Introduction
Cervical cancer is the third most common type of
cancer and the fourth most frequent cause of cancer-associated
mortality in women worldwide (1).
There are ~530,000 novel cervical cancer cases every year (1). Furthermore, the majority of these
cervical cancer cases occur in developing countries, including
China (1,2). Due to its recurrence and metastasis,
the prognosis for cervical cancer patients is unsatisfactory,
although great improvements have been made in surgery combined with
radiotherapy and/or chemotherapy (2). Therefore, novel therapeutic targets
for the treatment of cervical cancer are urgently required.
MicroRNAs (miRs), a class of small non-coding RNAs,
regulate gene expression by directly binding to the 3′-untranslated
region (UTR) of their target messeger (m)RNA, causing translation
repression or messenger (m)RNA degradation (3–5).
In the past decade, a large number of miRs have been identified to
performed important roles in various cellular biological processes,
such as cell proliferation, apoptosis, migration, invasion and
tumorigenesis (6–8). During the development and malignant
progression of cervical cancer, many miRs have been demonstrated to
be significantly deregulated and act as oncogenes or tumor
suppressors, such as miR-27 (9),
miR-124 (10), miR-140 (11) and miR-200 (12). Recently, miR-195 was identified to
exert a tumor suppressive role in various common types of human
cancer (13,14). For example, miR-195 was observed
to inhibit glucose uptake and proliferation of human bladder cancer
cells by targeting solute carrier family 2 member 3 (13). In addition, miR-195 was found to
inhibit the proliferation, migration and invasion of cervical
cancer cells by targeting SMAD family member 3 (Smad3), cyclin D2
(CCND2), MYB proto-oncogene, transcription factor (MYB) and cyclin
D1 (15–17). Thus, miR-195 may be key in the
development and progression of cervical cancer; however, whether
other target genes of miR-195 also contribute to cervical cancer
remains largely unclear.
In the current study, the clinical significance of
miR-195 expression levels in cervical cancer, as well as the
regulatory mechanism of miR-195 underlying cervical cancer
progression were investigated. The data indicate that miR-195
performs a suppressive role in cervical cancer by targeting
DCUN1D1.
Materials and methods
Clinical tissue samples
The present study was approved by the Ethics
Committee of Affiliated Qingdao Hiser Hospital of Qingdao
University (Qingdao, China). A total of 72 cervical cancer tissue
samples and matched adjacent normal tissue samples were obtained
from the Department of Gynecology at the Qingdao Hiser Hospital
between April 2010 and March 2012. Written informed consent was
obtained from each patient involved in the study. All patients had
not received radiation therapy or chemotherapy prior to surgery.
The survival time was counted from the date of surgery to the
follow-up at 5 years, or the date of death. The tissue samples were
stored immediately in liquid nitrogen following surgical resection
and stored at −80°C prior to use.
Cell culture and transfection
Certain common human cervical cancer cell lines
[HeLa (ATCC® CRM-CCL-2™); SiHa (ATCC®
HTb-35™); C33A (ATCC® HTb-31™)] were purchased from the
American Type Culture Collection (Manassas, VA, USA). Cells were
cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented
with 10% fetal bovine serum (FbS) (both from Thermo Fisher
Scientific, Inc., Waltham, MA, USA) and maintained at 37°C in a
humidified atmosphere containing 5% CO2. Cell
transfection was performed using Lipofectamine 2000 (Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions.
Following transfection for 48 h, the expression level of miR-195 or
DCUN1D1 was evaluated.
RNA extraction and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the tissue samples and
cells using TRIzol Reagent (Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. DNase (Takara
Biotechnology Co., Ltd., Dalian, China) treatment was used to
remove genomic DNA, according to the manufacturer's instructions.
For miR expression detection, a SYbR-Green PrimeScript miRNA RT-PCR
kit (cat. no. RR716; Takara biotechnology Co., Ltd.) was used to
perform the RT-qPCR, according to the manufacturer's instruction.
For mRNA expression detection, a SYbR-Green PrimeScript miRNA
RT-PCR kit (cat. no. RR716; Takara biotechnology Co., Ltd.) was
used to perform the RT-qPCR, according to the manufacturer's
instructions. U6 or GAPDH served as the internal reference for miR
or mRNA expression, respectively. The primer sequences were as
follows: Forward, 5′-AGGATCATTGGACAGGAAGAAGT-3′ and reverse,
5′-TGCCAGGTCATCACAGAACTG-3′ for DCUN1D1; and forward,
5′-GGAGCGAGATCCCTCCAAAAT-3′ and reverse,
5′-GGCTGTTGTCATACTTCTCATGG-3′ for GAPDH. The primers for miR-195
(HmiRQP0283) and U6 (HmiRQP9001) were purchased from Guangzhou
FulenGen, Co., Ltd. (Guangzhou, China). The reaction conditions
were as follows: 95°C for 10 min, and 45 cycles of 95°C for 15 sec
and 60°C for 15 sec. The relative expression was analyzed according
to the 2−ΔΔCq method (18). This experiment was repeated three
times.
MTT assay
The HeLa and SiHa cell suspension (5×104
cells/well) was plated in a 96-well plate and cultured at 37°C for
0, 24, 48 or 72 h, respectively. Subsequently, MTT (10 μl; 5
mg/ml) was added to each well and the cells were incubated at 37°C
for 4 h. The supernatant was then removed and 100 μl
dimethyl sulfoxide was added to each well. The absorbance at a
wavelength of 570 nm was determined using a Model 680 Microplate
Reader (bio-Rad Laboratories, Inc., Hercules, CA, USA). The
experiment was repeated three times.
Western blot analysis
Tissue samples and cells were lysed in cold
radioimmunoprecipitation assay buffer (Thermo Fisher Scientific,
Inc.). A bCA Protein Assay kit (cat. no. 23225; Pierce; Thermo
Fisher Scientific, Inc.) was used to determine the concentration of
protein according to the manufacturer's instructions. Protein (50
μg) was then separated using 10% SDS-PAGE (50 V for 1 h
followed by 400 mA for 4 h) and transferred to a polyvinylidene
fluoride (PVDF) membrane (Thermo Fisher Scientific, Inc.). After
blocking in 5% non-fat dried milk in Dulbecco's phosphate-buffered
saline (DPbS; Thermo Fisher Scientific, Inc.) at room temperature
for 3 h, the PVDF membrane was incubated with rabbit anti-human
DCUN1D1 monoclonal antibody (1:50; cat. no. ab181233), or rabbit
anti-human GAPDH monoclonal antibody (1:50; cat. no. ab181602)
(both from Abcam, Cambridge, MA, USA) for 3 h at room temperature.
After washing with DPbS three times, the membrane was incubated
with goat anti-rabbit IgG (1:5,000; cat. no. ab6721; Abcam) for 1 h
at room temperature. After washing with DPbS three times, an
Enhanced Chemiluminescence (ECL) Western blotting kit (cat. no.
32109; Pierce; Thermo Fisher Scientific, Inc.) was used to detect
the immune complex on the PVDF membrane according to the
manufacturer's instructions. Image-Pro Plus software 6.0 (Media
Cybernetics, Inc., Rockville, MD, USA) was used to analyze the
protein expression. GAPDH served as an internal control and the
experiment was repeated three times.
Wound healing assay
HeLa and SiHa cells (105 cells) were
seeded in a 6-well plate and cultured to confluence. The cell
monolayer was scraped using a 200 μl pipette tip to generate
a wound, which was then washed with DMEM two times. Cells were
cultured in DMEM with 3% FbS at 37°C for 48 h. Cells were
subsequently photographed under a light microscope (Olympus
Corporation, Tokyo, Japan) at 0 and 48 h after wounding. The
experiment was repeated three times.
Transwell assay
HeLa and SiHa cells were plated into the top side of
a Transwell chamber pre-coated with Matrigel (BD Biosciences, San
Jose, CA, USA). The lower chamber contained DMEM with 10% FBS as an
attractant. Following incubation at 37°C for 24 h, the upper
surface of the Transwell chamber was scraped using cotton swabs.
