Introduction
Multinucleated osteoclasts originate from bone
marrow myeloid precursor cells and serve a major role in bone
remodeling (1,2). Abnormally activated osteoclasts may
give rise to osteosclerosis, osteoporosis, rheumatoid arthritis,
Huppert’s disease, periodontal disease and metabolic bone disorders
(3). Therefore, sufficient
understanding of the osteoclast differentiation mechanisms is
crucial for the prevention and treatment of these diseases.
Cytokines that mediate the differentiation and maturation of
osteoclasts directly and/or indirectly regulate the expression of
critical genes through complex signal transduction pathways. At
present, the signal transduction terminal transcription factors
including nuclear factor kappa-light-chain-enhancer of activated B
cells (NF-κB), proto-oncogene c-Fos (c-Fos), Jun proto-oncogene
(c-Jun) and nuclear factor of activated T-cells (NFAT) have been
identified to be involved in these processes. Furthermore, these
pathways have been demonstrated to interact with each other
(4–6).
The Ca2+ signaling pathway exerts a
dominative role in osteoclast differentiation and activation.
Seales et al (7) and
Berridge et al (8)
suggested that Ca2+ oscillations occur in the process of
receptor activator of NF-κB ligand (RANKL)-induced osteoclast
formation. Calmodulin and its downstream factors are also activated
by RANKL. Ca2+/calcineurin, and calcineurin target NFAT,
are the key regulators of osteoclast formation (7,8).
Previous studies have indicated that Ca2+/calmodulin
dependent protein kinases (CaMKs) regulate the formation of
osteoclasts via the activated transcription factor activator
protein1 (AP-1), while the AP-1 family proteins members c-Fos and
c-Jun are involved in the regulation of bone metabolism (7,9).
In addition, CaMKs may inhibit the osteoclast differentiation by
negatively regulating the glycoprotein 130 (gp130) receptor pathway
in order to promote the formation of osteoclasts (7). RANKL initiates Ca2+
oscillations, which results in calcineurin-mediated activation of
NFAT, cytoplasmic 1 (NFATC1) and ultimately promotes the formation
of osteoclasts (10). KN93 is a
CaMK inhibitor that lacks subtype specificity (11). It was demonstrated that the CaMKs
inhibitor KN93 was involved in osteoclast differentiation, although
the mechanism remains unknown (12–14).
Odanacatian is a Cathepsin K kinase inhibitor and
Saraconine is an inhibitor of Src family kinases. These molecules
are required for normal osteoclastic functions and were suggested
to restrain osteoclast resorption activity (15–17). To the best of our knowledge, the
investigation of the CaMKs inhibitor KN93 as a biologically-active
osteoclast inhibitor has not yet been described. Our previous study
indicated that KN93 (10 ng/ml) effectively blocked CaMKs activation
in osteoclasts (18). However,
whether KN93 directly affects the differentiation and activation of
osteoclasts remains unknown.
TRAP staining, bone resorption activity assays,
filamentous (F)-actin staining, as well as the determination of
intracellular calcium ([Ca2+]i) levels,
osteoclast-specific gene expression levels monitoring and protein
levels of important transcription factors protein levels, were
investigated in the present study. The results suggested that KN93
inhibited the formation of TRAP-positive multinucleated cells, the
shaping of F-actin rings and the resorption activity of the cells.
Furthermore, the results revealed that KN93 decreased the
intracellular concentrations of [Ca2+]i,
expression levels of osteoclast-specific genes and protein
expression levels of important transcription factors associated
with the macrophage colony-stimulating factor (M-CSF) +
RANKL-induced osteoclast model.
Materials and methods
Cell culture and KN93 treatment
The murine mono-cyte/macrophage RAW 264.7 cell line
was obtained from the American Type Culture Collection (Manassas,
VA, USA). The cells were incubated in Dulbecco’s modified Eagle’s
medium (GE Healthcare, Chicago, IL, USA) containing 10% fetal
bovine serum (FBS; GE Healthcare), 2 mM/l L-glutamine, 100 U/ml
penicillin and 100 µg/ml streptomycin at 37°C in a
humidified atmosphere of 5% CO2. In order to induce
osteoclast differentiation, RAW264.7 cells were resuspended in
minimum essential medium (MEM)-α (GE Healthcare) and seeded into
6-well plates (5×104 cells/ml) and cultured for 4 h at
37°C in a humidified atmosphere of 5% CO2. The medium
was changed to MEM-α containing 25 ng/ml M-CSF + 30 ng/ml RANKL
(PeproTech, Inc., Rocky Hill, NJ, USA) + dimethyl sulfoxide
[(DMSO); control, KN93 was dissolved in DMSO), 25 ng/ml M-CSF + 30
ng/ml RANKL and 25 ng/ml M-CSF + 30 ng/ml RANKL + 10 µM KN93
(Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) + DMSO. The cells
were cultured for an additional 4 days.
Cell viability assay (MTT assay)
To examine the effect of KN93 on cell growth,
RAW264.7 cells were treated with various concentrations of KN93 and
cell growth was measured by the MTT assay (Beijing Solarbio Science
& Technology Co., Ltd., Beijing, China). In summary, the cells
were plated in 96-well plates in MEM-α containing 10% FBS and
cultivated for 4 h. Different concentrations of KN93 (1, 2, 5, 10
and 20 µM), and KN93 (1, 2, 5, 10 and 20 µM) + 25
ng/ml M-CSF + 30 ng/ml RANKL were added into the culture for an
additional 96 h. At the end of the incubation, the plates were
washed with PBS 3 times, and 100 µl 0.5 mg/ml MTT solution
in PBS was added into each well. Following an overnight incubation
at 37°C, the insoluble formazan crystals were dissolved in 100
µl DMSO. Following 10 sec of shaking, the optical densities
(OD) of the samples were immediately measured at 595 nm using a
Sunrise microplate reader (Tecan Group, Ltd., Männedorf,
Switzerland).
Osteoclast differentiation detection
Osteoclast differentiation was detected by counting
cells positively stained by tartrate-resistant acid phosphatase
(TRAP) using TRAP Acid Phosphatase kit 387-A (Sigma-Aldrich; Merck
KGaA). TRAP staining was performed in accordance with the protocol
provided by the manufacturer. The samples were monitored under a
phase-contrast microscope (original magnification, ×100) and
TRAP-positive multinucleated (3 nuclei) cells were selected in five
random visual fields in different areas of each well.
Osteoclast resorption activity assay
Osteoclast resorption activity was detected using
the Corning Osteo Assay Surface (COAS; Corning Inc., Corning, NY,
USA). RAW264.7 cells were seeded onto the COAS (1.5×104
cells/ml, 200 µl/well) and cultured at 37°C in a humidified
atmosphere of 5% CO2 for 4 h. The medium used was MEM-α
with or without KN93, and the cells were incubated at 37°C in a
humidified atmosphere of 5% CO2 for 4 days. The
operating methods were based on the manufacturer’s protocol, as
described previously (19). The
resorbing lacunae appeared as single or multiple clusters on the
bottom of the COAS wells and were visualized under an inverted
phase contrast microscope (original magnification, × 100). A total
of five visual fields were selected randomly and counted in each
sample. The resorption area was measured using morphological
microscopic image analysis system (version 1.0; Shenzhen Jeda
Technology Development Co., Ltd., Shenzhen, China).
