Introduction
The study of the interface between bone and the
immune system (termed osteoimmunology) focuses on shared niches,
mechanistic receptors and cytokines in physiological and
pathological conditions (1,2).
In recent years, intensive studies suggest that there is a close
relationship between the abnormal activation of immune cells and
the bone absorption function of osteoclasts (OCs) (3). OCs are stimulated to differentiate
and become activated by cytokines that are produced by immune
cells, such as dendritic cells and T cells, in vitro or
in vivo in experimental models (4–7).
Bone homeostasis is continuously regulated by two
processes: Bone formation and bone resorption by osteoblasts and
OCs, respectively (8,9). OCs are multinuclear cells that are
essential for bone remodeling, and their activity depends on the
high local concentration of the receptor activator of nuclear
factor-κB ligand (RANKL) and macrophage-colony stimulating factor
(M-CSF) (10). RANKL, a member of
the tumor necrosis factor (TNF) superfamily, interacts with
receptor activator of nuclear factor-κB (RANK) that is expressed on
OC progenitors, and then binds to TNF receptor-associated factor 6
(TRAF6) to activate downstream signaling cascades (11), such as the mitogen-activated
protein kinase (MAPK)/nuclear factor-κB (NF-κB) signal pathway, to
promote OC formation and bone resorption (12). Most studies have focused on
osteoclastogenesis from the monocytes. However, immature dendritic
cells (iDCs) derived from hematopoietic progenitors are also
crucial in osteoclastogenesis, especially in pathologies, such as
rheumatoid arthritis and lytic bone metastases of malignancies
(4,13,14). iDCs are alternate OC precursors
and have been reported to be more efficient in OC
transdifferentiation than monocytes (15). Suppressing iDC
transdifferentiation into OCs might be a novel strategy for the
treatment of bone disease, such as myeloma bone disease. γδ T cells
are a subset of T cells and serve an important role in antitumor
immunity. The effect of γδ T cells on osteoclastogenesis has rarely
been studied. The present study aimed to explore the role of human
γδ T cells in the process of iDC transdifferentiation into OCs.
Materials and methods
Human γδ T cells preparation
Peripheral blood mononuclear cells (PBMNCs) from ten
healthy male adult volunteers were isolated by Ficoll Paque
(Pharmacia Biotech, Piscataway, NJ, USA). The protocols were
approved by the Medical Ethics Committee of The First Affiliated
Hospital of Fujian Medical University, with the approval reference
number 2016[016]. The PBMNCs were stimulated with 1 µM zoledronate
acid (ZOL) and 100 U/ml recombinant human interleukin-2 (rh-IL-2;
Sigma-Alrdich; Merck KGaA, Darmstadt, Germany) in RPMI-1640 medium
(Gibco; Thermo Fisher Scientific, Inc., Waltham, MA, USA) at a
concentration of 1×106/ml and were incubated at 37°C
with 5% CO2 for 10 days. The medium was half replaced
with fresh medium containing 100 U/ml rh-IL-2 every three days. At
day 7, the cells were stained with anti-TCR Vγ9-fluorescein
isothiocyanate (cat. no. 130-107-487; 1:50: Miltenyi Biotec GmbH,
Bergisch Gladbach, Germany) and TCR Vδ2-phycoerythrin (cat. no.
130-111-010; 1:50: Miltenyi Biotec GmbH), or isotype controls, and
analyzed by flow cytometry for enrichment of Vγ9 and Vδ2-double
positive cells. At day 10, the cells were positively purified using
magnetic-activated cell sorting with anti-γδ TCR magnetic beads kit
(Miltenyi Biotec GmbH), as previously described (16).
CD14+ monocyte purification
and iDC culture
PBMNCs were purified using CD14 MicroBeads human
monocyte kit (Miltenyi Biotec GmbH). The phenotype of the monocytes
displayed >95% purity, as examined by flow cytometry analysis
for CD14 expression. To obtain iDCs, these CD14+
monocytes were suspended at 5×105 cells/ml in RPMI 1640
with 10% FBS (both Gibco; Thermo Fisher Scientific, Inc.), 100
ng/ml rh-GM-CSF (PeproTech, Inc., Rocky Hill, NJ, USA) and 10 ng/ml
rh-IL-4 (PeproTech, Inc.) for 6 days. The membrane marker CD1a and
isotype were assessed by flow cytometric analysis. Immunolabeling
of cell suspensions was performed in 1% bovine serum albumin (BSA)
and 3% human serum/PBS (both Gibco; Thermo Fisher Scientific,
Inc.). The purity of the isolated cell subsets was assessed by flow
cytometry on a FACSCalibur (BD Biosciences, San Jose, CA, USA) and
the data were analyzed using the Cell Quest program version 6.0
(FACScan; BD Biosciences).
Co-culture of γδ T cells and iDCs
γδ T cells and autologous iDCs from the same healthy
volunteer were co-cultured at different ratios in Transwell
inserts. The γδ T cells were added in the upper and the iDCs were
added in the lower compartment of the Transwells, with the medium
containing 10% FBS, 100 ng/ml rh-RANKL (PeproTech, Inc.) and 25
ng/ml rh-M-CSF (PeproTech, Inc.). To further confirm if the stage
of OC differentiation was important for the inhibitory effect of γδ
T cells, γδ T cells were co-cultured with iDCs at the ratio of 1:1
and iDCs were collected at different time-points of culture. The
day 0–1 group, day 3–4 group and the day 0–4 group were treated
with γδ T cells during days 0–1, day 3–4 and day 0–4,
respectively.
Tartrate resistant acid phosphatase
(TRAP) staining assay
At day 9 of co-culture, iDCs were collected, fixed
with 5% para-formaldehyde for 10 min and treated with
acetone/ethanol for 30 sec. Then the cells were washed with PBS,
plated on the slides and stained using the Leukocyte Acid
Phosphatase kit 387-A (Sigma-Aldrich; Merck KGaA) to TRAP activity.
Multinucleated cells with >3 nuclei were counted under
microscopy in ten different fields/well.
Bone resorption capacity assay
To assess the OC resorption capacity, iDCs from the
lower compartment of Transwelll inserts were collected and
resuspended in 96-well plates with a dentine disc at the bottom, at
a density of 2×104 cells/ml. After 24 h incubation so
that the cells attach, the culture was maintained in the medium
containing 25 ng/ml rh-M-CSF and 100 ng/ml rh-RANKL for 14 days.
Cultures had half of the medium changed with fresh cytokines and
conditioned medium every 2–3 days. Then, dentine was stained with
1% toluidine. Images of the resorption pit were captured using
reflected light microscopy and the pit area of dentine was measured
by Image-pro plus software version 6.0 (Media Cybernetics, Inc.,
Rockville, MD, USA).
