Introduction
Seaweed, including laver, is a standard food
consumed in Asian countries, including Korea and Japan. Several
studies have reported that seaweed has various bioactive substances
with anticancer, antiviral and anticoagulant effects (1-5).
The majority of studies on seaweed bioactivity have investigated
solvent extracts, rather than mono-constituent substances, and
there has been limited investigation of seaweed proteins and
peptides (6). Previous studies
have progressed our understanding of the bioactive components in
seaweed, including the partial creation of a seaweed protein
database, identification of a seaweed protein that changes
according to stress and environment, proteomic investigations on
its usefulness as a biomarker, and the development of a DNA marker
based on the genomic sequence of the laver Pyropia species
(7). However, there remains a
paucity of basic information describing the activity and function
of the bioactive components of seaweed.
The most prominent characteristic of seaweed is
photosynthesis, which occurs in semiautonomous organelles,
chloroplasts. Red algae contain a pigment termed phycobilin, which
forms a covalent bond with a water-soluble protein. Unlike
chlorophyll, a fat-soluble pigment that requires extraction with
organic solvents, phycobilin is a water-soluble pigment that can be
readily purified (8).
Phycobiliproteins (PBPs) are largely divided into three types. The
first type is phycoerythrin (PE), a complex between
phycoerythrobilin and a protein, which has a scarlet color. The
second type is phycocyanin (PC), a complex between phycocyanobilin
and a protein, which forms a blue color. The third type is the
indigo-blue allophycocyanin (APC), a complex between
phycocyanobilin and a protein with a distinct absorption spectrum.
These three complexes are all acidic, with isoelectric points of
~4.3. Each type of PBP is further divided into subtypes. PE has
three subtypes: B, R and C; PC has two subtypes: C and R; and APC
has only one subtype. In Pyropia yezoensis, the PBP type
depends on its color variation. PBPs have been reported to protect
hepatocytes and to have anti-inflammatory and antioxidant effects,
indicating their potential application as therapeutic agents
(9,10). Previous studies have extracted,
refined and functionally analyzed particular PBPs from
cyanobacteria and seaweed (8,10,11,12). According to a previous report, it
was possible to separate three types of PBPs simultaneously from
Lyngbya sp. (13).
However, there have been no reports on the separation of PBPs from
seaweed and their functional analysis. Therefore, the present study
aimed to identify proteins separated from laver, seaweed rich in
PBPs, using two-dimensional polyacrylamide gel electrophoresis
(2D-PAGE) and to analyze their antioxidant activities.
Reactive oxygen species (ROS) that are generated
in vivo are a major cause of human aging and disease. The
generation of ROS leads to disease through toxic effects on cells
and tissues. ROS include free radical species, including superoxide
anions, hydroxyl radicals and singlet oxygen, in addition to
non-radical species, including hydrogen peroxide
(H2O2). ROS levels are tightly controlled in
the body by antioxidant enzymes, including superoxide dismutase
(SOD), catalase (CAT) and glutathione peroxidase (GPx).
The liver is an important tissue responsible for
metabolism and the detoxification of nutrients in the human body.
Liver injury can be caused by alcohol, viral infection, drug abuse
and oxidative stress (14-16).
In the present study, HepG2 cells were used to examine the
antioxidant function of hepatocytes. HepG2 cells were first
reported as hepatocellular carcinoma cells, however, certain genes
were confirmed to be more similar to hepatoblastoma in 2009
(17,18). These cells have been used to
investigate liver metabolism, development, oncogenesis
(chemocarcinogenesis and mutagenesis), and hepatotoxicity in
thousands of cases in previous studies (17,19-26). HepG2 cells are cancer cells,
however, they do not differ from normal liver cells in their
detoxification and antioxidant mechanism (18). In the present study, the HepG2
cell line was used for the antioxidant function of hepatocytes.
PBPs have a variety of bonds between pigments and
proteins, and even the same PBP type can include different
phycobilins, with the composition influenced by the environment. As
reported previously, PBPs are suggested to have potential
antioxidant properties (27).
Therefore, the present study aimed to identify PBPs in laver,
elucidate their features and evaluate the potential of peptides
derived from these PBPs as antioxidant agents.
Materials and methods
Red algal materials and crude
extraction
P. yezoensis was obtained from Myeongji
(Busan, Korea) and stored at −70°C. The frozen samples were
lyophilized and homogenized prior to protein extraction. The crude
soluble fraction was extracted with 0.1 M sodium acetate buffer (pH
6.0). Samples of P. yezoensis (1 g) were mixed with 50 ml
sodium acetate buffer for 16 h with continuous shaking at 4°C, and
then filtered through Whatman No. 1 filter paper. The extract was
then precipitated with methanol and chloroform (28). Protein material was precipitated
to increase the purity and the resulting supernatants were treated
with a 2-D Clean-up kit (GE Healthcare Life Sciences, Logan, UT,
USA). Protein concentrations were determined using a bicinchoninic
acid assay with bovine serum albumin (Thermo Fisher Scientific,
Inc., Waltham, MA, USA) as the standard.
Identification of purified proteins
For 2D-PAGE analysis, isoelectric focusing was
performed as the first dimension and SDS-PAGE as the second
dimension, according to the manufacturer's protocols (GE Healthcare
Life Sciences). Protein samples (150 µg) containing 0.5%
immobilized pH gradient (IPG) buffer (pH 4-7) and 5% bromophenol
blue were applied to an immobilized IPG DryStrip (pH 4-7, 13 cm, GE
Healthcare Life Sciences) and rehydrated for 16 h in lysis solution
containing 4 M urea, 4% 3-(3-cholami-dopropyl dimethylammonio)
propanesulfonate, 2 M thiourea, 100 mM dithiothreitol and 1%
polyvinylpyrrolidone. Prior to performing the second dimension
SDS-PAGE, focused IPG strips were equilibrated for 20 min in a
solution of 50 mM Tris-HCl (pH 8.8), 6 M urea, 30% glycerol, 2% SDS
and 1% DTT, and then for an additional 20 min in the same solution,
but with the DTT replaced with 0.5% iodoacetamide. The equilibrated
strips were positioned on a 15% polyacrylamide gel (14×13 cm, 1.5
mm) and, following SDS-PAGE the gels were silver-stained. Analysis
of the partial amino acid sequences of the soluble proteins was
performed using electrospray ionization quadrupole time-of-flight
mass spectrometry/mass spectrometry (ESI Q TOF MS/MS), as described
previously (29).
