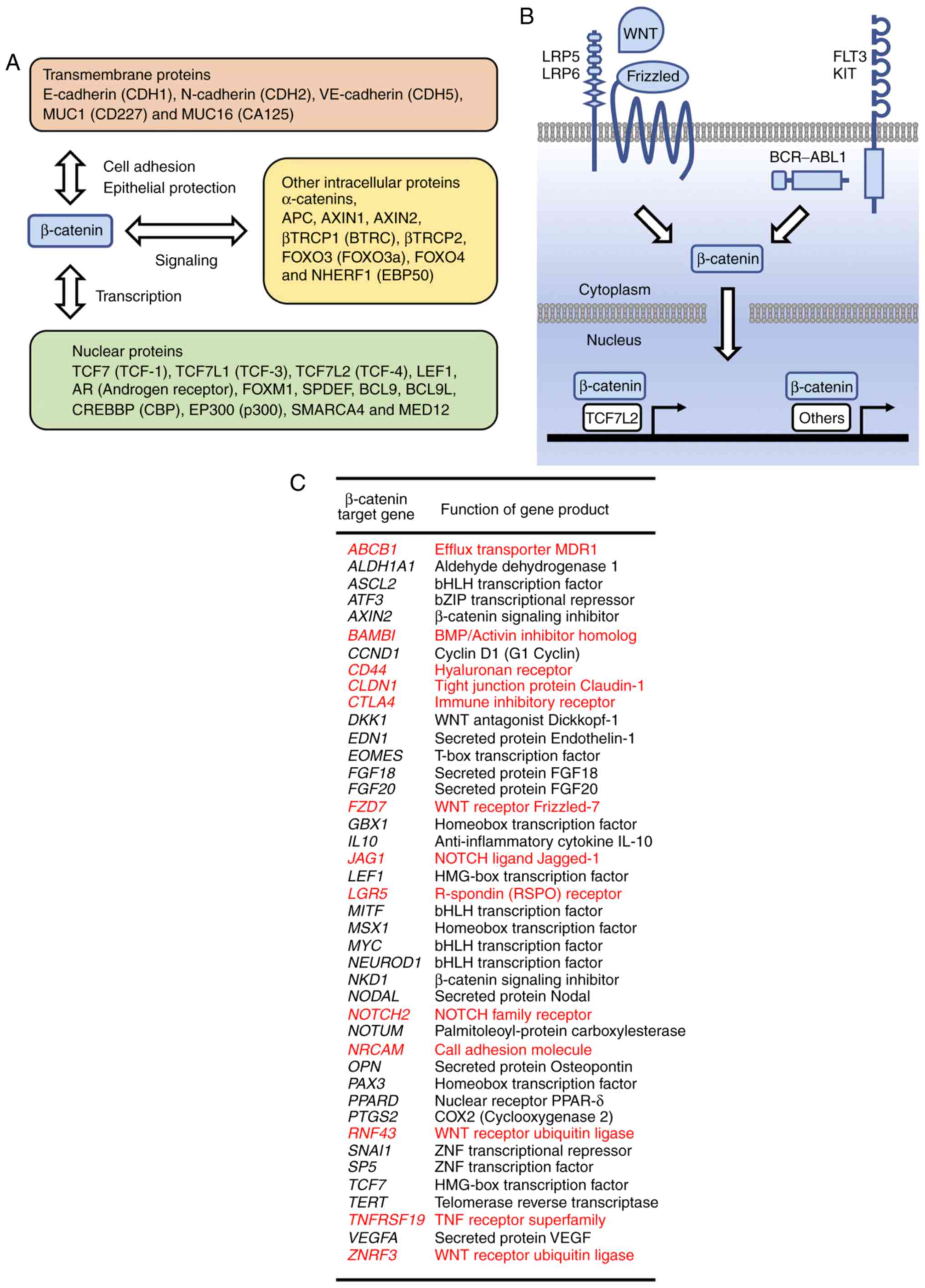
β-catenin undergoes post-translational
modifications, including acetylation, glycosylation, methylation,
phosphorylation and ubiquitylation. Oncogenic tyrosine kinases
phosphorylate β-catenin at Y654 to release β-catenin from cadherin
complexes, whereas canonical WNT signals prevent the
phosphorylation of β-catenin at S33, S37, T41 and S45 to release
β-catenin from ubiquitylation-mediated degradation (1). The β-TRCP complex is involved in the
poly-ubiquitylation of S33/S37/T41/S45-phosphorylated β-catenin and
subsequent proteasome-mediated degradation (9,11),
whereas USP7 is involved in the de-ubiquitylation and stabilization
of β-catenin (12). Stabilized
β-catenin is translocated into the nucleus to activate the
transcription of TCF/LEF target genes (Fig. 1B). The acetylation of β-catenin at
K49 leads to transcriptional activation, whereas the methylation of
β-catenin at K49 leads to transcriptional repression (13,14). The functions of β-catenin are
regulated by its localization, PPIs and stability based on
post-translational modifications.
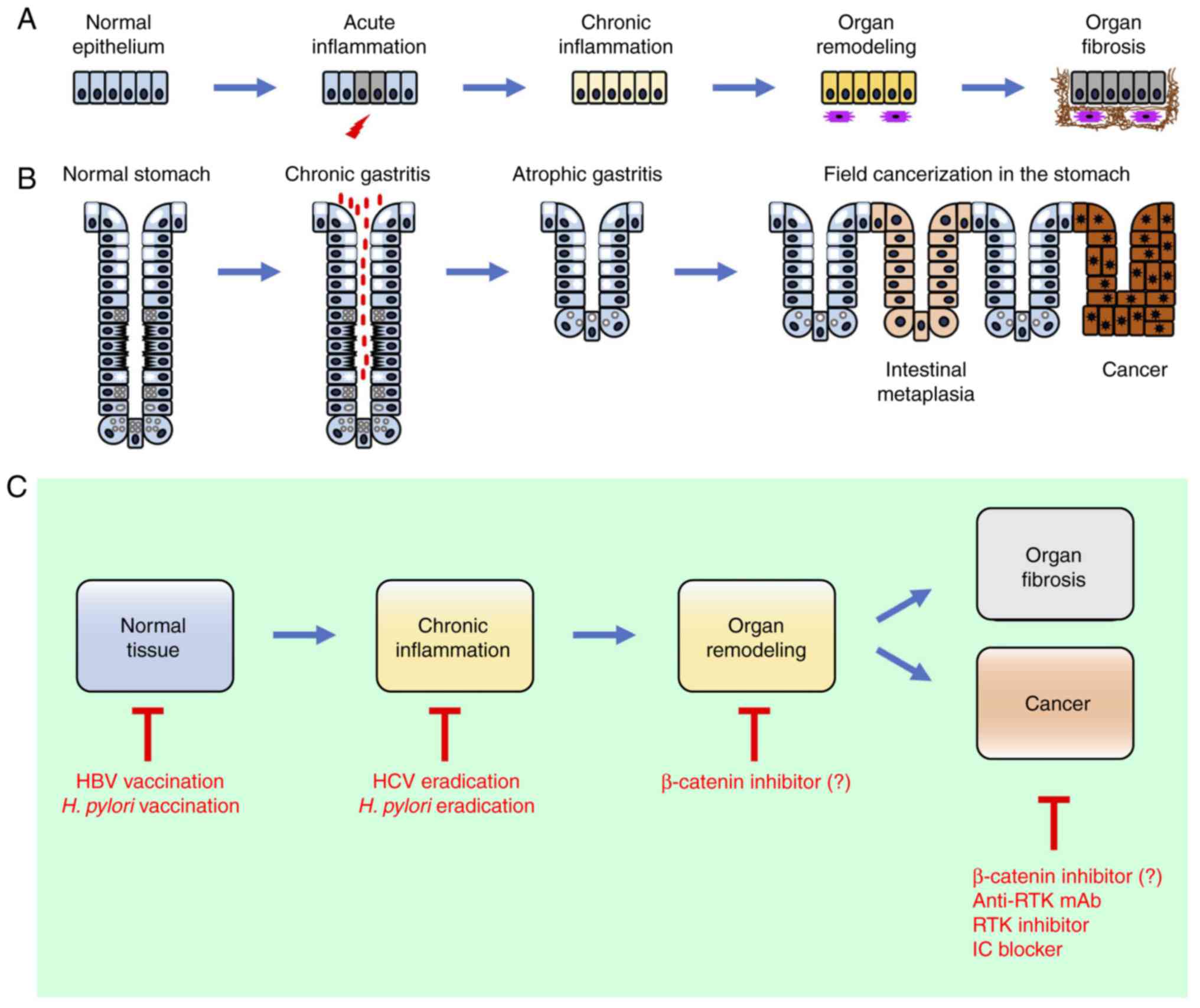
Inflammation is an immune response to repair tissue
damage caused by infectious agents, environmental stimuli and
endogenous irritants. The failure to resolve acute inflammation
leads to chronic inflammation characterized by the continuous
activation of macrophages and lymphocytes in the inflamed tissue
microenvironment and elevated levels of the pro-inflammatory
cytokines IL-1β, IL-6, IL-17 and TNF-α (31,32). Chronic persistent inflammation
then leads to the collapse of homeostatic interactions among
epithelial cells, stromal cells and immune cells in the tissue
microenvironment, which causes organ fibrosis through the
myofibroblast-like transition of tissue-resident fibroblasts,
stellate cells or bone marrow-derived fibrocytes, and the
subsequent deposition of extracellular matrix (ECM) components,
including collagen, fibronectin and hyaluronan (Fig. 2A).