Affymetrix GeneChip assays
IDCs were co-cultured with γδ T cells (1:1 ratio) in
the presence of rh-RANKL and rh-M-CSF. As a control, iDCs (without
γδ T cells) were cultured alone. Four samples were prepared from
two unrelated healthy volunteer donors. On day 9 of co-culture,
total RNA was extracted from the iDCs, using TRIzol reagent (Thermo
Fisher Scientific, Inc.), according to the manufacturer's
instructions. The samples were then purified with the RNeasy
MinElute cleanup kit (Qiagen GmbH, Hilden, Germany). Affymetrix
Human Genome U133 Plus 2.0 Arrays were used in the microarray
analysis and the data were analyzed using Affymetrix GeneChip
Command Console Software (TGCC; Affymetrix; Thermo Fisher
Scientific, Inc.). Afterwards, genes with ̔absent̓ scores were
filtered out and the remaining genes were analyzed. Microarray data
were normalized using the Robust Multiarray Average method.
Significance Analysis of Microarrays (17) was used to identify genes that are
differentially expressed. Furthermore, fold-change analysis, which
calculated the ratios of geometric means of expression intensities,
was performed. To select the differentially expressed genes (DEGs),
the criteria were: Threshold values of ≥2 and ≤−2-fold change; and
significance level <5%. Differentially expressed genes were
subjected to gene ontology (GO) and Kyoto Encyclopedia of Genes
Genomes (KEGG) functional analysis for biological processes using
Ingenuity Pathway Analysis (Qiagen GmbH).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR) analysis
RNA was extracted using RNeasy Mini kit (Qiagen
GmbH) from each group of iDCs, converted to cDNA using a
PrimeScript RT reagent kit (Takara Biotechnology Co., Ltd., Dalian,
China), and qPCR analysis was performed using the SYBR PreMix Ex
Taq II kit (Takara Bio, Inc., Otsu, Japan). The sequences of
primers used in this study were listed in Table I. qPCR reactions were run on an
ABI 7500 Sequence Detection System (Applied Biosystems; Thermo
Fisher Scientific, Inc.). The qPCR reaction was performed at 95°C
for 5 min, followed by 40 cycles of 95°C for 30 sec, 60°C for 30
sec, and 72°C for 30 sec. The data were analyzed using the
2−ΔΔCq relative expression method (18). qPCR reactions were performed in
triplicate.
 | Table ISequences of primers used in reverse
transcription-quantitative polymerase chain reaction. |
Table I
Sequences of primers used in reverse
transcription-quantitative polymerase chain reaction.
| Gene | Primer | Sequence
(5′-3′) |
|---|
| Cathepsin K | Forward |
GGAAGAAGACCCACAGGAAGCAATA |
| Reverse |
GAGAAGCCTCAAGGTTATGGATGGA |
| ATP6V0D2 | Forward |
GTCCCATTCTTGAGTTTGAGG |
| Reverse |
GGATAGAGTTTGCCGAAGGTT |
| RANK | Forward C |
AGTGAGAAGCATTATGAGCATC |
| Reverse |
ATTCCAGCTATCCAAGTATTCATCC |
| c-Fos | Forward |
TTGCTGCATAAAGTTTGTGATACAG |
| Reverse |
AGGAAAAGGCATCAGAGAAGTAGC |
| GAPDH | Forward CC |
AGCAAGAGCACAAGAGGAAGAG |
| Reverse |
GGTCTACATGGCAACTGTGAGGAG |
Western blot analysis
IDCs were seeded at 1.0×106 cells/well in
6-well plates with 100 ng/ml rh-RANKL and 25 ng/ml rh-M-CSF and
co-cultured with 1×105 γδ T cells for 12 h. As a
control, iDCs cultured alone (without γδ T cells) were used. At day
9, the cells were lysed with RIPA buffer. The protein
concentrations were determined using a Bio-Rad protein assay kit.
Then 20 µg of protein from each sample were resolved by 10% sodium
dodecyl sulfate-polyacrylamide gel electrophoresis and transferred
to polyvinylidene difluoride membranes (BD Biosciences). The
membranes were blocked with 5% non-fat milk in TBST buffer.
Membranes were incubated with the following primary antibodies:
β-actin (cat. no. bs-0061R; BIOSS, Beijing, China), RANK (cat.no.
4845; Cell Signaling Technology, Inc., Danvers, MA, USA), c-Fos
(cat. no. 4384; Cell Signaling Technology, Inc.) and ATP6V0D2 (cat.
no. ab194557; Abcam, Cambridge, MA, USA) at a dilution of 1:1,000
at 4°C overnight. The immunoreactive bands were then incubated with
the appropriate horseradish peroxidase (HRP)-conjugated secondary
antibodies (cat. no. A0208; 1:1,000; Beyotime Institute of
Biotechnology, Haimen, China) for 1 h at room temperature. The
bands were visualized using an enhanced chemiluminescence detection
system (Amersham; GE Healthcare Life Sciences, Piscataway, NJ,
USA). Densitometric analysis was performed using Quantity One
software (Bio-Rad Laboratories, Inc., Hercules, CA, USA).
Statistical analysis
Statistical analysis was conducted using SPSS 20.0
(IBM Corp., Armonk, NY, USA). All quantitative data were expressed
as mean ± standard deviation. Statistical differences were analyzed
by one-way analysis of variance, followed by Tukey post-hoc test
for multiple-group comparisons. P<0.05 was considered to
indicate a statistically significant difference.
