Introduction
Flavonoids are naturally occurring polyphenolic
metabolites in plants that serve significant roles in traditional
medicine and as food additives (1–3).
Glycosylated forms of numerous flavonoids are produced in order to
alter their chemical properties, such as their bioavailability and
water solubility (4). Flavonoids
without glycosylation are known as aglycones, which are often more
reactive forms of flavonoids and present greater antioxidant
capacity, as well as cellular toxicity, compared with the
glycosylated flavonoids (5,6).
Both aglycone and glycosylated forms of flavonoids are recognized
to be beneficial in regard to anti-tumorigenicity and
anti-mutagenicity, due to their antioxidant and radical scavenging
effects.
Numerous flavonoids, including quercetin, have been
previously reported to induce mutation frequency in bacterial
systems and chromosome aberrations in mammalian cells (7–10).
This mutagenic potential may be associated with the ability of
flavonoids to produce hydroxyl radicals, resulting in DNA breaks
(11,12). In the presence of specific metal
ions, flavonoids can cause DNA scission, but rarely DNA
double-strand breaks, resulting from the interaction of hydrogen
peroxide with metal ions (12).
This hydroxyl radical production is coupled with the reduction of
cupric ions to cuprous ions (from Cu2+ to
Cu1+). Quercetin forms chelating complexes with cupric
ions and portrays broad biological activities (13–16). This cupric ion and the quercetin
complex interact at the 3′ and 4′ hydroxyl groups on the B ring,
and the 3-hydroxyl group and 4-oxygen residue on the A ring
(17). Furthermore, the ability
to produce hydroxyl radicals and to induce DNA breaking activity
has also been reported for other compounds that are structurally
associated with quercetin, including fisetin, baicalein, taxifolin
and curcumin with cupric ions (18,19), as well as epigallocatechin gallate
and ferrous ions (20). However,
the exact mechanisms underlying the formation of hydroxyl radicals
from quercetin and its effects on glycosylation of flavonoids
remain unclear.
The present study utilizes a total of 7 natural and
5 recently semi-synthesized novel flavonoids to identify the
specific molecular mechanisms required to induce DNA scissions in
the presence of cupric ions. To analyze DNA scission formation in
an in vitro gel electrophoresis system, three aglycones and
their glycosylated flavonoids were reacted in the presence of
cupric ions at different temperatures.
Materials and methods
Chemicals
All natural and synthetic flavonoids were obtained
from Tokyo Sugar Refining Co., Ltd. (Tokyo, Japan). The tested
compounds, including three aglycone flavonoids (quercetin,
naringenin and hesperetin) and their glycosylated flavonoids are
summarized in Fig. 1. Quercetin
is an active aglycone form of isoquercetin, rutin,
maltooligosyl-isoquercetin, monoglucosyl-rutin and
maltooligosyl-rutin. The glycosylated form of quercetin loses the
3-position hydroxyl group on the B ring by glycosylation. Compared
with quercetin, naringenin does not contain a hydroxyl group at the
3-position on the B ring nor at the 3′-position on the C ring. In
addition, compared with quercetin, hesperetin does not contain a
hydroxyl group at the 3-position on the C ring nor at the
4′-position on the C ring. Glycosylated naringenin and hesperetin
lost the hydroxyl group at the 7-position on the A ring by