F-actin staining
Tetramethylrhodamine (TRITC)-conjugated phalloidin
(20 µM; Invitrogen; Thermo Fisher Scientific, Inc., Waltham,
MA, USA) was employed to stain the F-actin cytoskeleton. RAW264.7
cells were resuspended in MEM-α, seeded into 24-well plates and
cultured for 4 h at 37°C in a humidified atmosphere of 5%
CO2. Following this, the medium was replaced with MEM-α
containing 25 ng/ml M-CSF + 30 ng/ml RANKL+ DMSO, 25 ng/ml M-CSF +
30 ng/ml RANKL or 25 ng/ml M-CSF + 30 ng/ml RANKL + 10 µM
KN93 + DMSO. Cells were then cultured for an additional 4 days at
37°C in a humidified atmosphere of 5% CO2. Then medium
was discarded, and the cells were fixed with 4% paraformaldehyde
solution for 10 min at room temperature, permeabilized by 0.1%
Triton X-100, and stained for 20 min at room temperature according
to the manufacturer’s protocol. The stained cells were observed
under an inverted phase contrast fluorescence microscope (original
magnification, ×100) (DMI 3000B; Leica Microsystems GmbH, Wetzlar,
Germany).
[Ca2+]i
concentration quantification by flow cytometry
RAW264.7 cells were resuspended in MEM-α and seeded
into 24-well plates and cultured for 4 h at 37°C in a humidified
atmosphere of 5% CO2. The medium was changed to MEM-α
containing 25 ng/ml M-CSF + 30 ng/ml RANKL+ DMSO, 25 ng/ml M-CSF +
30 ng/ml RANKL and 25 ng/ml M-CSF + 30 ng/ml RANKL + 10 µM
KN93 + DMSO. The cells were cultured for an additional 4 days.
Following this, the medium was discarded, and the plates were
washed with PBS three times in order to remove the FBS and the
phenol red in the cell culture medium. The cells were collected and
transferred to microcentrifuge tubes. A total of 400 µl
Fluo-4-AM (Dojindo Molecular Technologies, Inc., Kumamoto, Japan)
was added to a final concentration of 5 µM. The cells were
incubated in the dark for 30 min at 37°C and rinsed three times
with PBS. A total of 400 µl PBS was added to the
microcentrifuge tubes. The cells were resuspended and incubated for
30 min at 37°C in a thermostatic water bath. The mean fluorescent
intensity from 1×104 randomly selected cells was
measured using a flow cytometer (Accuri C6, BD Biosciences,
Franklin Lakes, NJ, USA) at an emission wavelength of 516 nm with
alternate excitation wavelengths of 494 nm. The results were
analyzed by FlowJo 7.6 software (TreeStar, Inc., Ashland, OR,
USA).
Reverse transcription quantitative
polymerase chain reaction (RT-qPCR) analysis of TRAP, Cathepsin K,
and matrix metalloproteinase (MMP)-9 mRNA levels
RAW264.7 cells were resuspended in MEM-α at a
density of 5×104 cells/ml and then seeded into 6-well
culture plates at a volume of 2 ml/well. Following 4 h of culture,
the medium was changed to MEM-α with or without KN93 and the cells
were cultured for an additional 96 h at 37°C in a humidified
atmosphere of 5% CO2. Total RNA was extracted using a
Total RNA Kit I (Omega Bio-Tek, Inc., Norcross, GA, USA) and cDNA
synthesis was conducted with 2 µg RNA using the RevertAid
First strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.).
The Real-time PCR SYBR Premix Dimer Eraser kit was purchased from
Takara Bio., Inc. (Otsu, Japan), and qPCR was performed using the
Q6 Flex Real Time PCR system (Applied Biosystems; Thermo Fisher
Scientific, Inc.). The qPCR thermocycling conditions used for TRAP
were as follows: 95°C for 30 sec; followed by 40 cycles of 57°C for
1 min; and finally 72°C for 34 sec. The qPCR thermocycling
conditions used for MMP-9 were as follows: 95°C for 30 sec;
followed by 40 cycles of 55.6°C for 1 min; 72°C for 34 sec. The
qPCR thermocycling conditions used for Cathepsin K were as follows:
95°C for 30 sec; followed by 40 cycles of 56.8°C for 1 min; and
finally 72°C for 34 sec. The specificity of the PCR products was
verified by melting curve analysis. The 2−ΔΔCq method
was used to analyze relative gene expression data (20). The specific sequences of the PCR
primers for TRAP, MMP-9, Cathepsin K and GAPDH were as follows:
TRAP forward 5′-GGC TAC TTG CGG TTT CAC TAT-3′; TRAP reverse 5′-TCC
TTG GGA GGC TGG TCT-3′; MMP-9 forward 5′-GCC CTG GAA CTC ACA CGA
CA-3′; MMP-9 reverse 5′-TTG GAA ACT CAC ACG CCA GAA G-3′; Cathepsin
K forward 5′-CGC CTG CGG CAT TAC CAA-3′; Cathepsin K reverse 5′-TAG
CAT CGC TGC GTC CCT-3′; GAPDH forward 5′-AAA TGG TGA AGG TCG GTG
TG-3′ and GAPDH reverse 5′-TGA AGG GGT CGT TGA TGG-3′.
SDS-PAGE and western blot analysis
Cultured cells were collected and lysed in 100
µl radioimmunoprecipitation assay (Beyotime Institute of
Biotechnology, Haimen, China) buffer with 1 µl phenylmethyl
sulphonyl fluoride for 30 min on ice. Following ultrasonic
disruption (at 4°C, 2×10 sec and 20 kHz), the lysed cells were
centrifuged (12,000 × g for 15 min at 4°C) and the supernatants
containing the total protein were collected. The concentration of
total protein was determined by a bicinchoninic acid Protein Assay
kit (Beyotime Institute of Biotechnology) for each sample, and
equal amounts (50 µg) were separated on 12% SDS-PAGE gels at
110 V for 120 min. The separated protein bands were transferred by
electroblotting to nitrocellulose filter membranes (Beyotime
Institute of Biotechnology). The membranes were blocked in Blocking
buffer (Beyotime Institute of Biotechnology) for 60 min at room
temperature. The membranes were incubated overnight at 4°C with
primary anti-c-Fos (1:1,000; cat. no. 2250S), anti-phosphorylated
(phospho)-c-Fos (1:1,000; cat. no. 5348S), anti-c-Jun (1:1,000;
cat. no. 9165S), anti-phospho-c-Jun (1:1,000; cat. no. 3270S),
anti-NFATC1 (1:1,000; cat. no. 8032S) (all from Cell Signaling
Technology, Inc., Danvers, MA, USA), and anti-β-actin antibodies
(1:1,000; cat. no. sc-47778; Santa Cruz Biotechnology, Inc.,
Dallas, TX, USA). The antibodies were diluted in primary antibody
dilution buffer (Beyotime Institute of Biotechnology). The
membranes were incubated at room temperature for 60 min with goat
anti-rabbit IgG-horseradish peroxidase (HRP) secondary antibody
(cat. no. sc-2357; Santa Cruz Biotechnology, Inc.). The antibodies
were diluted (1:5,000) by secondary antibody dilution buffer
(Beyotime Institute of Biotechnology). The immunoreactive proteins
were visualized by electrochemiluminescence using Immobilon Western
Chemiluminescent HRP Substrate Detection reagent (EMD Millipore,
Billerica, MA, USA). The gray levels of the protein bands were
evaluated by Bio-Rad image lab analyzer v.5.2 software (Bio-Rad
Laboratories, Inc., Hercules, CA, USA).