Results
γδ T cells inhibit OC differentiation
from iDCs in vitro
At day 10 of culture of the PBMNCs in the medium
containing ZOL and rh-IL-2, the cells were collected and analyzed
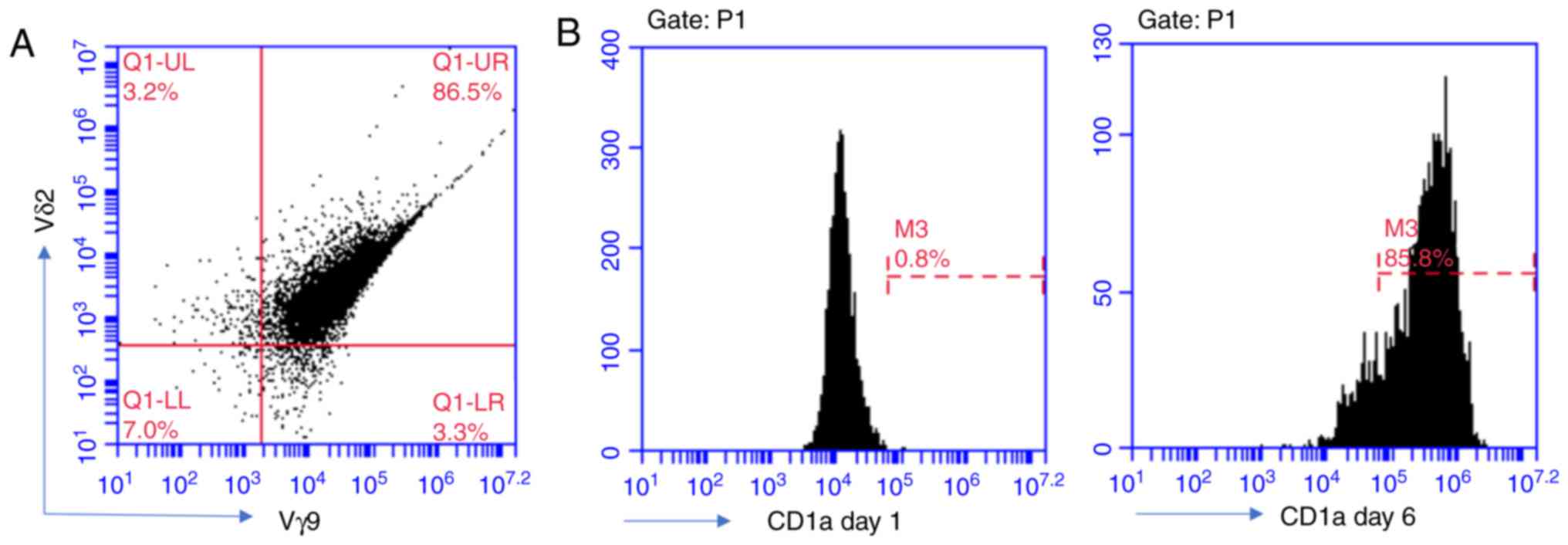
with flow cytometry. The results demonstrated that the ratio of Vγ9
and Vδ2-double positive cells was 86.5% of total (Fig. 1A). CD14+ PBMNCs were
cultured with rh-GM-CSF and rh-IL-4 in RPMI 1640 medium. After 6
days of incubation, these round mononuclear cells grew branched
projections. Flow cytometric analysis revealed that the
CD1a-positive rate was up to 86%, and therefore, these cells could
be regarded as iDCs (Fig. 1B).
After iDCs were co-cultured with γδ T cells, maintaining culture of
these iDCs in the presence of RANKL and M-CSF produced
significantly fewer TRAP+ OCs compared with the control
iDCs (without γδ T cell exposure; P<0.001). The higher the radio
of γδ T: iDCs in the culture, the fewer TRAP-positive OCs were
observed (Fig. 2A). The
difference between each adjacent ratio group was highly significant
(P<0.001). Furthermore, γδ T cells were added at different
time-points of OC differentiation. The number of TRAP+
OCs in the day 0–1 group was significantly lower compared with the
control group (P<0.001) and also significantly lower compared
with the day 3–4 group (P<0.001; Fig. 2B). No significant difference was
observed between the day 3–4 group and control (Fig. 2B). Thus, only the cells at the
early stage of OC differentiation (treatment during the 0–1 days)
were sensitive to the inhibitory effect of γδ T cells.
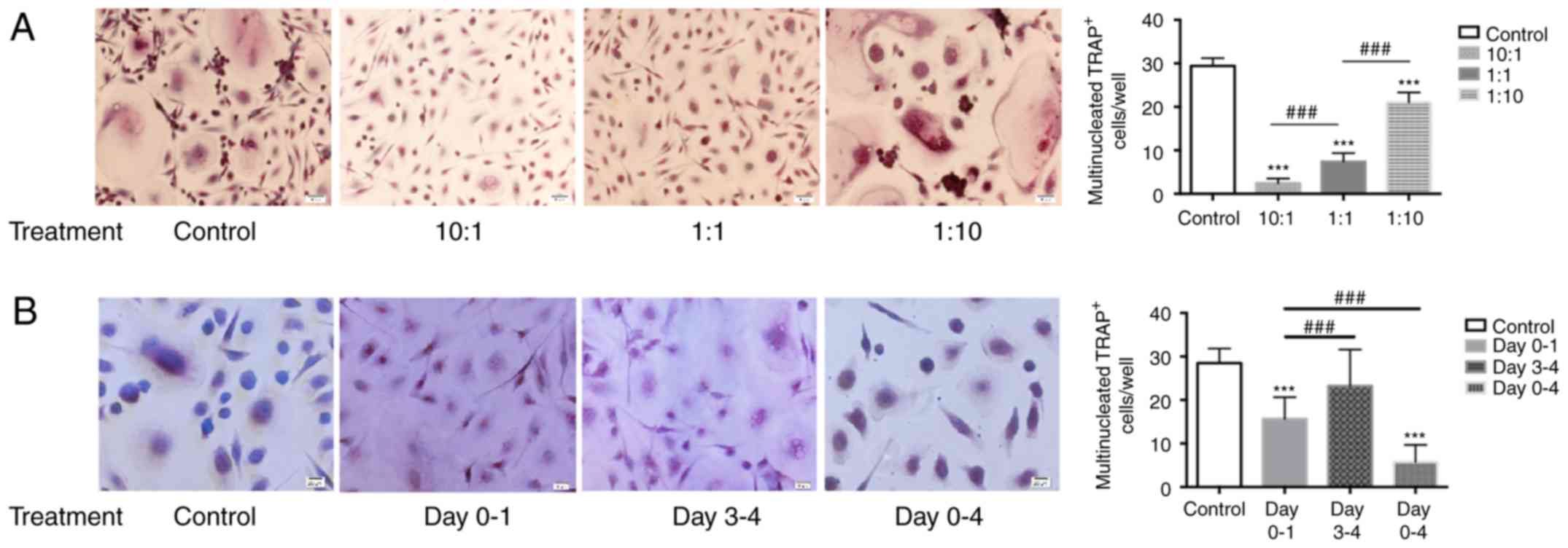 | Figure 2TRAP-positive OCs generated from iDCs
following co-culture with γδ T cells. iDCs were collected following
co-culture, maintained in α-MEM supplemented with rh-RANKL and
rh-M-CSF for 9 days and assayed for TRAP. (A) γδ T cells were
co-cultured with iDCs in the Transwell insert at the ratio of 10:1,
1:1 and 1:10 for 24 h. (B) γδ T cells were co-cultured with iDCs in
the Transwell insert at the ratio of 1:1 and iDCs were collected at
different time-points of culture (duration of treatment, days 0–1,
3–4 or 0–4). Representative images of the staining (magnification,
×200) and quantification of TRAP+ cells/well is shown.
Data are presented as the mean ± standard deviation from three
experiments from independent donors. ***P<0.001
compared with control; ###P<0.001 with comparisons
indicated by lines. TRAP, tartrate resistant acid phosphatase; OCs,
osteoclasts; iDCs, immature dendritic cells; rh, recombinant human;
RANKL, receptor activator of nuclear factor-κB ligand; M-CSF,
macrophage-colony stimulating factor. |
γδ T cells inhibit OC resorption capacity
in vitro
Toluidine blue staining revealed that the lacunar
resorption area on dentine slices of the co-culture group was
significantly decreased compared with the control group
(P<0.001; Fig. 3). The higher
the radio of γδT: iDCs, the smaller the lacunar resorption area.