glycosylation and are chemically inactive. All flavonoids were
prepared by dissolving in dimethyl sulfoxide (Thermo Fisher
Scientific, Inc., Waltham, MA, USA) at the concentration of 1 mM as
a stock solution. M13mp18 single-stranded DNA (250 μg/ml)
was purchased from New England BioLabs, Inc. (Ipswich, MA, USA).
Double-stranded lambda phage DNA (0.46 μg/μl) was
purchased from Nippon Gene Co., Ltd. (Tokyo, Japan). Copper
chloride dehydrate was purchased from Sigma-Aldrich (Merck KGaA,
Darmstadt, Germany).
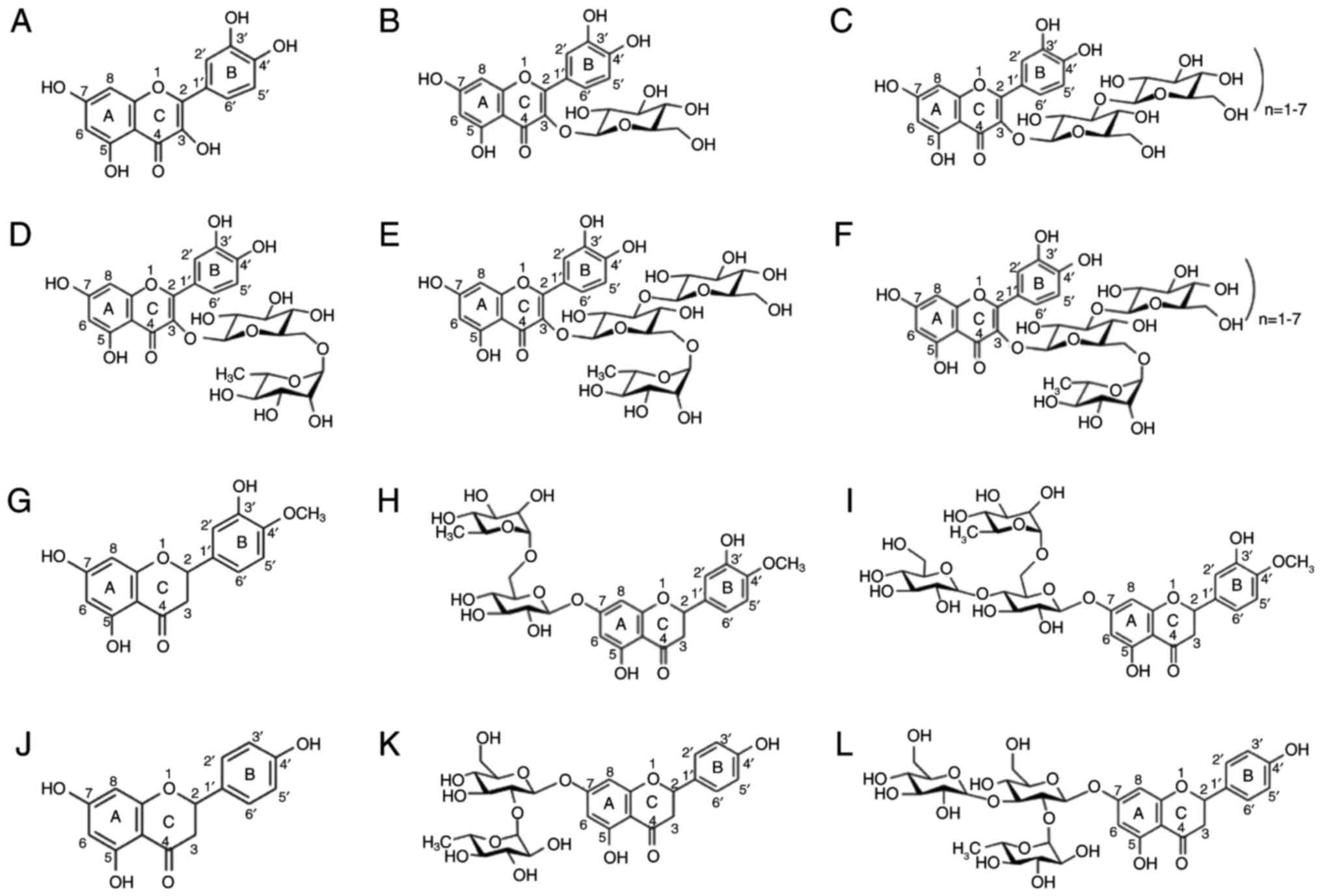 | Figure 1Structure of flavonoids. (A)
Quercetin, (B) isoquercetin, (C) rutin, (D) monoglucosyl-rutin, (E)
maltooligosyl-isoquercetin, (F) maltooligosyl rutin, (G)
hesperetin, (H) hesperidin, (I) monoglucosyl hesperidin, (J)
naringenin, (K) naringin, and (L) monoglucosyl naringin. |
DNA scission reaction
A total of 20 μl reaction solution was used,
containing 1.5 ng/μl single-stranded DNA or 1.25
ng/μl of double-stranded DNA, 10 mM Tris-HCl, 0.2 mM
CuCl2 and various concentrations of flavonoids (0.1, 1,
10 and 100 μM). The reaction mixtures were incubated at
different temperatures (4°C, 20°C, 37°C or 54°C) for 1 h. Next, 4
μl of 6X loading dye, containing 15% Ficoll, 10% glycerol,
0.25% bromophenol blue, and 0.25% xylene cyanol in water, was added
to the mixture and electrophoresis was conducted on 1% agarose gel
stained with ethidium bromide with 1X Tris-acetate-EDTA buffer at
100 V for 1 h. Subsequent to electrophoresis, destaining of
ethidium bromide was performed, and a gel image was obtained by
ChemiDoc XRS system (Bio-Rad Laboratories, Inc., Hercules, CA, USA)
via Image Lab software (Bio-Rad Laboratories, Inc.). Intact M13mp18
single-stranded DNA presented two bands, indicative of the circular
and linear form of DNA. The DNA intensity of the circular form was
obtained by Image Lab, while the fraction of intact DNA was
calculated as follows:
Half maximal inhibitory concentration
(IC50) values, which refer to the specific
concentrations required to induce 50% of DNA break, were obtained
from sigmoidal regression curves obtained by Prism 6 software
(GraphPad Software, Inc., La Jolla, CA, USA).
Absorption spectrum analysis
In order to investigate the cupric ion chelating
capability of flavonoids, the flavonoids and cupric ions were mixed
at rations of 1:2, 1:1 and 2:1, and absorbance values from 230 to
430 nm were obtained via a Nanodrop spectrophotometer (Thermo
Fisher Scientific, Inc.). According to the peak shifting and the
reduction of peak height, the chelating capacity of cupric ions was
estimated (14,15,17).
Oxidation of luminol
In order to investigate the potential mechanisms
underlying the induction of DNA damages, oxidative capacity
analysis was conducted. In total, 100 μl of reaction
solution, including 20 μl enhanced chemiluminescence
solution (Thermo Fisher Scientific, Inc.), 2 μl of 100
μM flavonoids and 2 μl of 100 μM
CuCl2, were mixed. Subsequently, the arbitrary relative
luminescence unit (RLU) was measured immediately until 3 min after
mixing via a Lumat LB9507 luminometer (Berthold Technologies, Oak
Ridge, TN, USA). RLU values were plotted with the exponential decay
model, and half time values were obtained.
Statistical analysis
Statistical calculations were preformed via Prism 6
software. Two-way analysis of variance (ANOVA) and Student’s t-test
were conducted. P-values of <0.05 were considered to indicate
differences that were statistically significant.