Statistical analysis
All experiments were repeated in triplicate and the
data are presented as the mean ± standard error of the mean.
Statistical differences between groups were evaluated by analysis
of variance followed by a Tukey’s post-hoc test using SPSS v.17.0
software (SPPS, Inc., Chicago, IL, USA). P<0.05 was considered
to indicate a statistically significant difference.
Results
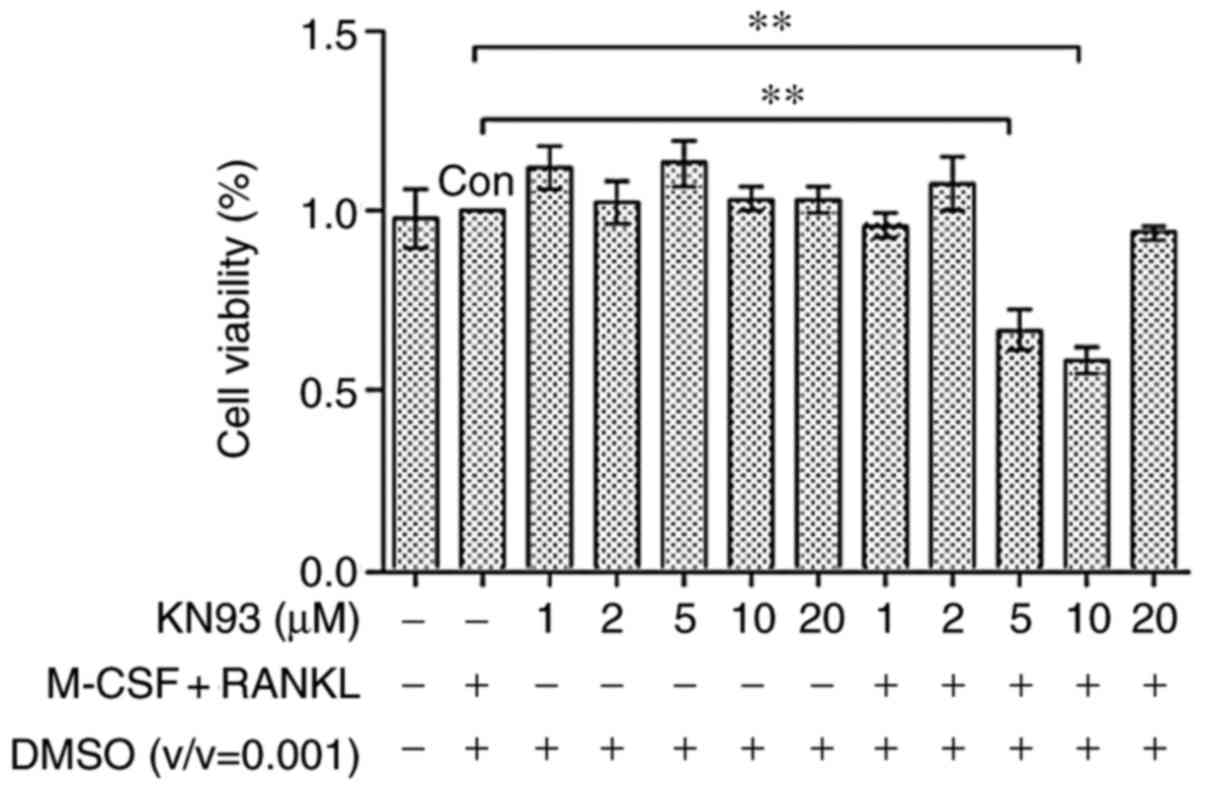
KN93 affects cell viability
KN93 did not affect RAW 264.7 cell growth rate
(Fig. 1), sustaining substantial
viability even at concentrations of 20 µM (102.95%). The
data demonstrated that KN93 was not cytotoxic to RAW264.7 cells.
However, KN93 affected the cell growth rate of 25 ng/ml M-CSF + 30
ng/ml RANKL-treated RAW 264.7 cells at concentrations of 5
µM (66.79%) and 10 µM (58.19%). Therefore, the
concentrations of 10 µM KN93 + 25 ng/ml M-CSF + 30 ng/ml
RANKL were used in subsequent experiments.
