burden on
postmenopausal women (4,5). Therefore, it is crucial to uncover
the mechanism and develop accurate diagnostic biomarkers of
PMOP.
Previous studies have indicated that osteoporosis
and BMD are heritable (6,7), and >60 susceptible loci have been
found to be associated with osteoporosis and BMD (6). Among these, polymorphisms of several
genes have been found to be involved in PMOP, including tumor
necrosis factor (TNF)-α, interleukin (IL)10, osteoprotegerin,
estrogen receptor 1 gene, estrogen receptor α, cannabinoid receptor
2, vitamin D receptor gene and LDL receptor related protein 5
(8–14).
Long non-coding RNAs (lncRNAs) are a set of
non-coding RNAs containing >200 nucleotides. There has been
increased interest focused on lncRNAs, which have been found to be
involved in diseases, including cancer and osteoporosis by
regulating their target genes at the transcriptional,
post-transcriptional and epigenetic levels (15,16). An lncRNA, DANCR was found to be
involved in PMOP by regulating TNF-α and IL6 (16). LncRNA MEG3 can suppress the
osteogenic differentiation of bone marrow mesenchymal stem cells
induced by PMOP (17). However,
reports of lncRNAs in PMOP remain limited.
In the present study, the lncRNA and mRNA expression
profile of blood samples from patients with PMOP and normal
controls (NCs) were identified by high-throughput RNA-sequencing.
To the best of our knowledge, the present study is the first to
obtain the lncRNA expression profiles of PMOP by RNA sequencing.
Based on the identified differentially expressed lncRNAs
(DElncRNAs) and differentially expressed mRNAs (DEmRNAs) in PMOP,
compared with NC, the DElncRNAs-DEmRNAs co-expression network was
constructed. The potential roles of these DElncRNAs were further
examined according to the functional annotation of their
co-expressed DEmRNAs. These findings may provide clues for
understanding the pathogenesis and novel insight for developing
diagnostic biomarkers of PMOP.
Materials and methods
Patients and samples
From April 2016 to March 2017, three women with PMOP
and two healthy women from Beijing Friendship Hospital were
enrolled in the present study. The inclusion criteria of patients
with PMOP were as follows: i) Postmenopausal women who were
diagnosed with osteoporosis. Osteoporosis was defined by the World
Health Organization criteria of a BMD T-score of −2.5 standard
deviations below the average for a young adult at peak bone density
in the femoral neck, total hip, or L1-L4; ii) clinically
symptomatic postmenopausal women with painful vertebral fractures
verified by X-ray and MRI within the last 6 months, who returned
for further examination and treatment. The patient characteristics
are listed in Table I. All
individuals provided written informed consent for use of their
samples in the present study. The present study was approved by the
Ethics Committee of Beijing Friendship Hospital, Capital Medical
University (Beijing, China; 2017-P2-084-01). From every
participant, a 2.5 ml peripheral whole blood was collected in
PAXgene® RNA blood tubes (PreAnalytiX GmbH,
Hombrechtikon, Switzerland) and stored at −80°C prior to
processing.
 | Table IPatient characteristics. |
Table I
Patient characteristics.
| Characteristic | Case 1 | Case 2 | Case 3 | Control 1 | Control 2 |
|---|
| Age (years) | 81 | 68 | 79 | 67 | 68 |
| Sex | Female | Female | Female | Female | Female |
| BMI
(kg/m2) | 23.1 | 15.6 | 15.6 | 28.6 | 22.5 |
| BMD-T score | −3 | −4.2 | −3.7 | 1.5 | 0.4 |
| History of
smoking | No | No | No | No | No |
| History of alcohol
intake | No | No | No | No | No |
| History of coffee
or carbonated drink intake | No | No | No | No | No |
| Family history of
matrilineal family | No | No | No | No | No |
| Lack of physical
activity | No | No | No | No | No |
| Bone
metabolism-associated disease | No | No | No | No | No |
| Bone
metabolism-associated drugs | No | No | No | No | No |
RNA isolation and sequencing
RNA isolation was performed using the PAXgene blood
RNA kit (PreAnalytiX GmbH) according to the manufacturer's
protocol. The concentration and purity of RNA were assessed using a
Nanodrop ND-2000 spectrophotometer (Thermo Fisher Scientific, Inc.,
Waltham, MA, USA). The integrity of RNA was assessed using a 2%
agarose gel. A RIN value was obtained using an Agilent 2100
bioanalyzer. The criteria for cDNA library construction were as
follows: i) Total RNA >5 µg; ii) concentration of RNA ≥200
ng/ml; iii) OD260/280 value 1.8-2.2.
Following removal of the ribosomal RNA using the
Ribo-Zero Magnetic kit (EpiCentre, Madison, WI, USA), the RNA was
purified and fragmented into 200-500-base pair fragments. The RNA
fragments were primed with random hexamer primers and the first
cDNA strand was synthe-sized, with the second cDNA strand
synthesized with dUTP instead of dTTP. The blunt ends of
double-stranded DNA were produced from cohesive ends of
double-stranded DNA using End Repair Enzyme mix (New England
BioLabs, Inc., Ipswich, MA, USA). Subsequently, 3′end adenylation
and adapter ligation were performed. When the second digested cDNA
strand was digested using the UNG enzyme (Illumina, Inc., San
Diego, CA, USA), polymerase chain reaction (PCR) was performed with
PCR Primer Cocktail (Illumina, Inc.) and PCR Master Mix (Illumina,
Inc.) to amplify the libraries. The following thermocycling
conditions were used for the PCR: Initial denaturation at 98°C for
30 sec; 15 cycles of 98°C for 10 sec, 65°C for 30 sec and 72°C for
30 sec, followed by a final extension step of 72°C for 5 min.
Certified Low Range Ultra Agarose (Bio-Rad Laboratories, Inc.,
Hercules, CA, USA) was used to purify the libraries, and the
libraries were quantified using Picogreen double-stranded DNA
quantitation kit (Molecular Probes; Thermo Fisher Scientific, Inc.)
on a TBS380 fluorometer (Promega Corporation, Madison, WI, USA).
The qualified libraries were amplified on cBot to generate the
cluster on the flowcell using TruSeq PE Cluster kit V3-cBot-HS
(Illumina, Inc.) according to the manufacturer's protocol.
Sequencing was performed on the Illumina Hiseq Xten platform
(Illumina, Inc.).
Quality control of raw sequence and
mapping of clean reads
The FASTQ sequence data were obtained from the
RNA-seq data using Base Calling V0.11.4 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/).
To obtain the high quality clean data, the low quality reads
including adaptor sequences, sequences with a quality score <20,
and sequences with an N base rate of raw reads >10% were removed
by using Cutadapt V1.9.1 (https://cutadapt.readthedocs.io/en/stable/). With
TopHat release 2.1.1 (http://tophat.cbcb.umd.edu/) and Ensemble gene
annotation, clean reads were aligned with the human reference
genome, Ensemble GRCh38.p7 (ftp://ftp.ncbi.nlm.nih.gov/genomes/Homo_sapiens). The
expression of mRNAs and lncRNAs was determined and outputted using
Cuffquant V2.2.1 (http://cufflinks.cbcb.umd.edu/).
Identification of DEmRNAs and DElncRNAs
in PMOP compared with NC
Fragments per Kilobase of exon per million fragments
mapped (FPKM) was used to determine the transcription abundance of
lncRNAs and mRNAs. The FPKMs of lncRNAs and mRNAs were calculated
using Cuffdiff (http://cole-trapnell-lab.github.io/cufflinks/cuff-diff/index.html).
A paired t-test was performed to obtain the DEmRNAs and DElncRNAs
in PMOP compared with NC. The thresholds of the DEmRNAs and
DElncRNAs was P<0.05.
DElncRNA-DEmRNA co-expression
network
To further examine the potential roles of DElncRNAs
and DEmRNAs in PMOP the DElncRNA-DEmRNA co-expression network was
constructed. Firstly, the Pearson's correlation coefficient (PCC)
between the expression levels of each DElncRNA-DEmRNA pair in the
PMOP and the NC group were calculated. Secondly, DElncRNA-DEmRNA
pairs with an absolute value of PCC ≥0.90 and P<0.05 were
defined as co-expressed DElncRNA-DEmRNA pairs. Those co-expressed
DElncRNA-DEmRNA pairs in which the expression level of DEmRNAs was
positively correlated with the expression level of DElncRNAs in
PMOP were defined as positively co-expressed DElncRNA-DEmRNA pairs.
Co-expressed DElncRNA-DEmRNA pairs in which the expression level of
DEmRNAs was negatively correlated with the expression level of
DElncRNAs in PMOP were defined as negatively co-expressed
DElncRNA-DEmRNA pairs. The positively and negatively co-expressed
DElncRNA-DEmRNA networks were visualized using Cytoscape 3.1
(http://cytoscape.org/).
Nearby DEmRNAs of the DElncRNAs
In order to identify the targeted DEmRNAs of
DElncRNAs by cis-regulatory effects, a search was performed
for the DEmRNAs transcribed within a 100-kb window upstream or
downstream of DElncRNAs, which served as nearby cis-targeted
DEmRNAs of DElncRNAs.
Functional annotation of DEmRNAs
co-expressed lncRNAs
Functional annotation, including Gene Ontology (GO)
function and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway
enrichment analyses of the DEmRNAs co-expressed with DElncRNAs was
performed using the GeneCoDis3 tool (http://genecodis.cnb.csic.es/analysis). A
hypergeometric test was used to obtain the P-value. The false
discovery rate (FDR; corrected P-value) of <0.05 was set as the
cut-off for significant GO terms and KEGG pathways.