Peptide synthesis
The P. yezoensis PBP-derived peptides (PBP
1-13) were synthesized by Peptron (Daejeon, Korea). Purification of
the 13 peptides was performed on a C18 column (Shiseido CAPCELL
PAK; Shiseido, Tokyo, Japan) attached to a high-performance liquid
chromatography apparatus with elution in 0.1% trifluoroacetic acid
and a gradient of 3-70% acetonitrile, a flow rate of 1 ml/min and
UV detection at 220 nm. The peptide molecular weights, as shown in
Table I, were determined by mass
analysis (HP 1100 series LC/MSD; Agilent Technologies, Inc., Santa
Clara, CA, USA).
 | Table IIdentification of two-dimensional
polyacrylamide gel electrophoresis spots and synthesis of peptides
based on phycobiliproteins from Pyropia yezoensis. |
Table I
Identification of two-dimensional
polyacrylamide gel electrophoresis spots and synthesis of peptides
based on phycobiliproteins from Pyropia yezoensis.
| Spot | Identification | Database/accession
no. | Taxonomy | Sequence | PI | Nominal mass
(Mr) | Score | Protein sequence
coverage (%) | Synthesized
peptide |
|---|
| 42 | R-phycoerythrin β
chain |
SwissProt/PHEB_PYRHA | Pyropia
haitanensis |
KAAAVAFITNTASQRK | 6.23 | 18,817 | 47 | 7 | PBP1 |
| 43 | Phycoerythrin β
subunit |
NCBI/gi|73761963 | Aglaothamnion
callophyllidicola |
KAAAVAFITNTASQRK | 5.41 | 17,570 | 71 | 8 | PBP1 |
| R-phycoerythrin β
chain |
SwissProt/PHEB_PORPU | Porphyra
purpurea |
RYVSYALLAGDPSVLEDRC | 6.23 | 18,833 | 63 | 9 | PBP2 |
| 47 | Allophycocyanin β
subunit |
NCBI/gi|11465727 | Porphyra
purpurea |
MQDAITSVINAADVQGKY | 5.17 | 17,589 | 85 | 10 | PBP3 |
| 48 | Allophycocyanin β
subunit |
NCBI/gi|11465727 | Porphyra
purpurea |
MQDAITSVINAADVQGKY
RAAATIAANAATIIKE
RYATYGMLAGDPSILEERV | 5.17 | 17,589 | 210 | 29 | PBP3
PBP4
PBP5 |
| 49 | Allophycocyanin α
chain | NCBI/gi|129987 | Aglaothamnion
neglectum |
RLVTYGIVAGDVTPIEEI
GLVGVKE | 4.78 | 17,648 | 81 | 14 | PBP6 |
| 51 | Phycoerythrin β
subunit |
NCBI/gi|73761963 | Aglaothamnion
callophyllidicola |
KAAAVAFITNTASQRK | 5.41 | 17,570 | 71 | 8 | PBP1 |
| Phycoerythrin α
subunit |
NCBI/gi|2317690 | Pyropia
yezoensis |
RFPSSSDLESVQGNIQRA | 5.40 | 17,587 | 103 | 9 | PBP7 |
| R-phycoerythrin α
chain |
NCBI/gi|3914333 | Pyropia
tenera |
KSVITTTISAADAAGRFPS
SSDLESVQGNIQRA | 5.40 | 17,887 | 91 | 18 | PBP8 |
| 52 | Phycoerythrin β
subunit |
NCBI/gi|73761963 | Aglaothamnion
callophyllidicola |
KAAAVAFITNTASQRK | 5.41 | 17,570 | 69 | 8 | PBP1 |
| Phycoerythrin α
subunit |
NCBI/gi|2317690 | Pyropia
yezoensis |
RFPSSSDLESVQGNIQRA
RTLNLPTSAYVASFAFARD | 5.40 | 17,857 | 94
78 | 9
10 | PBP7
PBP9 |
| 54 | Phycoerythrin α
subunit |
NCBI/gi|2317690 | Pyropia
yezoensis |
RFPSSSDLESVQGNIQRA | 5.40 | 17,857 | 77 | 9 | PBP7 |
| 61 | Phycocyanin α
subunit |
NCBI/gi|90994569 | Pyropia
yezoensis |
RFLSNGELQAINGRY
RLITGAAQSVYTKF | 7.71 | 17,569 | 90 | 15 | PBP10
PBP11 |
| 69 | Phycocyanin α
subunit |
NCBI/gi|11465844 | Porphyra
purpurea |
KTPITEAIASADSQGRF
KFPYVTQMPGPTYASSAIGKA | 7.71 | 17,555 | 96 | 20 | PBP12
PBP13 |
Measurements of cell viability and
ROS
The HepG2 human hepatoblastoma cell lines were
obtained from American Type Culture Collection (Rockville, MD,
USA). The cells were cultured at 37°C in a humidified 5%
CO2, 95% air equilibrated incubator in modified Eagle's
medium (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) supplemented
with heat-inactivated 10% fetal bovine serum (FBS; HyClone, Logan,
UT, USA), 100 U/ml penicillin and 100 µg/ml streptomycin.
The adherent cells were detached with trypsin-EDTA solution and
plated at 70-80% confluence.
Cell viability was measured with fluorescein
diacetate (FDA) and propidium iodide (PI), which stain viable cells
and dead cells, respectively. The cells were seeded at a density of
1.5×104 cells per well in a 96-well plate. PBP 1-13 (1
µg/ml) were added and the cells were treated with 5 mM
H2O2 for 1 h. The cell culture medium was
then removed and FDA (at a final concentration of 10 µg/ml)
or PI (at a final concentration of 5 µg/ml) was added. The
cells were incubated at 37°C for 30 min. The staining solutions
were removed and the samples were washed with phosphate-buffered
saline (PBS). In order to determine the experimental concentrations
of PBP peptides, a preliminary experiment was performed using 1-100
µg/ml of peptides with FDA and PI. The subsequent
experiments were performed using the range of 0.001-0.01-0.1-1
µg/ml, which showed the concentration-dependent changes and
suitability in measurement (data not shown).
Inhibition of ROS production by PBP 1-13 was
evaluated in the HepG2 cells using a cell permeable probe,
2′,7′-dichlorofluorescein diacetate (DCF-DA). The cells were seeded
onto 96-well plates (1.5×104 cells per well), grown
until confluence and pre-incubated with 50 µM DCF-DA for 1 h
at 37°C in the dark. Following washing with PBS away excess probe,
the cells were treated with PBS or PBP 1-13 (1 µg/ml) in the
presence or absence of 5 mM H2O2, for 1 h.
Ascorbic acid (1 µg/ml) was used as an antioxidant
control.
Fluorescence was measured using a FilterMax F5
Multi-Mode microplate reader (Molecular Devices LLC, Sunnyvale, CA,
USA) and an Epi-fluorescence microscope (Nikon Corporation, Tokyo,
Japan) with the following settings: Excitation 485 nm and emission
535 nm for FDA and DCF-DA, and excitation 535 nm and emission 615
nm for PI. The viability and ROS production of the synthetic
peptide-treated cells were calculated as a percentage of that in
the untreated control cells.
Quantitative analysis of living and dead cells
according to PBP2 concentrations were performed according to the
manufacturer's protocol of the FITC Annexin V Apoptosis Detection
Kit I (BD Biosciences, Franklin Lakes, NJ, USA) and measured with a
Muse™ Cell Analyzer (EMD Millipore, Billerica, MA, USA). To measure
the number of stained cells, ImageJ software (version 1.41;
National Institutes of Health, Bethesda, MD, USA) was used
alongside Cell Counting Macro 1 [version 1.0; developed by Kurt De
Vos (Academic Neurology, University of Sheffield, Shefield, UK) and
used by the GNU General Public License, https://github.com/grishagin/CellCounting/archive/master.zip].
The cells were seeded at a density of
1.5×104 cells per well in a 96-well plate. PBP2 (1
µg/ml) was added and the cells were treated with 5 mM
H2O2 for 1 h. The cell culture medium was
then removed, and the cells were washed with PBS. The cells were
then stained with Annexin V and PI for 15 min in the dark at
25°C.