β-catenin signaling dysregulation is involved in
chronic inflammation, organ fibrosis, and various types of human
cancer (1,33). However, β-catenin-targeted therapy
is not yet approved for the treatment of patients with
β-catenin-related diseases. As β-catenin is an intracellular
protein without intrinsic enzymatic activity, it is difficult to
target β-catenin for drug development. In this review, gain- and
loss-of-function β-catenin alterations in human cancer types are
summarized, and the pathophysiology of β-catenin-related chronic
inflammation and/or organ fibrosis are presented with emphases on
carcinogenesis in the stomach, liver and lungs. Finally, the
development of β-catenin inhibitors targeting its upstream
regulators, PPIs and downstream effectors are reviewed.
β-catenin-dependent transcription is aberrantly
activated in human cancer due to gain-of-function mutations in the
CTNNB1 gene itself, in addition to genetic alterations in
the APC, AXIN2, RNF43 and RSPO3 genes
involved in the canonical WNT/β-catenin signaling cascade,
upregulation of canonical WNT ligands in the tumor
microenvironment, or Y654 phosphorylation of β-catenin by oncogenic
tyrosine kinases, including BCR-ABL1 fusion, FLT3-ITD mutation and
overexpression of MET (34).
Missense mutations at or in-frame deletions around
S33, S37, T41 and S45 in β-catenin give rise to gain-of-function
β-catenin mutants that are resistant to ubiquitylation-mediated
proteasomal degradation and induce the upregulation of oncogenic
target genes, including CCND1 and MYC, in
adrenocortical tumors, bladder cancer, colorectal cancer, gastric
cancer, liver cancer, lung cancer, pancreatic cancer, prostate
cancer and uterine cancer (35-40). Aberrant β-catenin-dependent
transcriptional activation drives human carcinogenesis through the
induction of cancer stem cell (CSC) features, bulk tumor
proliferation and the epithelial-to-mesenchymal transition (EMT) in
the solid tumors mentioned above.
By contrast, nonsense or frame-shift mutations in
β-catenin, including R95*, K335fs, R449fs, E458fs, R474*, R535*,
E571* and E642fs, also occur in human cancer, including gastric
cancer and head and neck squamous cell carcinoma (37,41-43). In melanoma, decreased β-catenin
not only promotes invasion and metastasis through disrupted
cell-cell adhesion but also resistance to targeted therapy through
MITF/APE1 axis repression (44,45). Ctnnb1 haploinsufficiency
has been shown to promote aggressiveness and metastasis in a mouse
model of HER2-positive basal breast cancer (46). β-catenin exerts not only oncogenic
but also tumor-suppressor functions in a context-dependent
manner.
β-catenin mutations are classified as i)
gain-of-function mutations clustered at or around S33, S37, T41 and
S45, ii) loss-of-function mutations due to nonsense or frame-shift
mutations and iii) other mutations to be further characterized. In
addition to these coding mutations, copy number gain (47) and regulatory mutations in the
proximal promoter region (48) of
the CTNNB1 gene encoding β-catenin have been reported in
prostate cancer and breast cancer, respectively. Owing to the pro-
and anti-oncogenic roles of β-catenin, integrative omics analyses,
including whole-genome sequencing, transcriptome and
immunohistochemical analyses, are necessary to precisely prescribe
β-catenin-targeted therapeutics in personalized or precision
medicine in the future.
By contrast, atezolizumab, avelumab, durvalumab,
nivolumab and pembrolizumab are representative immune checkpoint
inhibitors that are approved for the treatment of patients with
certain types of cancer (34,68). As H. pylori eradication
eliminates H. pylori-related chronic inflammation and H.
pylori-specific immune evasion, and reduces non-specific immune
evasion caused by the DC-mediated Treg expansion and IL-10
elevation in the gastric microenvironment, H. pylori
eradication may synergize with immune checkpoint inhibitors for the
treatment of advanced gastric cancer with chronic active
gastritis.
Chronic liver inflammation associated with hepatitis
C virus (HCV) infection, hepatitis B virus (HBV) infection, alcohol
abuse, non-alcoholic fatty liver disease and other etiologies leads
to liver fibrosis due to the myofibroblast-like transition of
hepatic stellate cells or other mesenchymal cells and subsequent
accumulation of excessive ECM (31,33). Liver cirrhosis is the most
advanced stage of liver fibrosis, which is characterized by
impaired liver functions and complications, including ascites,
hepatic encephalopathy and upper gastrointestinal bleeding.
Persistent liver inflammation leads to field cancerization in the
liver through the sequential progression of chronic hepatitis,
liver fibrosis and hepatocellular carcinoma (HCC) (69), similar to H. pylori-related
field cancerization in the stomach (Fig. 2C).
The canonical WNT/β-catenin signaling cascade is
involved in the development and homeostasis of the liver (70,71). Canonical WNT/β-catenin signals
promote the proliferation of LGR5+ hepatocyte
progenitors in the peri-venous zone of hepatic lobules, whereas
non-canonical WNT and other signals promote the proliferation of
cholangiocyte progenitors in the peri-portal zone of hepatic
lobules (71-73). Canonical Wnt-dependent
Lgr5+ liver stem/progenitor cells in an organoid culture
have successfully been applied in transplantation therapy for liver
failure in a rat model (74).
HCV is a single-stranded RNA virus that infects
hepatocytes to produce Core, NS3/4A, NS5A, NS5B and other viral
proteins, and HCV upregulates the expression of β-catenin and MYC
in hepatocytes (69). HBV is a
partially double-stranded DNA virus that infects hepatocytes to
produce HBx, pre-S, S and other viral proteins, and HBx upregulates
the expression of EPCAM, β-catenin and MYC and activates NF-κB
signaling in hepatocytes (69).
The RSPO-dependent activation of the WNT/β-catenin signaling
cascade is involved in the activation of hepatic stellate cells to
promote liver fibrosis (75),
whereas a β-catenin inhibitor (PRI-724) has been shown to prevent
HCV-related liver fibrosis in a mouse model (76). By contrast, the WNT/β-catenin
signaling cascade is aberrantly activated in human HCC due to
gain-of-function mutations in the CTNNB1 gene and
loss-of-function mutations in the APC and AXIN1 genes
(35). Additionally,
gain-of-function β-catenin mutations (S33Y or S45Y) and the
overexpression of Met have been shown to synergistically promote
liver tumorigenesis in a mouse model (77), whereas an oncolytic adenovirus
Ad.wnt-E1A(Δ24 bp)-TSLC1 has been shown to effectively target liver
cancer cells with aberrant β-catenin-TCF/LEF signaling activation
and repress in vivo tumorigenesis and metastasis (78). β-catenin plays key roles in
multiple processes of chronic inflammation-related liver
pathophysiology, including hepatocyte proliferation, stellate-cell
activation, liver fibrosis and liver tumorigenesis.
The eradication of HCV is predicted to be an optimal
approach to prevent HCC; as such, the eradication of H.
pylori may be used to prevent gastric cancer. Direct-acting
antivirals, including HCV NS3/4A protease inhibitors (glecaprevir
and paritaprevir), HCV NS5A inhibitors (ledipasvir, ombitasvir,
pibrentasvir and velpatasvir) and HCV NS5B RNA polymerase
inhibitors (dasabuvir and sofosbuvir), have been developed for the
eradication of HCV. For example, glecaprevir/pibrentasvir and
velpatasvir/sofosbuvir are combination drugs that are approved for
the treatment of HCV genotypes 1, 2, 3, 4, 5 and 6 (79,80). Studies in Italy and Spain revealed
that the occurrence and recurrence of HCC were not prevented by HCV
eradication (81,82), whereas a study in Japan revealed
that the occurrence of HCC was successfully prevented by HCV
eradication (83). As
complications of non-viral etiologies of chronic liver
inflammation, ethnic or genetic backgrounds, stages of liver
fibrosis, and genomic or epigenetic alterations in premalignant
lesions may affect the outcomes of HCV eradication, further
investigations in larger cohorts are necessary to address the
controversy regarding the rate of HCC development following HCV
eradication.
Fibrosis is a common pathology of chronic
inflammation in the liver and other organs, including the lungs,
heart and kidneys (31,32). Irreversible pulmonary fibrosis,
cardiac fibrosis or renal fibrosis leads to organ destruction and
subsequent decompensation, which is the final serious condition in
patients with non-cancerous diseases. For example, cardiac fibrosis
is caused by the transformation of cardiac fibroblasts into
myofibroblasts and leads to myocardial stiffness and ventricular
dysfunction (32), whereas
pulmonary fibrosis is caused by chronic inflammation associated
with cancer therapy, cigarette smoking, connective tissue diseases,
environmental pollution, infection, pulmonary hypertension and
idiopathic pulmonary fibrosis (84,85).
Airway damage-induced canonical WNT/β-catenin
signaling activation in alveolar epithelial type II cells promotes
the activation and remodeling of interstitial fibroblasts;
transient remodeling leads to resolution, whereas persistent
remodeling leads to pulmonary fibrosis (31,85). Canonical WNT/β-catenin signaling
activation in pulmonary endothelial cells activates perivascular
fibroblasts to undergo a myofibroblast-like transition, which also
leads to ECM accumulation and increased tissue stiffness, further
promoting pulmonary fibrosis (86,87). Pulmonary injuries also induce the
chemokine-dependent recruitment of monocytes and their subsequent
transition into monocyte-derived alveolar macrophages that express
higher levels of pro-inflammatory and pro-fibrotic genes than
tissue-resident alveolar macrophages (88). However, β-catenin inhibitors,
including ICG-001 and XAV939, ameliorate chronic lung injury and
prevent the progression to severe pulmonary fibrosis (89,90). The canonical WNT/β-catenin
signaling cascade is involved in the pathogenesis of pulmonary
fibrosis.