Validation in the Gene Expression Omnibus
(GEO) dataset
The GSE56815 dataset was obtained from the GEO
(https://www.ncbi.nlm.nih.gov/geo/),
which consisted of 20 postmenopausal women with low hip BMD (case
group) and 20 postmenopausal women with high hip BMD (normal
group). All 40 females were Caucasian. The expression pattern of
selected DElncRNAs and DEmRNAs was verified using the GSE56815
dataset. GSE7158 was another dataset obtained from GEO, which
consisted of 12 women with a low peak bone mass (case group) and 14
women with a high peak bone mass (normal group). GSE7158 was also
used to validate the expression pattern of selected DElncRNAs.
Results
RNA-sequencing data
Total RNA extracted from each of the blood samples
met the criteria for cDNA library construction and RNA-sequencing.
Following trimming of the raw reads, 6.8×107,
6.8×107 and 6.7×107 clean reads were obtained
from the three respective blood samples from patients with
postmenopausal osteoporosis; 6.8×107 and
6.7×107 clean reads were obtained from the two
respective NCs. All of the clean reads were aligned to the human
reference genome (GRCh38.p7) and the mapped ratio of all samples
was >80%.
DEmRNAs and DElncRNAs in PMOP
A total of 185 significantly DEmRNAs (184
upregulated DEmRNAs and one downregulated DEmRNAs) were obtained
with P<0.05. The top 30 significant DEmRNAs are listed in
Table II. A total of 51
significantly DElncRNAs (25 upregulated DElncRNAs and 26
downregulated DElncRNAs) were obtained with P<0.05 (Table III). LOC105372321 was the most
markedly upregulated DElncRNA and LOC105374771 was the most
markedly downregulated DElncRNA in PMOP, compared with NC. NOD-like
receptor family pyrin domain containing 6 was the most
significantly upregulated DEmRNA and PAGE family member 2B was the
only downregulated DEmRNA. Furthermore, these DElncRNAs were
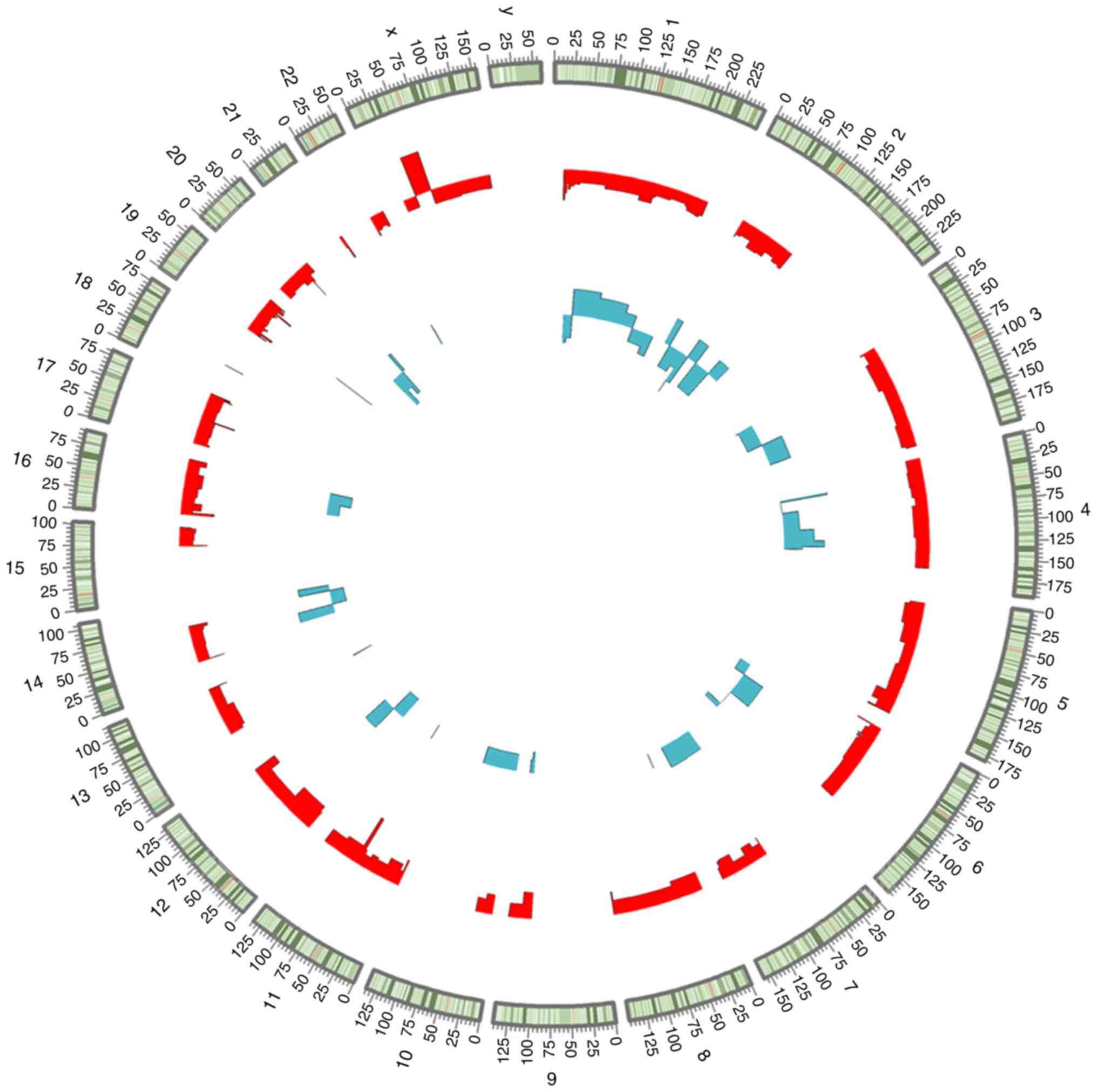
distributed in all chromosomes (chr.), with the exception of chr.
5, 15, 17, 18, 21 and sex chr. X and Y, whereas the DEmRNAs were
widely distributed in all chromosomes, except sex chr.Y (Fig. 1).
 | Table IITop 30 significantly DEGs in patients
with postmenopausal osteoporosis compared with normal controls. |
Table II
Top 30 significantly DEGs in patients
with postmenopausal osteoporosis compared with normal controls.
| DEG | Locus | log2
(fold-change) | Regulation | P-value |
|---|
| FOLR3 |
chr11:72135709-72139892 | −4.03 | Down |
5.00×10−05 |
| PI3 |
chr20:45174898-45176544 | −2.88 | Down |
5.00×10−05 |
| KRT23 |
chr17:40922695-40987135 | −2.76 | Down |
5.00×10−05 |
| CD177 |
chr19:43353658-43366075 | −2.57 | Down |
5.00×10−05 |
| REM2 |
chr14:22883164-22887680 | −2.15 | Down |
5.00×10−05 |
| NLRP6 |
chr11:278364-285942 | −2.14 | Down |
5.00×10−05 |
| MANSC1 |
chr12:12329262-12350541 | −2.06 | Down |
1.00×10−04 |
| LRG1 |
chr19:4537214-4540024 | −2.14 | Down |
1.50×10−04 |
| HCAR2 |
chr12:122695781-122710104 | −1.89 | Down |
2.00×10−04 |
| ADM |
chr11:10304979-10307402 | −1.67 | Down |
2.00×10−04 |
| MAK |
chr6:10762722-10838954 | −2.07 | Down |
2.50×10−04 |
| ABCG1 |
chr21:42199688-42304389 | −1.98 | Down |
4.50×10−04 |
| LGALSL |
chr2:64454192-64461383 | −1.69 | Down |
5.00×10−04 |
| BTNL8 |
chr5:180899076-180952166 | −1.89 | Down |
5.50×10−04 |
| CCNJL |
chr5:160251651-160339592 | −1.84 | Down |
6.50×10−04 |
| HIP1 |
chr7:75533297-75738976 | −1.51 | Down |
6.50×10−04 |
| SOCS3 |
chr17:78356776-78360079 | −1.85 | Down |
8.00×10−04 |
| PADI2 |
chr1:17066760-17119453 | −1.85 | Down |
8.00×10−04 |
| TGFA |
chr2:70402845-70554015 | −1.60 | Down |
1.05×10−03 |
| KIAA0226L |
chr13:46341999-46390042 | −1.35 | Down |
1.30×10−03 |
| OSM |
chr22:30262827-30266843 | −1.65 | Down |
1.40×10−03 |
| CRISPLD2 |
chr16:84819980-84909510 | −1.44 | Down |
1.55×10−03 |
| KCNS1 |
chr20:45091213-45101112 | −1.37 | Down |
1.55×10−03 |
| IL1B |
chr2:112829757-112836842 | −1.52 | Down |
1.70×10−03 |
| KAZN |
chr1:13893386-15220480 | −2.41 | Down |
2.10×10−03 |
| HLX |
chr1:220832762-220885059 | −1.31 | Down |
2.20×10−03 |
| ABCA1 |
chr9:104781001-104939096 | −2.05 | Down |
2.25×10−03 |
| HSPA1A |
chr6:31815513-31817942 | −1.45 | Down |
2.35×10−03 |
| KREMEN1 |
chr22:29058671-29168333 | −1.52 | Down |
2.45×10−03 |
 | Table IIISignificantly differentially
expressed lncRNAs in patients with postmenopausal osteoporosis
compared with normal controls. |
Table III
Significantly differentially
expressed lncRNAs in patients with postmenopausal osteoporosis
compared with normal controls.