Western blot analysis
Whole cell extracts for the detection of SOD, CAT
and GPx by immunoblotting were isolated from the
H2O2-treated HepG2 cells with or without PBP
1-13 peptide treatment. Separation of nuclei was performed using
the Nuclei EZ Prep nuclear isolation kit (Sigma-Aldrich; Merck
KGaA, Darmstadt, Germany) according to the manufacturer's
protocols. Protein concentration was determined by bicinchoninic
acid assay. The cell lysates (30 µg) and nuclei (20
µg) were separated by electrophoresis on a 15% SDS-PAGE gel
and transferred onto polyvinylidene difluoride membranes (EMD
Millipore) using a semi-dry transfer system. The membranes were
blocked with 1% BSA in TBST buffer (0.1% Tween 20 in TBS) overnight
at 4°C and incubated with the following primary antibodies: SOD2
(1:2,000; cat. no. OASE00357; Aviva Systems Biology, San Diego, CA,
USA), CAT (1:500; cat. no. OAAB05216; Aviva Systems Biology), GPx1
(1:1,000; cat. no. OAAF05777; Aviva Systems Biology), PCNA
(1:1,000; cat. no. sc-56, Santa Cruz Biotechnology, Inc.,
Heidelberg, Germany), β-actin (1:1,000; cat. no. sc-47778; Santa
Cruz Biotechnology Inc.), nuclear factor erythroid-derived 2-like 2
(Nrf2; 1:1,000; cat. no. sc-722; Santa Cruz Biotechnology, Inc.);
p-Nrf2 (1:5,000; cat. no. ab76026; Abcam, Cambridge, UK) in TBST
solution containing 1% BSA for 1 h at room temperature. Following
washing three times with TBST, the membranes were incubated for 1 h
at room temperature with anti-rabbit and anti-mouse secondary
antibodies (Thermo Fisher Scientific, Inc.) with horseradish
peroxidase and washed with TBST three times. The final detection
was performed with enhanced chemiluminescence western blotting
reagents (Santa Cruz Biotechnology, Inc.). GeneTools software
(version 4.03; Syngene, Cambridge, UK) was used for
densitometry.
Statistical analysis
Data are expressed as the mean ± standard deviation
and were evaluated by one-way analysis of variance using the
statistical package for social sciences version 10.0 (SPSS, Inc.,
Chicago, IL, USA). Values were compared with controls using one-way
analysis of variance followed by Duncan's multiple range tests.
P<0.05 was considered to indicate a statistically significant
difference.
Results
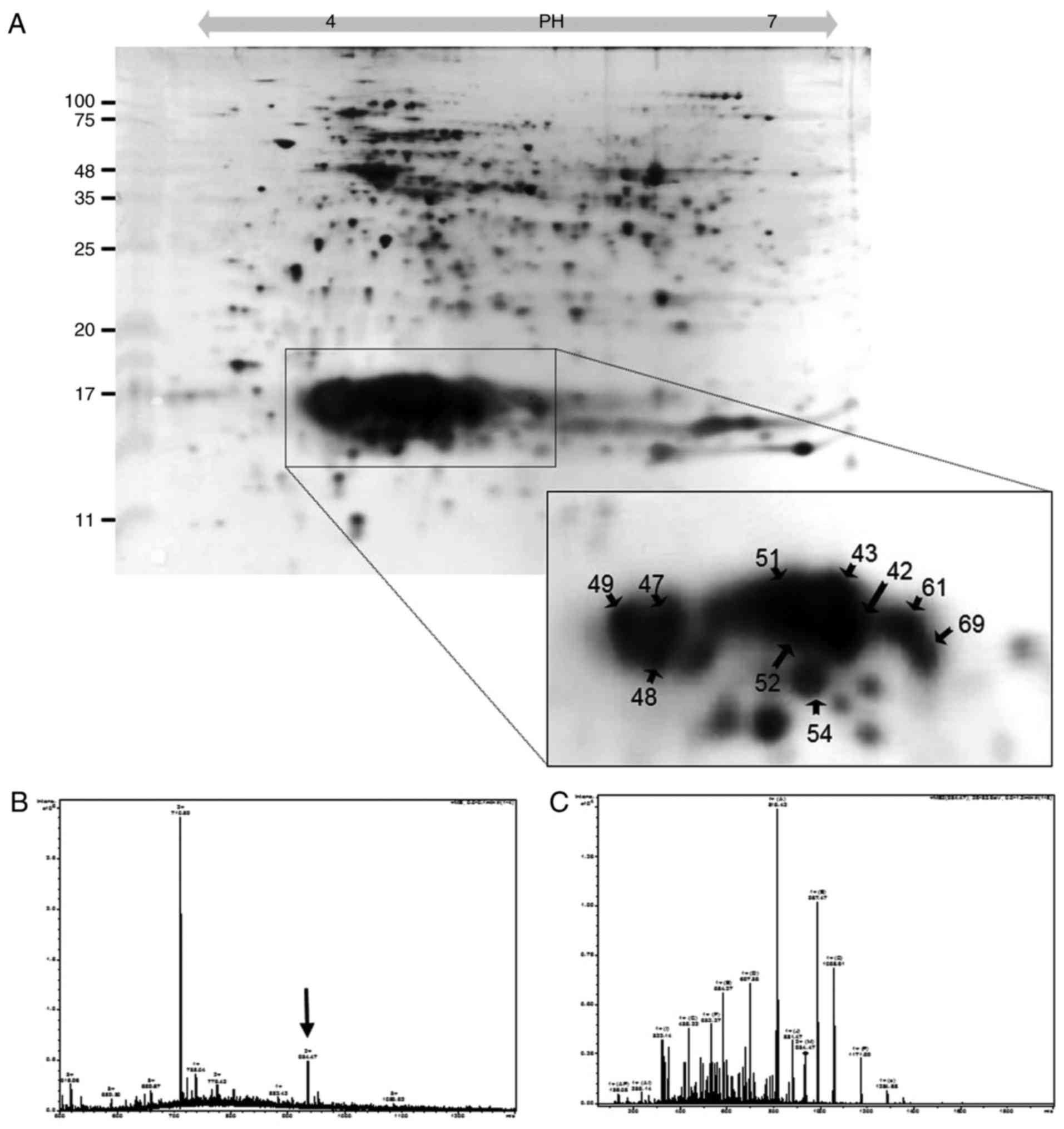
Identification of PBPs
Soluble proteins were extracted from P.
yezoensis and separated by 2D-PAGE. The soluble proteins were
classified by their isoelectric points (pIs) using a strip with a
pH range of 4-7. As shown in Fig.
1, ~200 spots were resolved. To characterize PBPs with pI
values of 4-6, 60 spots observed between 11 and 25 kDa were
analyzed by ESI-Q TOF MS/MS. As a result, 10 spots were identified
as PBPs (enclosed with circles in the tetragon in Fig. 1A). As shown in Table I, the identified PBPs were PE
(spots 42, 43, 51, 52 and 54), PC (spots 61 and 59) and APC (spots
47, 48 and 49). PE was identified as the α-type based on peptide
sequences analyzed from spots 51, 52 and 54, RFP SSS DLE SVQ GNI
QRA, RTL NLP TSA YVA SFA FAR D and KSV ITT TIS AAD AAG RFP SSS DLE
SVQ GNI QRA; and as the β-type based on sequences from spots 42, 43
(Fig. 1B and C), 51 and 52, KAA
AVA FIT NTA SQR K and RYV SYA LLA GDP SVL EDR C. PC was identified
as the α-type with the sequences analyzed from spots 61 and 69, RFL
SNG ELQ AIN GRY, RLI TGA AQS VYT KF, KTP ITE AIA SAD SQG RF and KFP
YVT QMP GPT YAS SAI GKA. APC-α was identified by the sequences of
spots 47 and 48, MQD AIT SVI NAA DVQ GKY, RAA ATI AAN AAT IIK E and
RYA TYG MLA GDP SIL EER V, whereas APC-β was identified by the
sequence of spot 49, RLV TYG IVA GDV TPI EEI GLV GVK E.