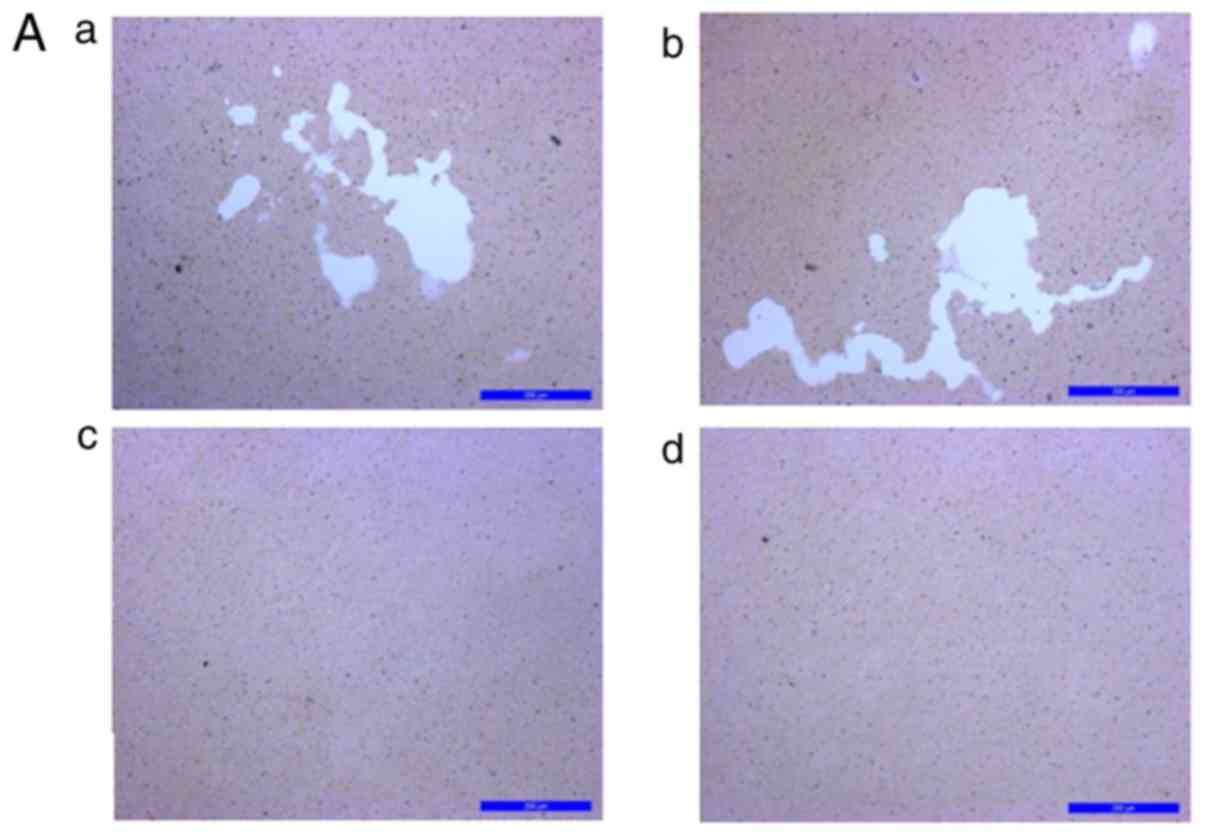
KN93 suppresses the formation of
TRAP-positive multi-nucleated cells
RAW 264.7 cells were treated with M-CSF + RANKL for
96 h and then differentiated into TRAP-positive multinucleated
cells (Fig. 2A). There was no
significant difference between the number of TRAP-positive cells in
the M-CSF + RANKL group (20.11±2.69), and in the KN93 (in DMSO) +
M-CSF + RANKL group (15.55±3.98). Concomitantly, 10 µM KN93
+ M-CSF + RANKL significantly suppressed osteoclast differentiation
(2.66±0.58), as indicated in Fig.
2B. The number of TRAP-positive cells between M-CSF + RANKL
group and the KN93 + M-CSF + RANKL group was significantly
different (P<0.01).
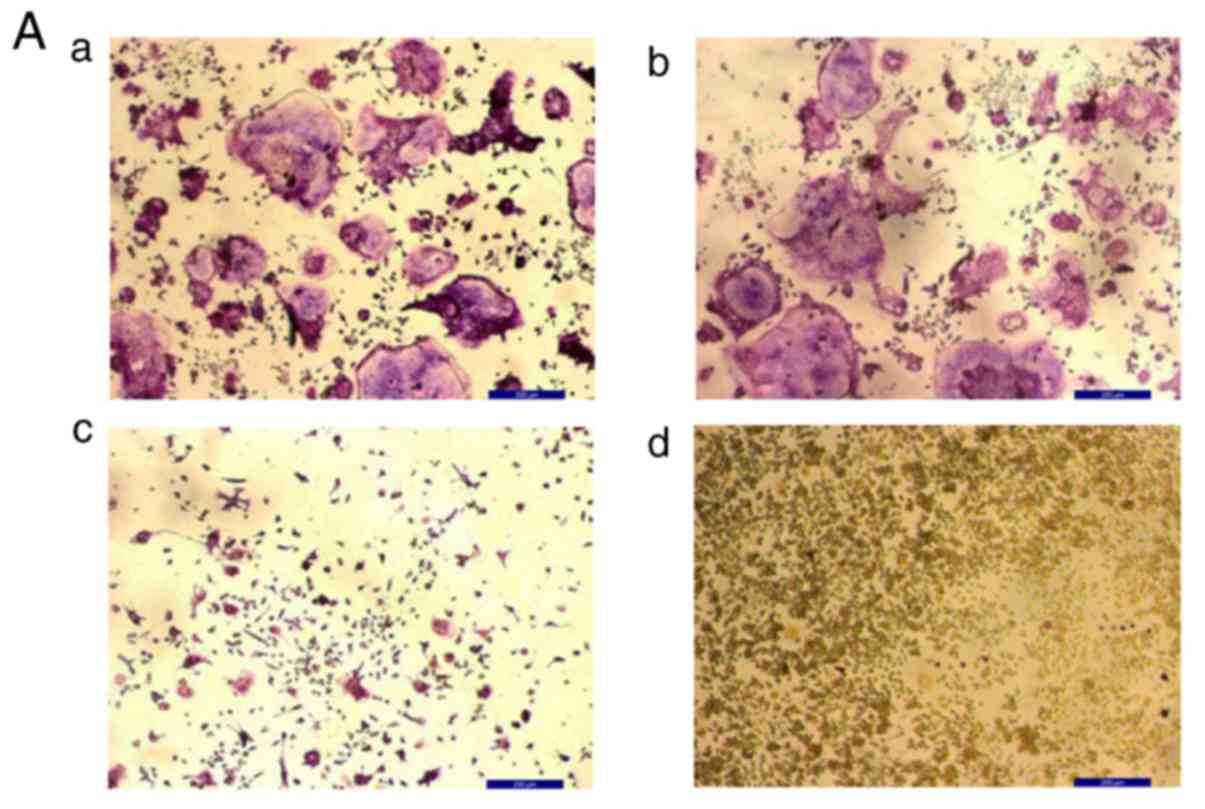 | Figure 2KN93 inhibits osteoclast
differentiation in M-CSF+RANKL-stimulated RAW 264.7 cells. (A)
M-CSF (25 ng/ml) + RANKL (30 ng/ml) with or without 10 µM
KN93 was added to RAW 264.7 cells. Following 96-h incubation, cells
were stained with TRAP and images were captured. a, M-CSF + RANKL +
DMSO; b, M-CSF + RANKL; c, M-CSF + RANKL + DMSO + KN93; and d,
RAW264.7 cells. (B) TRAP+ multinucleated cells were counted and
compared. The results are expressed as mean ± standard error of the
mean. **P<0.01 vs. M-CSF + RANKL-stimulated RAW 264.7
cells. Original magnification, ×100. Scale bar=200 µm.
M-CSF, macrophage colony-stimulating factor; RANKL, receptor
activator of nuclear factor kappa-light-chain-enhancer of activated
B cells ligand; DMSO, dimethyl sulfoxide; TRAP, tartrate-resistant
acid phosphatase; TRAP+, TRAP-positive. |
KN93 inhibits the activation of
osteoclasts
The effect of KN93 on M-CSF + RANKL-induced
osteoclast activation was additionally examined. RAW 264.7 cells
were stimulated with M-CSF + RANKL in order to induce
differentiation into functionally-active osteoclasts with
bone-resorbing capacities. The lacunae were shaped on the bottoms
of the COAS in the M-CSF + RANKL and the DMSO + M-CSF + RANKL
groups, whereas pit formation was evident in the KN93 + M-CSF +
RANKL group (Fig. 3A). The
resorption lacunae area is presented in Fig. 3B. The area of the KN93 + M-CSF +
RANKL group (4,978.26±2,270.20 µm2) was
significantly decreased compared with that of the M-CSF + RANKL
group (105,653.3±30,017.07 µm2) (P<0.01).