| LncRNA | Locus | Regulation | log2 (fold
change) | P-value |
|---|
| LOC105374771 |
chr2:64390955-64425399 | Down | −2.57 |
5.00×10−05 |
| LOC105372321 |
chr19:21444103-21464331 | Up | 3.58 |
5.00×10−05 |
| PSMD5-AS1 |
chr9:120843041-120854373 D | own | −1.70 |
1.00×10−04 |
| PAX8-AS1 |
chr2:113215996-113278950 D | own | −2.41 |
7.00×10−04 |
| LOC105372578 |
chr20:24919978-24932985 | Down | −2.70 |
1.65×10−03 |
| LINC00570 |
chr2:11393980-11403077 | Up | 2.20 |
1.95×10−03 |
| LOC105369213 |
chr16:81739026-81777351 | Down | −1.82 |
5.00×10−05 |
| LOC105378020 |
chr6:137943074-137957648 | Up | inf |
3.75×10−03 |
| SNHG5 |
chr6:85677006-85678733 | Up | 1.83 |
4.15×10−03 |
| LOC105378415 |
chr10:88061829-88104391 | Down | −1.37 |
4.80×10−03 |
| LOC105374150 |
chr3:148439991-148465791 | Up | 1.76 | 0.01 |
| LINC00282 |
chr13:51804681-51845150 | Down | −1.69 | 0.01 |
| LOC102724231 |
chr3:44421131-44424025 | Down | −1.46 | 0.01 |
| LINC00211 |
chr2:37826246-37875863 | Down | −1.92 | 0.01 |
| LOC101929638 |
chr22:29180622-29205834 | Up | 1.77 | 0.01 |
| JHDM1D-AS1 |
chr7:140177260-140179640 | Up | 1.77 | 0.01 |
| LOC105370449 |
chr14:34551436-34557529 | Up | 2.89 | 0.01 |
| LOC105372881 |
chr1:207365821-207373252 | Down | −1.14 | 0.01 |
| LOC105373262 |
chr1:244230505-244325182 | Down | −1.75 | 0.01 |
| LOC105371455 |
chr1:157225405-157283617 | Up | 1.68 | 0.01 |
| LOC100507487 |
chr4:128428015-128519398 | Up | 3.25 | 0.01 |
| LOC101929866 |
chr20:45178476-45191638 | Down | −1.37 | 0.01 |
| LINC00963 |
chr9:129488659-129513686 | Down | −1.08 | 0.02 |
| LOC101928143 |
chr14:73460934-73463642 | Down | −1.16 | 0.02 |
| LOC399715 |
chr10:6326543-6335982 | Down | −1.37 | 0.02 |
| LOC105373730 |
chr2:165821976-165848198 | Up | 1.43 | 0.02 |
| LOC100507639 |
chr4:141321123-141332617 | Up | 1.45 | 0.02 |
| LOC105374768 |
chr2:64299870-64344064 | Down | −1.34 | 0.02 |
| LOC100506159 |
chr12:9936578-9943495 | Down | −1.91 | 0.02 |
| LOC105376834 |
chr1:21585689-21591187 | Down | −2.23 | 0.02 |
| LINC01094 |
chr4:78645993-78684501 | Up | 1.29 | 0.02 |
| LOC105378085 |
chr6:159586906-159604657 | Down | −1.36 | 0.02 |
| LOC105377067 |
chr3:46130889-46190381 | Down | −1.26 | 0.02 |
| LOC105369823 |
chr12:69624414-69699416 | Up | 2.14 | 0.02 |
| LOC101929422 |
chr14:101120762-101123545 | Up | 2.50 | 0.02 |
| LOC105375328 |
chr7:64944845-64950665 | Up | 1.73 | 0.02 |
| LINC01271 |
chr20:50292719-50321342 | Down | −2.13 | 0.03 |
| LOC102723828 |
chr4:31997378-32155406 | Up | inf | 0.03 |
| LOC105377384 |
chr4:116344095-116355205 | Up | 2.43 | 0.03 |
| LOC100506113 |
chr11:75801640-75814797 | Down | −1.26 | 0.03 |
| LOC105377782 |
chr8:2199669-2206204 | Up | 1.20 | 0.03 |
| HCG27 |
chr6:31197759-31203968 | Down | −0.90 | 0.03 |
| LINC01137 |
chr1:37454878-37474443 | Down | −1.84 | 0.03 |
| LOC105374546 |
chr4:26859623-26860599 | Up | 3.84 | 0.03 |
| LOC105376995 |
chr20:62533992-62536728 | Up | 1.78 | 0.03 |
| LOC105374852 |
chr2:88016353-88021354 | Up | 1.72 | 0.04 |
| LOC105378701 |
chr1:47172216-47177080 | Up | 2.11 | 0.04 |
| LOC101928595 |
chr16:30096429-30113557 | Down | −0.98 | 0.04 |
| LOC105372991 |
chr22:30447958-30472047 | Up | 1.33 | 0.04 |
| LOC105374769 |
chr2:64299870-64344064 | Down | −3.63 | 0.05 |
| GAS5 |
chr1:173863247-173867987 | Up | 1.03 | 0.05 |
DElncRNA-DEmRNA co-expression
network
Based on the expression levels of DElncRNAs and
DEmRNAs, the PCC describing the co-expression association between
185 DElncRNAs and 51 DEmRNAs, was calculated. A total of 3,057
DElncRNA-DEmRNA co-expression pairs were obtained with an absolute
value of PCC ≥0.90 and P<0.05. Among these, a total of 2,756
lncRNA-mRNA pairs were identified as being positively co-expressed,
whereas 301 lncRNA-mRNA pairs were negatively co-expressed. The
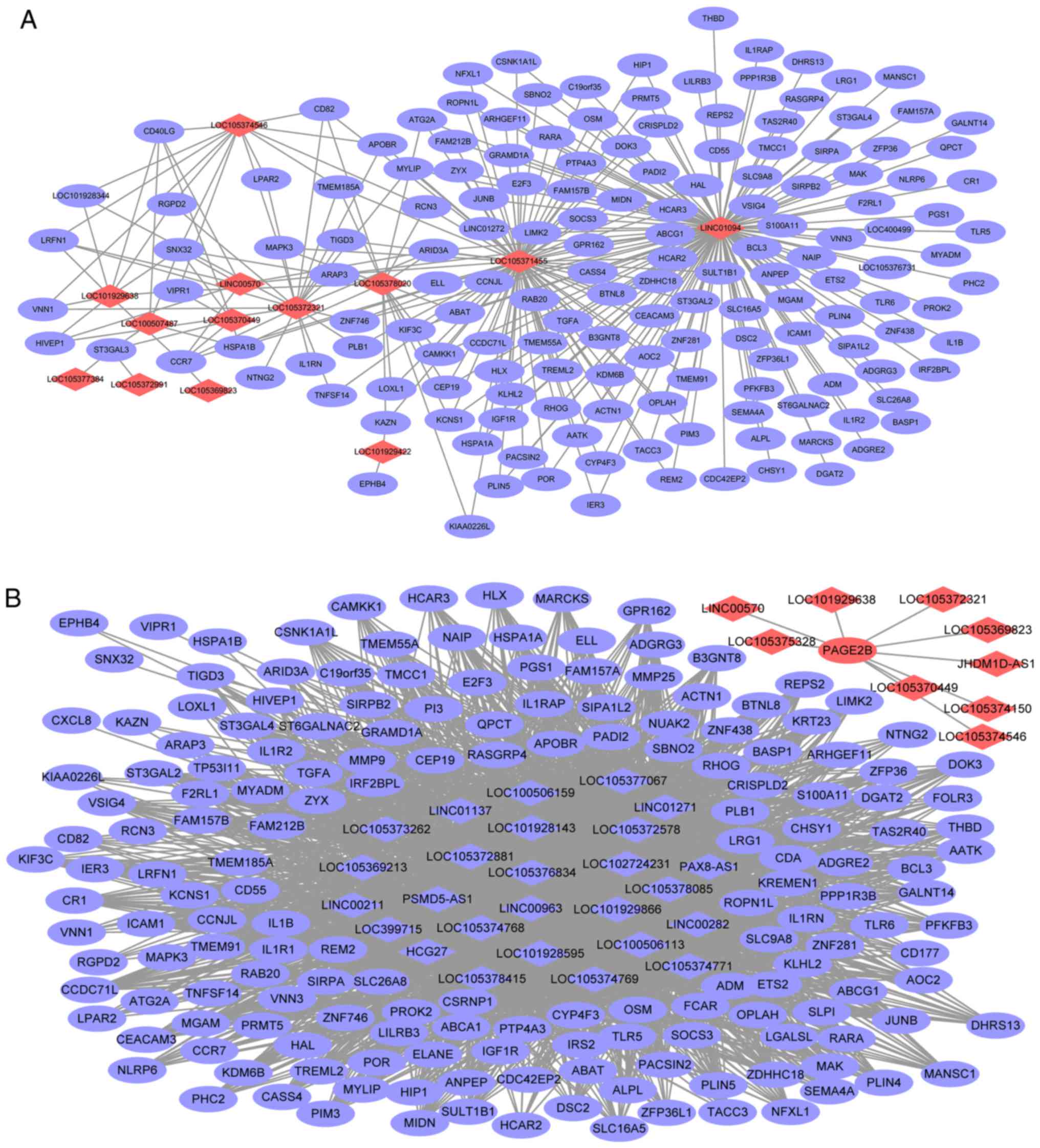
positively co-expressed DElncRNA-DEmRNA network (Fig. 2A) consisted of 215 nodes and 2,756
edges, and its hub lncRNAs were LOC105378415 (degree=159),
LOC105377067 (degree=157), HCG27 (degree=157), LOC101928143
(degree=154) and LINC00963 (degree=154). The negatively
co-expressed DElncRNA-DEmRNA network (Fig. 2B) consisted of 175 nodes and 301
edges, and its hub lncRNAs were LINC01094 (degree=135) and
LOC105371455 (degree=85).