Genetic alterations in the canonical WNT/β-catenin
signaling regulators are relatively rare in human lung cancer
(91). However, lung CSCs
differentiate into WNT-producing supporting cells to maintain the
stemness of CSCs and promote the expansion of bulk tumor cells
(92). The upregulation of RSPO2
or RSPO3 leads to canonical WNT/β-catenin signaling activation in
patient-derived xenograft (PDX) models of human lung cancer
(93), and nuclear β-catenin
staining is associated with poor prognosis in patients with lung
cancer (94,95). Despite relatively infrequent
genetic alterations, the canonical WNT/β-catenin signaling cascade
is involved in multi-step tumorigenesis in the human lungs through
WNT- and RSPO-dependent paracrine signaling.
Receptor tyrosine kinases (RTKs), including ALK,
DDR2, EGFR, FGFR1, FGFR2, HER2, MET, NTRK1, RET and ROS1, are
aberrantly activated in human lung cancer due to gene
amplification, gene fusions or point mutations (96-99). Although ALK inhibitors (alectinib
and ceritinib), an ALK/ROS1 inhibitor (crizotinib) and EGFR
inhibitors (afatinib, erlotinib, gefitinib and osimertinib) are
approved for the treatment of patients with lung cancer, drug
resistance and recurrence are difficult to avoid due to acquired
mutations in the targeted RTKs, the by-passed activation of other
RTKs and β-catenin signaling activation (34). Therapy-related chronic
inflammation, in addition to cancer-cell plasticity and intra-tumor
heterogeneity, lead to resistance to RTK-targeted therapeutics, in
part through canonical WNT signaling activation.
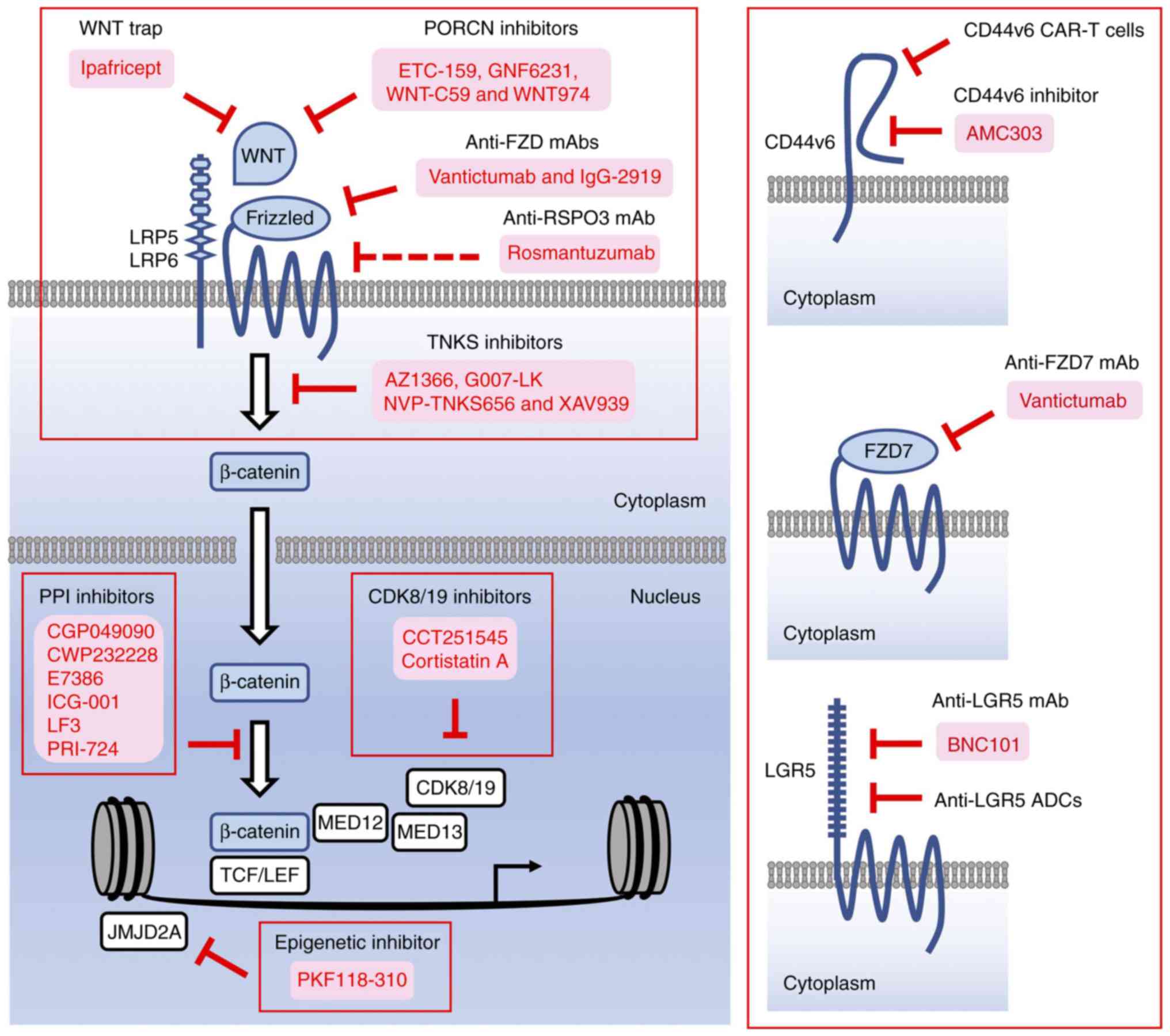
β-catenin is involved in chronic inflammation, organ
fibrosis and carcinogenesis; however, β-catenin lacking intrinsic
enzymatic activity is a difficult target for drug development.
Antibody-based or decoy-receptor drugs targeting ligands or
receptors involved in canonical WNT signaling and small-molecule
compounds targeting porcupine (PORCN), tankyrase (TNKS) and
β-catenin PPIs have been developed as β-catenin inhibitors for
preclinical studies and/or clinical trials (34). In addition to these
investigational drugs, small-molecule compounds targeting
epigenetic/transcriptional regulators involved in
β-catenin-dependent transcription and antibody- or peptide-based
drugs binding to β-catenin-target gene products have also been
suggested as β-catenin inhibitors. β-catenin-targeted therapeutics
are functionally classified as follows: i) β-catenin inhibitors
targeting upstream regulators, ii) β-catenin inhibitors targeting
PPIs, iii) β-catenin inhibitors targeting epigenetic regulators,
iv) β-catenin inhibitors targeting mediator complexes, and v)
β-catenin inhibitors targeting transcriptional outputs (Fig. 3). Follows is a discussion of the
pros and cons of each β-catenin inhibitor class.
Epigenetic components that permit access of the
β-catenin complex to the promoter and enhancer regions of its
target genes are downstream regulators of β-catenin-dependent
transcription (Fig. 3). JMJD2A
(KDM4A), JMJD2B (KDM4B) and JMJD2C (KDM4C) are Jumonji
domain-containing enzymes that demethylate histone H3 at K9 and K36
(H3K9 and H3K36) (117,118). JMJD2C associated with β-catenin
and chromatin is required for the expression of CCND1 and
cell growth in colorectal cancer cells (119). Mouse Jmjd2a and Jmjd2c are
required for the self-renewal of embryonic stem cells (ESCs)
(120), and Jmjd2c associated
with a mediator complex is required for the lineage-specific gene
expression and multi-lineage differentiation of ESCs (121). PKF118-310 was initially
identified as a compound that inhibits β-catenin-dependent
transcription via inhibition of the interaction between β-catenin
and TCF7L2, and PKF118-310 has been re-discovered as a JMJD2A
inhibitor (8,122,123). PKF118-310 has been shown to
exert antitumor effects on colorectal cancer and prostate cancer
in vitro and breast cancer and HCC in vivo (122,124,125). PKF118-310 also exerts
antifibrotic effects in mouse models of dermal fibrosis (126). Other epigenetic regulators,
including EZH2 (127,128), KDM1A (LSD1) (129) and PRMT5 (130), are also involved in canonical
WNT/β-catenin signaling activation in certain contexts. As
epigenetic regulators are desirable targets in the field of
clinical oncology (127,131-134), EZH2 inhibitors, including
GSK2816126 and tazemetostat/EPZ-6438), KDM1A inhibitors, including
GSK2879552 and pargyline, and PRMT5 inhibitors, including
GSK3235025/EPZ015666 and PJ-68, have been developed; however, the
mechanisms of action of these investigational drugs on
β-catenin-dependent transcription require further clarification to
identify biomarkers for patient selection. PKF118-310 is a
promising compound to be optimized for the treatment of patients
with β-catenin-dependent fibrosis and cancer.
Mediator complexes that assemble transcription
factors, cofactors and other regulators of RNA polymerase
II-mediated mRNA synthesis are involved in the β-catenin-dependent
transcription of oncogenic targets, including CCND1 and
MYC (135-137) (Fig. 3). β-catenin binds to MED12, which
associates with MED13, cyclin C (CCNC) and CDK8/19 to form the
kinase module of the mediator complex. As CDK8 and CDK19 are key
components of the mediator complex, CCT251545 (138,139) and cortistatin A (140,141) have been characterized as CDK8/19
inhibitors that suppress β-catenin-dependent transcription and the
in vivo tumorigenesis of colorectal cancer, breast cancer
and acute myeloid leukemia. CDK8/19 phosphorylates mediator complex
components (CCNC, MED12, MED13, MED14 and MED26), epigenetic
regulators (BCL9, BPTF, BRD9, KDM3A, MLL2, SETD1A and SIRT1) and
transcription factors or cofactors (ATF7, FOXC1, KLF12 and STAT1)
(141). Additionally, mediator
complexes are involved in β-catenin-dependent transcription and
transcription dependent on other transcription factors (136,137). Due to the unknown on-target
effects associated with the various functions of CDK9/18 and
mediator complexes, the application of CDK9/18 inhibitors for the
treatment of patients with WNT-driven cancer has been suspended at
the preclinical stage.
As transmembrane proteins expressed on tumor cells
are appropriate targets for the development of peptide-based drugs
(142), mAbs, antibody-drug
conjugates (ADCs), bi-specific antibodies (bsAbs) and chimeric
antigen receptor-modified T cells (150-155), drugs targeting CD44v6, FZD7 and
LGR5 are under development as functional β-catenin inhibitors.