Results
DNA damage formation
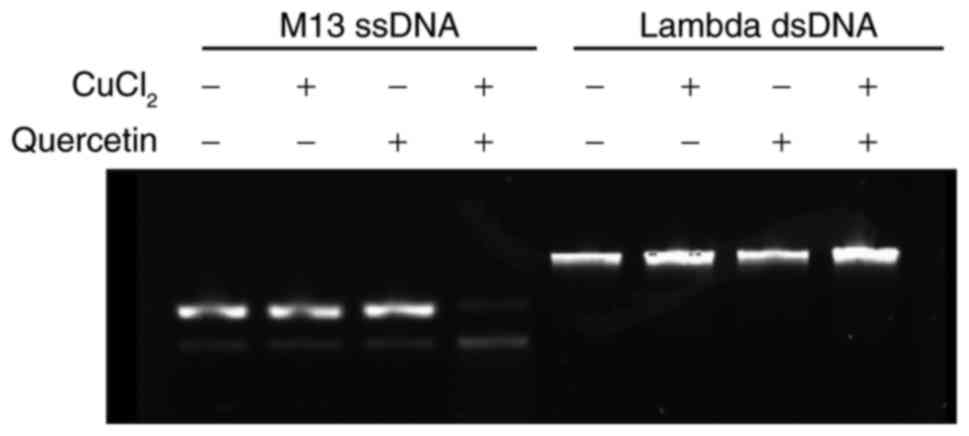
Fig. 2
demonstrates the DNA damage subsequent to reaction with quercetin
in the absence or presence of cupric ions at 37°C. Without cupric
ions in the reaction mixture, DNA damage was not detected from the
single-stranded and double-stranded DNA. Upon the incorporation of
cupric ions in the reaction mixture, single-stranded DNA was
degraded, whereas double-stranded DNA was not affected. Therefore,
quercetin without cupric ions did not produce DNA damage. In
addition, DNA damage produced by quercetin and cupric ions mainly
involved single-strand break rather than double-strand break.
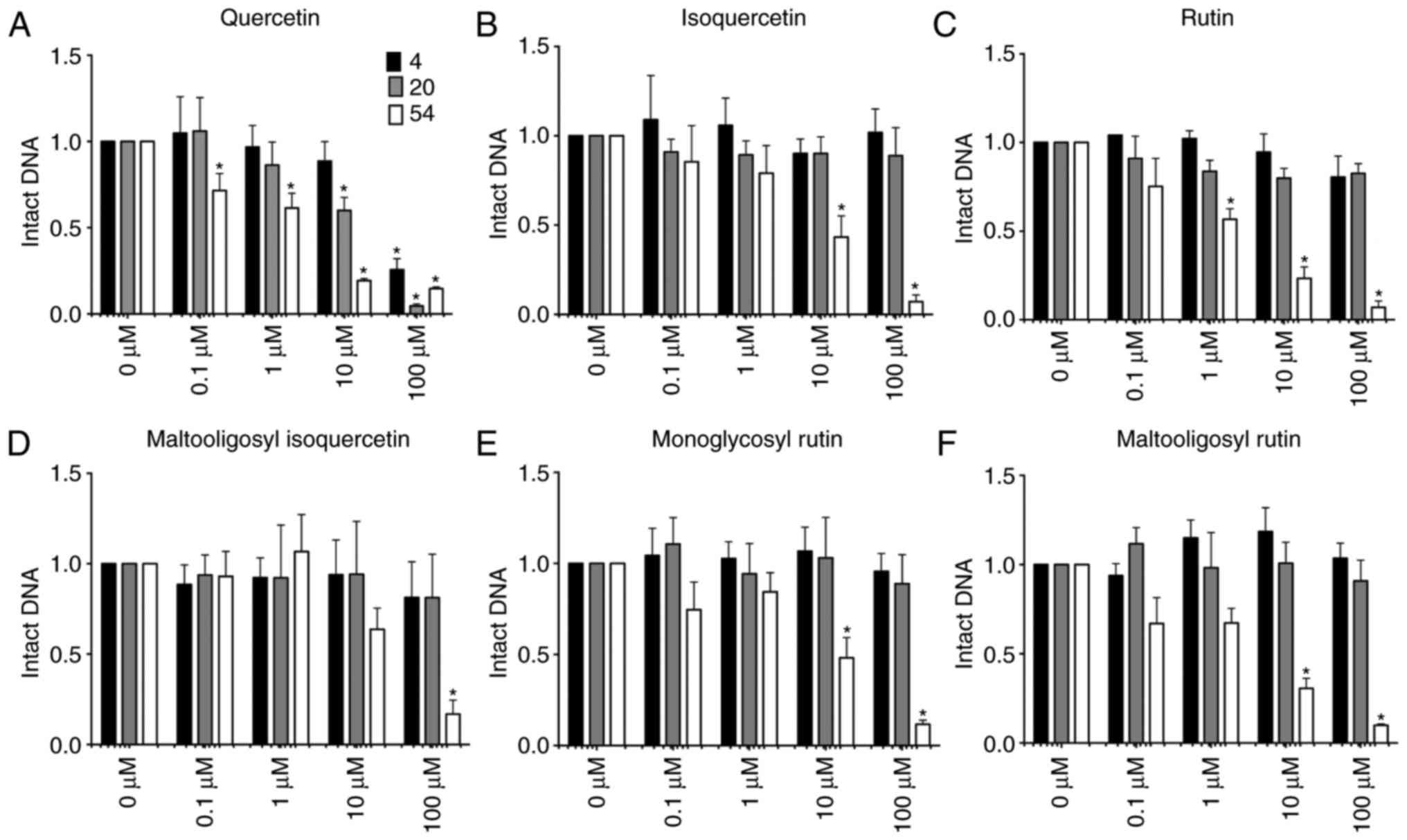
Fig. 3 shows the
fraction of intact DNA following reaction with glycosylated
flavonoids in the presence of cupric ions in the reaction mixture.