There was a significant difference between the adjacent ratio
groups (P<0.001; Fig. 3).
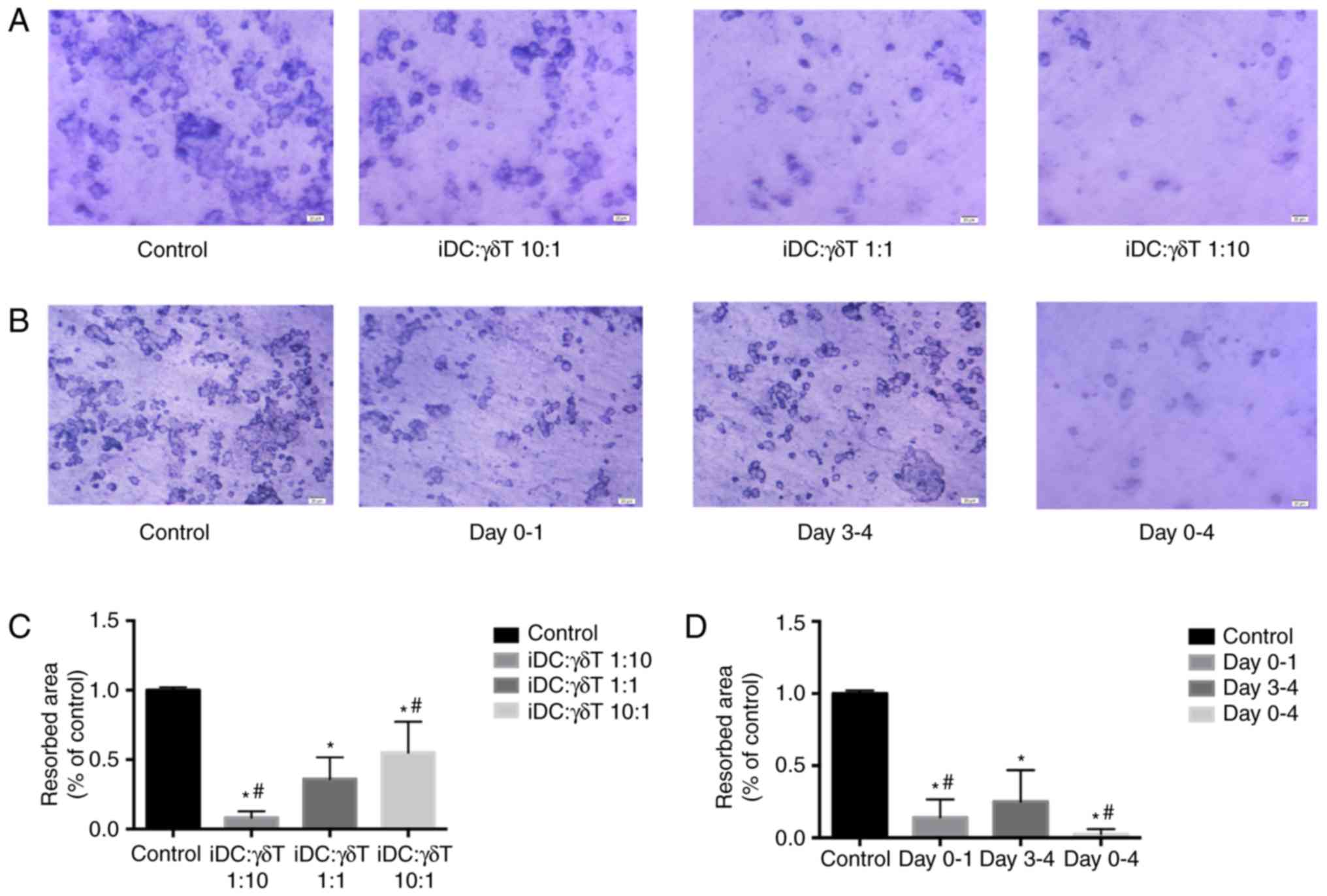 | Figure 3Resorption capacity of OCs generated
from iDCs following co-culture with γδ T cells. iDCs were collected
following co-culture, maintained in α-MEM supplemented with
rh-RANKL and rh-M-CSF for 14 days. Dentine was stained with 1%
toluidine. (A) γδ T cells were co-cultured with iDCs in the
Transwelll insert at the ratio of 10:1, 1:1 and 1:10 for 24 h. (B)
γδ T cells were co-cultured with iDCs in the Transwell insert at
the ratio of 1:1 and iDCs were collected at different time-points
of culture (duration of treatment, days 0–1, 3–4 or 0–4). (C and D)
The ratio of resorbed area of control in (A and B), respectively.