CD44v6 CAR-T cells (156) and
anti-LGR5 ADCs (157,158) and are in preclinical stages,
whereas the phase I clinical trial of anti-CD44v6 ADC (bivatuzumab
mertansine) for patients with head and neck squamous cell carcinoma
was terminated due to severe on-target skin toxicities (159). A peptide-based CD44v6 inhibitor
(AMC303) (160) is in a phase I
clinical trial for the treatment of patients with advanced solid
tumors (ClinicalTrials.gov Identifier:
NCT03009214); anti-FZD7 mAb (vantictumab), which cross-reacts with
FZD1, FZD2, FZD5 and FZD8 (100), is in phase I clinical trials for
cancer patients, as mentioned above (ClinicalTrials.gov Identifier: NCT01957007 and
NCT01973309); anti-LGR5 mAb (BNC101) (161) is also in a phase I clinical
trial for cancer patients (ClinicalTrials.gov Identifier: NCT02726334). Antibody-
or peptide-based drugs are promising options for the treatment of
β-catenin-driven human diseases; however, further evaluation of the
benefits, costs and on-target toxicities are necessary prior to
clinical application.
Clinical medicine, particularly clinical oncology,
is moving toward genomics-based personalized medicine due to the
development of nucleotide sequence technologies. Such personalized
medicine is expected to further evolve into omics- and clinical
record-based precision medicine with relatively inexpensive costs
due to increasing medical expenses in aging societies (162). Genomics-based testing platforms,
including MSK-IMPACT (43),
FoundationOne (163) and
Oncomine Comprehensive Panel (164), and organoid- or PDX-based drug
screening (165-167) are useful tools for the
prescription of targeted therapeutics; however, there remain unmet
medical needs for patients with refractory cancer driven by
gain-of-function mutations in non-enzymatic oncogenes, including
CTNNB1 and KRAS, or loss-of-function mutations in
tumor-suppressor genes, including APC and TP53.
β-catenin signaling is involved in myofibroblast
activation and subsequent organ fibrosis (Fig. 2A), whereas the activation of
β-catenin and NF-κB signaling is involved in field cancerization in
the liver associated with HBV, HCV and other etiologies, and field
cancerization in the stomach associated with H. pylori
infection (Fig. 2B). Vaccines
against HBV and H. pylori are available for the prevention
of cancer-associated infections (168,169). The eradication of HCV and H.
pylori are optimal choices for the first-line prevention of HCC
and gastric cancer, respectively (Fig. 2C); however, pathogen eradication
is not always successful and may occur too late to reverse the
process of field cancerization. β-catenin inhibitors are expected
to be applicable for organ fibrosis prevention, second-line HCC
prevention and treating β-catenin-driven cancer (Fig. 2C). Although β-catenin without
intrinsic enzymatic activity is difficult to target in drug
development, several classes of investigational β-catenin
inhibitors (Fig. 3) are in
preclinical stages or clinical trials for treating patients with
β-catenin-related diseases. The multi-layered prevention and
treatment strategy of β-catenin-related human diseases is realistic
at present and necessary for the implementation of precision
medicine in the future.
This study was financially supported in part by a
grant-in-aid for the Knowledgebase Project from M. Katoh's
Fund.
Not applicable.
MK analyzed the literatures, designed and wrote the
study, and produced the figures.
Not applicable.
Not applicable.
The author declares that he has no competing
interests.
Not applicable.
|
1
|
Katoh M and Katoh M: Molecular genetics
and targeted therapy of WNT-related human diseases (Review). Int J
Mol Med. 40:587–606. 2017.PubMed/NCBI
|
|
2
|
Takeichi M: Dynamic contacts: Rearranging
adherens junctions to drive epithelial remodelling. Nat Rev Mol
Cell Biol. 15:397–410. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
McCrea PD and Gottardi CJ: Beyond
β-catenin: Prospects for a larger catenin network in the nucleus.
Nat Rev Mol Cell Biol. 17:55–64. 2016. View Article : Google Scholar
|
|
4
|
Kufe DW: MUC1-C oncoprotein as a target in
breast cancer: Activation of signaling pathways and therapeutic
approaches. Oncogene. 32:1073–1081. 2013. View Article : Google Scholar
|
|
5
|
Liu Q, Cheng Z, Luo L, Yang Y, Zhang Z, Ma
H, Chen T, Huang X, Lin SY, Jin M, et al: C-terminus of MUC16
activates Wnt signaling pathway through its interaction with
β-catenin to promote tumorigenesis and metastasis. Oncotarget.
7:36800–36813. 2016.PubMed/NCBI
|
|
6
|
Klaus A and Birchmeier W: Wnt signalling
and its impact on development and cancer. Nat Rev Cancer.
8:387–398. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vaquero J, Nguyen Ho-Bouldoires TH,
Clapéron A and Fouassier L: Role of the PDZ-scaffold protein
NHERF1/EBP50 in cancer biology: From signaling regulation to
clinical relevance. Oncogene. 36:3067–3079. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Katoh M and Katoh M: WNT signaling pathway
and stem cell signaling network. Clin Cancer Res. 13:4042–4045.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Valenta T, Hausmann G and Basler K: The
many faces and functions of β-catenin. EMBO J. 31:2714–2736. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Lo YH, Noah TK, Chen MS, Zou W, Borras E,
Vilar E and Shroyer NF: SPDEF induces quiescence of colorectal
cancer cells by changing the transcriptional targets of β-catenin.
Gastroenterology. 153:205–218.e8. 2017. View Article : Google Scholar
|
|
11
|
Frescas D and Pagano M: Deregulated
proteolysis by the F-box proteins SKP2 and β-TrCP: Tipping the
scales of cancer. Nat Rev Cancer. 8:438–449. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Novellasdemunt L, Foglizzo V, Cuadrado L,
Antas P, Kucharska A, Encheva V, Snijders AP and Li VSW: USP7 is a
tumor-specific WNT activator for APC-mutated colorectal cancer by
mediating β-catenin deubiquitination. Cell Rep. 21:612–627. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Hoffmeyer K, Junghans D, Kanzler B and
Kemler R: Trimethylation and acetylation of β-catenin at Lysine 49
represent key elements in ESC pluripotency. Cell Rep. 18:2815–2824.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Alok A, Lei Z, Jagannathan NS, Kaur S,
Harmston N, Rozen SG, Tucker-Kellogg L and Virshup DM: Wnt proteins
synergize to activate β-catenin signaling. J Cell Sci.
130:1532–1544. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Herbst A, Jurinovic V, Krebs S, Thieme SE,
Blum H, Göke B and Kolligs FT: Comprehensive analysis of β-catenin
target genes in colorectal carcinoma cell lines with deregulated
Wnt/β-catenin signaling. BMC Genomics. 15:742014. View Article : Google Scholar
|
|
16
|
Watanabe K, Biesinger J, Salmans ML,
Roberts BS, Arthur WT, Cleary M, Andersen B, Xie X and Dai X:
Integrative ChIP-seq/microarray analysis identifies a CTNNB1 target
signature enriched in intestinal stem cells and colon cancer. PLoS
One. 9:e923172014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Funa NS, Schachter KA, Lerdrup M, Ekberg
J, Hess K, Dietrich N, Honoré C, Hansen K and Semb H: β-Catenin
regulates primitive streak induction through collaborative
interactions with SMAD2/SMAD3 and OCT4. Cell Stem Cell. 16:639–652.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Condello S, Morgan CA, Nagdas S, Cao L,
Turek J, Hurley TD and Matei D: β-Catenin-regulated ALDH1A1 is a
target in ovarian cancer spheroids. Oncogene. 34:2297–2308. 2015.
View Article : Google Scholar
|
|
19
|
Spranger S, Bao R and Gajewski TF:
Melanoma-intrinsic β-catenin signalling prevents anti-tumour
immunity. Nature. 523:231–235. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yun EJ, Zhou J, Lin CJ, Hernandez E, Fazli
L, Gleave M and Hsieh JT: Targeting cancer stem cells in
castration-resistant prostate cancer. Clin Cancer Res. 22:670–679.
2016. View Article : Google Scholar
|
|
21
|
Miwa N, Furuse M, Tsukita S, Niikawa N,
Nakamura Y and Furukawa Y: Involvement of Claudin-1 in the
β-catenin/Tcf signaling pathway and its frequent upregulation in
human colorectal cancers. Oncol Res. 12:469–476. 2001. View Article : Google Scholar
|
|
22
|
Shah KV, Chien AJ, Yee C and Moon RT:
CTLA-4 is a direct target of Wnt/β-catenin signaling and is
expressed in human melanoma tumors. J Invest Dermatol.
128:2870–2879. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yan KS, Janda CY, Chang J, Zheng GXY,
Larkin KA, Luca VC, Chia LA, Mah AT, Han A, Terry JM, et al:
Non-equivalence of Wnt and R-spondin ligands during
Lgr5+ intestinal stem-cell self-renewal. Nature.
545:238–242. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kaur A, Webster MR and Weeraratna AT: In
the Wnt-er of life: Wnt signalling in melanoma and ageing. Br J
Cancer. 115:1273–1279. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ravindranath A, Yuen HF, Chan KK, Grills
C, Fennell DA, Lappin TR and El-Tanani M: Wnt-β-catenin-Tcf-4
signalling-modulated invasiveness is dependent on osteopontin
expression in breast cancer. Br J Cancer. 105:542–551. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gnemmi V, Bouillez A, Gaudelot K, Hémon B,
Ringot B, Pottier N, Glowacki F, Villers A, Vindrieux D, Cauffiez
C, et al: MUC1 drives epithelial-mesenchymal transition in renal
carcinoma through Wnt/β-catenin pathway and interaction with SNAIL
promoter. Cancer Lett. 346:225–236. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Low KC and Tergaonkar V: Telomerase:
Central regulator of all of the hallmarks of cancer. Trends Biochem
Sci. 38:426–434. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Schön S, Flierman I, Ofner A, Stahringer
A, Holdt LM, Kolligs FT and Herbst A: β-catenin regulates NF-κB
activity via TNFRSF19 in colorectal cancer cells. Int J Cancer.