In the presence of 0.2 mM CuCl2 in the reaction mixture,
quercetin and its glycosylated flavonoids induced DNA breaks in a
concentration and temperature dependent manner. Less intact DNA was
observed following reaction with high concentration of flavonoids,
and high temperature efficiently induced DNA damages.
Quercetin was the only flavonoid capable of inducing
a DNA break in reactions performed at 4 and 20°C. Furthermore, the
reaction at 20°C induced a greater degree of DNA damage as compared
with the reaction at 4°C. The IC50 values of quercetin
at 4 and 20°C were 47.7 and 20.5 μM, respectively, while the
reaction at 54°C reduced the IC50 value to 2.6
μM. By contrast, all quercetin glycosides, including
isoquercetin, rutin, monoglucosyl rutin, maltooligosyl-rutin and
maltooligosyl-isoquercetin, failed to induce any DNA damage
following 4 and 20°C reactions under the tested conditions.
However, these glucosyl flavonoids did induce DNA damage when
reactions were conducted at 54°C for 1 h. The IC50
values were 7.7 μM for isoquercetin, 3.4 μM for
rutin, 9.5 μM for monoglucosyl rutin, 4.5 μM for
maltooligosyl rutin and 12.1 μM for maltooligosyl
isoquercetin. This indicates that glycosyl modifications at the
3-position on the C ring of quercetin suppressed the DNA scission
ability of quercetin at 4 and 20°C. However, DNA scission capacity
was not altered with glycosylation at the 3-position on the C ring
of quercetin at 54°C. Therefore, increased glucosyl modifications
did not contribute toward DNA degradation. In other words, the
hydroxyl groups of glucosyl residues did not portray any
contributions towards DNA damage.
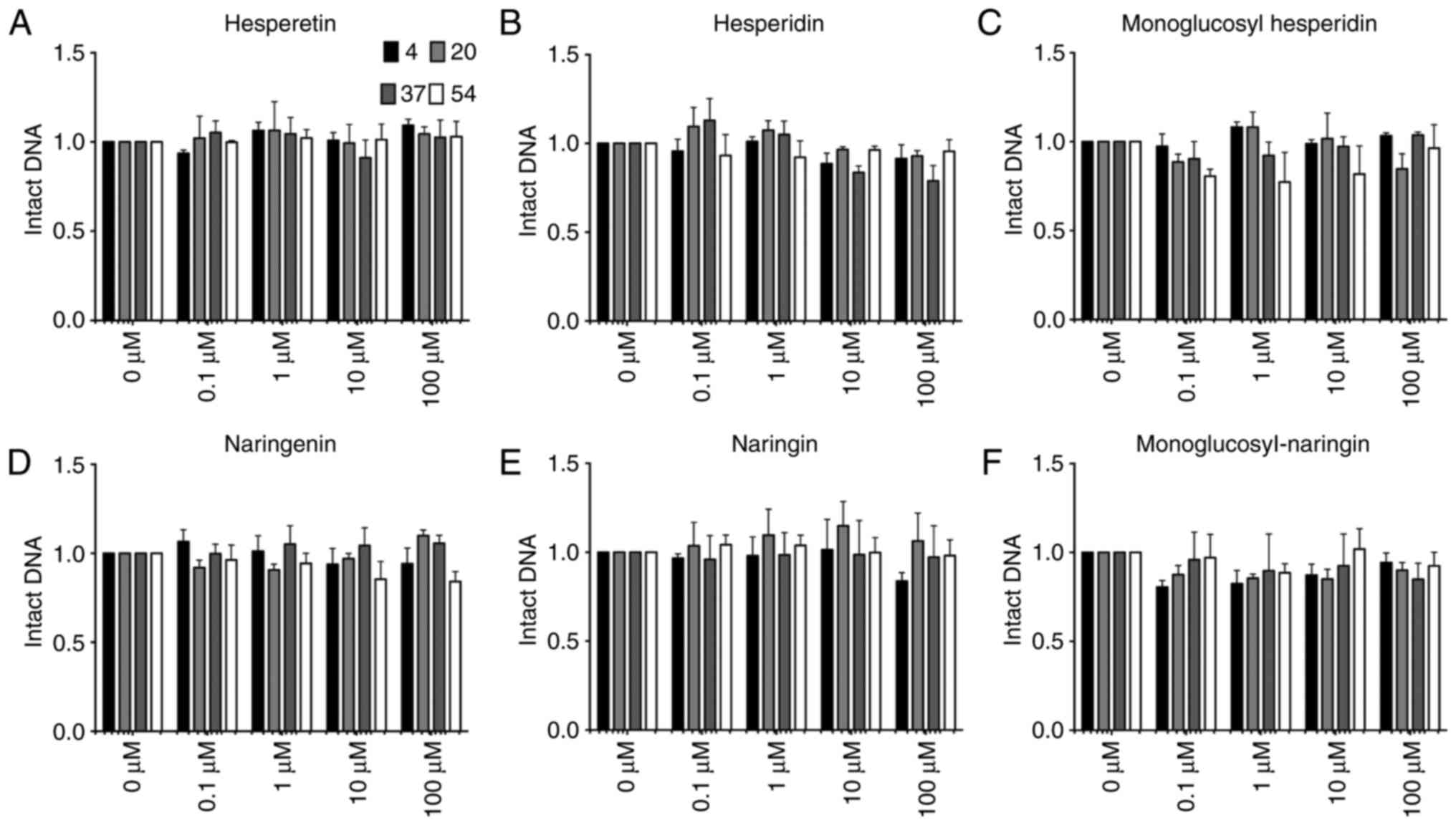
In order to clarify which position of the hydroxyl
group is responsible for inducing DNA damage, two other flavone
aglycones, hesperetin and naringenin, and their glycosylated
flavonoids were also tested in the same system to assess hydroxyl
radical formation by DNA damage observation (Fig. 4). Hesperetin, naringenin and their
glycosylated flavonoids, which do not have hydroxyl groups at the
3-position on the C ring nor at the 3′- or 4′-position on the B
ring, were incapable of producing any detectable single-stranded
DNA damage in the reaction at any tested temperature. Therefore,
this indicates that the presence of hydroxyl groups at the 3′- and
4′-positions is required for the induction of DNA damage at 54°C.