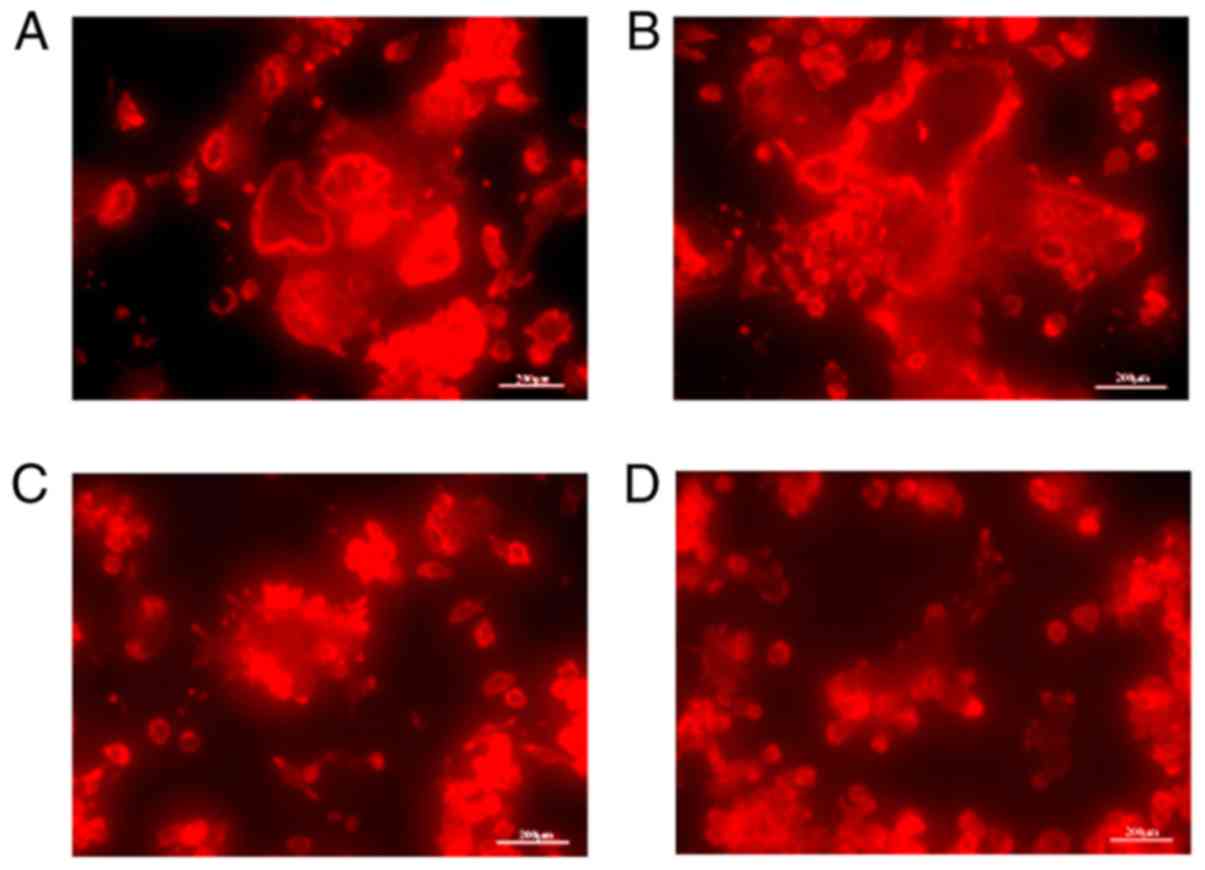
Effect of KN93 on the F-actin shape in
M-CSF + RANKL-treated RAW264.7 cells
The results revealed that intact and regular F-actin
rings were evident in the M-CSF + RANKL + DMSO group (Fig. 4A). Additionally, in the M-CSF +
RANKL-treated cell population, the majority of differentiated
osteoclasts were characterized by cells with a rounded morphology
that contained clearly visible F-actin rings (Fig. 4B). Nevertheless, treatment with 10
µM KN93 inhibited the formation of F-actin rings in the
M-CSF + RANKL-treated RAW264.7 cells (Fig. 4C).
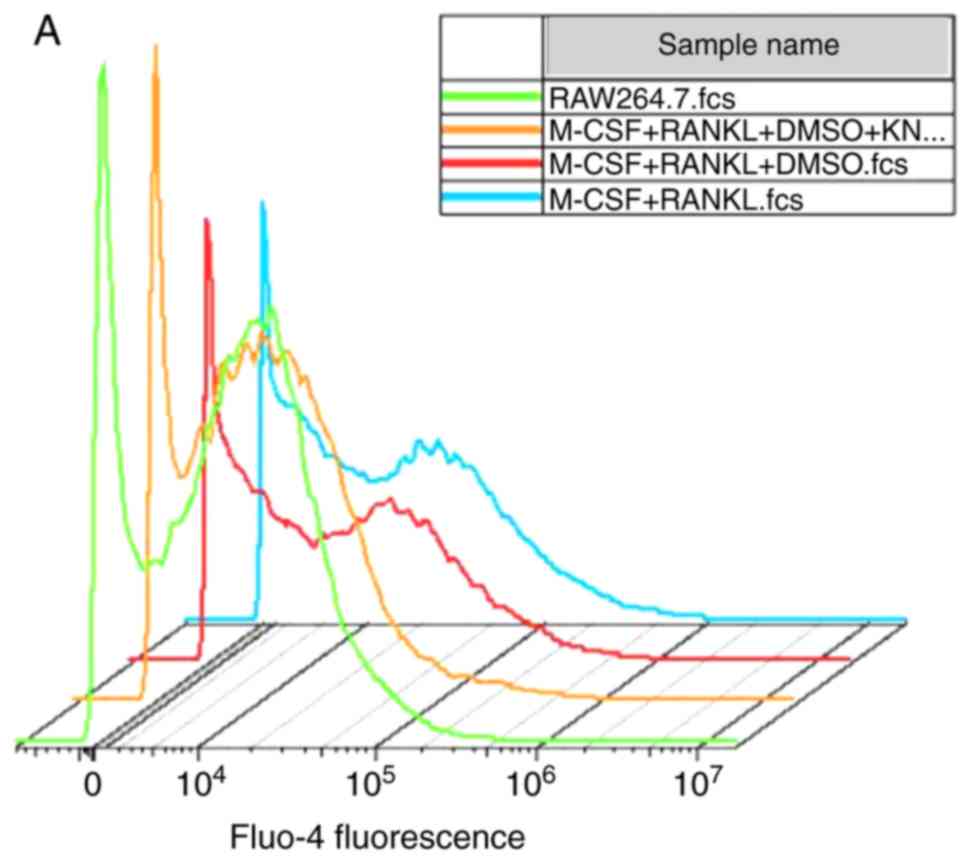
KN93 affects the levels of
[Ca2+]i in osteoclast differentiation
The concentration of [Ca2+]i
was indirectly elucidated according to the fluorescence intensity
of Fluo-4-AM. No significant difference in the concentration of
[Ca2+]i in M-CSF + RANKL-induced RAW264.7
cells compared with M-CSF + RANKL + DMSO-treated cells was
observed. Concomitantly, the level of [Ca2+]i
in the M-CSF + RANKL + DMSO + KN93 group was significantly
decreased compared with that of the M-CSF + RANKL group (P<0.05;
Fig. 5).