The invaded cells on the lower surface of the Transwell chamber
were fixed with ethanol, stained for 10 min at room temperature
using 0.05% Crystal Violet (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) and counted under a light microscope. The experiment was
repeated three times.
Bioinformatics prediction
TargetScan (www.targetscan.org), miRDB (www.mirdb.org) and DIANAmT (www.microrna.gr) were applied to predict the putative
target genes of miR-195, according to the manufacturer's
instruction.
Luciferase activity assay
The wild-type (WT) DCUN1D1 3′-UTR containing the
binding sites of miR-195 was amplified and subcloned into the
pmirGLO luciferase reporter vector (Promega Corporation, Madison,
WI, USA). The mutant-type (MuT) DCUN1D1 3′-UTR lacking
complementarity with miR-195 binding sites was generated using the
QuickChange Site-Directed Mutagenesis kit (cat. no. 200518
Stratagene; Agilent Technologies, Inc., Santa Clara, CA, USA)
according to the manufacturer's instructions, which was also
subcloned into the pmirGLO luciferase reporter vector. HeLa and
SiHa cells (5×104) were seeded in 24-well plates and
co-transfected with WT or MuT 3′-UTR vectors and miR-195 mimics or
miR-NC using Lipofectamine 2000. After 48 h, the cells were assayed
for luciferase activity using the Dual-Luciferase Reporter Assay
System (Promega Corporation), according to the manufacturer's
instructions. The firefly luciferase activities were normalized to
Renilla luciferase activity. This experiment was repeated
three times.
Statistical analysis
Data were presented as means ± standard deviation.
SPSS 17.0 statistical software (SPSS, Inc., Chicago, IL, USA) was
used to conduct the statistical analysis. An independent,
two-tailed Student's t test and one-way ANOVA were performed to
compare the difference. The association between the expression
level of miR-195 or DCUN1D1 and the clinicopathological
characteristics of cervical cancer patients were examined using the
χ2 test or Fisher's exact test. Survival analysis was
evaluated using the Kaplan-Meier method. The correlation between
the miR-195 and DCUN1D1 expression in cervical cancer was assessed
using Pearson's correlation analysis. P<0.05 was considered to
indicate a statistically significant difference.
Results
Downregulation of miR-195 is associated
with aggressive progression in cervical cancer
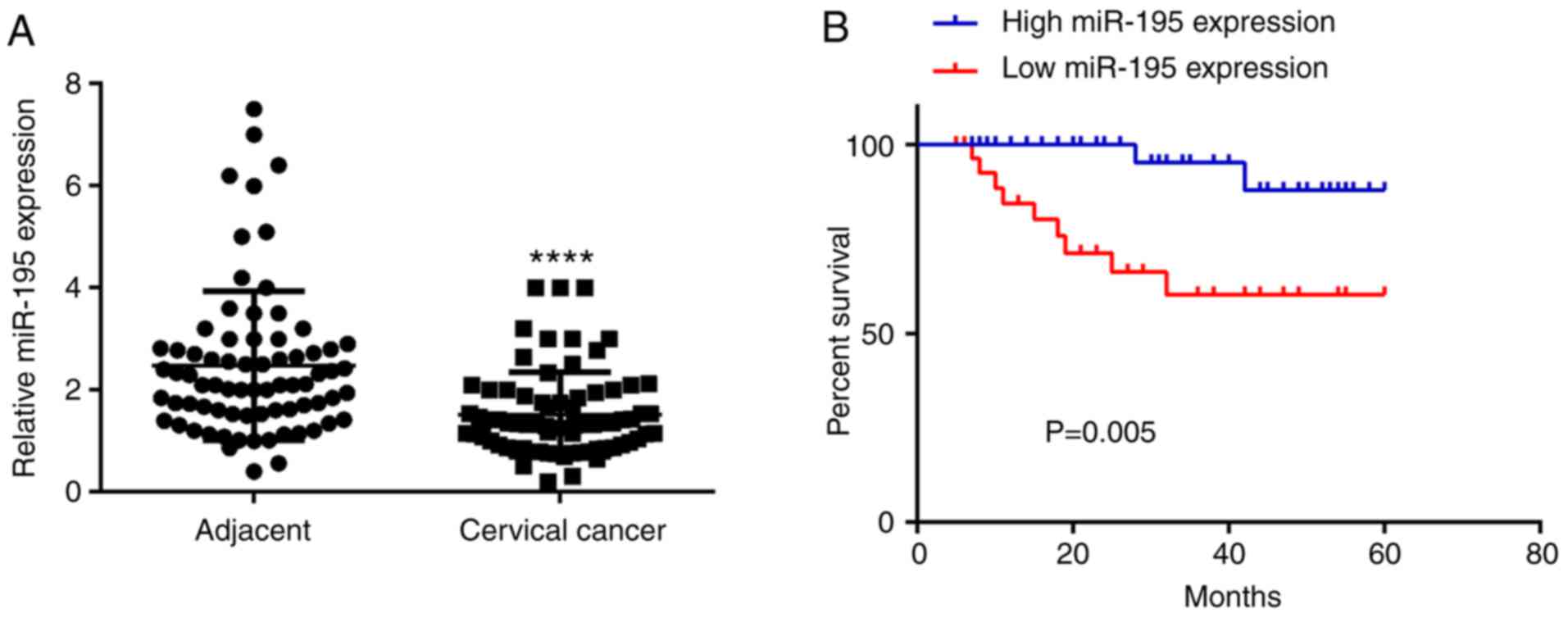
In the present study, the expression levels of
miR-195 were examined using RT-qPCR in cervical cancer and adjacent
non-tumor tissue samples. The data indicated that the expression
levels of miR-195 were significantly decreased in cervical cancer
tissue samples compared with adjacent non-tumor tissue samples
(Fig. 1A). The clinical
significance of miR-195 expression levels in cervical cancer was
then evaluated. According to the mean value of miR-195 expression
levels (1.52), these cervical cancer patients were divided into
high and low miR-195 expression groups. As presented in Table I, the low expression level of
miR-195 was significantly associated with node metastasis and an
advanced TNM clinical stage in cervical cancer. In addition, as all
of the cervical cancer patients involved in the present study were
high-risk human papilloma virus (HPV)-positive, no correlation was
observed between the miR-195 expression level and high-risk HPV
infection (data not shown).
 | Table IAssociation between miR-195
expression level and clinicopathological characteristics of
patients with cervical cancer. |
Table I
Association between miR-195
expression level and clinicopathological characteristics of
patients with cervical cancer.
| Variable | Patients
(n=72) | Low miR-195
(n=46) | High miR-195
(n=26) | P-value |
|---|
| Age (years) | | | | 1.000 |
| <55 | 27 | 19 | 8 | |
| ≥55 | 45 | 37 | 18 | |
| Tumor size
(cm) | | | | 0.623 |
| ≤4 | 44 | 27 | 17 | |
| >4 | 28 | 19 | 9 | |
|
Differentiation | | | | 0.165 |
| Well to
moderate | 53 | 31 | 22 | |
| Poor | 19 | 15 | 4 | |
| Clinical stage | | | | 0.043 |
| I-II | 47 | 26 | 21 | |
| III-IV | 25 | 20 | 5 | |
| Lymph node
metastasis | | | | 0.008 |
| No | 49 | 27 | 22 | |
| Yes | 23 | 20 | 3 | |
| Distant
metastasis | | | | 0.083 |
| No | 62 | 37 | 25 | |
| Yes | 10 | 9 | 1 | |
Downregulation of miR-195 predicts poor
prognosis of patients with cervical cancer
The prognostic value of miR-195 expression levels in
cervical cancer was then evaluated. The correlation between miR-195
expression and overall survival time of cervical cancer patients
was examined using the Kaplan-Meier method. As presented in
Fig. 1B, patients with low
expression levels of miR-195 demonstrated shorter survival times
compared with those with high expression levels of miR-195.