Furthermore, flavone glycosides did not cause any DNA damage,
confirming that the hydroxyl groups of glucosyl residue have no
effect on DNA damage.
Chelating capacity
Absorption spectrum alterations, shown in Fig. 5A-L, occurred when flavonoids and
cupric ions were mixed in different ratios (1:2, 1:1 and 2:1).
Quercetin and its glycosides displayed peaks at ~250 and 370 nm, as
previously described (14,15,17).
Upon addition of cupric ions, the first peak at 250 nm skewed to
the left and the size of the second peak was reduced. The reduction
in the second peak was the greatest for quercetin. Naringenin,
hesperetin and their glycosides presented the first peak at 250 nm
and the second peak at 290 nm. As observed in quercetin and its
glycosides, the first peak was shifted toward the left and the
second peak size was decreased in the presence of cupric ions. The
reduction ratio of the second peak was used for assessment of
cupric ion chelating capacity, and the results are summarized in
Fig. 5M. Quercetin demonstrated
>6-fold reduction in the absorbance peak height in the presence
of cupric ions. The peak heights of other flavonoids were reduced
by 2-4-fold. Furthermore, quercetin showed statistically
significant differences compared with the other flavonoids (ANOVA;
P<0.0001).
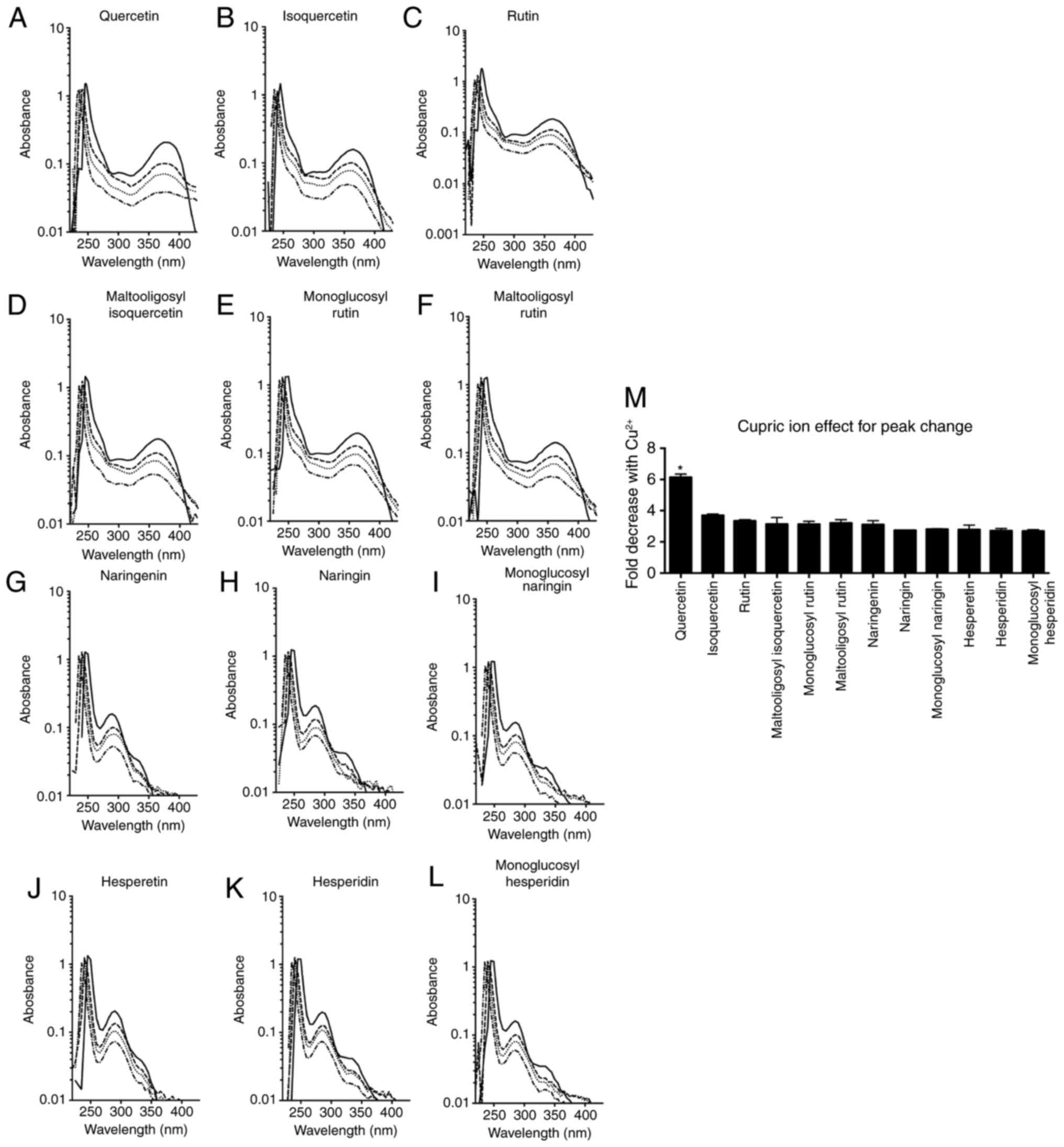 | Figure 5Absorption spectrum changes in the
presence of cupric ions. Solid lines indicate the flavonoid only,
and the dashed, dotted and dash-dotted lines indicate a flavonoid
and cupric ion mixture at a ratio of 2:1, 1:1 and 1:2,
respectively. (A) Quercetin, (B) isoquercetin, (C) rutin, (D)
maltooligosyl isoquercetin, (E) monoglucosyl rutin, (F)
maltooligosyl rutin, (G) naringenin, (H) naringin, (I) monoglucosyl
naringin, (J) hesperetin, (K) hesperidin and (L) monoglucosyl
hesperidin spectrum changes are shown. (M) Effect of cupric ion on
peak changes is displayed. *P<0.05 vs. other
flavonoids. Error bars indicate the standard error of the mean.