TRAP, MMP-9 and Cathepsin K expression in
KN93 + M-CSF + RANKL-treated osteoclast cells The gene expression
levels of TRAP, MMP-9 and Cathepsin K were examined by RT-qPCR
No significant difference in the expression levels
of TRAP, MMP-9 and Cathepsin K mRNA between the M-CSF + RANKL +
DMSO and the M-CSF + RANKL groups was observed. Furthermore, the
expression levels of TRAP in the M-CSF + RANKL + DMSO +
KN93-treated cells were significantly decreased compared with the
M-CSF + RANKL groups (P<0.05). The expression levels of MMP-9 in
the M-CSF + RANKL + DMSO + KN93-treated cells were significantly
decreased compared with the M-CSF + RANKL groups (P<0.05). The
expression levels of Cathepsin K in the M-CSF + RANKL + DMSO +
KN93-treated cells were significantly decreased compared with the
M-CSF + RANKL groups (P<0.01). Treatment of the cells with 10
µM KN93 decreased the expression levels of TRAP, MMP-9 and
Cathepsin mRNA to 0.57±0.07, 0.34±0.16 and 0.97±0.05-fold,
respectively, compared with the M-CSF + RANKL groups (Fig. 6).
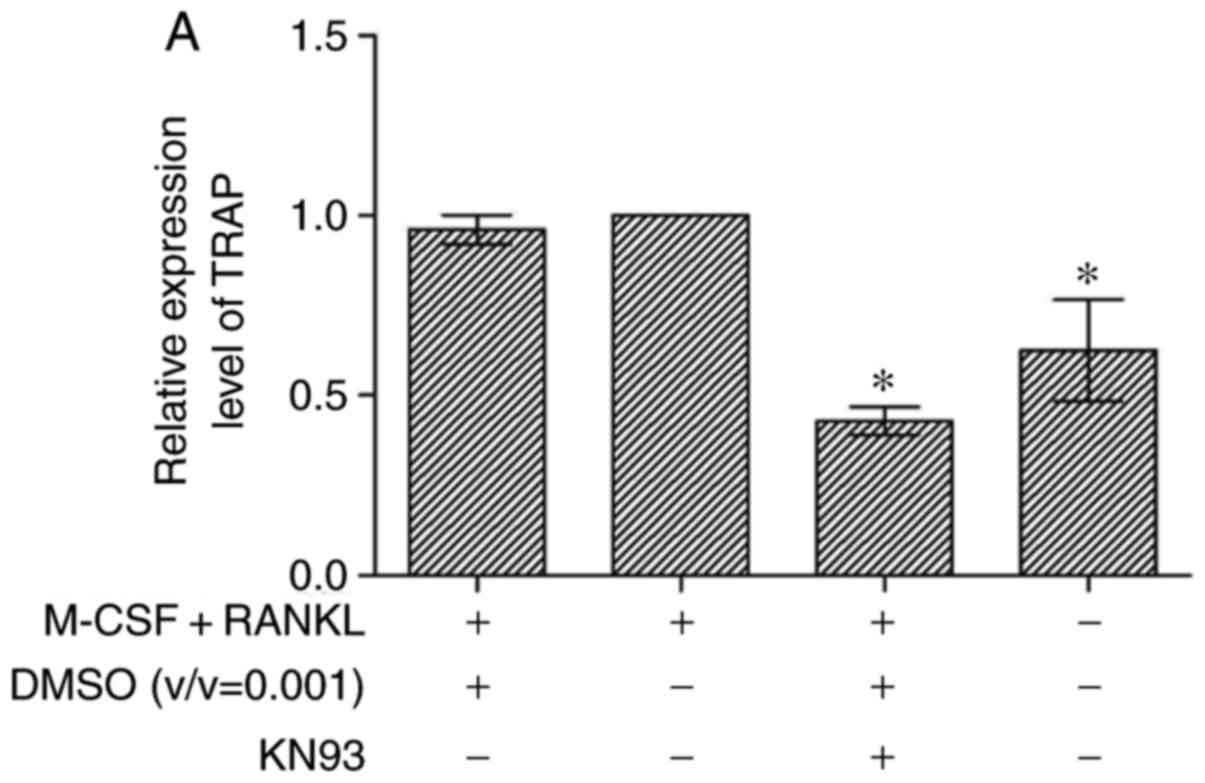 | Figure 6Effect of KN93 on the expression of
TRAP, MMP-9 and Cathepsin K genes. RAW264.7 cells were cultured in
minimum essential medium-α, and then seeded in 6-well culture
plates. After 4 h of culture, M-CSF + RANKL with or without KN93
were added. Following additional culture steps, total RNA were
collected, and mRNA expression levels of (A) TRAP, (B) MMP-9 and
(C) Cathepsin K genes were measured. The results are expressed as
mean ± standard error of the mean. *P<0.05 and
**P<0.01 vs. M-CSF + RANKL groups. M-CSF, macrophage
colony-stimulating factor; RANKL, receptor activator of nuclear
factor kappa-light-chain-enhancer of activated B cells ligand;
DMSO, dimethyl sulfoxide; TRAP, tartrate-resistant acid
phosphatase; MMP-9, matrix metalloproteinase 9. |
Effects of KN93 on the expression of
osteoclast-specific transcription factors
NFAT c1 (NFATc1), and AP-1 protein family members
c-Jun and c-Fos are considered to be the most crucial
osteoclast-specific transcription factors (21). The present study examined whether
these transcription factors were modulated by KN93 using western
blot analysis. Treatment of RAW264.7 cells with 10 µM KN93 +
M-CSF + RANKL significantly decreased the M-CSF + RANKL-stimulated
upregulation of the NFATc1, p-C-Jun, and p-C-Fos transcription
factors compared with the M-CSF + RANKL-treated cells (Fig. 7).