Therefore, downregulation of miR-195 may predict poor prognosis of
patients with cervical cancer.
miR-195 exerts suppressive effects on the
proliferation, migration and invasion of cervical cancer cells
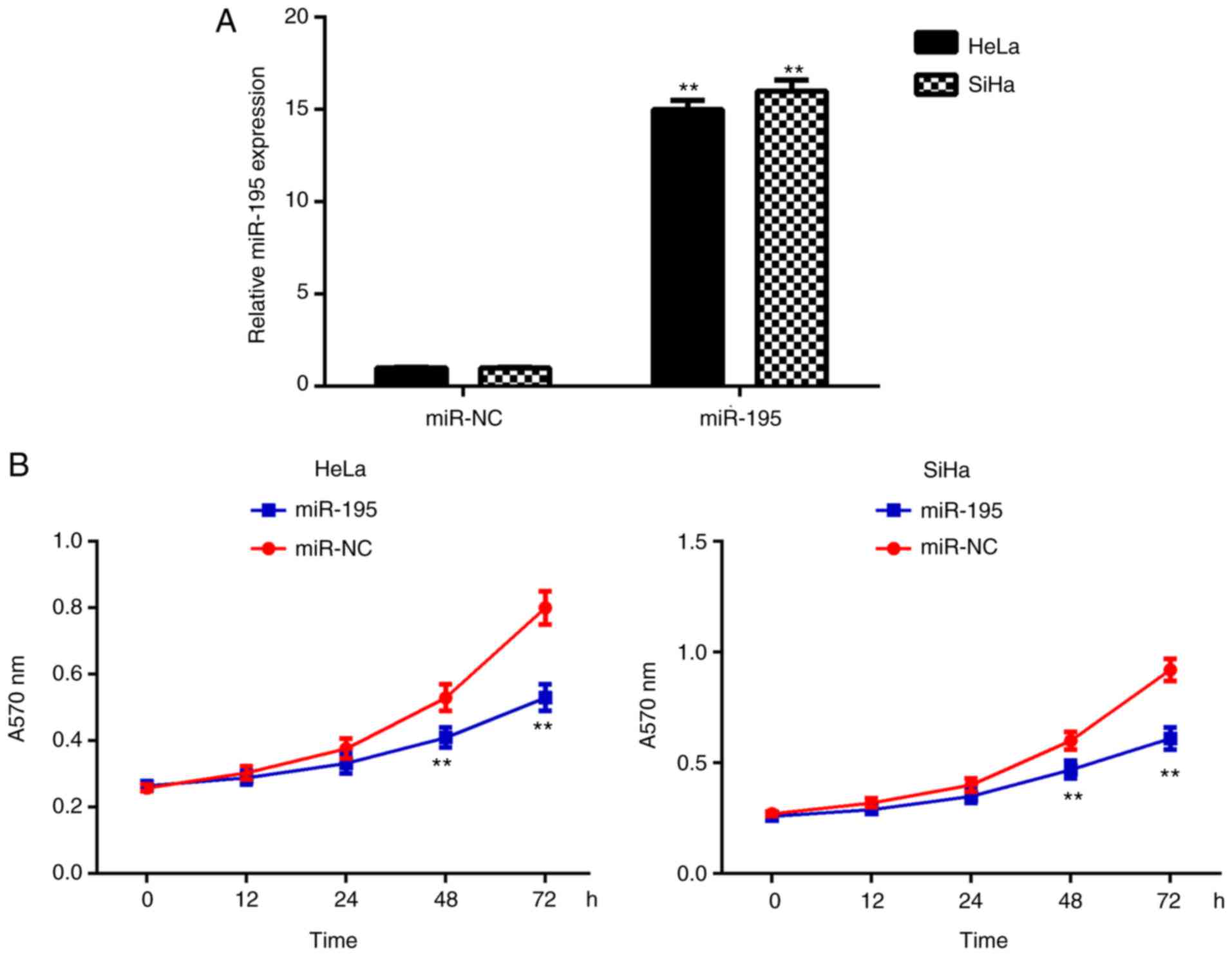
Subsequently, the effects of miR-195 on the
malignant phenotypes of cervical cancer cells were investigated. An
miR-195 mimic was used to transfect HeLa and SiHa cells. Following
transfection, the miR-195 expression levels were significantly
upregulated compared with the miR-negative control (NC) group
(Fig. 2A). The MTT assay data
further indicated that the miR-195-overexpressing cells
demonstrated reduced proliferation when compared with those in the
miR-NC group (Fig. 2B). These
findings indicate that miR-195 may exert suppressive effects on
cervical cancer growth. The role of miR-195 in the regulation of
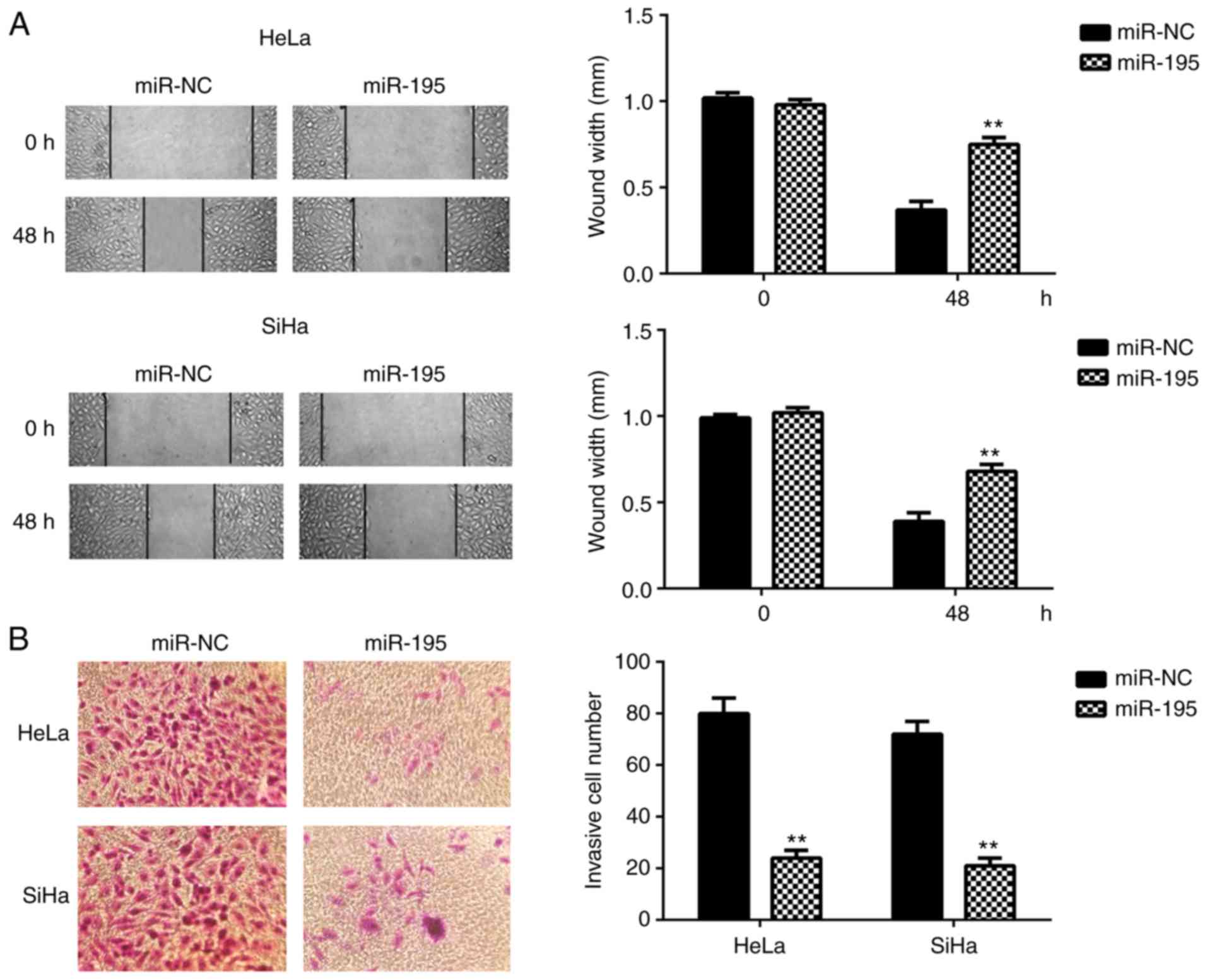
cervical cancer cell migration and invasion was then evaluated.