Three independent experiments were performed. |
Oxidation of luminol
In addition to the interaction between flavonoids
and cupric ions, oxidative capacity analysis was conducted with
luminol as a substrate in the presence of flavonoids and cupric
ions. Subsequent to mixing, the reduction of the luminescence
signal was observed (Fig. 6).
Emitted glow signals are associated with not only cupric ion
induction, but also generated hydrogen peroxide by flavonoids in
the presence of cupric ions. The luminescence signal (arbitrary
RLU) was rapidly decreased with an exponentially decreased model
for all flavonoids. Quercetin and isoquercetin exhibited the
fastest reduction of RLU with a half-life of 0.22 min. In addition,
rutin, monoglucosyl-rutin, maltooligosyl-rutin and
maltooligosyl-isoquercetin presented intermediate reduction in
kinetics with a half-life between 0.28-0.33 min. Naringenin,
naringin, monoglucosyl-naringin, hesperetin, hesperidin and
monoglucosyl-hesperidin also displayed the slowest kinetics with a
half-life of 0.35-0.45 min.
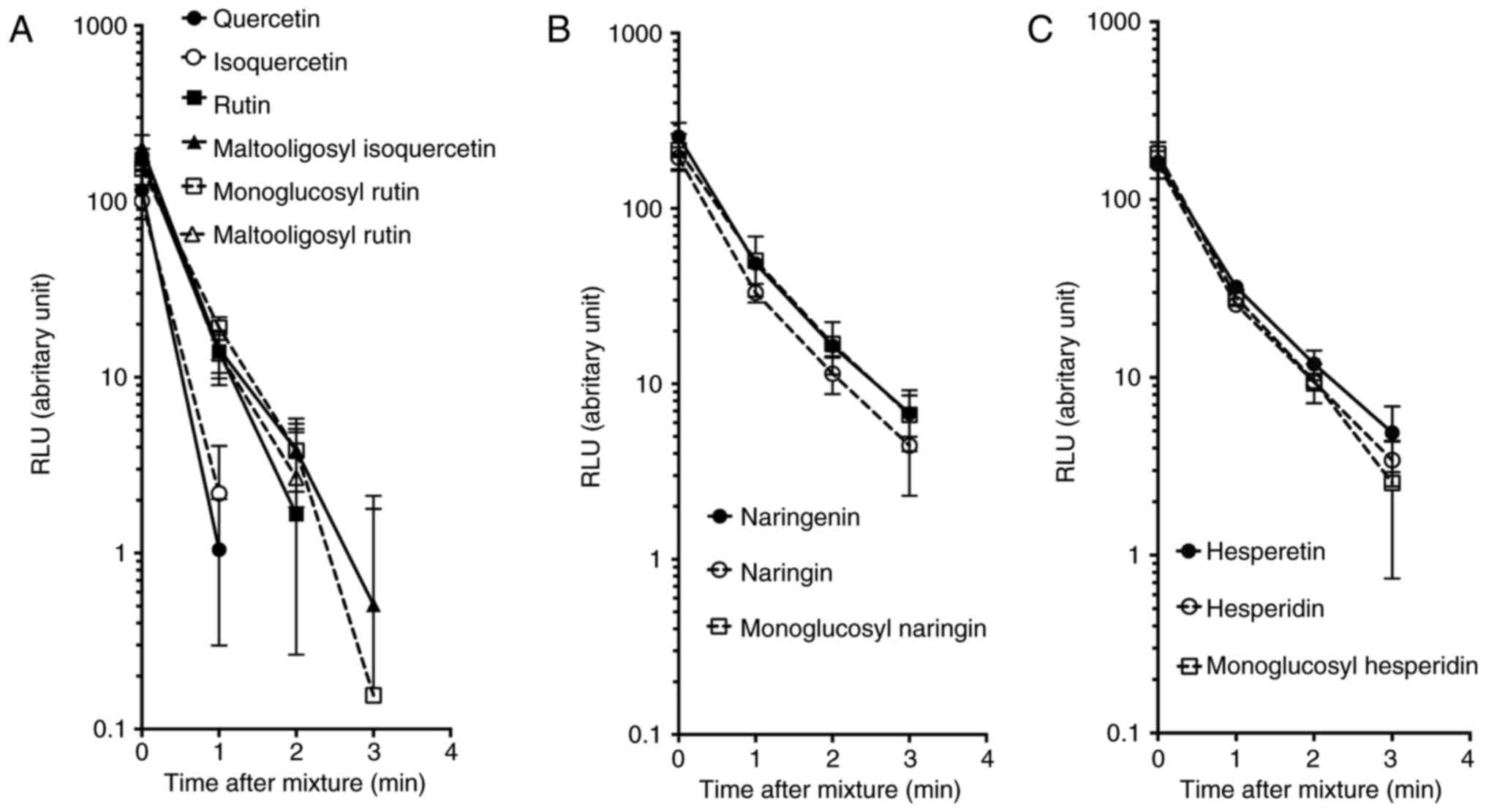 | Figure 6Oxidation of luminol in the presence
of hydrogen peroxide, CuCl2 and flavonoids. (A)
Quercetin, isoquercetin, rutin, maltooligosyl isoquercetin,
monoglucosyl rutin, and maltooligosyl rutin. (B) Naringenin,
naringin, and monoglucosyl naringin. (C) Hesperetin, hesperidin,
and monoglucosyl hesperidin. Error bars indicate the standard error
of the mean values. Three independent experiments were performed.