135:1800–1811. 2014. View Article : Google Scholar
|
|
29
|
De Jaime-Soguero A, Aulicino F, Ertaylan
G, Griego A, Cerrato A, Tallam A, Del Sol A, Cosma MP and Lluis F:
Wnt/Tcf1 pathway restricts embryonic stem cell cycle through
activation of the Ink4/Arf locus. PLoS Genet. 13:e10066822017.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ring A, Kim YM and Kahn M: Wnt/catenin
signaling in adult stem cell physiology and disease. Stem Cell Rev
Rep. 10:512–525. 2014. View Article : Google Scholar
|
|
31
|
Bataller R and Brenner DA: Liver fibrosis.
J Clin Invest. 115:209–218. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wynn TA and Ramalingam TR: Mechanisms of
fibrosis: Therapeutic translation for fibrotic disease. Nat Med.
18:1028–1040. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Monga SP: β-catenin signaling and roles in
liver homeostasis, injury, and tumorigenesis. Gastroenterology.
148:1294–1310. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Katoh M: Canonical and non-canonical WNT
signaling in cancer stem cells and their niches: Cellular
heterogeneity, omics reprogramming, targeted therapy and tumor
plasticity. Int J Oncol. 51:1357–1369. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Guichard C, Amaddeo G, Imbeaud S, Ladeiro
Y, Pelletier L, Maad IB, Calderaro J, Bioulac-Sage P, Letexier M,
Degos F, et al: Integrated analysis of somatic mutations and focal
copy-number changes identifies key genes and pathways in
hepatocellular carcinoma. Nat Genet. 44:694–698. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Kandoth C, McLellan MD, Vandin F, Ye K,
Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, et al:
Mutational landscape and significance across 12 major cancer types.
Nature. 502:333–339. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Cancer Genome Atlas Research Network:
Comprehensive molecular characterization of gastric adenocarcinoma.
Nature. 513:202–209. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Robinson D, Van Allen EM, Wu YM, Schultz
N, Lonigro RJ, Mosquera JM, Montgomery B, Taplin ME, Pritchard CC,
Attard G, et al: Integrative clinical genomics of advanced prostate
cancer. Cell. 161:1215–1228. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Teo AE, Garg S, Shaikh LH, Zhou J, Karet
Frankl FE, Gurnell M, Happerfield L, Marker A, Bienz M, Azizan EA
and Brown MJ: Pregnancy, primary aldosteronism, and adrenal CTNNB1
mutations. N Engl J Med. 373:1429–1436. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bailey P, Chang DK, Nones K, Johns AL,
Patch AM, Gingras MC, Miller DK, Christ AN, Bruxner TJ, Quinn MC,
et al: Genomic analyses identify molecular subtypes of pancreatic
cancer. Nature. 531:47–52. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Cancer Genome Atlas Network: Comprehensive
genomic characterization of head and neck squamous cell carcinomas.
Nature. 517:576–582. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Cancer Genome Atlas Research Network; Asan
University; BC Cancer Agency; Brigham and Women's Hospital; Broad
Institute; Brown University; Case Western Reserve University;
Dana-Farber Cancer Institute; Duke University; et al: Integrated
genomic characterization of oesophageal carcinoma. Nature.
541:169–175. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zehir A, Benayed R, Shah RH, Syed A,
Middha S, Kim HR, Srinivasan P, Gao J, Chakravarty D, Devlin SM, et
al: Mutational landscape of metastatic cancer revealed from
prospective clinical sequencing of 10,000 patients. Nat Med.
23:703–713. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Kaur A, Webster MR, Marchbank K, Behera R,
Ndoye A, Kugel CH III, Dang VM, Appleton J, O'Connell MP, Cheng P,
et al: sFRP2 in the aged microenvironment drives melanoma
metastasis and therapy resistance. Nature. 532:250–254. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Webster MR, Kugel CH III and Weeraratna
AT: The Wnts of change: How Wnts regulate phenotype switching in
melanoma. Biochim Biophys Acta. 1856:244–251. 2015.PubMed/NCBI
|
|
46
|
Bui T, Schade B, Cardiff RD, Aina OH,
Sanguin-Gendreau V and Muller WJ: β-Catenin haploinsufficiency
promotes mammary tumorigenesis in an ErbB2-positive basal breast
cancer model. Proc Natl Acad Sci USA. 114:E707–E716. 2017.
View Article : Google Scholar
|
|
47
|
Beltran H, Prandi D, Mosquera JM, Benelli
M, Puca L, Cyrta J, Marotz C, Giannopoulou E, Chakravarthi BV,
Varambally S, et al: Divergent clonal evolution of
castration-resistant neuroendocrine prostate cancer. Nat Med.
22:298–305. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Rheinbay E, Parasuraman P, Grimsby J, Tiao
G, Engreitz JM, Kim J, Lawrence MS, Taylor-Weiner A,
Rodriguez-Cuevas S, Rosenberg M, et al: Recurrent and functional
regulatory mutations in breast cancer. Nature. 547:55–60. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Bugaytsova JA, Björnham O, Chernov YA,
Gideonsson P, Henriksson S, Mendez M, Sjöström R, Mahdavi J,
Shevtsova A, Ilver D, et al: Helicobacter pylori adapts to chronic
infection and gastric disease via pH-responsive BabA-mediated
adherence. Cell Host Microbe. 21:376–389. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Javaheri A, Kruse T, Moonens K,
Mejías-Luque R, Debraekeleer A, Asche CI, Tegtmeyer N, Kalali B,
Bach NC, Sieber SA, et al: Helicobacter pylori adhesin HopQ engages
in a virulence-enhancing interaction with human CEACAMs. Nat
Microbiol. 2:161892016. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Salama NR, Hartung ML and Müller A: Life
in the human stomach: Persistence strategies of the bacterial
pathogen Helicobacter pylori. Nat Rev Microbiol. 11:385–399. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Yamaoka Y and Graham DY: Helicobacter
pylori virulence and cancer pathogenesis. Future Oncol.
10:1487–1500. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Käbisch R, Mejías-Luque R, Gerhard M and
Prinz C: Involvement of Toll-like receptors on Helicobacter
pylori-induced immunity. PLoS One. 9:e1048042014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
McCracken KW, Catá EM, Crawford CM,
Sinagoga KL, Schumacher M, Rockich BE, Tsai YH, Mayhew CN, Spence
JR, Zavros Y and Wells JM: Modelling human development and disease
in pluripotent stem cell-derived gastric organoids. Nature.
516:400–404. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Bertaux-Skeirik N, Feng R, Schumacher MA,
Li J, Mahe MM, Engevik AC, Javier JE, Peek RM Jr, Ottemann K,
Orian-Rousseau V, et al: CD44 plays a functional role in
Helicobacter pylori-induced epithelial cell proliferation. PLoS
Pathog. 112:e10046632015. View Article : Google Scholar
|
|
56
|
Song X, Xin N, Wang W and Zhao C:
Wnt/β-catenin, an oncogenic pathway targeted by H. pylori in
gastric carcinogenesis. Oncotarget. 6:35579–35588. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Sigal M, Logan CY, Kapalczynska M,
Mollenkopf HJ, Berger H, Wiedenmann B, Nusse R, Amieva MR and Meyer
TF: Stromal R-spondin orchestrates gastric epithelial stem cells
and gland homeostasis. Nature. 548:451–455. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Katoh M, Hirai M, Sugimura T and Terada M:
Cloning, expression and chromosomal localization of Wnt-13, a novel
member of the Wnt gene family. Oncogene. 13:873–876.
1996.PubMed/NCBI
|
|
59
|
Katoh M, Kirikoshi H, Terasaki H and
Shiokawa K: WNT2B2 mRNA, up-regulated in primary gastric cancer, is
a positive regulator of the WNT-β-catenin-TCF signaling pathway.
Biochem Biophys Res Commun. 289:1093–1098. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Jiang X and Cong F: Novel regulation of
Wnt signaling at the proximal membrane level. Trends Biochem Sci.
41:773–783. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Milne AN, Carneiro F, O'Morain C and
Offerhaus GJ: Nature meets nurture: Molecular genetics of gastric
cancer. Hum Genet. 126:615–628. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Leodolter A, Alonso S, González B, Ebert
MP, Vieth M, Röcken C, Wex T, Peitz U, Malfertheiner P and Perucho
M: Somatic DNA hypomethylation in H. pylori-associated high-risk
gastritis and gastric cancer: Enhanced somatic hypomethylation
associates with advanced stage cancer. Clin Transl Gastroenterol.
6:e852015. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Ajani JA, Lee J, Sano T, Janjigian YY, Fan
D and Song S: Gastric adenocarcinoma. Nat Rev Dis Primers.
3:170362017. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Huh CW, Youn YH, Jung da H, Park JJ, Kim
JH and Park H: Early attempts to eradicate Helicobacter pylori
after endoscopic resection of gastric neoplasm significantly
improve eradication success rates. PLoS One. 11:e01622582016.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Dang BN and Graham DY: Helicobacter pylori
infection and antibiotic resistance: A WHO high priority? Nat Rev
Gastroenterol Hepatol. 14:383–384. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Osumi H, Fujisaki J, Suganuma T, Horiuchi
Y, Omae M, Yoshio T, Ishiyama A, Tsuchida T and Miki K: A
significant increase in the pepsinogen I/II ratio is a reliable
biomarker for successfulHelicobacter pylori eradication. PLoS One.