Cell viabilities and ROS inhibition
activities of synthetic peptides
The HepG2 human hepatoblastoma cell line derived
from the HepG2 cell line can be used for investigations of
xenobiotic metabolism as it maintains the synthesis and secretion
of plasma proteins and cell surface receptors, which are specific
functions of normal liver parenchymal cells (18,19). This was performed in the present
study to confirm the protective effect of PBP peptides on oxidative
stress and liver injury induced by the H2O2
in vitro system. As the liver is central for detoxification
in vivo, HepG2 cell line, which is a human liver cell, was
used as a target to observe the protective effect of oxidative
stress.
The synthetic peptides, listed in Table I, were administered at 1
µg/ml to the HepG2 human liver cells, and their viabilities
and ROS inhibition activities were determined. The fluorescent
indicator dyes DCF-DA, FAD and PI load readily into guard cells,
and their optical properties make them amenable to analysis using a
fluorescence spectrophotometer. As shown in Fig. 2A, the number of viable cells was
reduced to 47% by 5 mM H2O2. PBP1, 2 and 7-9
increased the number of viable cells by 23-32%, whereas dead cells
stained with PI increased 2.3-fold by 5 mM
H2O2 (Fig.
2B). PBP 1-13 reduced dead cells by 30-143%. In particular,
PBP2 decreased to the negative control level. The ratio of viable
cells to dead cells was reduced to 0.23 by 5 mM
H2O2, but recovered in the presence of
PBP1-4, 7-9 and 12 (Fig. 2C). In
particular, PBP1-2 led to recovery with a ratio over three times
higher than that of the positive control. Treatment with 5 mM
H2O2 increased the production of ROS ~2-fold
compared with that of the negative control (Fig. 2D). PBP 1, 2, 7-9 and 11-13 led to
a 17-56% reduction in the production of ROS compared with that of
the positive control. As a result, PBP 1, 2, 7-9 and 12 protected
cells from oxidative stress by H2O2 and
exhibited an antioxidative effect.
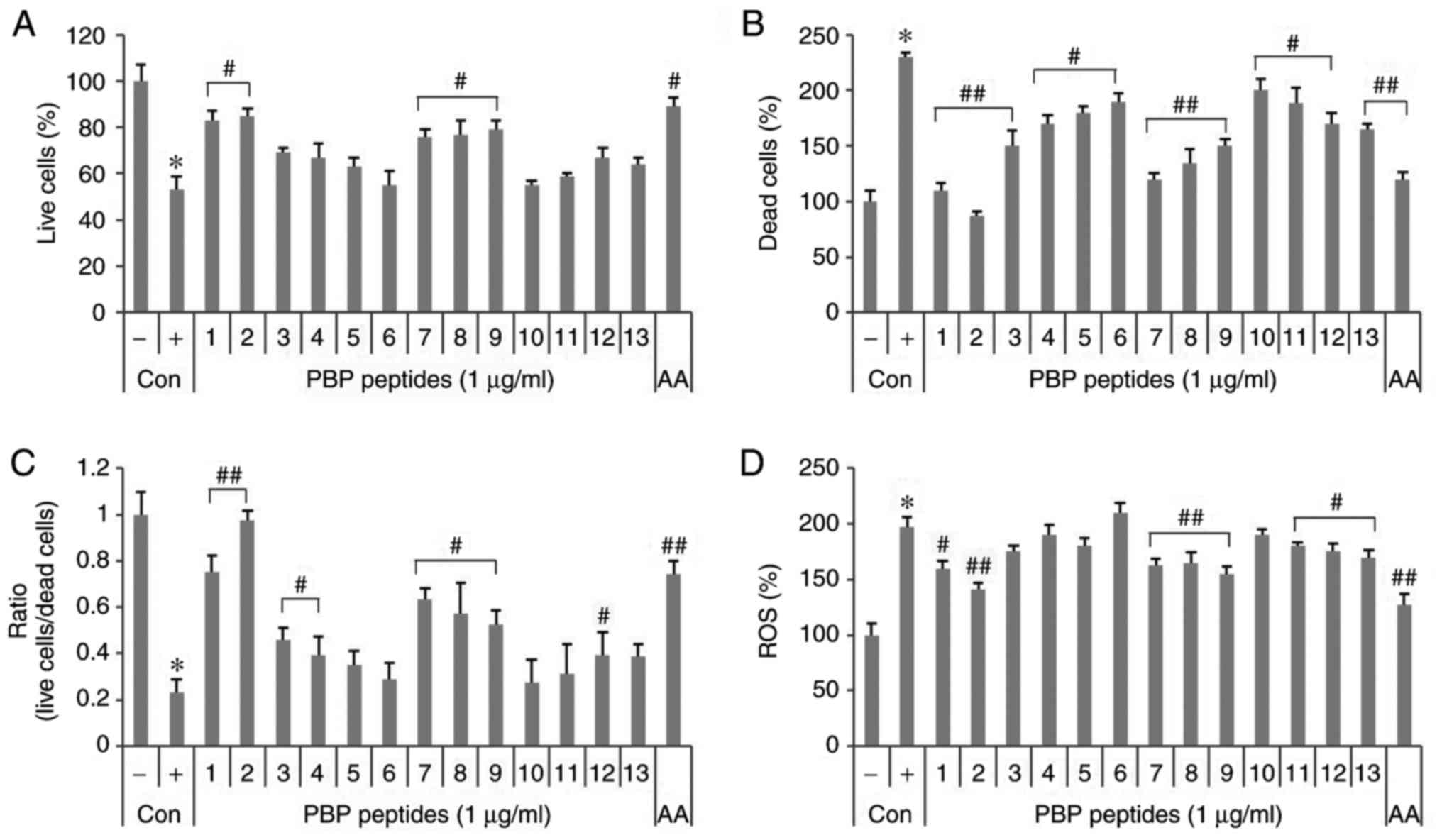 | Figure 2Effects of synthetic PBP peptides in
HepG2 cells. Induction of 5 mM H2O2-induced
cell toxicity and the effects of synthetic peptides. Live cells and
dead cells were stained with (A) fluorescein diacetate and (B)
propidium iodide. (C) Ratio of live cells/dead cells. (D) Effects
of the synthetic peptides on the formation of
H2O2-induced ROS. Cells were pretreated with
2′,7′-dichlorofluorescin diacetate. Values are expressed as the
mean ± standard deviation (n=3). *P<0.05, vs.
untreated control; #P<0.05, vs. 5 mM
H2O2-treated control. ##P<0.01,
vs. 5 mM H2O2-treated control. -, control
cells without 5 mM H2O2; +, control cells
treated with 5 mM H2O2; PBP,
phycobiliprotein; AA, 1 µg/ml ascorbic acid;
H2O2, hydrogen peroxide; ROS, reactive oxygen
species; Con, control. |
In particular, PBP2 showed good efficacy in terms of
cell survival, prevention of death and inhibition of ROS production
compared with 1 µg/ml ascorbic acid, a well-known
antioxidant.
Effects of PBP2 under
H2O2-induced oxidative stress in HepG2
cells
The HepG2 cells were stained with DCF-DA, PI,
Annexin V and FDA, and observed with a fluorescence microscope and
a fluorescence absorbance spectrophotometer to determine
cytoprotective activity and antioxidant activity according to the
concentration of PBP2 (0.001-1 µg/ml). The viable cell
number decreased in the presence of 5 mM
H2O2, whereas the number of dead cells and
ROS increased (Fig. 3A and B).