Nearby DEmRNAs of DElncRNAs
A total of 97 DElncRNAs nearby DEmRNA pairs were
obtained. LOC101928595, LOC101929866 and HCG27 had 13, 8 and 7
nearby DEmRNAs, respectively, and were the top three DElncRNAs with
the most nearby DEmRNAs (Table
IV). DElncRNAs nearby DEmRNA pairs in which the expression
levels of DEmRNAs were co-expressed with DElncRNAs are listed in
Table V.
 | Table IVNearby DEmRNAs of DElncRNAs in
postmenopausal osteoporosis. |
Table IV
Nearby DEmRNAs of DElncRNAs in
postmenopausal osteoporosis.
| Count | DElncRNA | lncRNA
location | mRNA | mRNA location |
|---|
| 1 | LOC105376834 |
chr1:21585689-21591187 | ALPL |
chr1:21508981-21578412 |
| 4 | LINC01137 |
chr1:37454878-37474443 | ZC3H12A |
chr1:37474517-37484377 |
| SNIP1 |
chr1:37531436-37554344 |
| DNALI1 |
chr1:37556918-37595985 |
| GNL2 |
chr1:37556918-37595985 |
| 2 | LOC105378701 |
chr1:47172216-47177080 | CYP4Z1 |
chr1:47067487-47118320 |
| CYP4A22 |
chr1:47137424-47149738 |
| 1 | LOC105371455 |
chr1:157225405-157283617 | ETV3 |
chr1:157121190-157138591 |
| 4 | GAS5 |
chr1:173863247-173867987 C | ENPL |
chr1:173799549-173824639 |
| ZBTB37 |
chr1:173868094-173891122 |
| SERPINC1 |
chr1:173903803-173917378 |
| RC3H1 |
chr1:173931083-173993072 |
| 2 | LOC105372881 |
chr1:207365821-207373252 CD | 55 |
chr1:207321471-207360966 |
| CR2 |
chr1:207454299-207489895 |
| 1 | LOC105373262 |
chr1:244230505-244325182 C | 1orf100 |
chr1:244352062-244389896 |
| 1 | LINC00570 |
chr2:11393980-11403077 | E2F6 |
chr2:11444374-11466177 |
| 1 | LOC105374771 |
chr2:64390955-64425399 | LGALSL |
chr2:64454192-64461383 |
| 2 | LOC105374852 |
chr2:88016353-88021354 | RGPD2 |
chr2:87748086-87992864 |
| SMYD1 |
chr2:88067779-88113384 |
| 1 | LOC105373730 |
chr2:165821976-165848198 | GALNT3 |
chr2:165747802-165796352 |
| 1 | LOC102724231 | | TOPAZ1 |
chr3:44241885-44338010 |
| 2 | LOC105377067 |
chr3:46130889-46190381 | XCR1 |
chr3:46016989-46086803 |
| CCR1 |
chr3:46201708-46208341 |
| 1 | LOC105374546 |
chr4:26859623-26860599 | STIM2 |
chr4:26860690-27025381 |
| 1 | LINC01094 |
chr4:78645993-78684501 | ANXA3 |
chr4:78551587-78610451 |
| 1 | LOC100507639 |
chr4:141321123-141332617 | ZNF330 |
chr4:141220293-141234697 |
| 7 | HCG27 |
chr6:31197759-31203968 C | 6orf15 |
chr6:31111222-31112555 |
| PSORS1C1 |
chr6:31114830-31140092 |
| CDSN |
chr6:31114830-31140092 |
| PSORS1C2 |
chr6:31114830-31140092 |
| CCHCR1 |
chr6:31142438-31158238 |
| POU5F1 |
chr6:31164336-31170693 |
| HLA-C |
chr6:31268748-31272136 |
| 1 | LOC105378020 |
chr6:137943074-137957648 | TNFAIP3 |
chr6:137823668-137883314 |
| 1 | LOC105378085 |
chr6:159586906-159604657 | SOD2 |
chr6:159679063-159789703 |
| 4 | LOC105375328 |
chr7:64944845-64950665 | ZNF138 |
chr7:64794387-64853800 |
| LOC441239 |
chr7:64882492-64937316 |
| ZNF117 |
chr7:64974451-65006746 |
| ERV3-1 |
chr7:64974451-65006746 |
| 2 | LINC00963 |
chr9:129488659-129513686 | NTMT1 |
chr9:129608883-129642169 |
| C9orf50 |
chr9:129608883-129642169 |
| 2 | LOC399715 |
chr10:6326543-6335982 | PFKFB3 |
chr10:6144801-6254648 |
| PRKCQ |
chr10:6393037-6585361 |
| 2 | LOC105378415 |
chr10:88061829-88104391 | PTEN |
chr10:87863437-87975287 |
| RNLS |
chr10:88131897-88583860 |
| 2 | LOC100506113 |
chr11:75801640-75814797 | MOGAT2 |
chr11:75701595-75732958 |
| DGAT2 |
chr11:75768732-75801536 |
| 4 | LOC100506159 |
chr12:9936578-9943495 | KLRF2 |
chr12:9881488-9932430 |
| CLEC2A |
chr12:9881488-9932430 |
| CLEC12B |
chr12:10006137-10030606 |
| CLEC9A |
chr12:10030676-10066030 |
| 2 | LOC105369823 |
chr12:69624414-69699416 | LRRC10 |
chr12:69608563-69611162 |
| BEST3 |
chr12:69624414-69699416 |
| 4 | LINC00282 |
chr13:51804681-51845150 | WDFY2 |
chr13:51584193-51804206 |
| DHRS12 |
chr13:51584193-51804206 |
| CCDC70 |
chr13:51861980-51866236 |
| ATP7B |
chr13:51932668-52012130 |
| 3 | LOC101928143 |
chr14:73460934-73463642 | NUMB |
chr14:73275209-73458580 |
| HEATR4 |
chr14:73478483-73634418 |
| C14orf169 |
chr14:73478483-73634418 |
| 13 | LOC101928595 |
chr16:30096429-30113557 | INO80E |
chr16:29996208-30023280 |
| DOC2A |
chr16:29996208-30023280 |
| C16orf92 |
chr16:30023333-30053026 |
| FAM57B |
chr16:30023333-30053026 |
| ALDOA |
chr16:30053089-30070420 |
| PPP4C |
chr16:30075975-30085377 |
| TBX6 |
chr16:30085792-30091919 |
| GDPD3 |
chr16:30096429-30113557 |
| MAPK3 |
chr16:30114104-30123309 |
| CORO1A |
chr16:30183392-30189076 |
| BOLA2B |
chr16:30192929-30206927 |
| SLX1A |
chr16:30192929-30206927 |
| SULT1A3 |
chr16:30192929-30206927 |
| 2 | LOC105369213 |
chr16:81739026-81777351 | CMIP |
chr16:81445169-81711762 |
| PLCG2 |
chr16:81779257-81962693 |
| 3 | LOC105372578 |
chr20:24919978-24932985 | CST7 |
chr20:24949229-24959928 |
| APMAP |
chr20:24962924-24992974 |
| ACSS1 |
chr20:25006229-25058182 |
| 8 | LOC101929866 |
chr20:45178476-45191638 | STK4 |
chr20:44966473-45079977 |
| KCNS1 |
chr20:45091213-45101112 |
| WFDC5 |
chr20:45109451-45116321 |
| WFDC12 |
chr20:45123425-45124465 |
| PI3 |
chr20:45174898-45176544 |
| SEMG1 |
chr20:45206963-45209773 |
| SEMG2 |
chr20:45221368-45224458 |
| SLPI |
chr20:45230820-45290352 |
| 1 | LINC01271 |
chr20:50292719-50321342 | LINC01272 |
chr20:50267467-50279795 |
| 2 | LOC105376995 |
chr20:62533992-62536728 | GATA5 |
chr20:62463496-62475970 |
| MIR1-1HG |
chr20:62543069-62570764 |
| 3 | LOC101929638 |
chr22:29180622-29205834 | KREMEN1 |
chr22:29058671-29168333 |
| RHBDD3 |
chr22:29259854-29300525 |
| EWSR1 |
chr22:29259854-29300525 |
| 5 | LOC105372991 |
chr22:30447958-30472047 | SF3A1 |
chr22:30331987-30378655 |
| SEC14L2 |
chr22:30396940-30436501 |
| SEC14L3 |
chr22:30447958-30472047 |
| SEC14L6 |
chr22:30522796-30546717 |
| GAL3ST1 |
chr22:30554634-30574588 |
 | Table VDElncRNA-nearby DEmRNA pairs in which
DEmRNAs are co-expressed with DElncRNAs. |
Table V
DElncRNA-nearby DEmRNA pairs in which
DEmRNAs are co-expressed with DElncRNAs.