MO, maltooligosyl; MG, monoglucosyl. |
Discussion
Flavonoids with a variety of chemical structures
with modified residues exist; for instance, a number of hydroxyl
groups are attached to the different positions of benzene rings
(21). Depending on the hydroxyl
groups present at specific locations, metal ions can bind to
flavonoids and induce the Fenton-like reaction, resulting in DNA
damage (11,12). DNA double-strand breaks are rare
in this event, but single-strand scission and oxidative damage are
induced (11,22). This DNA damage may be associated
with mutagenesis in the cell culture system following flavonoid
reaction with cells in the presence of metals (23).
The present study clearly demonstrated the
importance of the specific positions of hydroxyl groups on the
flavonoids to induce DNA damage. The results revealed that
quercetin was the only flavonoid capable of inducing DNA damage at
any of the tested experimental temperatures (Fig. 3). Naringenin and hesperetin, which
do not possess a hydroxyl group at the specific positions (the
3-position on the C ring, and the 3′- and 4′-positions on the B
ring), were incapable of inducing DNA damage (Fig. 4). Furthermore, at low
temperatures, the glycosylated flavonoids of quercetin were
incapable of inducing DNA breaks (Fig. 3). This suggests that a hydroxyl
group at the 3-position on the C ring of quercetin is the most
reactive, inducing DNA damage in a Fenton-like reaction with cupric
ions. These findings are in agreement with the previous studies
that refer to the possibly significant role of this location
(11) in forming a complex with
cupric ions to induce DNA damage. It is also worth noting that the
hydroxyl group at the 3-position on the C ring of quercetin has the
ability to scavenge radicals (24). The high cupric ion chelating
capacity of quercetin (Fig. 5)
and fast oxidation of luminol (Fig.
6) were also in agreement with these previous findings.
The structural differences among quercetin,
naringenin and hesperetin helped to investigate the impact of
hydroxyl groups at the 3′- and 4′-positions on the B ring in the
induction of DNA damage. With two hydroxyl groups at the 3′- and
4′-positions on the B ring, flavonoids induced DNA damage at a high
temperature, as observed with glycosylated quercetins (including
isoquercetin, rutin, maltooligosyl-isoquercetin, monoglucosyl-rutin
and maltooligosyl-rutin) (Fig.
3). When one of the two hydroxyl groups was replaced with other
residues, high temperature-specific DNA damage was not observed in
naringenin and hesperetin. Additionally, hydroxyl groups at the
7-positions on the A ring did not contribute to DNA damage
induction based on the observation of the glycosylated naringenin
and hesperetin. Previously, it has been reported that the hydroxyl
groups of quercetin at the 3′- and 4′-positions on the B ring have
higher radical scavenging effects in comparison with the hydroxyl
group at the 3-position on the A ring (24). This may be associated with the
temperature-dependent pro- and anti-oxidant properties of
quercetin.
The DNA scission observed in the present study is a
result of radical formation from a Fenton-like reaction between
specific hydroxyl groups and cupric ions. Temperature-dependent DNA
damage between a hydroxyl group at the 3-position on the C ring and
the hydroxyl groups at the 3′- and 4′-positions on the B ring may
be associated with the amount of radical formation at the different
temperatures. In this case, the hydroxyl group at the 3-positions
on the C ring can efficiently produce more radicals compared with
the hydroxyl groups at 3′- and 4′-positions on the B ring in the
presence of cupric ions. Another possible mechanism of differential
DNA damage is an interaction between the quercetin-cupric ion
complex and DNA. The proposed models of the quercetin-cupric ion
complex involve a hydroxyl group at the 3-position on the C ring
and hydroxyl groups at the 3′- and 4′-positions on the B ring. It
is possible that the complex formation between quercetin-cupric ion
at the 3′- and 4′-positions on the B ring is
temperature-dependent.
In conclusion, the present study reported that
quercetin induces DNA scissions in the presence of cupric ions in a
broad range of temperatures. The hydroxyl group at the 3-position
on the C ring contributes to the temperature-independent DNA
scission due to high chelating capacity and oxidative reaction. In
addition, the hydroxyl groups at the 3′- and 4′-positions on the B
ring contribute to DNA scission formation in the presence of cupric
ions at high temperature.
Acknowledgments
Not applicable.