PBP2 at concentrations of 1 µg/ml increased the number of
living cells and restored apoptotic cells and dead cells to
negative control levels. In addition, 0.1-1 µg/ml PBP2
reduced the production of ROS by >50% when compared with that of
the positive control.
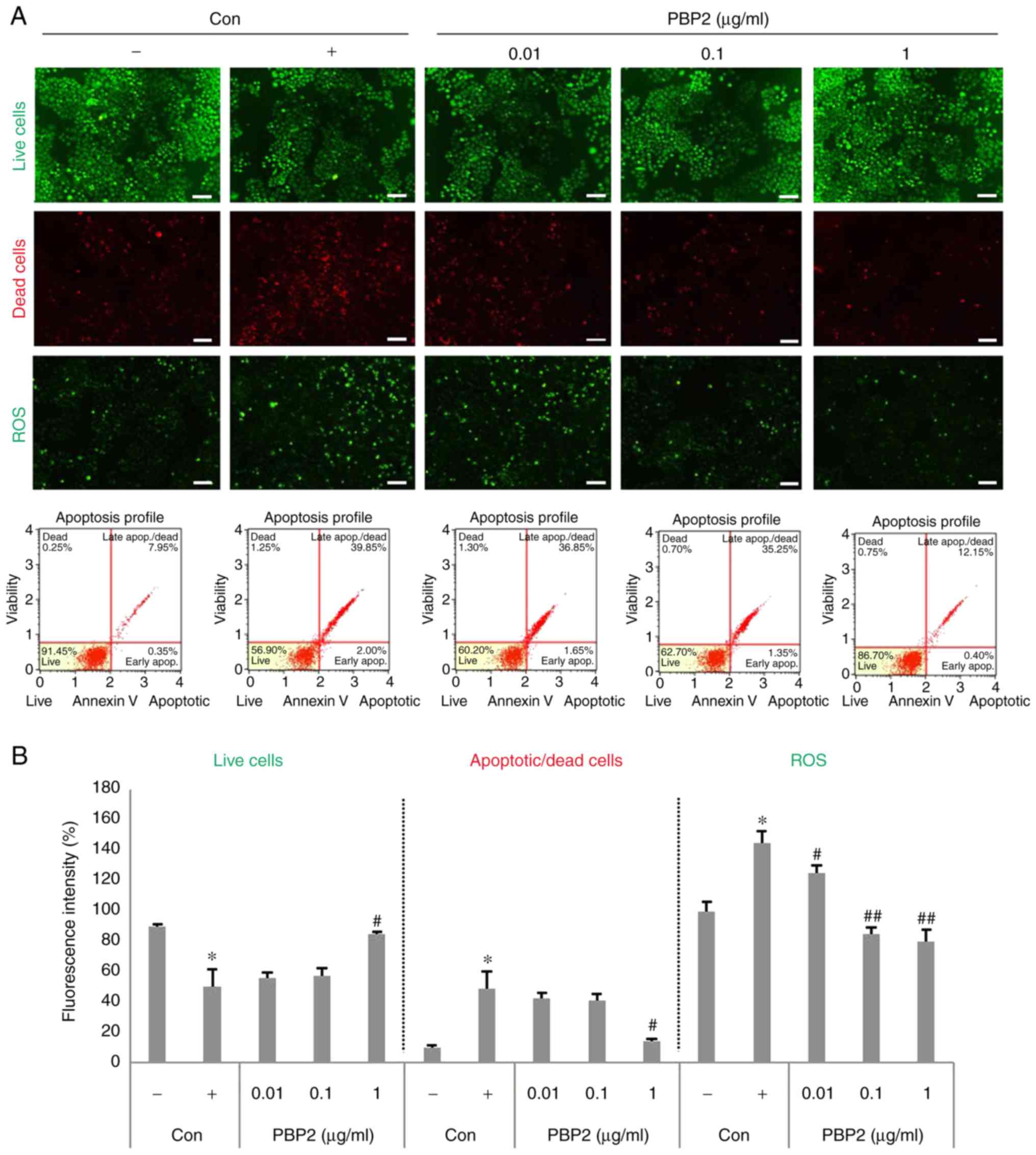 | Figure 3PBP2 resorted 5 mM
H2O2-induced oxidative stress in HepG2 cells.
(A) Live cells, apoptotic cells, dead cells and ROS were detected
by fluorescein diacetate, propidium iodide, Annexin V and
2′,7′-dichlorofluorescein diacetate staining. Scale bar=100
µm. (B) Fluorescence intensity was analyzed using ImageJ
software and the Muse™ cell analyzer. Values are expressed as the
mean ± standard deviation (n=3). *P<0.05, vs.
untreated control; #P<0.05, vs. 5 mM
H2O2-treated control; ##P<0.01,
vs. 5 mM H2O2-treated control. -, control
cells without 5 mM H2O2; +, control cells
treated with 5 mM H2O2; PBP2,
phycobiliprotein 2; H2O2, hydrogen peroxide;
ROS, reactive oxygen species; Con, control. |
PBP2 upregulates the protein expression
of p-Nrf2 and antioxidant enzymes
To examine the possible mechanisms underlying the
antioxidative effect, the present study investigated the effects of
1 µg/ml PBP2 on the p-Nrf2/antioxidant enzyme (CAT, SOD and
GPx) pathway in HepG2 cells. As shown in Fig. 4A and B, HepG2 cells exposed to 5
mM H2O2 showed a significant decrease in the
level of p-Nrf2. However, a marked increase in the level of p-Nrf2
was detected in the 0.1-1 µg/ml PBP2-treated groups. The
expression levels of CAT and GPX were not significantly affected by
5 mM H2O2, however, SOD was decreased by 5 mM
H2O2 and increased by treatment with PBP2
(Fig. 4C and D). GPX was not
affected by 5 mM H2O2, but was increased by
the effect of PBP2, suggesting that it was used to reduce the
increased ROS. This increase was not concentration-dependent in the
range of 0.01-1 µg/ml, suggesting that the removal of ROS
used antioxidant enzymes other than SOD and GPX. The present study
did not examine all the antioxidant enzymes and mechanisms, but
confirmed the possibility. Therefore, the results of the present
study may assist in elucidating the more specific mechanism of
PBP2.