| DEmRNA D | ElncRNA | PCC | P-value |
|---|
| DGAT2 | LOC100506113 |
9.77×10−01 |
4.05×10−03 |
| KCNS1 | LOC101929866 |
9.02×10−01 |
3.64×10−02 |
| PI3 | LOC101929866 |
9.65×10−01 |
7.89×10−03 |
| SLPI | LOC101929866 |
9.48×10−01 |
1.41×10−02 |
| LGALSL | LOC105374771 |
9.93×10−01 |
6.56×10−04 |
| ALPL | LOC105376834 |
9.60×10−01 |
9.51×10−03 |
Functional annotation
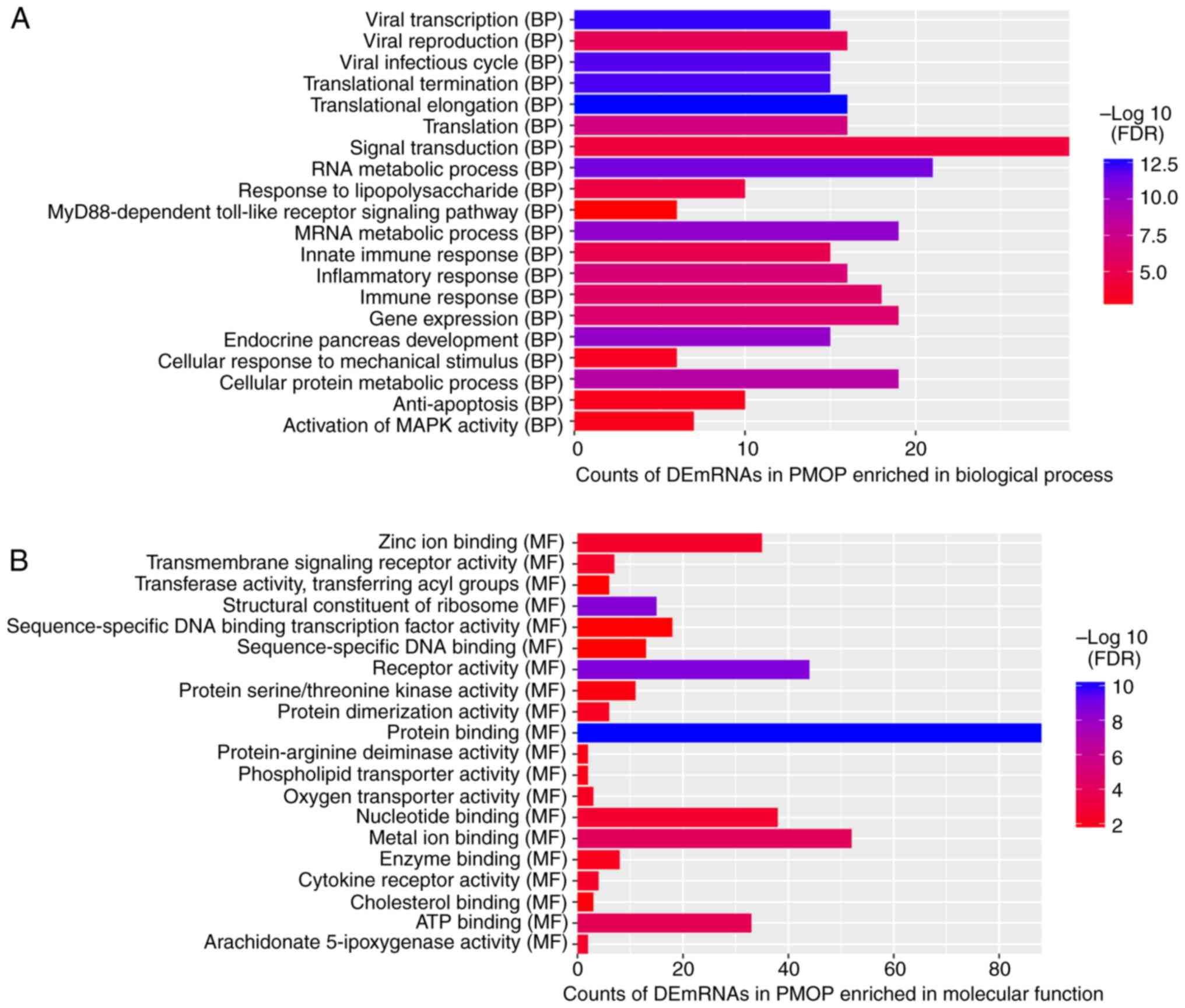
Based on the GO enrichment analysis of DEmRNAs
co-expressed with DElncRNAs, inflammatory response (FDR=3.14E-5),
anti-apoptosis (FDR=5.44E-05), plasma membrane (FDR=1.48E-11),
integral to membrane (FDR=6.06E-10), protein binding
(FDR=1.39E-06), and receptor activity (FDR=8.92E-06) were the most
significantly enriched GO terms in PMOP (Fig. 3A–C). Hematopoietic cell lineage
(FDR=0.000244565), Osteoclast differentiation (FDR=0.000438367) and
Cytokine-cytokine receptor interaction (FDR=0.00212347) were the
most significantly enriched KEGG pathways in PMOP (Fig. 3D).
Validation in the GEO dataset
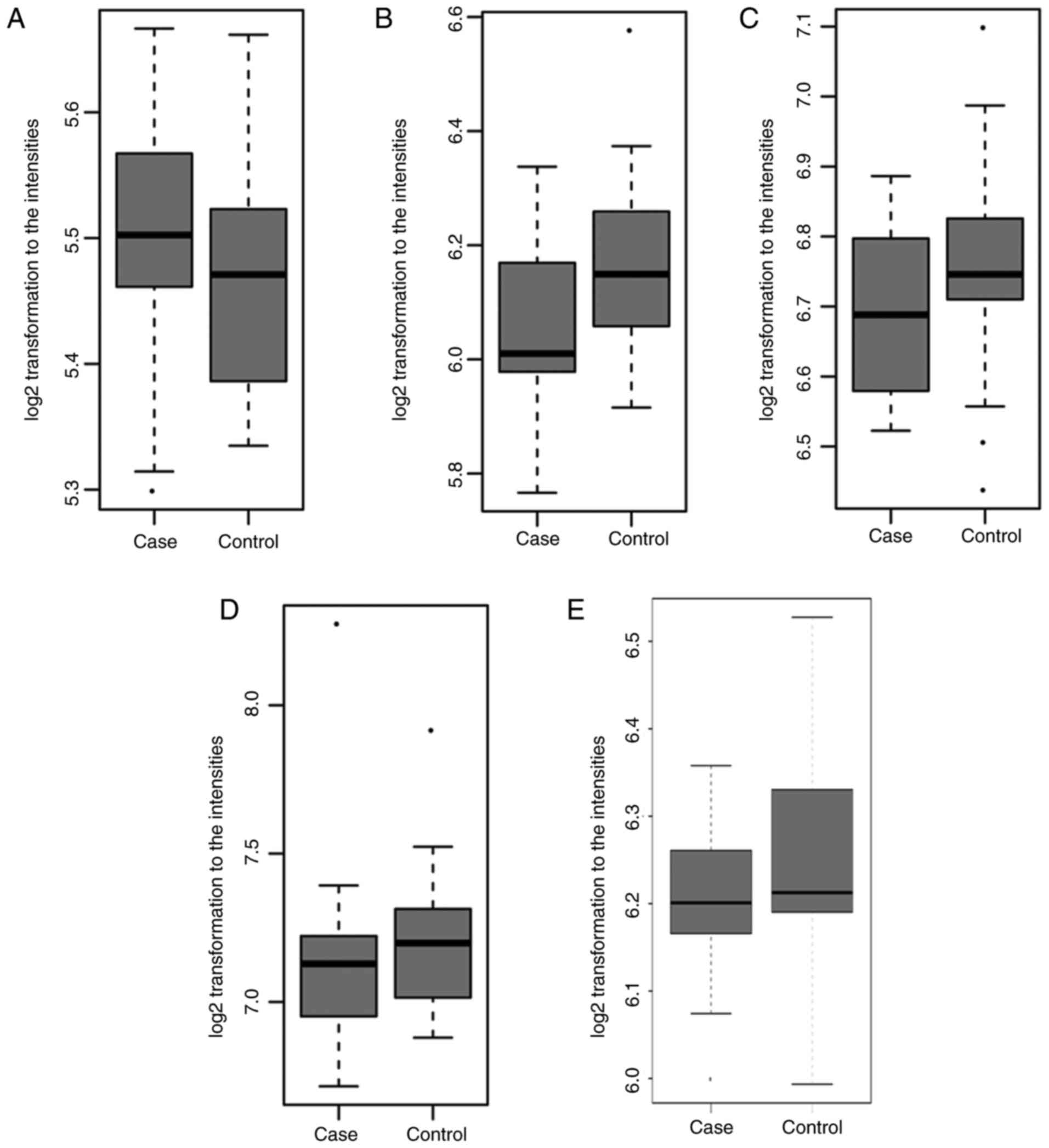
The expression patterns of selected DElncRNAs
(LINC00963, LOC105376834, LOC101929866, LOC105374771 and
LOC100506113) and DEmRNAs [alkaline phosphatase, liver/bone/kidney
(ALPL), suppressor of cytokine signaling 3 (SOCS3), secretory
leukocyte peptidase inhibitor (SLPI) and CD177] were verified using
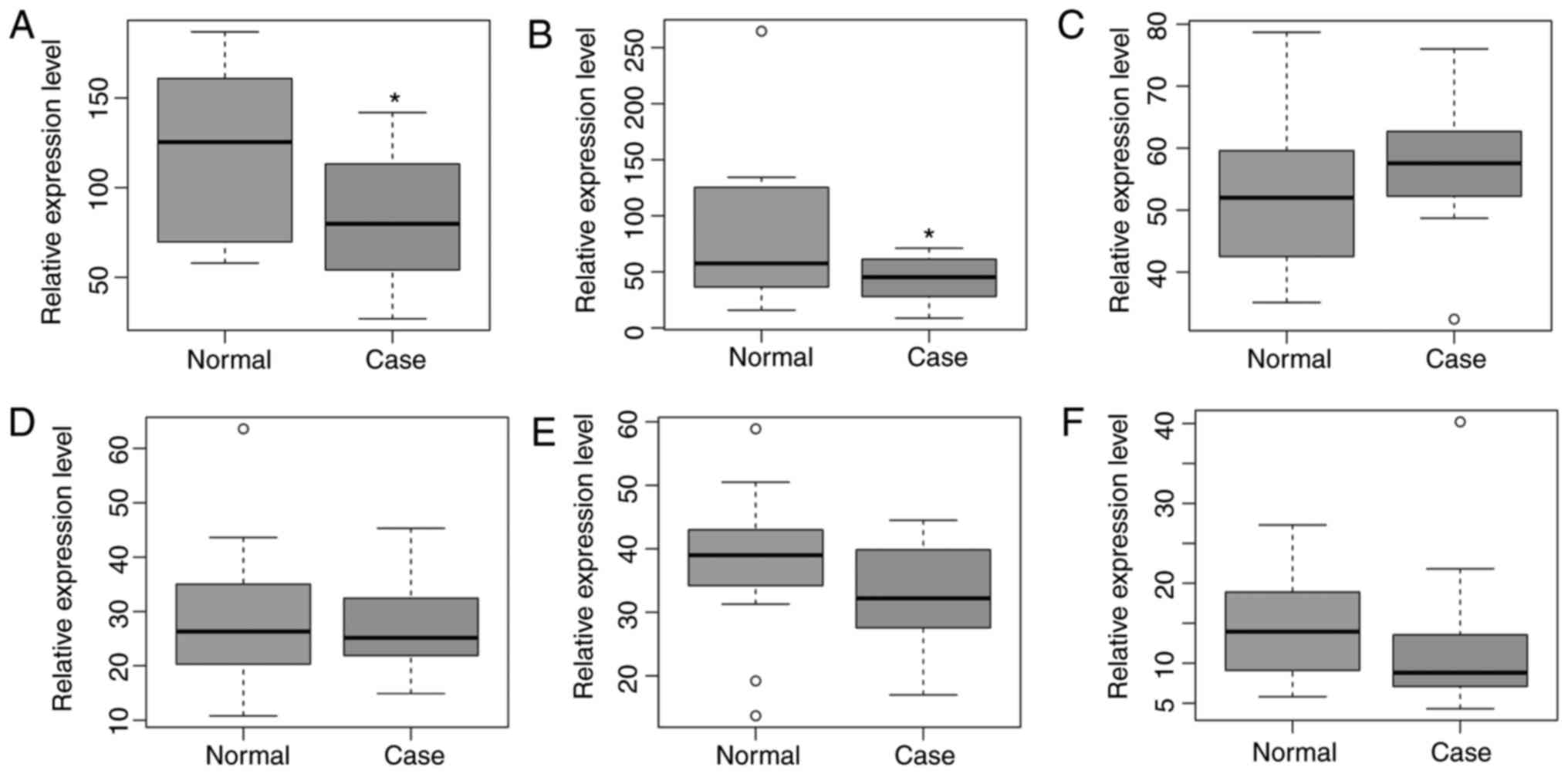
the GSE56815 dataset. As shown in Fig. 4A–D, SOCS3, SLPI and CD177 were
upregulated in PMOP, which was consistent with the RNA-sequencing
results. ALPL was downregulated in PMOP, which was inconsistent
with the RNA-sequencing results. However, only one of these five
DElncRNAs, LINC00963, was detected in GSE56815, which may be due to
the restriction of the microarray. LINC00963 was downregulated in
PMO, which showed the same pattern with that in the RNA-sequencing
results (Fig. 4E).
The expression pattern of six DElncRNAs (PSMD5-AS1,
PAX8-AS1, JHDM1D-AS1, LINC00963, LOC100506113 and HCG27) was
validated by GSE7158. Five DElncRNAs (PSMD5-AS1, PAX8-AS1,
LINC00963, LOC100506113 and HCG27) were downregulated, whereas
JHDM1D-AS1 was upregulated, in PMOP compared with NC (Fig. 5), which was the same pattern found
in the RNA-sequencing results.
Discussion
Although the function of the majority of lncRNAs
remains to be elucidated, previous studies have indicated that
lncRNAs may be involved in the pathogenesis of PMOP. Identifying
the key DElncRNAs in PMOP not only provides novel clues for
understanding the function of lncRNAs, but also contributes to
developing novel biomarkers of PMOP.
In the present study, the landscape of lncRNAs in
PMOP was obtained and a total of 51 DElncRNAs in PMOP were
identified. With the exception of LINC00963 and GAS5, no previous
study has reported on the function of the remaining 49 DElncRNAs.
In addition, the present study is the first, to the best of our
knowledge, to show that these 51 DElncRNAs may be associated with
PMOP.
LINC00963 is reported to be involved in cell
viability, motility and invasiveness in prostate cancer cells by
affecting the expression of epidermal growth factor receptor
(18). In the present study,
LINC00963 was a significantly downregulated lncRNA in PMOP. Whether
LINC00963 is involved in PMOP by regulating the viability, motility