Data from the wound healing and Transwell assays indicated that
ectopic expression of miR-195 significantly reduced the migration
and invasion of HeLa and SiHa cells (Fig. 3). These findings indicate that
miR-195 may have a suppressive role in cervical cancer
metastasis.
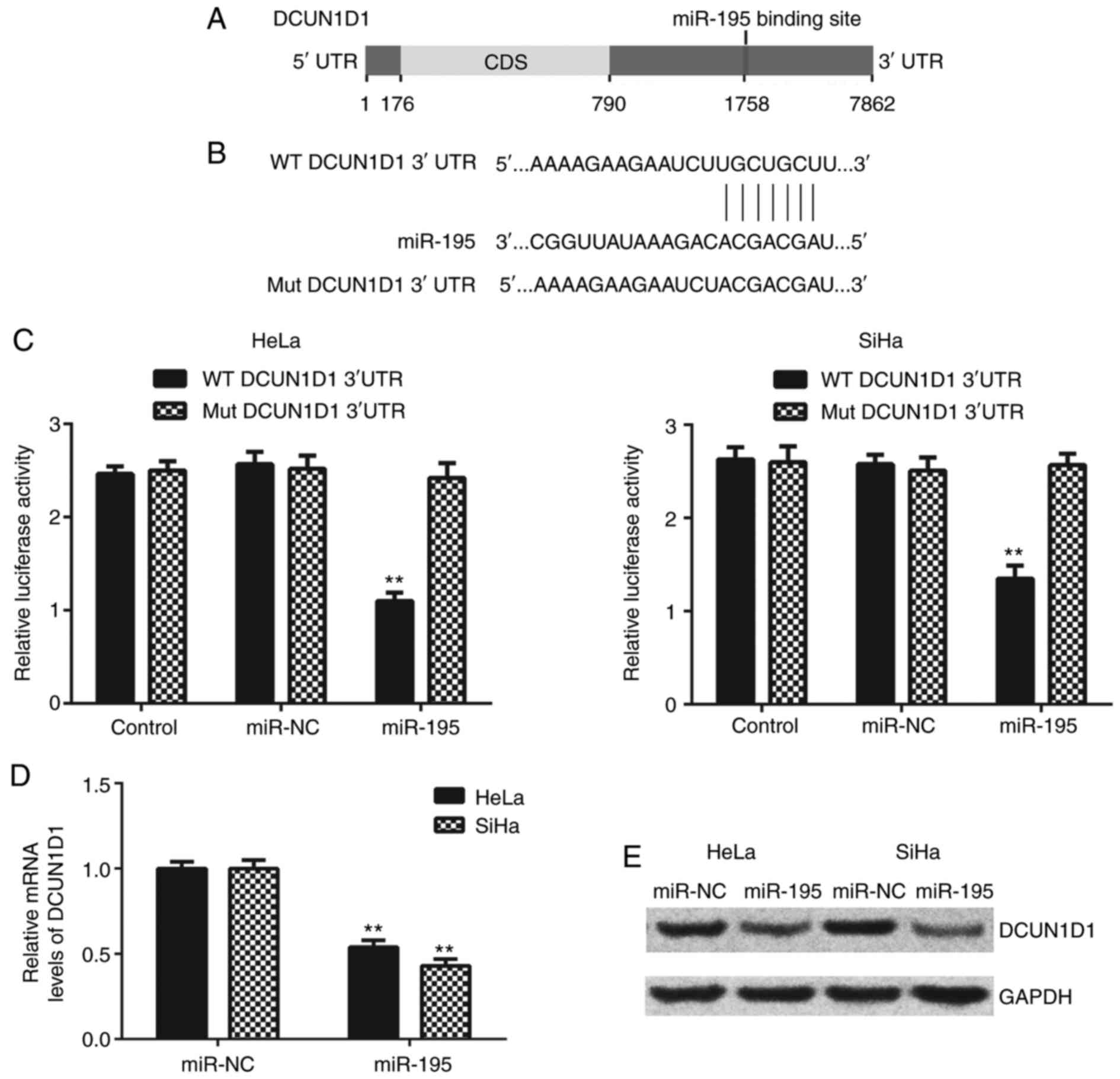
DCUN1D1 is a target gene of miR-195 in
cervical cancer cells
DCUN1D1 was predicted as a putative target gene of
miR-195 using TargetScan, miRDB, and DIANAmT (Fig. 4A). To confirm this prediction, the
WT and MuT DCUN1D1 3′-UTR luciferase reporter plasmids were
generated (Fig. 4B). The
luciferase reporter gene assay was performed in HeLa and SiHa
cells. As presented in Fig. 4C,
luciferase activity was significantly reduced in the presence of
miR-195 in the HeLa and SiHa cells transfected with WT DCUN1D1
3′-UTR luciferase reporter plasmid, but not those transfected with
MuT DCUN1D1 3′-UTR luciferase reporter plasmid.
The effects of miR-195 on the expression levels of
DCUN1D1 in cervical cancer cells were evaluated. The data indicated
a significant reduction in the mRNA and protein expression levels
of DCUN1D1 in miR-195-overexpressing HeLa and SiHa cells (Fig. 4D and E). Thus, DCUN1D1 appears to
be the direct target of miR-195 in cervical cancer cells.