Representative images (magnification, ×400) and quantification of
the resorbed are is shown. Data are presented as the mean ±
standard deviation from five experiments. *P<0.001
compared with the control; #P<0.001 compared with the
1:1 group. OCs, osteoclasts; iDCs, immature dendritic cells; rh,
recombinant human; RANKL, receptor activator of nuclear factor-κB
ligand; M-CSF, macrophage-colony stimulating factor. |
γδ T cells negatively regulate the
RANK/c-Fos/ATP6V0D2 pathway
Fig. 4A
illustrates the changed in mRNA expression patterns between the
co-culture of γδ T cells and iDCs and the individual culture of
iDCs. A total of 293 mRNAs, whose expression change was >2-fold,
were identified. Among them, 123 mRNAs were upregulated and 170
mRNAs were downregulated in iDCs co-cultured with γδ T cells,
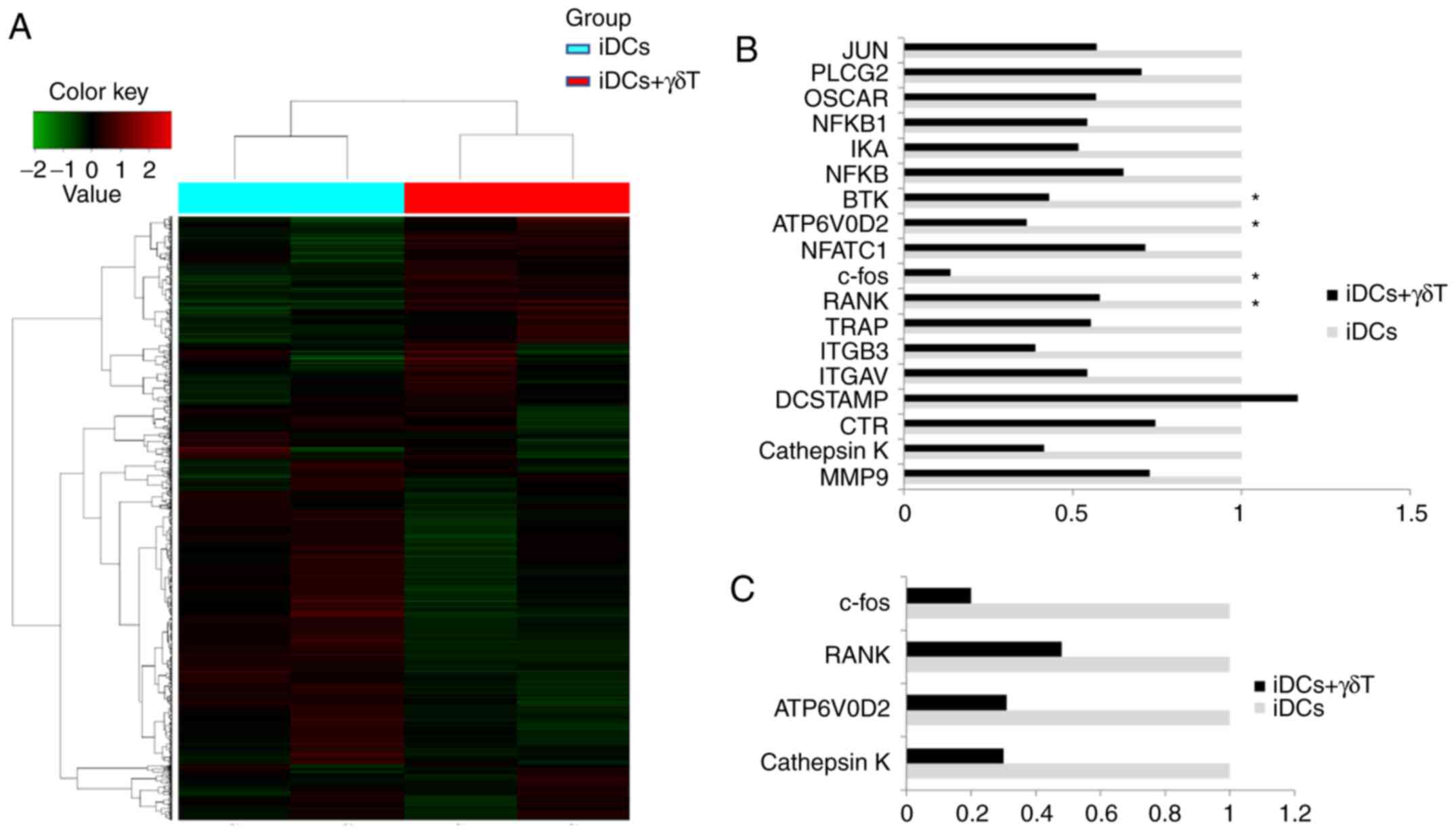
compared with iCDs cultured alone (Fig. 4A). The DEGs, identified from the
microarray analysis as associated with OC differentiation are shown
in Fig. 4B. RT-qPCR was then used
to confirm the changes in mRNA expression for several of these
genes. The mRNA expression levels of RANK, c-Fos, ATP6V0D2,
cathepsin K and CTR were decreased in iDCs co-cultured with γδ T
cells, compared with iDCs alone, and these changes were consistent
with the microarray results (Fig.
4C). The significantly differentially expressed genes were
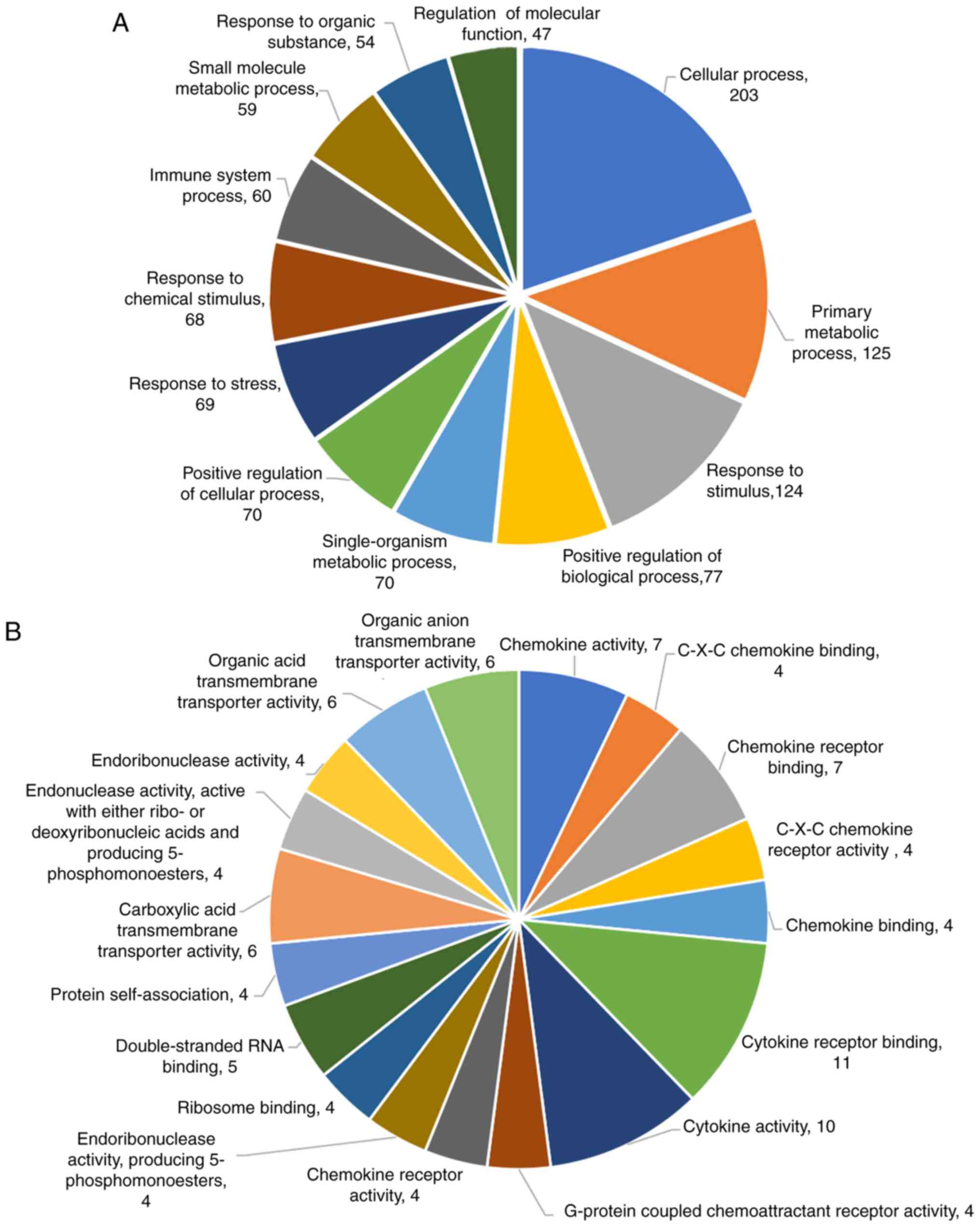
examined further by GO and KEGG functional analysis. The GO
enrichment analysis revealed that the DEGs in biological processes
were mainly related to cellular process, primary metabolic process
and response to stimulus positive regulation of biological process
(Fig. 5A). The DEGs in molecular
function were involved in chemokine activity, chemokine receptor
binding, C-X-C chemokine binding and cytokine receptor binding
activity (Fig. 5B). The results
from KEGG functional enrichment analysis are listed in Table II. Western blot analysis also
confirmed that the protein expression levels of RANK, c-Fos,
ATP6V0D2 and cathepsin K in the co-culture groups (γδ T cells: iDCs
co-cultures ratios, 1:1 and 10:1) were significantly decreased
compared with the control group (iDC alone; Fig. 6).