and invasiveness of osteo-clasts and osteoblast requires further
investigation.
Although the functions of lncRNAs remain to be fully
elucidated, previous studies have indicated that lncRNAs are
important in regulating the expression levels of genes and
proteins, and are involved in a variety of biochemical processes
and diseases (19–21). To date, calculating the
correlation coefficients between the expression levels of lncRNAs
and genes has been the most popular approach to identify potential
target genes of lncRNAs (22,23). Accumulated evidence has indicated
that lncRNA-mRNA co-expression analysis can be used to examine the
biological functions of lncRNAs in various diseases by examining
their co-expressed mRNAs (24–26). In addition, several lncRNA-gene
pairs have been validated by in vitro experiments (27).
In the present study, LINC00963 was a hub lncRNA of
the positively co-expressed DElncRNA-DEmRNA network. Among its 154
co-expressed DEmRNAs, SOCS3 and adrenomedullin (ADM) were two of
the top 20 DEmRNAs in PMOP. ADM is a 52-amino acid peptide with
several biological functions. Previous studies have demonstrated
that ADM is closely associated with regulating bone formation
(28). The expression of ADM has
been detected in chondrocytes and osteoblasts (29). ADM can promote growth of
chondrocytes and osteoblasts in vitro (29). Additionally, apoptotic cell death
in serum-starved osteoblasts can be reduced by ADM (30). In the present study, the
expression of ADM was significantly downregulated in the blood
samples of patients with PMOP. It was hypothesized that
downregulated ADM may be involved in PMOP through reducing the bone
formation induced by the reduced proliferation of osteoblasts. The
SOCS family includes cytokine-inducible negative regulators of
cytokine signaling. As a member of the SOCS family, SOCS3 can be
regulated by various cytokines (31). Previous studies have reported that
increased SOCS3 elevated transforming growth factor-β, TNF-α and
RANK ligand (RANKL)-induced osteoclast formation, and promoted
precursors to the osteoclast lineage through the inhibition of
specific anti-osteoclastic Janus kinase/signal transducer and
activator of transcription signals (31). In addition, increased SOCS3 is
closely associated with inflammation-induced bone loss (32). SOCS3 is also involved in
RANKL-mediated dendritic cell-derived osteoclastogenesis by
regulating associated cytokine signaling (32). Diabetes-associated
inflammation-induced alveolar bone loss can also be regulated by
SOCS3. In the present study, downregulated SOCS3 was detected in
blood samples of patients with PMOP, which suggested that SOCS3 may
also be a regulator of PMOP. It was hypothesized that LINC00963-ADM
and LINC00963-SOCS3 interactions may be key in PMOP.
Another lncRNA, GAS5, has been reported to regulate
apoptosis in prostate cancer, breast cancer, renal cell carcinoma
and gastric cancer (33–36). In the present study, GAS5 was a
significantly upregulated DElncRNA in POMP, which had four nearby
DEmRNAs (centromere protein L, zinc finger and BTB domain
containing 37, serpin family C member 1, and ring finger and
CCCH-type domains 1). It was hypothesized that GAS5 may be involved
in PMOP by regulating apoptosis and these four genes.