Upregulation of DCUN1D1 is associated
with the aggressive progression and poor prognosis in cervical
cancer
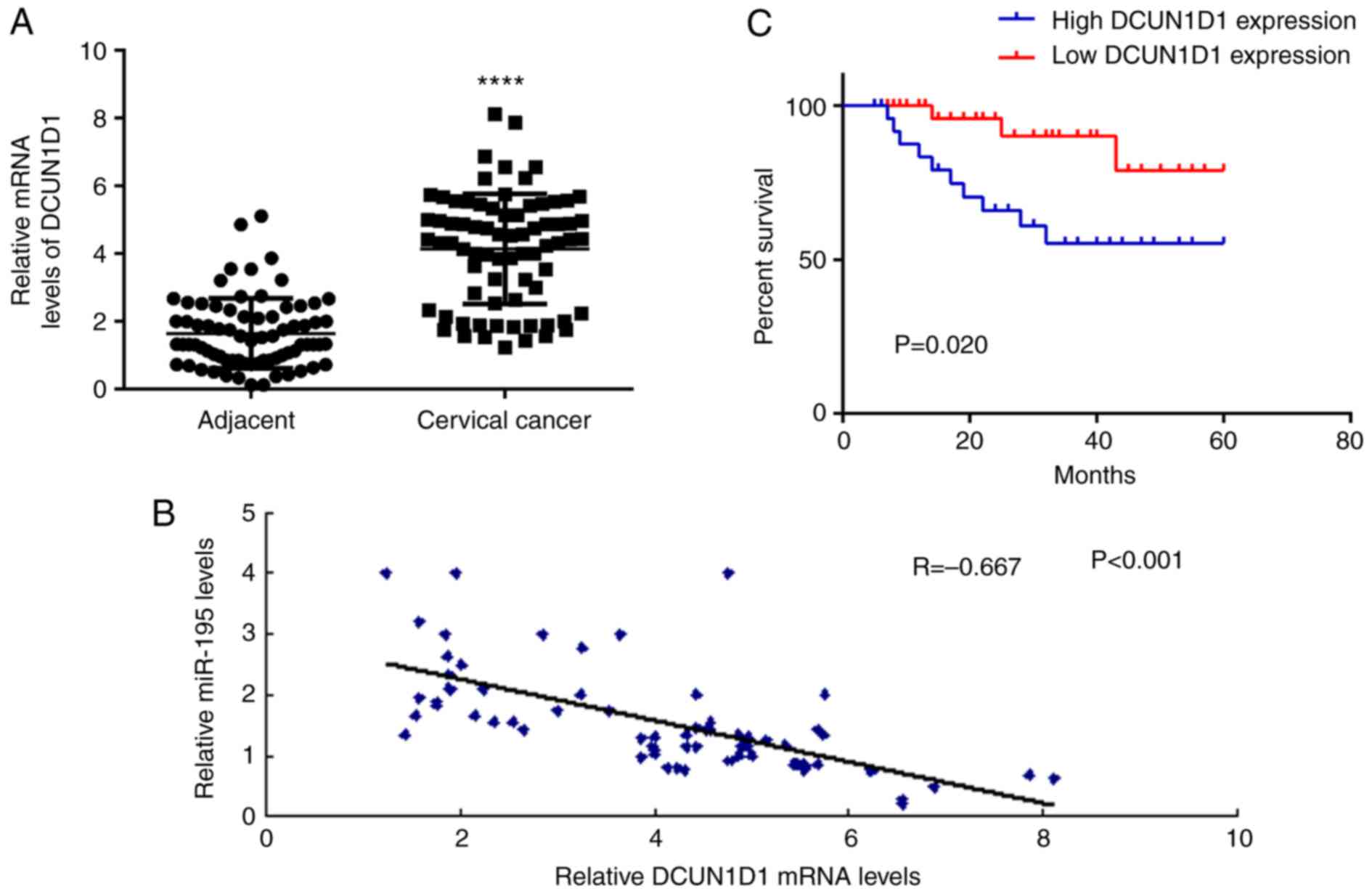
The expression level of DCUN1D1 in cervical cancer
and adjacent non-tumor tissue samples was investigated in the
current study. The data demonstrate that DCUN1D1 was significantly
upregulated in cervical cancer tissue samples when compared with
that in adjacent non-tumor tissue samples (Fig. 5A). The correlation between miR-195
and DCUN1D1 expression levels was then evaluated using Pearson's
correlation analysis. As presented in Fig. 5B, a significant negative
correlation was observed between the expression level of miR-195
and DCUN1D1 in cervical cancer tissue samples. These data indicate
that the upregulation of DCUN1D1 may be due to the downregulation
of miR-195 in cervical cancer. The clinical significance of DCUN1D1
expression levels was evaluated in cervical cancer. According to
the mean value of DCUN1D1 mRNA expression levels (4.14), the
cervical cancer patients were divided into a high DCUN1D1
expression level group and a low DCUN1D1 expression level group. As
presented in Table II, the
upregulation of DCUN1D1 was associated with lymph node metastasis,
distant metastasis, and an advanced clinical stage in cervical
cancer. Furthermore, the patients with high expression levels of
DCUN1D1 exhibited shorter survival times when compared with those
with low expression levels of DCUN1D1 (Fig. 5C). Therefore, an increased
expression level of DCUN1D1 may predict poor prognosis of patients
with cervical cancer.
 | Table IIAssociation between DCUN1D1
expression levels and clinicopathological characteristics of
patients with cervical cancer. |
Table II
Association between DCUN1D1
expression levels and clinicopathological characteristics of
patients with cervical cancer.
| Variable | Patients
(n=72) | Low DCUN1D1
(n=32) | High DCUN1D1
(n=40) | P-value |
|---|
| Age (years) | | | | 0.807 |
| <55 | 27 | 11 | 16 | |
| ≥55 | 45 | 21 | 24 | |
| Tumor size
(cm) | | | | 0.331 |
| ≤4 | 44 | 22 | 22 | |
| >4 | 28 | 10 | 18 | |
|
Differentiation | | | | 0.105 |
| Well to
moderate | 53 | 27 | 26 | |
| Poor | 19 | 5 | 14 | |
| Clinical stage | | | | 0.014 |
| I–II | 47 | 26 | 21 | |
| III–IV | 25 | 6 | 19 | |