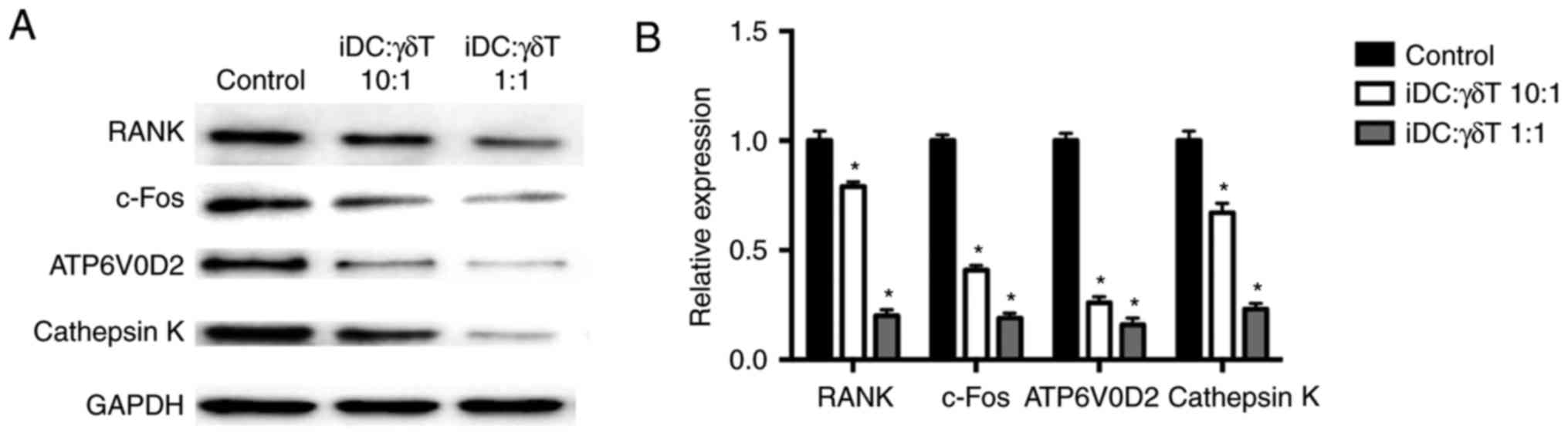 | Figure 6(A and B) Effect of γδ T cell
co-culture on protein expression of RANK, c-Fos, ATP6V0D2 and
cathepsin K in iDCs. iDCs were seeded at a density of
1.0×106 cells/well and cultured with or without γδ T
cells in the upper Transwell inserts for 24 h. The ratios of iDCs
and γδ T cells were 10:1 and 1:1. After maintaining the cultures in
RANKL- and M-CSF-containing medium for 9 days, the protein
expression levels of RANK, c-Fos, ATP6V0D2 and cathepsin K of iDCs
were detected by western blot analysis. Data are presented as the
mean ± standard deviation of three independent experiments.
*P<0.05 compared with the control group. RANK,
receptor activator of nuclear factor-κB; c-Fos, Fos proto-oncogene
AP-1 transcription factor subunit; ATP6V0D2, ATPase H+
transporting V0 subunit d2; iDCs, immature dendritic cells; RANKL,
receptor activator of nuclear factor-κB ligand; M-CSF,
macrophage-colony stimulating factor. |
 | Table IIKEGG enrichment analysis. |
Table II
KEGG enrichment analysis.
| KEGG pathway | Genes involved |
|---|
| [ko04060]
Cytokine-cytokine receptor interaction | CCL5; TNFSF10;
CXCR5; CXCL3; CCL17; CXCR4; CCL18; VEGFA; IL8; VEGFA; CXCR4;
TNFSF15 |
| [ko04620] Toll-like
receptor signaling pathway | CCL5; CEP170; IL8;
FTSJD1; TLR4 |
| [ko05323]
Rheumatoid arthritis | CCL5; ATP6V0D2;
CCL5; MMP1; VEGFA; IL8; VEGFA; TRBC1; TLR4 |
| [ko04623] Cytosolic
DNA-sensing pathway | CCL5; GOLGA7;
FTSJD1; DDX58; FCGR3A; FCGR3B; |
| [ko04380]
Osteoclast differentiation | BTK; FOSL2;
ATP6V0D2; BTUK; CTSK |
| [ko04621] NOD-like
receptor signaling pathway | CCL5; IL8;
PLD1 |
| [ko04666] Fc Gamma
R-mediated phagocytosis | ARPC1B; CEP170;
ARPC3; PPAP2B; PPAP2B; PLD1 |
Discussion
Previous studies have demonstrated that activated T
lymphocytes suppress osteoclastogenesis in vitro and in
vivo (7,19). Amongst the best understood
unconventional T cells, human γδ T cells that can readily be
expanded in vitro, contribute to anti-infection and tumor
immune responses and regulate local immune surveillance (20,21). Previous studies have demonstrated
that γδ T cells inhibit OC transdifferentiation from monocytes.
iDCs are involved in the OC differentiation process and can also
give rise to OCs in inflammatory conditions (22). However, the effects and the
mechanisms of γδ T cells on iDC osteoclastogenesis have not been
clearly elucidated.
In the present study, it was confirmed that γδ T
cells inhibited RANKL-mediated OC differentiation from human iDCs
and their resorptive function in vitro. Previous studies
have focused on effects of the T cells on osteoclastogenesis from
monocytes (23), while the
effects on the transdifferentiation from the iDCs into OCs have
seldom been discussed, particularly in humans. In the present
study, a co-culture Transwelll system of γδ T cells and iDCs was
established and the results demonstrated that γδ T cells, amplified
with Zol and rhIL-2 stimulation, suppressed iDC
transdifferentiation into OCs. The inhibitory effect of the γδ T
cells depended on the radio of γδ T: iDCs in the cultures, which
was consistent with previous studies (24,25). The current study also investigated
the inhibitory effect of γδ T cells on osteoclastogenesis at
different iDC differentiation stages. Notably, the results revealed
that there were significantly fewer TRAP+ OCs in the day
0–1 group compared with the day 3–4 group and the days 0–4 group.