LncRNAs have also been shown to regulate gene
expression in cis. LOC105376834-ALPL and LOC101929866-SLPI
were two DElncRNA-DEmRNA co-expression pairs in PMOP. In addition,
ALPL and SLPI were nearby DEmRNAs of LOC105376834 and LOC101929866,
respectively. It was hypothesized that LOC105376834 and
LOC101929866 may regulate the expression of ALPL and SLPI by a
cis-effect, in which the DElncRNAs were also co-expressed
with DElncRNAs. ALPL is an osteoblast marker and is reported to be
closely associated with the development of osteoporosis (37,38). Downregulated ALPL can reflect
decreased activity of osteoblasts, bone formation and extracellular
matrix mineralization (37).
Previous studies have detected downregulated ALPL in bone tissue
samples of patients with PMOP and ovariectomized mice, a model of
postmenopausal osteoporosis (37,39,40). In the present study, the
downregulation of ALPL was detected in blood samples from patients
with PMOP, which confirmed the importance of ALPL in PMOP and may
serve as a diagnostic marker of PMOP. As ALPL was a nearby
co-expressed DEmRNA of LOC105376834, it was hypothesized that
LOC105376834 may be involved in PMOP by cis-regulating the
expression of ALPL.
SLPI encodes a serine protease inhibitor, which
protects epithelial tissues from serine proteases. Additionally,
SLPI is an anti-inflammatory mediator (41). SLPI can contribute to wound
healing by decreasing the excessive inflammatory response,
elevating keratinocyte proliferation and increasing collagen
deposition by suppressing the activity of protease (42). To the best of our knowledge, the
association between SLPI and PMOP has not been reported previously.
A significant downregulation of SLPI was detected in patients with
PMOP in the present study. As accumulated evidence has indicated
that various inflammatory conditions are involved with osteoporosis
(43), the present study
hypothesized that SLPI may be involved in PMOP by regulating the
inflammatory condition. In addition, estrogen treatment has been
shown to increase the expression of SPLI in alveolar epithelial
cells in ovariectomized mice (44). The same result was found in the
rat uterus following treatment with estrogen (45). It was hypothesized that reduced
estrogen may be involved in PMOP by regulating SLPI. SLPI was the
nearby co-expressed DEmRNA of LOC101929866, which suggested that
LOC101929866 may be associated with PMOP. The other two nearby
co-expressed DEmRNAs (potassium voltage-gated channel subfamily S
member 1, and peptidase inhibitor 3) of LOC101929866 may also be
involved in PMOP.
CD177 was the third significant DEmRNA in PMOP,
which may also be an estrogen-associated gene. CD177 encodes a
glycosyl-phosphatidylinositol-linked cell surface glycoprotein
associated with neutrophil activation. Although there was no
previous report on the association between CD177 and PMOP, a low
expression of CD177 was found to be involved in clonal myeloid
disorders, particularly myelodysplasia (46). A significantly upregulated level
CD177 was previously detected in breast cancer cells following
treatment with estrogen receptors-β agonists (47), which suggested that CD177 was
closely associated with estrogen. It was hypothesized that reduced
CD177 may also be involved in PMOP by regulating estrogen. The
precise role of CD177 in PMOP requires further investigation.
Besides LOC105376834 a nd LOC10192986, LOC105374771
and LOC100506113 were two downregulated DElncRNAs in PMOP, which
had nearby co-expressed DEmRNAs. Therefore, LOC100506113 and
LOC105374771 may be involved in PMOP by regulating the expression
of diacylglycerol O-acyltransferase 2 and LGALSL, respectively. In
addition, LOC105374771 was the most markedly downregulated lncRNA,
which was co-expressed with 130 DEmRNAs, including ALPL, SOCS3,
ADM, CD177 and SLPI. LOC105374771 may affect the pathogenesis of
PMOP by regulating the expression of these DEmRNAs.
Besides LINC00963, the other hub lncRNAs of the
positively and negatively co-expressed DElncRNAs-DEmRNAs network
were LOC105378415, LOC105377067, HCG27, LOC101928143 and LINC01094.
Three PMOP-associated DEmRNAs, including ALPL, SOCS3 and ADM, were
common co-expressed DEmRNAs of these hub DElncRNAs, which indicated
the importance of these DElncRNAs in PMOP.
As hematopoietic cell lineage and osteoclast
differentiation are two well-known pathways in PMOP. DEmRNAs
enriched in these two pathways and their co-expressed DElncRNAs may
be involved in PMOP by regulating hematopoietic cell lineage or
osteoclast differentiation.
In conclusion, the present study identified five
DEmRNAs (ALPL, SOCS3, ADM, SLPI and CD177) co-expressed with
DElncRNAs, which may be involved in PMOP. DElncRNAs in PMOP,
including LINC00963, LOC105376834, LOC101929866, LOC105374771 and
LOC100506113, may be involved in the pathogenesis of PMOP by
regulating the expression of their nearby and co-expressed DEmRNAs
and the pathway of osteoclast differentiation. The results of the
present study may provide a foundation for future investigations of
lncRNAs in PMOP and contribute in developing novel diagnostic
biomarkers and drug design for PMOP. However, the sample size for
RNA sequencing in the present study was small, and the difference
in body mass index between the PMOP and NC groups may have affected
the results of RNA-sequencing, which were limitations of the study.
Although the validation based on GSE56815 and GSE7158 suggested
that the RNA-sequencing results were generally reliable,
investigations with a larger sample size are required to confirm
this conclusion. In addition, further experiments are required to
address the biological significance of key lncRNAs and genes in
PMOP.
Acknowledgments
Not applicable.
Notes
[1]
Funding
This study was supported by the Beijing Health
System High Level Health Technical Personnel Training Project
(grant no. 2015-3-009).
[2] Availability
of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
[3] Authors'
contributions
QF and AG were responsible for conceptionand design
of the experiments. QF and XDB performed the experiments. XDB and
JSL analyzed the data. HM and YY supplied reagents, materials and
analysis tools. All named authors wrote this manuscript and have
agreed to the publication of this manuscript, and it does not
infringe on any copyright or property rights.
[4] Ethics
approval and consent to participate
All individuals provided written informed consent
for use of their samples in the present study. The present study
was approved by the Ethics Committee of Beijing Friendship
Hospital, Capital Medical University (Beijing, China;
2017-P2-084-01).
[5] Consent for
publication
Not applicable.
[6] Competing
interests
The authors declare that they have no competing
interests.
References
|
1
|
Zhang Y, Liu H, Zhang C, Zhang T, Zhang B,
Li L, Chen G, Fu D and Wang K: Endochondral ossification pathway
genes and postmenopausal osteoporosis: Association and specific
allele related serum bone sialoprotein levels in Han Chinese. Sci
Rep. 5:167832015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ma M, Chen X, Lu L, Yuan F, Zeng W, Luo S,
Yin F and Cai J: Identification of crucial genes related to
postmenopausal osteoporosis using gene expression profiling. Aging
Clin Exp Res. 28:1067–1074. 2016. View Article : Google Scholar
|
|
3
|
Taguchi A, Ohtsuka M, Nakamoto T, Naito K,
Tsuda M, Kudo Y, Motoyama E, Suei Y and Tanimoto K: Identification
of post-menopausal women at risk of osteoporosis by trained general
dental practitioners using panoramic radiographs. Dentomaxillofac
Radiol. 36:149–154. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sasser AC, Taylor M, Birnbaum HG,
Schoenfeld MJ, Oster EF and Rousculp M: Assessing the economic
impact of chronic conditions in postmenopausal women. Expert Opin
Pharmacother. 6:1803–1814. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sasser AC, Rousculp MD, Birnbaum HG, Oster
EF, Lufkin E and Mallet D: Economic burden of osteoporosis, breast
cancer, and cardiovascular disease among postmenopausal women in an
employed population. Womens Health Issues. 15:97–108. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Richards JB, Zheng HF and Spector TD:
Genetics of osteoporosis from genome-wide association studies:
Advances and challenges. Nat Rev Genet. 13:576–588. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu YJ, Zhang L, Papasian CJ and Deng HW:
Genome-wide association studies for osteoporosis: A 2013 update. J
Bone Metab. 21:99–116. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kotrych D, Dziedziejko V, Safranow K,
Sroczynski T, Staniszewska M, Juzyszyn Z and Pawlik A: TNF-α and
IL10 gene polymorphisms in women with postmenopausal osteoporosis.
Eur J Obstet Gynecol Reprod Biol. 199:92–95. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shang DP, Lian HY, Fu DP, Wu J, Hou SS and
Lu JM: Relationship between estrogen receptor 1 gene polymorphisms
and postmenopausal osteoporosis of the spine in Chinese women.
Genet Mol Res. 15:2016. View Article : Google Scholar
|
|
10
|
Zhang C, Ma J, Chen G, Fu D, Li L and Li
M: Evaluation of common variants in CNR2 gene for bone mineral
density and osteoporosis susceptibility in postmenopausal women of
Han Chinese. Osteoporos Int. 26:2803–2810. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Luo D, Liu Y, Zhou Y, Chen Z, Yang L, Liu
Y, Xu Q, Xu H, Kuang H, Huang Q, et al: Association between dietary
phytoestrogen intake and bone mineral density varied with estrogen
receptor alpha gene polymorphisms in southern Chinese
postmenopausal women. Food Funct. 6:1977–1983. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Singh M, Singh P, Singh S, Juneja PK and
Kaur T: Vitamin D receptor (VDR) gene polymorphism influences the
risk of osteoporosis in postmenopausal women of Northwest India.