12:e01839802017. View Article : Google Scholar
|
|
67
|
Seta T, Takahashi Y, Noguchi Y, Shikata S,
Sakai T, Sakai K, Yamashita Y and Nakayama T: Effectiveness of
Helicobacter pylori eradication in the prevention of primary
gastric cancer in healthy asymptomatic people: A systematic review
and meta-analysis comparing risk ratio with risk difference. PLoS
One. 12:e01833212017. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Smyth MJ, Ngiow SF, Ribas A and Teng MW:
Combination cancer immunotherapies tailored to the tumour
microenvironment. Nat Rev Clin Oncol. 13:143–158. 2016. View Article : Google Scholar
|
|
69
|
Arzumanyan A, Reis HM and Feitelson MA:
Pathogenic mechanisms in HBV- and HCV-associated hepatocellular
carcinoma. Nat Rev Cancer. 13:123–135. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Touboul T, Chen S, To CC, Mora-Castilla S,
Sabatini K, Tukey RH and Laurent LC: Stage-specific regulation of
the WNT/β-catenin pathway enhances differentiation of hESCs into
hepatocytes. J Hepatol. 64:1315–1326. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Planas-Paz L, Orsini V, Boulter L,
Calabrese D, Pikiolek M, Nigsch F, Xie Y, Roma G, Donovan A, Marti
P, et al: The RSPO-LGR4/5-ZNRF3/RNF43 module controls liver
zonation and size. Nat Cell Biol. 18:467–479. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Okabe H, Yang J, Sylakowski K, Yovchev M,
Miyagawa Y, Nagarajan S, Chikina M, Thompson M, Oertel M, Baba H,
et al: Wnt signaling regulates hepatobiliary repair following
cholestatic liver injury in mice. Hepatology. 64:1652–1666. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Li J, Hu SB, Wang LY, Zhang X, Zhou X,
Yang B, Li JH, Xiong J, Liu N, Li Y, et al: Autophagy-dependent
generation of Axin2+ cancer stem-like cells promotes
hepatocarcinogenesis in liver cirrhosis. Oncogene. 36:6725–6737.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Kuijk EW, Rasmussen S, Blokzijl F, Huch M,
Gehart H, Toonen P, Begthel H, Clevers H, Geurts AM and Cuppen E:
Generation and characterization of rat liver stem cell lines and
their engraftment in a rat model of liver failure. Sci Rep.
6:221542016. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Yin X, Yi H, Wang L, Wu W, Wu X and Yu L:
RSPOs facilitated HSC activation and promoted hepatic fibrogenesis.
Oncotarget. 7:63767–63778. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Tokunaga Y, Osawa Y, Ohtsuki T, Hayashi Y,
Yamaji K, Yamane D, Hara M, Munekata K, Tsukiyama-Kohara K, Hishima
T, et al: Selective inhibitor of Wnt/β-catenin/CBP signaling
ameliorates hepatitis C virus-induced liver fibrosis in mouse
model. Sci Rep. 7:3252017. View Article : Google Scholar
|
|
77
|
Tao J, Xu E, Zhao Y, Singh S, Li X, Couchy
G, Chen X, Zucman-Rossi J, Chikina M and Monga SP: Modeling a human
hepatocellular carcinoma subset in mice through coexpression of Met
and point-mutant β-catenin. Hepatology. 64:1587–1605. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Zhang J, Lai W, Li Q, Yu Y, Jin J, Guo W,
Zhou X, Liu X and Wang Y: A novel oncolytic adenovirus targeting
Wnt signaling effectively inhibits cancer-stem like cell growth via
metastasis, apoptosis and autophagy in HCC models. Biochem Biophys
Res Commun. 491:469–477. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Lamb YN: Glecaprevir/pibrentasvir: First
global approval. Drugs. 77:1797–1804. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Nehra V, Rizza SA and Temesgen Z:
Sofosbuvir/velpatasvir fixed-dose combination for the treatment of
chronic hepatitis C virus infection. Drugs Today (Barc).
53:177–189. 2017. View Article : Google Scholar
|
|
81
|
Conti F, Buonfiglioli F, Scuteri A, Crespi
C, Bolondi L, Caraceni P, Foschi FG, Lenzi M, Mazzella G, Verucchi
G, et al: Early occurrence and recurrence of hepatocellular
carcinoma in HCV-related cirrhosis treated with direct-acting
antivirals. J Hepatol. 65:727–733. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Reig M, Mariño Z, Perelló C, Iñarrairaegui
M, Ribeiro A, Lens S, Díaz A, Vilana R, Darnell A, Varela M, et al:
Unexpected high rate of early tumor recurrence in patients with
HCV-related HCC undergoing interferon-free therapy. J Hepatol.
65:719–726. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Kobayashi M, Suzuki F, Fujiyama S,
Kawamura Y, Sezaki H, Hosaka T, Akuta N, Suzuki Y, Saitoh S, Arase
Y, et al: Sustained virologic response by direct antiviral agents
reduces the incidence of hepatocellular carcinoma in patients with
HCV infection. J Med Virol. 89:476–483. 2017. View Article : Google Scholar
|
|
84
|
Selman M, López-Otín C and Pardo A:
Age-driven developmental drift in the pathogenesis of idiopathic
pulmonary fibrosis. Eur Respir J. 48:538–552. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Knudsen L, Ruppert C and Ochs M: Tissue
remodelling in pulmonary fibrosis. Cell Tissue Res. 367:607–626.
2017. View Article : Google Scholar
|
|
86
|
Cao Z, Lis R, Ginsberg M, Chavez D, Shido
K, Rabbany SY, Fong GH, Sakmar TP, Rafii S and Ding BS: Targeting
of the pulmonary capillary vascular niche promotes lung alveolar
repair and ameliorates fibrosis. Nat Med. 22:154–162. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Andersson-Sjöland A, Karlsson JC and
Rydell-Törmänen K: ROS-induced endothelial stress contributes to
pulmonary fibrosis through pericytes and Wnt signaling. Lab Invest.
96:206–217. 2016. View Article : Google Scholar
|
|
88
|
Misharin AV, Morales-Nebreda L, Reyfman
PA, Cuda CM, Walter JM, McQuattie-Pimentel AC, Chen CI, Anekalla
KR, Joshi N, Williams KJN, et al: Monocyte-derived alveolar
macrophages drive lung fibrosis and persist in the lung over the
life span. J Exp Med. 214:2387–2404. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Henderson WR Jr, Chi EY, Ye X, Nguyen C,
Tien YT, Zhou B, Borok Z, Knight DA and Kahn M: Inhibition of
Wnt/β-catenin/CREB binding protein (CBP) signaling reverses
pulmonary fibrosis. Proc Natl Acad Sci USA. 107:14309–14314. 2010.
View Article : Google Scholar
|
|
90
|
Chen X, Shi C, Meng X, Zhang K, Li X, Wang
C, Xiang Z, Hu K and Han X: Inhibition of Wnt/β-catenin signaling
suppresses bleomycin-induced pulmonary fibrosis by attenuating the
expression of TGF-β1 and FGF-2. Exp Mol Pathol. 101:22–30. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Campbell JD, Alexandrov A, Kim J, Wala J,
Berger AH, Pedamallu CS, Shukla SA, Guo G, Brooks AN, Murray BA, et
al: Distinct patterns of somatic genome alterations in lung
adenocarcinomas and squamous cell carcinomas. Nat Genet.
48:607–616. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Tammela T, Sanchez-Rivera FJ, Cetinbas NM,
Wu K, Joshi NS, Helenius K, Park Y, Azimi R, Kerper NR, Wesselhoeft
RA, et al: A Wnt-producing niche drives proliferative potential and
progression in lung adenocarcinoma. Nature. 545:355–359. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Chartier C, Raval J, Axelrod F, Bond C,
Cain J, Dee-Hoskins C, Ma S, Fischer MM, Shah J, Wei J, et al:
Therapeutic targeting of tumor-derived R-Spondin attenuates
β-catenin signaling and tumorigenesis in multiple cancer types.
Cancer Res. 76:713–723. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
94
|
Yang Y, Shen J, He J, He J and Jiang G: A
meta-analysis of abnormal β-catenin immunohistochemical expression
as a prognostic factor in lung cancer: Location is more important.
Clin Transl Oncol. 18:685–692. 2016. View Article : Google Scholar
|
|
95
|
Jin J, Zhan P, Katoh M, Kobayashi SS, Phan
K, Qian H, Li H and Wang X and Wang X: Prognostic significance of
β-catenin expression in patients with non-small cell lung cancer: A
meta-analysis. Transl Lung Cancer Res. 6:97–108. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Mano H: ALKoma: A cancer subtype with a
shared target. Cancer Discov. 2:495–502. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Seo JS, Ju YS, Lee WC, Shin JY, Lee JK,
Bleazard T, Lee J, Jung YJ, Kim JO, Shin JY, et al: The
transcriptional landscape and mutational profile of lung
adenocarcinoma. Genome Res. 22:2109–2119. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Hirsch FR, Suda K, Wiens J and Bunn PA Jr:
New and emerging targeted treatments in advanced non-small-cell
lung cancer. Lancet. 388:1012–1024. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Katoh M: Therapeutics targeting FGF
signaling network in human diseases. Trends Pharmacol Sci.
37:1081–1096. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Gurney A, Axelrod F, Bond CJ, Cain J,
Chartier C, Donigan L, Fischer M, Chaudhari A, Ji M, Kapoun AM, et
al: Wnt pathway inhibition via the targeting of Frizzled receptors
results in decreased growth and tumorigenicity of human tumors.