These results suggest that the early stage of transdifferentiation
from iDCs into OCs was more vulnerable to γδ T cell inhibition.
A microarray analysis was performed for further
insight into the inhibitory roles of the γδ T cells on
osteoclastogenesis. Two different random/unrelated healthy donors
were used for sample collection, in order to eliminate genetic
variation in the samples. GO and KEGG functional analysis
demonstrated that the γδ T cell inhibition on osteoclastogenesis
might be associated with the RANK/RANKL signaling pathway,
toll-like receptors and the cytokine-cytokine receptor interaction,
among other pathway (Table II).
In the context of amino-bisphosphonate drugs, such as ZOL, the γδ T
cells can produce interferon (IFN)-γ, TNF-α and GM-CSF, which
mediate monocyte differentiation to iDCs and induce iDCs to further
mature (26,27). Pappalardo and Thompson (24) demonstrated that activated human γδ
T cells were capable of inhibiting OC formation and the resorption
capacity of mature OCs, via the production of GM-CSF and IFN-γ. In
the present study, the GO enrichment analysis of the data also
revealed that chemokine and chemokine receptor activity, which was
inhibited by γδ T cells, may have important roles in the process of
osteoclastogenesis. The current findings enrich our understanding
of T cell function in the regulation of OC differentiation.
Earlier studies have indicated that T lymphocytes
suppress osteoclastogenesis by diverting early monocyte/macrophage
progenitor lineage commitment towards dendritic cell
differentiation (19), the
mechanism for which so far remained undiscovered. In the present
experiments, an Affymetrix mRNA microarray was used to screen out
the differential mRNA expression and then RT-qPCR was used to
confirm the changes in mRNA expression of RANK, cathepsin K, c-Fos
and ATP6V0D2 that are associated with osteoclastogensis (Fig. 4). The results demonstrated that γδ
T cells decreased RANK, c-Fos and ATP6V0D2 expression in iDCs,
indicating that γδ T cells suppressed the RANK/RANKL pathway,
through downregulating RANK expression, which is essential for OC
differentiation and activation.
Previous studies have demonstrated that c-Fos is the
switch differentiation mechanism between OC and DC lineages
(19,28). c-Fos is a downstream molecule of
the NF-κB signaling pathway (29), and c-Fos regulates several
transcription factors that are essential for OC formation (30). c-Fos gene knockout mice exhibit
severe osteopetrosis because the OC differentiation process is
completely inhibited (31).
Furthermore, the gene expression of ATP6V0D2 was demonstrated to be
suppressed when iDCs were co-cultured with γδ T cells. As ATP6V0D2
is involved in the fusion of osteoclastic precursors (32) and ATP6V0D2 deficiency results in
OC precursor cell fusion dysfunction (33), inhibition of ATP6V0D2 might be
involved in the process that the γδ T cells use to suppress OC
differentiation and decrease their bone resorption activity.
In conclusion, the present results demonstrated that
γδ T cells were capable of inhibiting OC formation from iDCs and
their resorption capacity. The potential mechanism of the action of
γδ T cells might be by suppressing the gene expression of RANK,
c-Fos and ATP6V0D2 and the RANK/RANKL pathway.
Acknowledgments
We would like to thank Spandidos publications
(www.spandidos-publications.com) for English language
editing.
Abbreviations:
|
α-MEM
|
α-inimal essential medium
|
|
FBS
|
fetal bovine serum
|
|
PBMNCs
|
peripheral blood mononuclear cells
|
|
iDCs
|
immature dendritic cells
|
|
OC
|
osteoclast
|
|
rh-IL-2
|
recombinant human interleukin-2
|
|
rh-IL-4
|
recombinant human interleukin-4
|
|
GM-CSF
|
granulocyte macrophage-colony
stimulating factor
|
|
M-CSF
|
macrophage-colony stimulating
factor
|
|
RANK
|
receptor activator of nuclear
factor-κB
|
|
RANKL
|
receptor activator of nuclear
factor-κB ligand
|
Funding
This study was supported by grants from the National
Natural Science Foundation of China (grant no. 81400160), the
National Science Foundation of Fujian Province (grant no.
2015J01389) and the Medical Innovation Project of Fujian Province
(grant no. 2016-CX-34).
Availability of data and materials
The analyzed datasets generated during the study are
available from the corresponding author on reasonable request.
Authors' contributions
XZ performed the experiments and was the major
contributor in writing the manuscript. ZZ contributed to the
conception and design of the experiments. DQ participated in the
study design and performed the statistical analysis. JC analyzed
and interpreted the data. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The ethics committee of the First Affiliated
Hospital of Fujian Medical University approved the protocols
included in the present study [reference number, 2016 (016)].
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Long CL and Humphrey MB: Osteoimmunology:
The expanding role of immunoreceptors in osteoclasts and bone
remodeling. Bonekey Rep. 1:pii2012. View Article : Google Scholar
|
|
2
|
Takayanagi H: Osteoimmunology in 2014:
Two-faced immunology-from osteogenesis to bone resorption. Nat Rev
Rheumatol. 11:74–76. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Arron JR and Choi Y: Bone versus immune
system. Nature. 408:535–536. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Maitra R, Follenzi A, Yaghoobian A,
Montagna C, Merlin S, Cannizzo ES, Hardin JA, Cobelli N, Stanley ER
and Santambrogio L: Dendritic cell-mediated in vivo bone
resorption. J Immunol. 185:1485–1491. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wakkach A, Mansour A, Dacquin R, Coste E,
Jurdic P, Carle GF and Blin-Wakkach C: Bone marrow microenvironment
controls the in vivo differentiation of murine dendritic cells into
osteo-clasts. Blood. 112:5074–5083. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Rifas L and Weitzmann MN: A novel T cell
cytokine, secreted osteoclastogenic factor of activated T cells,
induces osteoclast formation in a RANKL-independent manner.
Arthritis Rheum. 60:3324–3335. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Karieb S and Fox SW: Suppression of T
cell-induced osteoclast formation. Biochem Biophys Res Commun.