References
|
1
|
Havsteen BH: The biochemistry and medical
significance of the flavonoids. Pharmacol Ther. 96:67–202. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yao LH, Jiang YM, Shi J, Tomás-Barberán
FA, Datta N, Singanusong R and Chen SS: Flavonoids in food and
their health benefits. Plant Foods Hum Nutr. 59:113–122. 2004.
View Article : Google Scholar
|
|
3
|
Amin A and Buratovich M: The anti-cancer
charm of flavonoids: A cup-of-tea will do! Recent Pat Anticancer
Drug Discov. 2:109–117. 2007. View Article : Google Scholar
|
|
4
|
Yu H, Haskins JS, Su C, Allum A, Haskins
AH, Salinas VA, Sunada S, Inoue T, Aizawa Y, Uesaka M and Kato TA:
In vitro screening of radioprotective properties in the novel
glucosylated flavonoids. Int J Mol Med. 38:1525–1530. 2016.
View Article : Google Scholar
|
|
5
|
Maeda J, Roybal EJ, Brents CA, Uesaka M,
Aizawa Y and Kato TA: Natural and glucosyl flavonoids inhibit
poly(ADP-ribose) polymerase activity and induce synthetic lethality
in BRCA mutant cells. Oncol Rep. 31:551–556. 2014. View Article : Google Scholar :
|
|
6
|
Engen A, Maeda J, Wozniak DE, Brents CA,
Bell JJ, Uesaka M, Aizawa Y and Kato TA: Induction of cytotoxic and
genotoxic responses by natural and novel quercetin glycosides.
Mutat Res Genet Toxicol Environ Mutagen. 784–785:15–22. 2015.
View Article : Google Scholar
|
|
7
|
Gaspar J, Rodrigues A, Laires A, Silva F,
Costa S, Monteiro MJ, Monteiro C and Rueff J: On the mechanisms of
genotoxicity and metabolism of quercetin. Mutagenesis. 9:445–449.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bjeldanes LF and Chang GW: Mutagenic
activity of quercetin and related compounds. Science. 197:577–578.
1977. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Brown JP and Dietrich PS: Mutagenicity of
plant flavonols in the Salmonella/mammalian microsome test:
Activation of flavonol glycosides by mixed glycosidases from rat
cecal bacteria and other sources. Mutat Res. 66:223–240. 1979.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
MacGregor JT and Jurd L: Mutagenicity of
plant flavonoids: Structural requirements for mutagenic activity in
Salmonella typhimurium. Mutat Res. 54:297–309. 1978. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rahman A, Shahabuddin, Hadi SM, Parish JH
and Ainley K: Strand scission in DNA induced by quercetin and
Cu(II): Role of Cu(I) and oxygen free radicals. Carcinogenesis.
10:1833–1839. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rahman A, Fazal F, Greensill J, Ainley K,
Parish JH and Hadi SM: Strand scission in DNA induced by dietary
flavonoids: Role of Cu(I) and oxygen free radicals and biological
consequences of scission. Mol Cell Biochem. 111:3–9. 1992.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu Y and Guo M: Studies on transition
metal-quercetin complexes using electrospray ionization tandem mass
spectrometry. Molecules. 20:8583–8594. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pekal A, Biesaga M and Pyrzynska K:
Interaction of quercetin with copper ions: Complexation, oxidation
and reactivity towards radicals. Biometals. 24:41–49. 2011.
View Article : Google Scholar
|
|
15
|
Ríha M, Karlícková J, Filipský T, Jahodár
L, Hrdina R and Mladenka P: In vitro copper-chelating properties of
flavonoids. Free Radic Biol Med. 75(Suppl 1): S462014. View Article : Google Scholar
|
|
16
|
Cherrak SA, Mokhtari-Soulimane N,
Berroukeche F, Bensenane B, Cherbonnel A, Merzouk H and Elhabiri M:
In vitro antioxidant versus metal ion chelating properties of
flavonoids: A structure-activity investigation. PLoS One.
11:e01655752016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bukhari SB, Memon S, Mahroof-Tahir M and
Bhanger MI: Synthesis, characterization and antioxidant activity
copper-quercetin complex. Spectrochim Acta A Mol Biomol Spectrosc.
71:1901–1906. 2009. View Article : Google Scholar
|
|
18
|
Yoshino M, Haneda M, Naruse M and Murakami
K: Prooxidant activity of flavonoids: Copper-dependent strand
breaks and the formation of 8-hydroxy-2′-deoxyguanosine in DNA. Mol
Genet Metab. 68:468–472. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ahsan H and Hadi SM: Strand scission in
DNA induced by curcumin in the presence of Cu(II). Cancer Lett.
124:23–30. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ohashi Y, Yoshinaga K and Yoshioka H and
Yoshioka H: Kinetic analysis of the effect of (-)-epigallocatechin
gallate on the DNA scission induced by Fe(II). Biosci Biotechnol
Biochem. 66:770–776. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kumar S and Pandey AK: Chemistry and
biological activities of flavonoids: An overview.
ScientificWorldJournal. 2013:1627502013. View Article : Google Scholar
|
|
22
|
Yamashita N, Tanemura H and Kawanishi S:
Mechanism of oxidative DNA damage induced by quercetin in the
presence of Cu(II). Mutat Res. 425:107–115. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Carver JH, Carrano AV and MacGregor JT:
Genetic effects of the flavonols quercetin, kaempferol, and
galangin on Chinese hamster ovary cells in vitro. Mutat Res.
113:45–60. 1983. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen JW, Zhu ZQ, Hu TX and Zhu DY:
Structure-activity relationship of natural flavonoids in hydroxyl
radical-scavenging effects. Acta Pharmacol Sin. 23:667–672.
2002.PubMed/NCBI
|