| Lymph node
metastasis | | | | 0.011 |
| No | 49 | 27 | 22 | |
| Yes | 23 | 5 | 18 | |
| Distant
metastasis | | | | 0.035 |
| No | 62 | 31 | 31 | |
| Yes | 10 | 1 | 9 | |
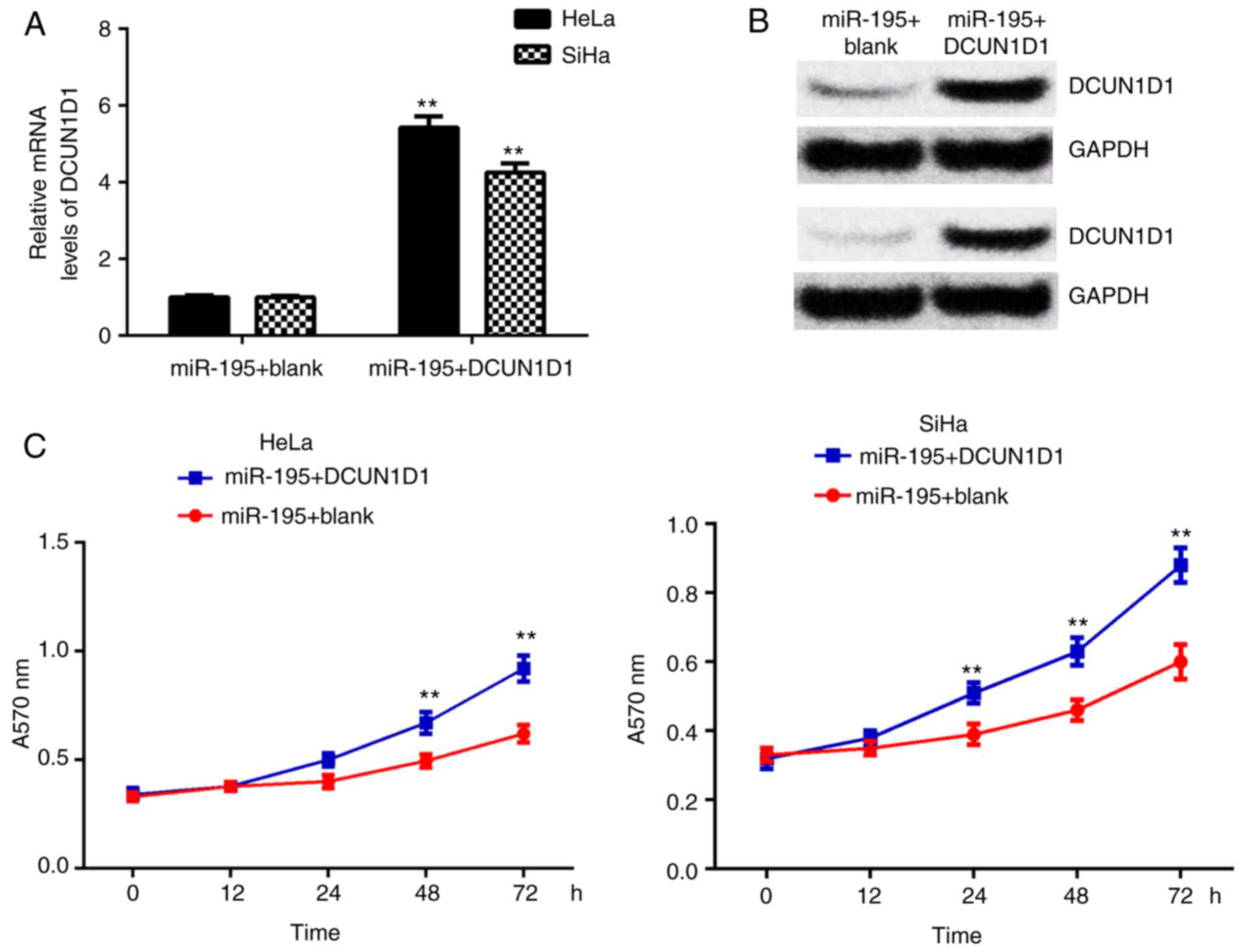
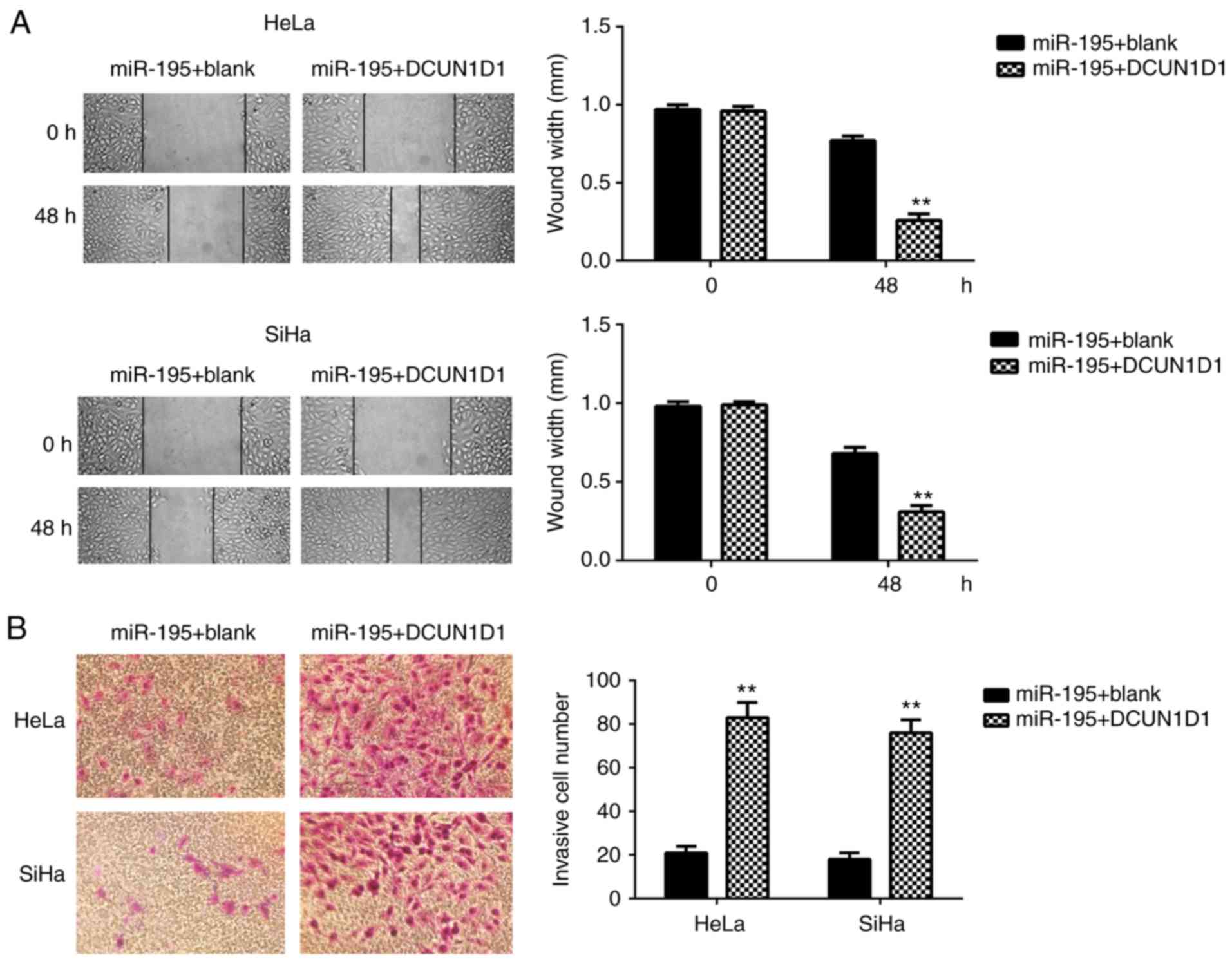
DCUN1D1 restored miR-195-mediated cell
proliferation, migration and invasion in cervical cancer
To confirm miR-195 exerts suppressive effects on
cervical cancer cells by targeting DCUN1D1, the
miR-195-overexpressing cells were transfected with pcDNA3.1-DCUN1D1
expression plasmid or with blank pcDNA3.1 vector as the control
group. Following transfection, the mRNA and protein expression
levels of DCUN1D1 were significantly increased in the miR-195 +
DCUN1D1 group compared with the miR-195 + blank group (Fig. 6A and B). Furthermore, MTT, wound
healing and Transwell assays demonstrated that overexpression of
DCUN1D1 restored the suppressive effects of miR-195 on the
proliferation, migration and invasion of cervical cancer cells
(Figs. 6C and 7). These data confirm that miR-195
exerts inhibitory effects on cervical cancer cells by directly
targeting DCUN1D1.
HPV16 E6 regulates the expression levels
of miR-195 and DCUN1D1 in cervical cancer cells
In the present study, three common cervical cancer
cell lines were used to perform in vitro experiments. C33A
cells are HPV−, SiHa cells are HPV16+, and
HeLa cells are HPV18+. Notably, the qPCR data
demonstrated that the expression levels of miR-195 were
significantly lower in the HeLa (HPV18+) and SiHa
(HPV16+) cells, when compared with those in C33A
(HPV−) cells (Fig.
8A). As HPVs contain two oncogenes (E6 and E7) (18), HPV16 E6, HPV16 E7, HPV18 E6 and
HPV18 E7 were knocked down in SiHa or HeLa cells using small
interfering (si)RNA. As presented in Fig. 8B, the expression level of miR-195
was significantly increased in SiHa and HeLa cells transfected with
E6 siRNA, indicating that E6 is the most important HPV oncoprotein
responsible for the inhibition of miR-195 in cervical cancer cells.
Consistently, knockdown of E6 significantly reduced the protein
expression of DCUN1D1 in SiHa and HeLa cells (Fig. 8C). Therefore, the oncogenic
effects of HPV E6 on cervical cancer may be partly via regulating
the expression level of miR-195 and, thus, its target gene,
DCUN1D1.