Proc Natl Acad Sci USA. 109:11717–11722. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Steinhart Z, Pavlovic Z, Chandrashekhar M,
Hart T, Wang X, Zhang X, Robitaille M, Brown KR, Jaksani S,
Overmeer R, et al: Genome-wide CRISPR screens reveal a Wnt-FZD5
signaling circuit as a druggable vulnerability of RNF43-mutant
pancreatic tumors. Nat Med. 23:60–68. 2017. View Article : Google Scholar
|
|
102
|
Bendell J, Eckhardt GS, Hochster HS,
Morris VK, Strickler J, Kapoun AM, Wang M, Xu L, McGuire K, Dupont
J, et al: Initial results from a phase 1a/b study of OMP-131R10, a
first-in-class anti-RSPO3 antibody, in advanced solid tumors and
previously treated metastatic colorectal cancer (CRC). Eur J
Cancer. 69(Suppl 1): S29–S30. 2016. View Article : Google Scholar
|
|
103
|
Le PN, McDermott JD and Jimeno A:
Targeting the Wnt pathway in human cancers: Therapeutic targeting
with a focus on OMP-54F28. Pharmacol Ther. 146:1–11. 2015.
View Article : Google Scholar
|
|
104
|
Madan B, Ke Z, Harmston N, Ho SY, Frois
AO, Alam J, Jeyaraj DA, Pendharkar V, Ghosh K, Virshup IH, et al:
Wnt addiction of genetically defined cancers reversed by PORCN
inhibition. Oncogene. 35:2197–2207. 2016. View Article : Google Scholar
|
|
105
|
Chen CW, Beyer C, Liu J, Maier C, Li C,
Trinh-Minh T, Xu X, Cole SH, Hsieh MH, Ng N, et al: Pharmacological
inhibition of porcupine induces regression of experimental skin
fibrosis by targeting Wnt signalling. Ann Rheum Dis. 76:773–778.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Blyszczuk P, Müller-Edenborn B, Valenta T,
Osto E, Stellato M, Behnke S, Glatz K, Basler K, Lüscher TF,
Distler O, et al: Transforming growth factor-β-dependent Wnt
secretion controls myofibroblast formation and myocardial fibrosis
progression in experimental autoimmune myocarditis. Eur Heart J.
38:1413–1425. 2017.
|
|
107
|
Liu J, Pan S, Hsieh MH, Ng N, Sun F, Wang
T, Kasibhatla S, Schuller AG, Li AG, Cheng D, et al: Targeting
Wnt-driven cancer through the inhibition of Porcupine by LGK974.
Proc Natl Acad Sci USA. 110:20224–20229. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Quackenbush KS, Bagby S, Tai WM,
Messersmith WA, Schreiber A, Greene J, Kim J, Wang G, Purkey A,
Pitts TM, et al: The novel tankyrase inhibitor (AZ1366) enhances
irinotecan activity in tumors that exhibit elevated tankyrase and
irinotecan resistance. Oncotarget. 7:28273–28285. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Lau T, Chan E, Callow M, Waaler J, Boggs
J, Blake RA, Magnuson S, Sambrone A, Schutten M, Firestein R, et
al: A novel tankyrase small-molecule inhibitor suppresses APC
mutation-driven colorectal tumor growth. Cancer Res. 73:3132–3144.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Shultz MD, Cheung AK, Kirby CA, Firestone
B, Fan J, Chen CH, Chen Z, Chin DN, Dipietro L, Fazal A, et al:
Identification of NVP-TNKS656: The use of structure-efficiency
relationships to generate a highly potent, selective, and orally
active tankyrase inhibitor. J Med Chem. 56:6495–6511. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
111
|
Huang SM, Mishina YM, Liu S, Cheung A,
Stegmeier F, Michaud GA, Charlat O, Wiellette E, Zhang Y, Wiessner
S, et al: Tankyrase inhibition stabilizes Axin and antagonizes Wnt
signalling. Nature. 461:614–620. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
Trautmann M, Sievers E, Aretz S, Kindler
D, Michels S, Friedrichs N, Renner M, Kirfel J, Steiner S, Huss S,
et al: SS18-SSX fusion protein-induced Wnt/β-catenin signaling is a
therapeutic target in synovial sarcoma. Oncogene. 33:5006–5016.
2014. View Article : Google Scholar
|
|
113
|
Jang GB, Hong IS, Kim RJ, Lee SY, Park SJ,
Lee ES, Park JH, Yun CH, Chung JU, Lee KJ, et al: Wnt/β-catenin
small-molecule inhibitor CWP232228 preferentially inhibits the
growth of breast cancer stem-like cells. Cancer Res. 75:1691–1702.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Yamada K, Hori Y, Yamaguchi A, Matsuki M,
Tsukamoto S, Yokoi A, Semba T, Ozawa Y, Inoue S, Yamamoto Y, et al:
Abstract 5177: E7386: First-in-class orally active CBP/β-catenin
modulator as an anticancer agent. Proceedings of the American
Association for Cancer Research Annual Meeting 2017; 2017 Apr 1–5;
Washington, DC. Philadelphia. AACR;
Cancer Res. 77(Suppl 13): 51772017.
View Article : Google Scholar
|
|
115
|
Fang L, Zhu Q, Neuenschwander M, Specker
E, Wulf-Goldenberg A, Weis WI, von Kries JP and Birchmeier W: A
small-molecule antagonist of the β-catenin/TCF4 interaction blocks
the self-renewal of cancer stem cells and suppresses tumorigenesis.
Cancer Res. 76:891–901. 2016. View Article : Google Scholar
|
|
116
|
Zhou H, Mak PY, Mu H, Mak DH, Zeng Z,
Cortes J, Liu Q, Andreeff M and Carter BZ: Combined inhibition of
β-catenin and Bcr-Abl synergistically targets tyrosine kinase
inhibitor-resistant blast crisis chronic myeloid leukemia blasts
and progenitors in vitro and in vivo. Leukemia. 31:2065–2074. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
117
|
Katoh M and Katoh M: Identification and
characterization of JMJD2 family genes in silico. Int J Oncol.
24:1623–1628. 2004.PubMed/NCBI
|
|
118
|
Berry WL and Janknecht R: KDM4/JMJD2
histone demethylases: Epigenetic regulators in cancer cells. Cancer
Res. 73:2936–2942. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Kim TD, Fuchs JR, Schwartz E, Abdelhamid
D, Etter J, Berry WL, Li C, Ihnat MA, Li PK and Janknecht R:
Pro-growth role of the JMJD2C histone demethylase in HCT-116 colon
cancer cells and identification of curcuminoids as JMJD2
inhibitors. Am J Transl Res. 6:236–247. 2014.PubMed/NCBI
|
|
120
|
Pedersen MT, Kooistra SM, Radzisheuskaya
A, Laugesen A, Johansen JV, Hayward DG, Nilsson J, Agger K and
Helin K: Continual removal of H3K9 promoter methylation by Jmjd2
demethylases is vital for ESC self-renewal and early development.
EMBO J. 35:1550–1564. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Tomaz RA, Harman JL, Karimlou D, Weavers
L, Fritsch L, Bou-Kheir T, Bell E, Del Valle Torres I, Niakan KK,
Fisher C, et al: Jmjd2c facilitates the assembly of essential
enhancer-protein complexes at the onset of embryonic stem cell
differentiation. Development. 144:567–579. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Lepourcelet M, Chen YN, France DS, Wang H,
Crews P, Petersen F, Bruseo C, Wood AW and Shivdasani RA:
Small-molecule antagonists of the oncogenic Tcf/β-catenin protein
complex. Cancer Cell. 5:91–102. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Franci G, Sarno F, Nebbioso A and Altucci
L: Identification and characterization of PKF118-310 as a KDM4A
inhibitor. Epigenetics. 12:198–205. 2017. View Article : Google Scholar :
|
|
124
|
Wei W, Chua MS, Grepper S and So S: Small
molecule antagonists of Tcf4/β-catenin complex inhibit the growth
of HCC cells in vitro and in vivo. Int J Cancer. 126:2426–2436.
2010.
|
|
125
|
Hallett RM, Kondratyev MK, Giacomelli AO,
Nixon AML, Girgis-Gabardo A, Ilieva D and Hassell JA: Small
molecule antagonists of the Wnt/β-catenin signaling pathway target
breast tumor-initiating cells in a Her2/Neu mouse model of breast
cancer. PLoS One. 7:e339762012. View Article : Google Scholar
|
|
126
|
Beyer C, Reichert H, Akan H, Mallano T,
Schramm A, Dees C, Palumbo-Zerr K, Lin NY, Distler A, Gelse K, et
al: Blockade of canonical Wnt signalling ameliorates experimental
dermal fibrosis. Ann Rheum Dis. 72:1255–1258. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Katoh M: Mutation spectra of histone
methyltransferases with canonical SET domains and EZH2-targeted
therapy. Epigenomics. 8:285–305. 2016. View Article : Google Scholar
|
|
128
|
Chen JF, Luo X, Xiang LS, Li HT, Zha L, Li
N, He JM, Xie GF, Xie X and Liang HJ: EZH2 promotes colorectal
cancer stem-like cell expansion by activating p21cip1-Wnt/β-catenin
signaling. Oncotarget. 7:41540–41558. 2016.PubMed/NCBI
|
|
129
|
Huang M, Chen C, Geng J, Han D, Wang T,
Xie T, Wang L, Wang Y, Wang C, Lei Z and Chu X: Targeting KDM1A
attenuates Wnt/β-catenin signaling pathway to eliminate
sorafenib-resistant stem-like cells in hepatocellular carcinoma.