Arch Osteoporos. 8:1472013. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kitjaroentham A, Hananantachai H, Phonrat
B, Preutthipan S and Tungtrongchitr R: Low density lipoprotein
receptor-related protein 5 gene polymorphisms and osteoporosis in
Thai menopausal women. J Negat Results Biomed. 15:162016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cvijetic S, Grazio S, Kosovic P, Uremovic
M, Nemcic T and Bobic J: Osteoporosis and polymorphisms of
osteoprotegerin gene in postmenopausal women-a pilot study.
Reumatologia. 54:10–13. 2016. View Article : Google Scholar :
|
|
15
|
Terashima M, Tange S, Ishimura A and
Suzuki T: MEG3 Long Noncoding RNA contributes to the epigenetic
regulation of epithelial-mesenchymal transition in lung cancer cell
lines. J Biol Chem. 292:82–99. 2017. View Article : Google Scholar :
|
|
16
|
Rajpathak SN, Vellarikkal SK, Patowary A,
Scaria V, Sivasubbu S and Deobagkar DD: Human 45, X fibroblast
transcriptome reveals distinct differentially expressed genes
including long noncoding RNAs potentially associated with the
pathophysiology of Turner syndrome. PLoS One. 9:e1000762014.
View Article : Google Scholar
|
|
17
|
Wang Q, Li Y and Zhang Y, Ma L, Lin L,
Meng J, Jiang L, Wang L, Zhou P and Zhang Y: LncRNA MEG3 inhibited
osteogenic differentiation of bone marrow mesenchymal stem cells
from postmenopausal osteoporosis by targeting miR-133a-3p. Biomed
Pharmacother. 89:1178–1186. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang L, Han S, Jin G, Zhou X, Li M, Ying
X, Wang L, Wu H and Zhu Q: Linc00963: A novel, long non-coding RNA
involved in the transition of prostate cancer from
androgen-dependence to androgen-independence. Int J Oncol.
44:2041–4449. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Costa FF: Non-coding RNAs: New players in
eukaryotic biology. Gene. 357:83–94. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Fitzgerald KA and Caffrey DR: Long
noncoding RNAs in innate and adaptive immunity. Curr Opin Immunol.
26:140–146. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liao Q, Liu C, Yuan X, Kang S, Miao R,
Xiao H, Zhao G, Luo H, Bu D, Zhao H, et al: Large-scale prediction
of long non-coding RNA functions in a coding-non-coding gene
co-expression network. Nucleic Acids Res. 39:3864–3878. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Dong R, Jia D, Xue P, Cui X, Li K, Zheng
S, He X and Dong K: Genome-wide analysis of long noncoding RNA
(lncRNA) expression in hepatoblastoma tissues. PLoS One.
9:e855992014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Wang P, Fu H, Cui J and Chen X:
Differential lncRNA-mRNA co-expression network analysis revealing
the potential regulatory roles of lncRNAs in myocardial infarction.
Mol Med Rep. 13:1195–1203. 2016. View Article : Google Scholar :
|
|
25
|
Fang M, Zhang P, Zhao Y and Liu X:
Bioinformatics and co-expression network analysis of differentially
expressed lncRNAs and mRNAs in hippocampus of APP/PS1 transgenic
mice with Alzheimer disease. Am J Transl Res. 9:1381–1391.
2017.PubMed/NCBI
|
|
26
|
Yang L, Yi K, Wang H, Zhao Y and Xi M:
Comprehensive analysis of lncRNAs microarray profile and
mRNA-lncRNA co-expression in oncogenic HPV-positive cervical cancer
cell lines. Oncotarget. 7:49917–49929. 2016.PubMed/NCBI
|
|
27
|
Wan ZY, Song F, Sun Z, Chen YF, Zhang WL,
Samartzis D, Ma CJ, Che L, Liu X, Ali MA, et al: Aberrantly
expressed long noncoding RNAs in human intervertebral disc
degeneration: A microarray related study. Arthritis Res Ther.
16:4652014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Martinez-Herrero S, Larrayoz IM,
Ochoa-Callejero L, Fernández LJ, Allueva A, Ochoa I and Martínez A:
Prevention of bone loss in a model of postmenopausal osteoporosis
through adrenomedullin inhibition. Front Physiol. 7:2802016.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Montuenga LM, Martínez A, Miller MJ,
Unsworth EJ and Cuttitta F: Expression of adrenomedullin and its
receptor during embryogenesis suggests autocrine or paracrine modes
of action. Endocrinology. 138:440–451. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Uzan B, Villemin A, Garel JM and Cressent
M: Adrenomedullin is anti-apoptotic in osteoblasts through CGRP1
receptors and MEK-ERK pathway. J Cell Physiol. 215:122–128. 2008.
View Article : Google Scholar
|
|
31
|
Sun GJ, Guo T, Chen Y, Xu B, Guo JH and
Zhao JN: Significant pathways detection in osteoporosis based on
the bibliometric network. Eur Rev Med Pharmacol Sci. 17:1–7.
2013.PubMed/NCBI
|
|
32
|
Zhang X, Alnaeeli M, Singh B and Teng YT:
Involvement of SOCS3 in regulation of CD11c+ dendritic
cell-derived osteoclastogenesis and severe alveolar bone loss.
Infect Immun. 77:2000–2009. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Pickard MR, Mourtada-Maarabouni M and
Williams GT: Long non-coding RNA GAS5 regulates apoptosis in
prostate cancer cell lines. Biochim Biophys Acta. 1832:1613–1623.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sun M, Jin F, Xia R, Kong R, Li JH, Xu TP,
Liu YW, Zhang EB, Liu XH and De W: Decreased expression of long
noncoding RNA GAS5 indicates a poor prognosis and promotes cell
proliferation in gastric cancer. BMC Cancer. 14:3192014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Qiao HP, Gao WS, Huo JX and Yang ZS: Long
non-coding RNA GAS5 functions as a tumor suppressor in renal cell
carcinoma. Asian Pac J Cancer Prev. 14:1077–1082. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mourtada-Maarabouni M, Pickard MR, Hedge
VL, Farzaneh F and Williams GT: GAS5, a non-protein-coding RNA,
controls apoptosis and is downregulated in breast cancer. Oncogene.
28:195–208. 2009. View Article : Google Scholar
|
|
37
|
Balla B, Kosa JP, Kiss J, Borsy A, Podani
J, Takács I, Lazáry A, Nagy Z, Bácsi K, Speer G, et al: Different
gene expression patterns in the bone tissue of aging postmenopausal
osteoporotic and non-osteoporotic women. Calcif Tissue Int.
82:12–26. 2008. View Article : Google Scholar
|
|
38
|
Rodríguez JP, Montecinos L, Ríos S, Reyes
P and Martínez J: Mesenchymal stem cells from osteoporotic patients
produce a type I collagen-deficient extracellular matrix favoring
adipogenic differentiation. J Cell Biochem. 79:557–565. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Hsiao HB, Lin H, Wu JB and Lin WC:
Kinsenoside prevents ovariectomy-induced bone loss and suppresses
osteoclastogenesis by regulating classical NF-κB pathways.
Osteoporos Int. 24:1663–1676. 2013. View Article : Google Scholar
|
|
40
|
de Castro LF, Lozano D, Portal-Núñez S,
Maycas M, De la Fuente M, Caeiro JR and Esbrit P: Comparison of the
skeletal effects induced by daily administration of PTHrP (1-36)
and PTHrP (107-139) to ovariectomized mice. J Cell Physiol.
227:1752–1760. 2012. View Article : Google Scholar
|
|
41
|
McKiernan PJ, McElvaney NG and Greene CM:
SLPI and inflammatory lung disease in females. Biochem Soc Trans.
39:1421–1426. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Park JJ, Bae CS, Choi BD, Jeong SJ, Wang
G, Lim DS, Kim BO, Cho YS, Kim SJ and Jeong MJ: Induction of
secretory leukocyte protease inhibitor (SLPI) in estradiol valerate
(EV) induced polycystic ovary. Arch Pharm Res. 34:1389–1397. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Al-Daghri NM, Aziz I, Yakout S, Aljohani
NJ, Al-Saleh Y, Amer OE, Sheshah E, Younis GZ and Al-Badr FB:
Inflammation as a contributing factor among postmenopausal Saudi
women with osteoporosis. Medicine (Baltimore). 96:e57802017.
View Article : Google Scholar
|
|
44
|
Draijer C, Hylkema MN, Boorsma CE, Klok
PA, Robbe P, Timens W, Postma DS, Greene CM and Melgert BN: Sexual
maturation protects against development of lung inflammation
through estrogen. Am J Physiol Lung Cell Mol Physiol.
310:L166–L174. 2016. View Article : Google Scholar
|
|
45
|
Chen D, Xu X, Cheon YP, Bagchi MK and
Bagchi IC: Estrogen induces expression of secretory leukocyte
protease inhibitor in rat uterus. Biol Reprod. 71:508–514. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Meyerson HJ, Osei E, Schweitzer K, Blidaru
G, Edinger A and Balog A: CD177 expression on neutrophils: In
search of a clonal assay for myeloid neoplasia by flow cytometry.
Am J Clin Pathol. 140:658–669. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Lattrich C, Schüler S, Häring J,
Skrzypczak M, Ortmann O and Treeck O: Effects of a combined
treatment with tamoxifen and estrogen receptor β agonists on human
breast cancer cell lines. Arch Gynecol Obstet. 289:163–171. 2014.
View Article : Google Scholar
|