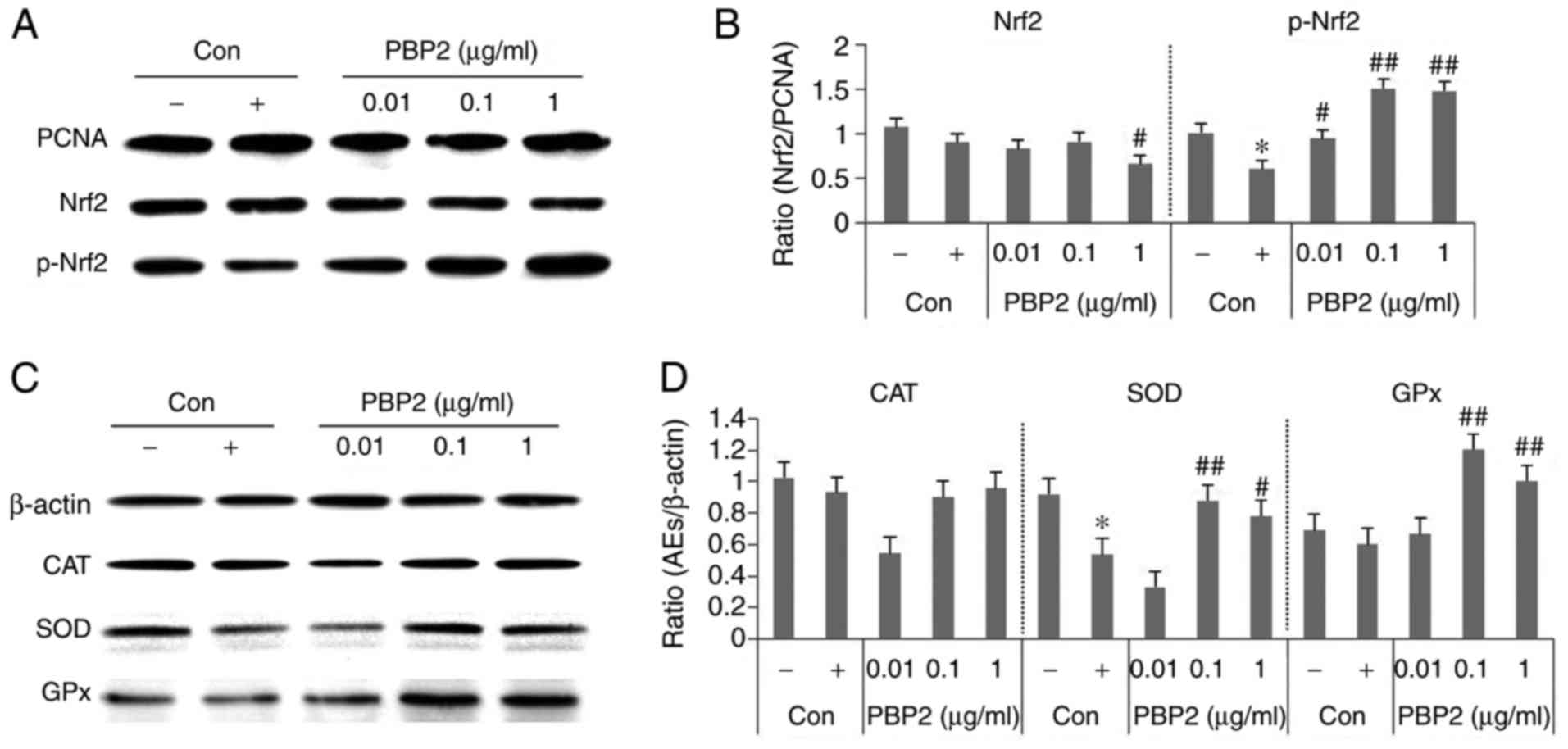 | Figure 4Effects of PBP2 on protein expression
levels of CAT, SOD and GPx in H2O2-treated
HepG2 cells. (A) Western blot analysis of protein levels of p-Nrf2.
(B) Data quantification of protein expression of Nrf2 and p-Nrf2.
(C) Western blot analysis of antioxidant enzyme levels. (D) Data
quantification of antioxidant enzyme/β-actin protein expression.
Values are expressed as the mean ± standard deviation (n=3).
*P<0.05, vs. untreated control;
#P<0.05, vs. 5 mM H2O2-treated
control; ##P<0.01, vs. 5 mM
H2O2-treated control. -, control cells
without 5 mM H2O2; +, control cells treated
with 5 mM H2O2; PBP2, phycobiliprotein 2;
Nrf2, nuclear factor erythroid-derived 2-like 2; p-Nrf2,
phosphorylated Nrf2; CAT, catalase; SOD, superoxide dismutase; GPx,
glutathione peroxidase; Con, control; AEs, antioxidant enzymes. |
The results showed that treatment with 5 mM
H2O2 led to decreases in the expression of
p-Nrf2 and SOD, which were reversed by treatment with 0.1-1
µg/ml PBP2. However, no significant difference in the
protein level of CAT was observed among these groups. These data
suggested that PBP2 controlled H2O2-induced
oxidative stress by regulating the expression of p-Nrf2/SOD.
Discussion
PBPs have been described as having potent
antioxidant activities and are used as pharmaceutical agents based
on such reported activities (9,10,12,13,30-32). In the present study, various
physiological activities of PBP were isolated and identified from
P. yezoensis.
PE is a hexamer in dilute acidic solutions. The
monomer has two peptide chains, α and β, each with a molecular
weight of 19.5 kDa. Therefore, the total molecular weight of the
monomer is 39 kDa. PE acquires a scarlet color by binding to
phycoerythrobilin, a pigment that normally has a minimal absorption
spectrum of 498, 540 and 565 nm, and, typically, two or three
molecules bind with one another. Red algae contain
phycoerythrobilin and phycourobilin, with a maximum absorbance of
490 nm and, typically, two molecules bind to the protein. Certain
types of PE show different intensities in the absorption bands and
have different absorption spectra; however, in the majority of
cases, PE has a maximum band of fluorescence emission at 552 nm
(33,34). In the present study, P.
yezoensis tissue underwent a lysis protocol, resulting in the
majority of protein complexes dissociating to yield proteins in
only the monomer form, which were then analyzed by 2D-PAGE. As a
result, two types of PE were observed at 19 kDa. However, PC-β was
not observed in the protein samples analyzed in the present study.
PC in seaweed is usually a monomer with a molecular weight of 40
kDa, and is composed of an α-chain with a molecular weight of 18.5
kDa and β-chain with a molecular weight of 20.5 kDa, and two or
three blue-colored phycocyanobilin molecules bind with one
phycoerythrobilin molecule (35,36). PC in red algae is the R-type with
an absorption band between 615 and 620 nm, however, certain types
show an absorption band at 550 nm, caused by phycoerythrobilin
(33).
In APC, a monomer with a molecular weight of 15.5
kDa forms a hexamer (35,36). APC is less abundant than PE or PC
and, like PC, has a bond between phycocyanobilin and the protein.
However, it differs from PE and PC in its absorption spectrum,
showing maximum absorption bands at 620 and 650 nm (33).
Color mutations in P. yezoensis yield, in
addition to the wild-type, a red type, a green type and a yellow
type, all of which have different types of phycobilin (37). The wild-type consists of R-PE and
C-PC; the red type consists of R-PE and R-PC; the green type
consists of B-PE and C-PC; and the yellow type consists of B-PE and
R-PC. APC occurs in all these genotypes. The present study used the
red-type P. yezoensis and its R-PE was identified in spots
5, 42, 43 and 51. However, PC could not be classified in present
study. Presumably, PC was dissolved or dissociated by the reagent
used in the pretreatment process for 2D-PAGE and ESI Q TOF, and
thus was not classified or detected.
Marine organisms, including seaweeds, contain
various chemical agents, including fatty acids, carotenoids,
polysaccharides and pigment proteins, all showing diverse
biological activities (1-5,31).
In particular, several studies have focused on the potential
biochemical applications of peptides originating from marine
organism-derived proteins (32).
The diversity of such applications is based potentially on the
essential amino acid compositions of these proteins.
In the present study, the antioxidant activity was
confirmed by synthesizing the identified PBP. No cytotoxicity was
observed for the 13 synthetic peptides assessed, as shown in
Fig. 2C. The percentage of cells
that survived oxidative stress was observed to increase when the
cells were treated with the synthesized peptides. Therefore, the
peptides showed no toxicity in these cell culture experiments. The
intracellular ROS levels, measured with DCF-DA, increased 2-3-fold
in cells treated with H2O2 and these values
decreased when treated with certain peptides. The ROS levels were
found to decrease by up to 56% in the presence of PBP2 (synthesized
from PE), as shown in Fig. 2D.