436:619–624. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Adamopoulos IE and Mellins ED: Alternative
pathways of osteoclastogenesis in inflammatory arthritis. Nat Rev
Rheumatol. 11:189–194. 2015. View Article : Google Scholar :
|
|
9
|
Criscitiello C, Viale G, Gelao L, Esposito
A, De Laurentiis M, De Placido S, Santangelo M, Goldhirsch A and
Curigliano G: Crosstalk between bone niche and immune system:
Osteoimmunology signaling as a potential target for cancer
treatment. Cancer Treat Rev. 41:61–68. 2015. View Article : Google Scholar
|
|
10
|
Ishii M and Saeki Y: Osteoclast cell
fusion: Mechanisms and molecules. Mod Rheumatol. 18:220–227. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Mochizuki A, Takami M, Miyamoto Y,
Nakamaki T, Tomoyasu S, Kadono Y, Tanaka S, Inoue T and Kamijo R:
Cell adhesion signaling regulates RANK expression in osteoclast
precursors. PLoS One. 7:e487952012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tomomura M, Suzuki R, Shirataki Y,
Sakagami H, Tamura N and Tomomura A: Rhinacanthin C inhibits
osteoclast differentiation and bone resorption: Roles of
TRAF6/TAK1/MAPKs/NF-κB/NFATc1 Signaling. PLoS One. 10:e01301742015.
View Article : Google Scholar
|
|
13
|
Tucci M, Ciavarella S, Strippoli S,
Brunetti O, Dammacco F and Silvestris F: Immature dendritic cells
from patients with multiple myeloma are prone to osteoclast
differentiation in vitro. Exp Hematol. 39:773–783.e1. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Rivollier A, Mazzorana M, Tebib J, Piperno
M, Aitsiselmi T, Rabourdin-Combe C, Jurdic P and Servet-Delprat C:
Immature dendritic cell transdifferentiation into osteoclasts: A
novel pathway sustained by the rheumatoid arthritis
microenvironment. Blood. 104:4029–4037. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gallois A, Lachuer J, Yvert G, Wierinckx
A, Brunet F, Rabourdin-Combe C, Delprat C, Jurdic P and Mazzorana
M: Genome-wide expression analyses establish dendritic cells as a
new osteoclast precursor able to generate bone-resorbing cells more
efficiently than monocytes. J Bone Miner Res. 25:661–672. 2010.
View Article : Google Scholar
|
|
16
|
Uchida R, Ashihara E, Sato K, Kimura S,
Kuroda J, Takeuchi M, Kawata E, Taniguchi K, Okamoto M, Shimura K,
et al: Gamma delta T cells kill myeloma cells by sensing mevalonate
metabolites and ICAM-1 molecules on cell surface. Biochem Biophys
Res Commun. 354:613–618. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Virginia GT, Robert T and Gilbert C:
Significance analysis of microarrays applied to the ionizing
radiation response. PNAS. 98:5116–5121. 2001. View Article : Google Scholar
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
19
|
Grcevic D, Lukić IK, Kovacić N, Ivcević S,
Katavić V and Marusić A: Activated T lymphocytes suppress
osteoclastogenesis by diverting early monocyte/macrophage
progenitor lineage commitment towards dendritic cell
differentiation through down-regulation of receptor activator of
nuclear factor-kappaB and c-Fos. Clin Exp Immunol. 146:146–158.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Tyler CJ, Doherty DG, Moser B and Eberl M:
Human Vgamma9/Vdelta2 T cells: Innate adaptors of the immune
system. Cell Immunol. 296:10–21. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kabelitz D: γδ T-cells: Cross-talk between
innate and adaptive immunity. Cell Mol Life Sci. 68:2331–2333.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Oh Y, Oh I, Morimoto J, Uede T and
Morimoto A: Osteopontin has a crucial role in osteoclast-like
multinucleated giant cell formation. J Cell Biochem. 115:585–595.
2014. View Article : Google Scholar
|
|
23
|
Bozec A, Zaiss MM, Kagwiria R, Voll R,
Rauh M, Chen Z, Mueller-Schmucker S, Kroczek RA, Heinzerling L,
Moser M, et al: T cell costimulation molecules CD80/86 inhibit
osteoclast differentiation by inducing the IDO/tryptophan pathway.
Sci Transl Med. 6:235ra602014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Pappalardo A and Thompson K: Activated γδ
T cells inhibit osteoclast differentiation and resorptive activity
in vitro. Clin Exp Immunol. 174:281–291. 2013.PubMed/NCBI
|
|
25
|
Cui Q, Shibata H, Oda A, Amou H, Nakano A,
Yata K, Hiasa M, Watanabe K, Nakamura S, Miki H, et al: Targeting
myeloma-osteoclast interaction with Vγ9Vδ2 T cells. Int J Hematol.
94:63–70. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Nace G, Evankovich J, Eid R and Tsung A:
Dendritic cells and damage-associated molecular patterns:
Endogenous danger signals linking innate and adaptive immunity. J
Innate Immun. 4:6–15. 2012. View Article : Google Scholar
|
|
27
|
Desch AN, Gibbings SL, Clambey ET, Janssen
WJ, Slansky JE, Kedl RM, Henson PM and Jakubzick C: Dendritic cell
subsets require cis-activation for cytotoxic CD8 T-cell induction.
Nat Commun. 5:46742014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xu X, Liu N, Wang Y, Pan LC, Wu D, Peng Q,
Zhang M, Wang HB and Sun WC: Tatarinan O, a lignin-like compound
from the roots of Acorus tatarinowii Schott inhibits osteoclast
differentiation through suppressing the expression of c-Fos and
NFATc1. Int Immunopharmacol. 34:212–219. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Baek JM, Park SH, Cheon YH, Ahn SJ, Lee
MS, Oh J and Kim JY: Esculetin attenuates receptor activator of
nuclear factor kappa-B ligand-mediated osteoclast differentiation
through c-Fos/nuclear factor of activated T-cells c1 signaling
pathway. Biochem Biophys Res Commun. 461:334–341. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wagner EF: Bone development and
inflammatory disease is regulated by AP-1 (Fos/Jun). Ann Rheum Dis.
69(Suppl 1): i86–i88. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Matsuo K, Owens JM, Tonko M, Elliott C,
Chambers TJ and Wagner EF: Fosl1 is a transcriptional target of
c-Fos during osteoclast differentiation. Nat Genet. 24:184–187.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kim K, Lee SH, Ha Kim J, Choi Y and Kim N:
NFATc1 induces osteoclast fusion via up-regulation of Atp6v0d2 and
the dendritic cell-specific transmembrane protein (DC-STAMP). Mol
Endocrinol. 22:176–185. 2008. View Article : Google Scholar
|
|
33
|
Park SJ, Park DR, Bhattarai D, Lee K, Kim
J, Bae YS and Lee SY: 2-(trimethylammonium) ethyl
(R)-3-methoxy-3-oxo-2-stearamidopropyl phosphate suppresses
osteoclast maturation and bone resorption by targeting
macrophage-colony stimulating factor signaling. Mol Cells.
37:628–635. 2014. View Article : Google Scholar : PubMed/NCBI
|