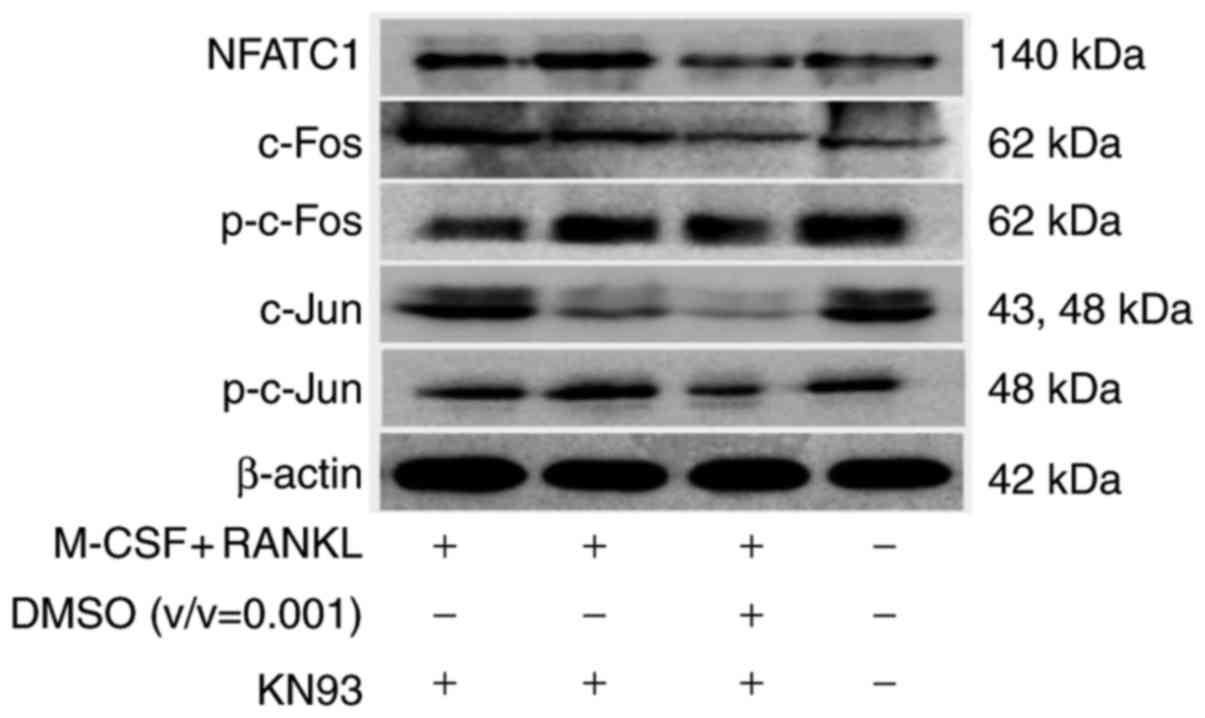 | Figure 7KN93 downregulates the expression of
osteoclast-specific transcription factors in M-CSF +
RANKL-stimulated RAW264.7 cells. RAW264.7 cells were cultured in
the presence of M-CSF + RANKL, and with or without 10 µM
KN93. Following culture for 96 h, total proteins were extracted and
equal amount of proteins were assayed by western blot analysis.
NFATC1, nuclear factor of activated T-cells, cytoplasmic 1; c-Fos,
proto-oncogene c-Fos; p, phosphorylated; c-Jun, Jun proto-oncogene;
M-CSF, macrophage colony-stimulating factor; RANKL, receptor
activator of nuclear factor kappa-light-chain-enhancer of activated
B cells ligand; DMSO, dimethyl sulfoxide. |
Discussion
M-CSF is critical in the survival and
differentiation of osteoclast precursor cells. M-CSF exerts its
anti-apoptotic function via the anti-apoptotic gene B-cell lymphoma
2 (Bcl-2). M-CSF may act through the stimulation of extracellular
signal regulated kinase (ERK) via growth factor receptor bound
protein 2, and the activation of protein kinase B (Akt) by the
phosphatidylinositol 3-kinase pathway and consequent promotion of
osteoclast precursor cell survival (22,23). In addition, M-CSF may induce the
expression of RANK in osteoclast precursor cells, and the
RANKL/RANK axis may serve an important role in the differentiation
of osteoclasts (24).
Phospholipase Cγ (PLCγ), MAP kinases and Akt are triggered via the
binding of RANKL to RANK. Activated PLCγ subsequently mediates the
expression of osteoclast-specific genes through downstream factors
(25).
The present study established a M-CSF +
RANKL-induced osteoclast model, which was characterized by
TRAP-positive staining, regular and intact F-actin rings and
vigorous resorption activity, as described previously (19). The concentration of
[Ca2+]i varies in the process of osteoclast
differentiation, and CaMKII kinase participates in osteoclast
differentiation (18). Based on
these data, the major role of the Ca2+ signaling
pathway, which is involved in osteoclast differentiation and
activation, was of interest. Therefore, whether the CaMKs inhibitor
KN93 affected the differentiation and activation of osteoclasts was
additionally investigated. The present study indicated that in the
M-CSF + RANKL-induced osteoclast system, KN93 inhibited the
differentiation of osteoclasts from their precursors, weakened the
resorption capacity of mature osteoclasts and caused disassembly of
the cytoskeletal structure. Concomitantly, KN93 downregulated the
M-CSF + RANKL-induced increase in [Ca2+]i
concentration in RAW264.7 cells and altered the expression levels
of the key genes TRAP, MMP-9 and Cathepsin K, and the key
transcription factors NFATc1, c-Fos and c-Jun, which are associated
with osteoclast differentiation and activation.
NFATc1 is a major osteoclastogenic transcription
factor, which is critical for osteoclast terminal differentiation.
It is indirectly activated by RANKL. RANK and its costimulatory
receptors, namely immunoreceptor tyrosine-based activation motif
proteins, coordinate to evoke PLCγ. Subsequently,
inositol-1,4,5-trisphosphate (IP3), which is produced by PLCγ, acts
on IP3-sensitive Ca2+ channels, which are located on the
surface of the endoplasmic reticulum. This give rise to
Ca2+ excretions from the endoplasmic reticulum and
intracellular Ca2+ levels increase rapidly in a short
time period. Subsequently, calcineurin is activated, and thereby
NFATc1 is stimulated and translocated to the nucleus (26). NFATc1 binds to the
osteoclast-specific gene promoter region and promotes gene
expression. Eventually, the osteoclast differentiation and
activation properties are attributed to the osteoclast-specific
gene expression (27–29). Ca2+ is a secondary
messenger, which is mediated by Ca2+-binding proteins,
including calmodulin. Following transient increases in
[Ca2+]i, Ca2+ interacts with
calmodulin and undergoes a conformational change, which leads to
the activation of CaMKs and the phosphatase calcineurin (26).
CaMKs act via the Ca2+-CaMK-cyclic
AMP-responsive element-binding protein (CREB) pathway and the
downstream factor AP-1 in order to regulate the early stages of
osteoclast differentiation (7).
Concomitantly, activated CaMKs activate the ERK-MAPK signaling
pathway in order to mediate the differentiation and activation of
osteoclasts (18). KN93 is an
inhibitor of CaMKs, and a previous study demonstrated that KN93
inhibited the activation of CaMKs in differentiated osteoclasts
(18). KN93 decreased the number
of TRAP-positive multinuclear cells (14). However, to the best of our
knowledge, a detailed and comprehensive investigation has not been
conducted to date.