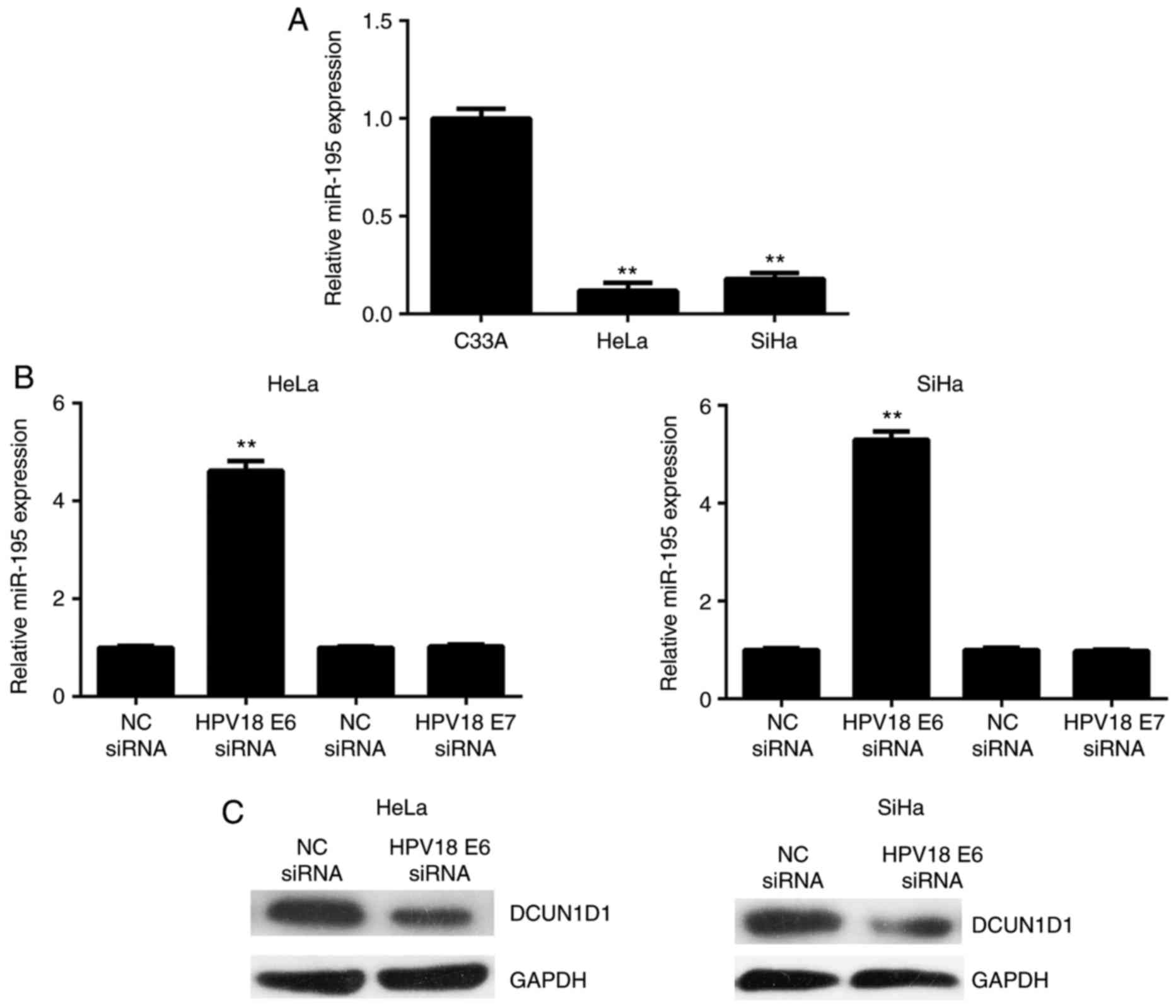 | Figure 8HPV16 E6 regulates the expression
levels of miR-195 and DCUN1D1 in cervical cancer cells. (A) Reverse
transcription-quantitative polymerase chain reaction was used to
examine the miR-195 expression levels in (A) cervical cancer cell
lines, including HeLa (HPV18+), SiHa (HPV16+)
and C33A (HPV−), and (b) HeLa and SiHa cells transfected
with HPV16 E6 siRNA, HPV16 E7 siRNA, HPV18 E6 siRNA and HPV18 E7
siRNA, respectively. **P<0.01 vs. C33A. (C) Western
blot analysis was performed to examine the protein expression level
of DCUN1D1 in SiHa and HeLa cells transfected with HPV16 E6 siRNA
or HPV18 E6 siRNA, respectively. The experiments were repeated
three times. HPV, human papilloma virus; miR, microRNA; DCUN1D1,
defective in cullin neddylation 1 domain containing 1; siRNA, small
interfering RNA; NC, negative control. |
Discussion
The exact regulatory mechanism of miR-195 underlying
cervical cancer progression remains largely unclear. In the present
study, miR-195 was significantly downregulated in cervical cancer,
and a low expression level of miR-195 was associated with malignant
progression and poor prognosis in cervical cancer. Ectopic
expression of miR-195 inhibited the proliferation, migration and
invasion of cervical cancer cells. DCUN1D1, which is significantly
upregulated in cervical cancer, was identified as a novel target
gene of miR-195, and upregulation of DCUN1D1 was associated with
the malignant progression and poor prognosis in cervical cancer.
DCUN1D1 demonstrated a negative correlation with miR-195 expression
levels in cervical cancer, and overexpression of DCUN1D1 restored
the suppressive effects of miR-195 on the malignant phenotypes of
cervical cancer cells. Notably, miR-195 was downregulated in HeLa
(HPV18+) and SiHa (HPV16+) cells compared
with that in C33A (HPV−) cells, and knockdown of HPV E6
increased the miR-195 expression level while it reduced the DCUN1D1
expression in HeLa and SiHa cells.
In recent years, accumulating evidence has
demonstrated that miRs are key in tumor initialization, growth, and
metastasis by regulating the expression of their specific target
genes (11,20,21). Among these cancer-associated miRs,
miR-195 typically acts as a tumor suppressor in certain common
cancer types, such as gastric (14), colorectal (22) and renal (23) cancer, osteosarcoma (24), and liver cancer (25). Furthermore, recent studies have
revealed a role of miR-195 in cervical cancer (15,17,26). Wang et al (26) reported that miR-195 was
downregulated in primary cervical cancer tissue samples, and may
inhibit the proliferation of cervical cancer cells via targeting
cyclin D1a and thus inducing G1 phase arrest. Similarly,
Li et al (17) identified
that miR-195 suppressed cervical cancer cell proliferation and
promoted cell apoptosis by targeting cyclin D1 (17). In the present study, miR-195 was
significantly downregulated in cervical cancer tissue samples when
compared with adjacent non-tumor tissue samples, and its
downregulation was associated with node metastasis and an advanced
clinical stage. In addition, Zhou et al (15) observed that the low miR-195
expression level was significantly correlated with a higher
clinical stage, node metastasis and deep stromal invasion (15). Furthermore, miR-195 inhibited
cervical cancer proliferation and repressed cancer cell migration
and invasion in the present study, which was consistent with the
findings of previous studies (15,16).
As miRs function by regulating the expression levels
of their target genes (7), the
target genes of miR-195 were then predicted in cervical cancer
cells using bioinformatics analysis. DCUN1D1 was selected as the
potential target of miR-195, as a recent study demonstrated that
miR-218 inhibits the migration, invasion and epithelial-mesenchymal
transition in cervical cancer cells by targeting DCUN1D1 (27). However, whether other miRs may
also directly target DCUN1D1 in cervical cancer remains unknown.
DCUN1D1, also termed squamous cell carcinoma-related oncogene, may
promote nuclear translocation and assembly of the neddylation E3
complex (28,29). Recently, DCUN1D1 was identified to
be involved in tumor progression and development of brain
metastasis in patients with non-small cell lung cancer (30). Furthermore, DCUN1D1 has been
reported to be activated by amplification in squamous cell
carcinomas (29). In the current
study, DCUN1D1 was significantly upregulated in cervical cancer
tissue samples when compared with adjacent normal tissue samples,
and its upregulation was significantly associated with node
metastasis, distant metastasis, an advanced clinical stage and
shorter survival time of patients with cervical cancer. As it was
observed that the expression level of DCUN1D1 was negatively
correlated to miR-195 expression levels in cervical cancer tissue
samples, it was hypothesized that its upregulation may be due to
the downregulation of miR-195. Further investigation indicated that
DCUN1D1 overexpression restored the suppressive effects of miR-195
on the proliferation, migration and invasion of cervical cancer
cells, deonstrating that DCUN1D1 functions as a downstream effecter
of miR-195 in cervical cancer cells. In addition to DCUN1D1,
various other target genes of miR-195 have been identified in other
studies, including Smad3, CCND2, MYB and cyclin D1 (15–17). These target genes have also been
proposed to be involved in the miR-195-mediated malignant
phenotypes of cervical cancer cells (15–17). For example, Du et al
(16) found that miR-195 exerted
suppressive effects on cervical cancer cell proliferation,
migration and invasion in vitro, and identified CCND2 and
MYb as two direct targets of miR-195 in cervical cancer HeLa cells
(16).
Notably, the HPV E6 affected the expression levels
of miR-195 and DCUN1D1 in cervical cancer cells in the present
study. These findings provide a novel potential molecular mechanism
in HPV-associated cervical cancer. In addition, the 72 cervical
cancer patients that were involved in the present study were
high-risk HPV+, and thus no correlation was observed
between the miR-195 expression level and HPV infection in these
cervical cancer patients.