Cancer Lett. 398:12–21. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Jin Y, Zhou J, Xu F, Jin B, Cui L, Wang Y,
Du X, Li J, Li P, Ren R and Pan J: Targeting methyltransferase
PRMT5 eliminates leukemia stem cells in chronic myelogenous
leukemia. J Clin Invest. 126:3961–3980. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Feinberg AP, Koldobskiy MA and Göndör A:
Epigenetic modulators, modifiers and mediators in cancer aetiology
and progression. Nat Rev Genet. 17:284–299. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Morera L, Lübbert M and Jung M: Targeting
histone methyltransferases and demethylases in clinical trials for
cancer therapy. Clin Epigenetics. 8:572016. View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Allis CD and Jenuwein T: The molecular
hallmarks of epigenetic control. Nat Rev Genet. 17:487–500. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Jones PA, Issa JP and Baylin S: Targeting
the cancer epigenome for therapy. Nat Rev Genet. 17:630–641. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Kim S, Xu X, Hecht A and Boyer TG:
Mediator is a transducer of Wnt/beta-catenin signaling. J Biol
Chem. 281:14066–14075. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Hnisz D, Abraham BJ, Lee TI, Lau A,
Saint-André V, Sigova AA, Hoke HA and Young RA: Super-enhancers in
the control of cell identity and disease. Cell. 155:934–947. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Yin JW and Wang G: The Mediator complex: A
master coordinator of transcription and cell lineage development.
Development. 141:977–987. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Mallinger A, Crumpler S, Pichowicz M,
Waalboer D, Stubbs M, Adeniji-Popoola O, Wood B, Smith E, Thai C,
Henley AT, et al: Discovery of potent, orally bioavailable,
small-molecule inhibitors of WNT signaling from a cell-based
pathway screen. J Med Chem. 58:1717–1735. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Dale T, Clarke PA, Esdar C, Waalboer D,
Adeniji-Popoola O, Ortiz-Ruiz MJ, Mallinger A, Samant RS,
Czodrowski P, Musil D, et al: A selective chemical probe for
exploring the role of CDK8 and CDK19 in human disease. Nat Chem
Biol. 11:973–980. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Pelish HE, Liau BB, Nitulescu II,
Tangpeerachaikul A, Poss ZC, Da Silva DH, Caruso BT, Arefolov A,
Fadeyi O, Christie AL, et al: Mediator kinase inhibition further
activates super-enhancer-associated genes in AML. Nature.
526:273–276. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
141
|
Poss ZC, Ebmeier CC, Odell AT,
Tangpeerachaikul A, Lee T, Pelish HE, Shair MD, Dowell RD, Old WM
and Taatjes DJ: Identification of Mediator kinase substrates in
human cells using cortistatin A and quantitative phosphoproteomics.
Cell Rep. 15:436–450. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Todaro M, Gaggianesi M, Catalano V,
Benfante A, Iovino F, Biffoni M, Apuzzo T, Sperduti I, Volpe S,
Cocorullo G, et al: CD44v6 is a marker of constitutive and
reprogrammed cancer stem cells driving colon cancer metastasis.
Cell Stem Cell. 14:342–356. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Schmitt M, Metzger M, Gradl D, Davidson G
and Orian-Rousseau V: CD44 functions in Wnt signaling by regulating
LRP6 localization and activation. Cell Death Differ. 22:677–689.
2015. View Article : Google Scholar :
|
|
144
|
Jiang WG, Sanders AJ, Katoh M, Ungefroren
H, Gieseler F, Prince M, Thompson SK, Zollo M, Spano D, Dhawan P,
et al: Tissue invasion and metastasis: Molecular, biological and
clinical perspectives. Semin Cancer Biol. 35(Suppl): S244–S275.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Hira VVV, Van Noorden CJF, Carraway HE,
Maciejewski JP and Molenaar RJ: Novel therapeutic strategies to
target leukemic cells that hijack compartmentalized continuous
hematopoietic stem cell niches. Biochim Biophys Acta. 1868:183–198.
2017.PubMed/NCBI
|
|
146
|
Vincan E, Flanagan DJ, Pouliot N, Brabletz
T and Spaderna S: Variable FZD7 expression in colorectal cancers
indicates regulation by the tumour microenvironment. Dev Dyn.
239:311–317. 2010.
|
|
147
|
Simmons GE Jr, Pandey S,
Nedeljkovic-Kurepa A, Saxena M, Wang A and Pruitt K: Frizzled 7
expression is positively regulated by SIRT1 and β-catenin in breast
cancer cells. PLoS One. 9:e988612014. View Article : Google Scholar
|
|
148
|
Qiu X, Jiao J, Li Y and Tian T:
Overexpression of FZD7 promotes glioma cell proliferation by
upregulating TAZ. Oncotarget. 7:85987–85999. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Carmon KS, Gong X, Yi J, Wu L, Thomas A,
Moore CM, Masuho I, Timson DJ, Martemyanov KA and Liu QJ: LGR5
receptor promotes cell-cell adhesion in stem cells and colon cancer
cells via the IQGAP1-Rac1 pathway. J Biol Chem. 292:14989–15001.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Ayyar BV, Arora S and O'Kennedy R:
Coming-of-age of antibodies in cancer therapeutics. Trends
Pharmacol Sci. 37:1009–1028. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Beck A, Goetsch L, Dumontet C and Corvaïa
N: Strategies and challenges for the next generation of
antibody-drug conjugates. Nat Rev Drug Discov. 16:315–337. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
152
|
Kontermann RE and Brinkmann U: Bispecific
antibodies. Drug Discov Today. 20:838–847. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
153
|
Stadler CR, Bähr-Mahmud H, Celik L, Hebich
B, Roth AS, Roth RP, Karikó K, Türeci O and Sahin Y: Elimination of
large tumors in mice by mRNA-encoded bispecific antibodies. Nat
Med. 23:815–817. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Jackson HJ, Rafiq S and Brentjens RJ:
Driving CAR T-cells forward. Nat Rev Clin Oncol. 13:370–383. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Dai H, Wang Y, Lu X and Han W: Chimeric
antigen receptors modified T-cells for cancer therapy. J Natl
Cancer Inst. 108:djv4392016. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Casucci M, Nicolis di Robilant B, Falcone
L, Camisa B, Norelli M, Genovese P, Gentner B, Gullotta F, Ponzoni
M, Bernardi M, et al: CD44v6-targeted T cells mediate potent
antitumor effects against acute myeloid leukemia and multiple
myeloma. Blood. 122:3461–3472. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Junttila MR, Mao W, Wang X, Wang BE, Pham
T, Flygare J, Yu SF, Yee S, Goldenberg D, Fields C, et al:
Targeting LGR5+ cells with an antibody-drug conjugate
for the treatment of colon cancer. Sci Transl Med. 7:314ra1862015.
View Article : Google Scholar
|
|
158
|
Gong X, Azhdarinia A, Ghosh SC, Xiong W,
An Z, Liu Q and Carmon KS: LGR5-targeted antibody-drug conjugate
eradicates gastrointestinal tumors and prevents recurrence. Mol
Cancer Ther. 15:1580–1590. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Riechelmann H, Sauter A, Golze W, Hanft G,
Schroen C, Hoermann K, Erhardt T and Gronau S: Phase I trial with
the CD44v6-targeting immunoconjugate bivatuzumab mertansine in head
and neck squamous cell carcinoma. Oral Oncol. 44:823–829. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Al-Rawi V, Laeufer T, Glocker K, Heneka Y
and Matzke-Ogi A: Abstract 4911: Allosteric inhibition of the
receptor tyrosine kinases c-MET, RON and VEGFR-2 via the
co-receptor CD44v6 by the novel compound AMC303. In: Proceedings of
the American Association for Cancer Research Annual Meeting 2017;
Apr 1-5, 2017; Washington, DC. Philadelphia. AACR;
Cancer Res. 77(Suppl 13): 49112017.
View Article : Google Scholar
|
|
161
|
Inglis DJ, Beaumont DM and Lavranos TC:
Abstract 4695: Targeting the LGR5 complex with BNC101 to improve
check- point inhibitor therapy in colorectal cancer. In:
Proceedings of the American Association for Cancer Research Annual
Meeting 2017; Apr 1-5, 2017; Washington, DC. Philadelphia.
AACR;
Cancer Res. 77(Suppl 13): 46952017.
View Article : Google Scholar
|
|
162
|
Katoh M: The integration of genomics
testing and functional proteomics in the era of personalized
medicine. Expert Rev Proteomics. 14:1055–1058. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Frampton GM, Fichtenholtz A, Otto GA, Wang
K, Downing SR, He J, Schnall-Levin M, White J, Sanford EM, An P, et
al: Development and validation of a clinical cancer genomic
profiling test based on massively parallel DNA sequencing. Nat
Biotechnol. 31:1023–1031. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Hovelson DH, McDaniel AS, Cani AK, Johnson
B, Rhodes K, Williams PD, Bandla S, Bien G, Choppa P, Hyland F, et
al: Development and validation of a scalable next-generation
sequencing system for assessing relevant somatic variants in solid
tumors. Neoplasia. 17:385–399. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Friedman AA, Letai A, Fisher DE and
Flaherty KT: Precision medicine for cancer with next-generation
functional diagnostics. Nat Rev Cancer. 15:747–756. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Pauli C, Hopkins BD, Prandi D, Shaw R,
Fedrizzi T, Sboner A, Sailer V, Augello M, Puca L, Rosati R, et al:
Personalized in vitro and in vivo cancer models to guide precision
medicine. Cancer Discov. 7:462–477. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
167
|
Whorehouses D and Caldas C: Of mice and
men: Patient-derived xenografts in cancer medicine. Ann Oncol.
28:2330–2331. 2017. View Article : Google Scholar
|
|
168
|
Singal AG and El-Serag HB: Hepatocellular
carcinoma from epidemiology to prevention: Translating knowledge
into practice. Clin Gastroenterol Hepatol. 13:2140–2151. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
169
|
Zeng M, Mao XH, Li JX, Tong WD, Wang B,
Zhang YJ, Guo G, Zhao ZJ, Li L, Wu DL, et al: Efficacy, safety, and
immunogenicity of an oral recombinant Helicobacter pylori vaccine
in children in China: A randomised, double-blind,
placebo-controlled, phase 3 trial. Lancet. 386:1457–1464. 2015.
View Article : Google Scholar : PubMed/NCBI
|