The ROS levels were reduced by PBP 1, 2 (synthesized from the PE β
subunit), 7-9 (synthesized from PE α subunit) and 10-12
(synthesized from PC), demonstrating the antioxidant properties of
PE and PC. For several decades, several in vitro and in
vivo studies have been performed on the potential removal of
free radicals by PBPs. Sonani et al reported on external
antioxidant and free radical removal activities of three types of
PBP extracted from Lyngbya sp., a cyanobacterium (13). PE exhibited antioxidant activity
by removing ROS through direct oxidation-reduction reactions,
whereas PC and APC exhibited antioxidant activities through
indirect mechanisms, including metal ion chelation. Such reports
indicate that the antioxidant activities of PBPs occur by various
mechanisms and the mechanism is dependent on amino acid side chains
of these PBPS. Here, it was hypothesized that the distribution of
the amino acids on the surface of PBPs defines their diverse
antioxidant activities. The peptides synthesized from PE types of
P. yezoensis also showed activity that reduced ROS levels.
In the present study, the antioxidant activities attributed to PBPs
were evaluated by synthesizing shorter peptide fragments of PBPs
rather than full-length proteins in an effort to identify novel
candidate peptides for experiments on antioxidant and anti-aging
activities.
Amino acids with hydrophobic branch chains are good
proton donors and metal ion chelation agents. Similarly, acidic,
basic and aromatic amino acids facilitate the removal of metal ions
(38). PBPs have been reported to
be metal ion chelation agents, and thus effective at removing
metals. It has been suggested that certain antioxidant activities
of PBPs are attributable to proton donation or metal ion chelation,
both dependent on amino acid composition (13,39). In particular, it was reported that
PE has low chelation capability but a high reduction capacity when
compared with that of PC or APC, indicating that PE and PC may be
important in oxidation-reduction reactions and, therefore, in
antioxidant activities (27). The
peptides synthesized from PE exhibited higher ROS removal
activities compared with the peptides synthesized from PC or
APC.
When oxidative stress occurs in a cell, the cell
activates several processes to reduce this stress (40). One of these processes is to
increase antioxidant enzyme levels through the Nrf2/antioxidant
responsive element (ARE) pathway. Nrf2 enters the nucleus and
converts to p-Nrf2, which activates ARE. This allows the expression
of antioxidant enzymes, including CAT, SOD and GPx, and is used to
eliminate oxidative stress. In the present study, PBP2 increased
the expression of p-Nrf2, a typical antioxidant stress reliever. In
addition, PBP2 increased the expression of antioxidant enzymes SOD
and GPx. PBP2 was able to inhibit ROS production directly by
increasing the activity of p-Nrf2 and SOD. The subsequent
experiments corroborated these findings.
Various biomolecules with potent antioxidant
activities have been examined for the prevention of aging. Aging is
defined as systematic decreases in physiological functions,
including biochemical functions, occurring in the majority of
organisms. According to the free radical theory of aging, a major
cause is activated oxygen species and, therefore, removal of such
species is being investigated for the prevention of aging. The
present study was performed to address whether certain peptides,
derived from PBPs, with identified antioxidant activities may be
useful for the prevention of aging. The findings identified several
peptides as candidate antioxidant agents. The aim of the present
study was to analyze the signaling pathways involved in the
mechanism of action of peptides synthesized from PBPs,
demonstrating their antioxidant activities in cells and addressing
their potential value as antioxidant agents.
In the present study, PBPs from P. yezoensis
were identified by proteomics and several PBP peptides were
synthesized to examine their inhibition of ROS generation. Several
synthesized PBPs peptides showed antioxidant activities in HepG2
cells. In particular, the PBP2 peptide was identified as a
potential antioxidant as this peptide downregulated ROS, and
appeared to function through the p-Nrf2/SOD pathways. In
conclusion, PBP2 attenuated H2O2-induced
oxidative stress in the HepG2 cells.
Acknowledgments
The authors would like to express sincere gratitude
to Dr Dong-Gyun Kim and Dr Young-Ok Kim (Korean National Fisheries
Science Research Institute) for helping to carry out 2D-PAGE.
Funding
This study was supported by the Basic Science
Research Program through the National Research Foundation of Korea,
which is funded by the Ministry of Education (grant no.
2012R1A6A1028677).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
The design of the experiment was done by EYK and
TJN. Experimentation and drafting of the manuscript was performed
by EYK. YHC assisted with revising the manuscript critically for
important intellectual content.
Ethics approval and consent to
participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Pádua D, Rocha E, Gargiulo D and Ramos AA:
Bioactive compounds from brown seaweeds: Phloroglucinol,
fucoxanthin and fucoidan as promising therapeutic agents against
breast cancer. Phytochem Lett. 14:91–98. 2015. View Article : Google Scholar
|
|
2
|
Pérez MJ, Falqué E and Dominguez H:
Antimicrobial action of compounds from marine seaweed. Mar Drugs.
14:e522016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Magalhaes KD, Costa LS, Fidelis GP,
Oliveira RM, Nobre LT, Dantas-Santos N, Camara RB, Albuquerque IR,
Cordeiro SL, Sabry DA, et al: Anticoagulant, antioxidant and
antitumor activities of heterofucans from the seawee Dictyopteris
delicatula. Int J Mol Sci. 12:3352–3365. 2011. View Article : Google Scholar :
|
|
4
|
Cunha L and Grenha A: Sulfated seaweed
polysaccharides as multifunctional materials in drug delivery
applications. Mar Drugs. 14:E422016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Michalak I and Chojnacka K: Algae as
production systems of bioactive compounds. Eng Life Sci.
15:160–176. 2015. View Article : Google Scholar
|
|
6
|
Harnedy PA and FitzGerald RJ: Bioactive
proteins, peptides, and amino acids from macroalgae(1). J Phycol.
47:218–232. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
López-Cristoffanini C, Zapata J, Gaillard
F, Potin P, Correa JA and Contreras-Porcia L: Identification of
proteins involved in desiccation tolerance in the red seaweed
Pyropia orbicularis (Rhodophyta, Bangiales). Proteomics.
15:3954–3968. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cai C, Li C, Wu S, Wang Q, Guo Z and He P:
Large scale preparation of phycobiliproteins from Porphyra
yezoensis using co-precipitation with ammonium sulfate. Nat Sci.
4:536–543. 2012.
|
|
9
|
Jensen GS, Attridge VL, Beaman JL, Guthrie
J, Ehmann A and Benson KF: Antioxidant and anti-inflammatory
properties of an aqueous cyanophyta extract derived from
Arthrospira platensis: Contribution to bioactivities by the
non-phycocyanin aqueous fraction. J Med Food. 18:535–541. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Manirafasha E, Ndikubwimana T, Zeng X, Lu
Y and Jing K: Phycobiliprotein: Potential microalgae derived
pharmaceutical and biological reagent. Biochem Eng J. 109:282–296.
2016. View Article : Google Scholar
|
|
11
|
Hemlata and Fatma T: Screening of
cyanobacteria for phycobiliproteins and effect of different
environmental stress on its yield. Bull Environ Contam Toxicol.
83:509–515. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Reis A, Mendes A, Lobo-Fernandes H, Empis
JA and Novais JM: Production, extraction and purification of
phycobiliproteins from Nostoc sp. Bioresour Technol. 66:181–187.