In conclusion, to the best of our knowledge, this is
the first study demonstrating that miR-195 inhibits the
proliferation, migration and invasion of cervical cancer cells by
directly targeting DCUN1D1. based on these findings, it is proposed
that the miR-195/DCUN1D1 axis may serve as a potential candidate
target for the treatment of cervical cancer. A limitation of the
present study is that the number of cervical cancer patients is
small. Further studies are required to establish the exact role of
miR-195 in cervical cancer growth and metastasis in vivo, as
well as the downstream signaling pathways.
Acknowledgments
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets generated/analysed during the current
study are available.
Authors' contributions
JZ did most of the experiments and was a major
contributor in writing the paper. HY collected clincial tissues and
performed the PCR assay. XX performed the statistical analysis.SK
designed the study and was also a major contributor in writing the
paper. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Affiliated Qingdao Hiser Hospital of Qingdao
University (Qingdao, China). Written informed consent was obtained
from each patient involved in the study.
Consent for publication
Consent for publication was obtained from the
participants.
Competing interests
The authors declare they have no competing
interests.
References
|
1
|
Siegel R and Naishadham D; Jemal A: Cancer
statistics: Cancer statistics, 2012. CA Cancer J Clin. 62:10–29.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Siegel RL and Miller KD; Jemal A: Cancer
statistics: 2015.CA Cancer J Clin. 65:5–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Li H, Xiang Z, Liu Y, Xu B and Tang J:
MicroRNA-133b inhibits proliferation, cellular migration, and
invasion via targeting LASP1 in hepatocarcinoma cells. Oncol Res.
25:1269–1282. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhu K, He Y, Xia C, Yan J, Hou J, Kong D,
Yang Y and Zheng G: MicroRNA-15a inhibits proliferation and induces
apoptosis in CNE1 nasopharyngeal carcinoma cells. Oncol Res.
24:145–151. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Jiang Z, Zhang Y, Cao R, Li L, Zhong K,
Chen Q and Xiao J: miR-5195-3p inhibits proliferation and invasion
of human bladder cancer cells by directly targeting oncogene KLF5.
Oncol Res. 25:1081–1087. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang G, Fu Y, Liu G, Ye Y and Zhang X:
miR-218 inhibits proliferation, migration, and EMT of gastric
cancer cells by targeting WASF3. Oncol Res. 25:355–364. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yao J, Deng B, Zheng L, Dou L, Guo Y and
Guo K: miR-27b is upregulated in cervical carcinogenesis and
promotes cell growth and invasion by regulating CDH11 and
epithelial-mesenchymal transition. Oncol Rep. 35:1645–1651. 2016.
View Article : Google Scholar
|
|
10
|
Zhang X, Cai D, Meng L and Wang B:
MicroRNA-124 inhibits proliferation, invasion, migration and
epithelial-mesenchymal transition of cervical carcinoma cells by
targeting astrocyte-elevated gene-1. Oncol Rep. 36:2321–2328. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Su Y, Xiong J, Hu J, Wei X, Zhang X and
Rao L: MicroRNA-140-5p targets insulin like growth factor 2 mRNA
binding protein 1 (IGF2bP1) to suppress cervical cancer growth and
metastasis. Oncotarget. 7:68397–68411. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zeng F, Xue M, Xiao T, Li Y, Xiao S, Jiang
B and Ren C: MiR-200b promotes the cell proliferation and
metastasis of cervical cancer by inhibiting FOXG1. biomed
Pharmacother. 79:294–301. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Fei X, Qi M, Wu B, Song Y, Wang Y and Li
T: MicroRNA-195-5p suppresses glucose uptake and proliferation of
human bladder cancer T24 cells by regulating GLUT3 expression. FEbS
Lett. 586:392–397. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang J, Li L, Jiang M and Li Y:
MicroRNA-195 inhibits human gastric cancer by directly targeting
basic fibroblast growth factor. Clin Transl Oncol. 19:1320–1328.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhou Q, Han LR, Zhou YX and Li Y: MiR-195
suppresses cervical cancer migration and invasion through targeting
Smad3. Int J Gynecol Cancer. 26:817–824. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Du X, Lin LI, Zhang L and Jiang J:
MicroRNA-195 inhibits the proliferation, migration and invasion of
cervical cancer cells via the inhibition of CCND2 and MYB
expression. Oncol Lett. 10:2639–2643. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Li Z, Wang H, Wang Z and Cai H: MiR-195
inhibits the proliferation of human cervical cancer cells by
directly targeting cyclin D1. Tumour biol. 37:6457–6463. 2016.
View Article : Google Scholar
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−delta delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
19
|
Gómez-Gómez Y, Organista-Nava J and
Gariglio P: Deregulation of the miRNAs expression in cervical
cancer: Human papilloma-virus implications. Biomed Res Int.
2013:4070522013. View Article : Google Scholar
|
|
20
|
Zhou W, Zou B, Liu L, Cui K, Gao J, Yuan S
and Cong N: MicroRNA-98 acts as a tumor suppressor in
hepatocellular carcinoma via targeting SALL4. Oncotarget.
7:74059–74073. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xu X, Zhang Y, Jasper J, Lykken E,
Alexander Pb, Markowitz GJ, McDonnell DP, Li QJ and Wang XF:
MiR-148a functions to suppress metastasis and serves as a
prognostic indicator in triple-negative breast cancer. Oncotarget.
7:20381–20394. 2016.PubMed/NCBI
|
|
22
|
Sun M, Song H, Wang S, Zhang C, Zheng L,
Chen F, Shi D, Chen Y, Yang C, Xiang Z, et al: Integrated analysis
identifies microRNA-195 as a suppressor of Hippo-YAP pathway in
colorectal cancer. J Hematol Oncol. 10:792017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wang K, Sun Y, Tao W, Fei X and Chang C:
Androgen receptor (AR) promotes clear cell renal cell carcinoma
(ccRCC) migration and invasion via altering the
circHIAT1/miR-195-5p/29a-3p/29c-3p/CDC42 signals. Cancer Lett.
394:1–12. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Qu Q, Chu X and Wang P: MicroRNA-195-5p
suppresses osteosarcoma cell proliferation and invasion by
suppressing naked cuticle homolog 1. Cell Biol Int. 41:287–295.
2017. View Article : Google Scholar
|
|
25
|
Zhang H, Zhou D, Ying M, Chen M, Chen P,
Chen Z and Zhang F: Expression of long non-coding RNA (lncRNA)
small nucleolar RNA host gene 1 (SNHG1) exacerbates hepatocellular
carcinoma through suppressing miR-195. Med Sci Monit. 22:4820–4829.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang N, Wei H, Yin D, Lu Y, Zhang Y, Zhang
Q, Ma X and Zhang S: MicroRNA-195 inhibits proliferation of
cervical cancer cells by targeting cyclin D1a. Tumour biol.
37:4711–4720. 2016. View Article : Google Scholar
|
|
27
|
Jiang Z, Song Q, Zeng R, Li J, Li J, Lin
X, Chen X, Zhang J and Zheng Y: MicroRNA-218 inhibits EMT,
migration and invasion by targeting SFMBT1 and DCUN1D1 in cervical
cancer. Oncotarget. 7:45622–45636. 2016.PubMed/NCBI
|
|
28
|
Huang G, Kaufman AJ, Ramanathan Y and
Singh B: SCCRO (DCUN1D1) promotes nuclear translocation and
assembly of the neddylation E3 complex. J biol Chem.
286:10297–10304. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Sarkaria I, O-charoenrat P, Talbot SG,
Reddy PG, Ngai I, Maghami E, Patel KN, Lee B, Yonekawa Y, Dudas M,
et al: Squamous cell carcinoma related oncogene/DCUN1D1 is highly
conserved and activated by amplification in squamous cell
carcinomas. Cancer Res. 66:9437–9444. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yoo J, Lee SH, Lym KI, Park SY, Yang SH,
Yoo CY, Jung JH, Kang SJ and Kang CS: Immunohistochemical
expression of DCUN1D1 in non-small cell lung carcinoma: Its
relation to brain metastasis. Cancer Res Treat. 44:57–62. 2012.
View Article : Google Scholar : PubMed/NCBI
|