1998. View Article : Google Scholar
|
|
13
|
Sonani RR, Singh NK, Kumar J, Thakar D and
Madamwar D: Concurrent purification and antioxidant activity of
phycobiliproteins from Lyngbya sp. A09DM: An antioxidant and
anti-aging potential of phycoerythrin in Caenorhabditis elegans.
Process Biochem. 49:1757–1766. 2014. View Article : Google Scholar
|
|
14
|
Dey A and Cederbaum AI: Alcohol and
oxidative liver injury. Hepatology. 43:S63–S74. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Loguercio C and Federico A: Oxidative
stress in viral and alcoholic hepatitis. Free Radic Biol Med.
34:1–10. 2003. View Article : Google Scholar
|
|
16
|
Mayoral W and Lewis JH: Drug-induced liver
disease. Curr Opin Gastroenterol. 16:231–238. 2000. View Article : Google Scholar
|
|
17
|
López-Terrada D, Cheung SW, Finegold MJ
and Knowles BB: Hep G2 is a hepatoblastoma-derived cell line. Hum
Pathol. 40:1512–1515. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Nowrouzi A, Meghrazi K, Golmohammadi T,
Golestani A, Ahmadian S, Shafiezadeh M, Shajary Z, Khaghani S and
Amiri AN: Cytotoxicity of subtoxic AgNP in human hepatoma cell line
(HepG2) after long-term exposure. Iran Biomed J. 14:23–32.
2010.PubMed/NCBI
|
|
19
|
Yook JS, Kim M, Pichiah PB, Jung SJ, Chae
SW and Cha YS: The antioxidant properties and inhibitory effects on
HepG2 cells of chicory cultivated using three different kinds of
fertilizers in the absence and presence of pesticides. Molecules.
20:12061–12075. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Martín MA, Ramos S, Mateos R,
Izquierdo-Pulido M, Bravo L and Goya L: Protection of human HepG2
cells against oxidative stress by the flavonoid epicatechin.
Phytother Res. 24:503–509. 2010.
|
|
21
|
Salla S, Sunkara R, Ogutu S, Walker LT and
Verghese M: Antioxidant activity of papaya seed extracts against
H2O2 induced oxidative stress in HepG2 cells.
LWT-Food Sci Tech. 66:293–297. 2016. View Article : Google Scholar
|
|
22
|
Zhao J, Ma D, Luo M, Wang W, Zhao C, Zu Y,
Fu Y and Wink M: In vitro antioxidant activities and antioxidant
enzyme activities in HepG2 cells and main active compounds of
endophytic fungus from pigeon pea [Cajanus cajan (L.) Millsp]. Food
Res Int. 56:243–251. 2014. View Article : Google Scholar
|
|
23
|
Song JS, Kim EK, Choi YW, Oh WK and Kim
YM: Hepatocyte-protective effect of nectandrin B, a nutmeg lignan,
against oxidative stress: Role of Nrf2 activation through ERK
phosphorylation and AMPK-dependent inhibition of GSK-3β. Toxicol
Appl Pharmacol. 307:138–149. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Tsai TH, Yu CH, Chang YP, Lin YT, Huang
CJ, Kuo YH and Tsai PJ: Protective effect of caffeic acid
derivatives on tertbutyl hydroperoxide-induced oxidative
hepatotoxicity and mitochondrial dysfunction in HepG2 cells.
Molecules. 22:E7022017. View Article : Google Scholar
|
|
25
|
Martín MA, Ramos S, Mateos R, Granado
Serrano AB, Izquierdo-Pulido M, Bravo L and Goya L: Protection of
human HepG2 cells against oxidative stress by cocoa phenolic
extract. J Agric Food Chem. 56:7765–7772. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Martín MA, Ramos S, Granado-Serrano AB,
Rodríguez-Ramiro I, Trujillo M, Bravo L and Goya L: Hydroxytyrosol
induces antioxidant/detoxificant enzymes and Nrf2 translocation via
extracellular regulated kinases and
phosphatidylinositol-3-kinase/protein kinase B pathways in HepG2
cells. Mol Nutr Food Res. 54:956–966. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sonani RR, Rastogi RP and Madamwar D:
Antioxidant potential of phycobiliproteins: Role in anti-aging
research. Biochem Anal Biochem. 4:2015. View Article : Google Scholar
|
|
28
|
Wessel D and Flugge UI: A method for the
quantitative recovery of protein in dilute solution in the presence
of detergents and lipids. Anal Biochem. 138:141–143. 1984.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Abu-Reidah IM, Arráez-Román D,
Quirantes-Piné R, Fernández- Arroyo S, Segura-Carretero A and
Fernández-Gutiérrez A: HPLC- ESI-Q-TOF-MS for a comprehensive
characterization of bioactive phenolic compounds in cucumber whole
fruit extract. Food Res Int. 46:108–117. 2012. View Article : Google Scholar
|
|
30
|
Sonani RR, Rastogi RP, Patel R and
Madamwar D: Recent advances in production, purification and
applications of phycobiliproteins. World J Biol Chem. 7:100–109.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cornish ML and Garbary DJ: Antioxidants
from macroalgae: Potential applications in human health and
nutrition. Algae. 25:155–171. 2010. View Article : Google Scholar
|
|
32
|
Cheung RC, Ng TB and Wong JH: Marine
peptides: Bioactivities and applications. Mar Drugs. 13:4006–4043.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sobiechowska-Sasim M, Stoń-Egiert J and
Kosakowska A: Quantitative analysis of extracted phycobilin
pigments in cyanobacteria-an assessment of spectrophotometric and
spectrofluorometric methods. J Appl Phycol. 26:2065–2074. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Galland-Irmouli AV, Pons L, Luçon M,
Villaume C, Mrabet NT, Guéant JL and Fleurence J: One-step
purification of R-phycoerythrin from the red macroalga Palmaria
palmata using preparative polyacrylamide gel electrophoresis. J
Chromatogr B Biomed Sci Appl. 739:117–123. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang L, Qu Y, Fu X, Zhao M, Wang S and Sun
L: Isolation, purification and properties of an R-phycocyanin from
the phycobilisomes of a marine red macroalg Polysiphonia urceolata.
PLoS One. 9:e878332014. View Article : Google Scholar
|
|
36
|
Wang Y, Gong X, Wang S, Chen L and Sun L:
Separation of native allophycocyanin and R-phycocyanin from marine
red macroalg Polysiphonia urceolata by the polyacrylamide gel
electrophoresis performed in novel buffer system. PLoS One.
9:e1063692014. View Article : Google Scholar
|
|
37
|
Hwang MS, Kim SO, Lee YS, Park EJ, Kim SC,
Ha DS, Gong YG, Baek J and Choi HG: Isolation and characterization
of pure lines of pigmentation and morphological mutants in Porphyra
tenera Kjellman (Bangiales, Rhodophyta). Kor J Fish Aquat Sci Doi.
2010.
|
|
38
|
Khantaphant S, Benjakul S and Ghomi MR:
The effects of pretreatments on antioxidative activities of protein
hydrolysate from the muscle of brownstripe red snapper (Lutjanus
vitta). LWT-Food Sci Technol. 4:1139–1148. 2011. View Article : Google Scholar
|
|
39
|
Sarmadi BH and Ismail A: Antioxidative
peptides from food proteins: A review. Peptides. 31:1949–1956.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kensler TW, Wakabayashi N and Biswal S:
Cell survival responses to environmental stresses via the
Keap1-Nrf2-ARE pathway. Annu Rev Pharmacol Toxicol. 47:89–116.
2007. View Article : Google Scholar
|