TRAP staining is one of the most frequently used
methods to distinguish the differentiation degree of osteoclasts.
The results of the present study indicated that KN93 markedly
inhibited the M-CSF + RANKL-induced osteoclastogenesis. These data
were in accordance with a previous study (14). Additionally, KN93 restricted the
activation of M-CSF + RANKL-induced RAW264.7 cells, which
manifested in weak bone resorption ability and inhibited the
formation of an intact F-actin ring. Healthy F-actin rings are
considered to be crucial for osteoclast bone eroding capacity
(21). In addition, the property
of resorbing mineralized calcium apatite and/or carbonate
substrates including bone, dentin and nacre is the most reliable
criterion for the identification of osteoclast function (30). During osteoclastogenesis,
Ca2+ oscillation is long-lasting, which may ensure the
sustained induction of NFATc1 autoamplification. Therefore,
intracellular Ca2+ is a crucial factor in
osteoclastogenesis (26).
NFATc1 is activated by sustained and low
Ca2+ activation (31),
and a low Ca2+ threshold with regard to the constructive
stimulation of transcription factors is decreased by
Ca2+ oscillations (32,33). Long-lasting Ca2+
oscillation is considered to contribute to the prolonged nuclear
localization of NFATc1 and ensures its sustained transcriptional
activation (34). This process is
required for cell terminal differentiation, including the
differentiation of osteoclasts (34). In the present study,
[Ca2+]i levels in M-CSF + RANKL induced RAW
264.7 cells were decreased by KN93. KN93 may downregulate
[Ca2+]i levels and inhibit Ca2+
oscillation, therefore leading to an inhibition of osteoclast
differentiation.
The product of TRAP gene expression is considered to
be the phenotypic marker of differentiated osteoclasts (35). In addition, TRAP serves a pivotal
role in the process of osteoclast bone resorption (36). Previously, the mechanism of
osteoclastic bone resorption activity has been investigated more
clearly. Mineralized bone matrix degradation primarily includes two
processes, namely disintegration of crystalline hydroxyapatite and
dissolution of organic matrix proteolytic activity (37). Carbonic anhydrase II is primarily
responsible for the process of inorganic matrix degradation and the
maintenance of the normal intracellular pH in the resorption area
between the osteoclast and the bone matrix (38,39). Subsequently, proteolytic enzymes
of the cysteine proteinase family, including Cathepsin K and the
MMPs family are activated for the organic matrix degradation
(40). In the present study, KN93
inhibited the expression of TRAP, MMP-9 and Cathepsin K, which
demonstrated that KN93 mediated osteoclast formation and function
at the molecular level. KN93 may exert its effects via the
Ca2+/calmodulin-dependent protein kinase signaling
pathway. As aforementioned, NFATc1 localizes in the nucleus and
regulates the expression of osteoclast-specific genes, including
TRAP, Cathepsin K, MMP-9, osteoclast-associated receptor and
calcitonin receptor.
AP-1 is crucial for osteoclastogenesis and consists
of 3 protein family members, namely Fos (c-Fos, FosB, Fra-1,
Fra-2), Jun (c-Jun, JunB, JunD), and activating transcription
factor (ATF; ATFa, ATF2, ATF4, B-ATF) (41). Notably, c-Fos in the Fos family
contributes to osteoclastogenesis to a major extent. The expression
of c-Fos is inhibited by CaMKs and CREB (13,26). During osteoclastogenesis, the Jun
family members may replace each other (42). During the differentiation of the
osteoclasts, NFATc1 is activated by c-Fos, and the sensitization of
the Ca2+-Calcineurin-NFATc1 pathway is critical for this
process (43). In the present
study, western blot analysis suggested that KN93 decreased the
levels of NFATc1, p-C-Jun and p-C-Fos during the process of
osteoclastogenesis. Therefore, we hypothesize that KN93 negatively
regulates osteoclastogenesis via the
Ca2+/calmodulin-dependent protein kinase signaling
pathway.
Apart from KN93, there are other potential factors
associated with the regulation of osteoclast differentiation and
activation, including the mitogen-activated protein kinase pathway
inhibitors SP600125, U0126 and SB203580, which will be the focus of
subsequent investigations.
Taken collectively, the data from the present study
suggest that KN93 inhibited the formation of TRAP-positive
multi-nucleated cells, restricted the shaping of F-actin rings and
decreased the resorption activity of osteoclasts. Additional
analyses indicated that KN93 may act through the
Ca2+/calmodulin-dependent protein kinase signaling
pathway, which regulates osteoclastogenesis. The data also
demonstrated that KN93 decreased the concentration of
[Ca2+]i, the expression levels of
osteoclast-specific genes and the protein levels of critical
transcription factors in the M-CSF + RANKL-induced osteoclast
model.
To the best of our knowledge, the present study
demonstrated for the first time that KN93, an inhibitor of CaMKs,
may directly affect the differentiation and activation of
osteoclasts via the Ca2+/calmodulin-dependent protein kinase
signaling pathway. Additional studies are required to elucidate the
detailed mechanisms of these processes. The results therefore
suggested that KN93 may represent a promising therapeutic agent for
the treatment of diseases associated with abnormal osteoclast
activity, such as osteoporosis and osteopetrosis.
Acknowledgments
Not applicable.
Funding
This study was supported by the Project of Natural
Science Foundation of Anhui Province (grant no., 1508085QH172), the
Natural Science Research Project of the Universities in Anhui
(grant nos., KJ2017A225 and KJ2017A237), the Natural Science
Foundation of the Bengbu Medical College (grant no. BYKY1614ZD) and
the National Training Programs of the Innovation and
Entrepreneurship for Undergraduate (grant nos. 201410367029 and
201610367004).
Availability of data and materials
The datasets used and/or analysed during the current
study are available from the corresponding author on reasonable
request.
Authors’ contributions
FX cultured the cells, helped to design the study,
analysed and interpreted the results, and was a major contributor
in the writing of the manuscript. DN and WS performed the MTT assay
and tartrate-resistant acid phosphatase staining. QY, WW and BT
conducted the osteoclast resorption activity assay and (F)-actin
staining. ZL, DZ and YM performed the intracellular Ca2+
concentration quantification and reverse
transcription-quantification polymerase chain reaction. CL, XL, SY
and SXu performed the western blot analysis. YF and XSun cultured
the cells. CC provided experimental ideas, analysed and interpreted
the results, and was a major contributor in the writing of the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
pNot applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kikuta J and Ishii M: Intravital bone
imaging: osteoclast. Clin Calcium. 28:211–216. 2018.In
Japanese.
|
|
2
|
Verma SK, Leikin E, Melikov K, Gebert C,
Kram V, Young MF, Uygur B and Chernomordik LV: Cell-surface
phosphatidylserine regulates osteoclast precursor fusion. J Biol
Chem. 293:254–270. 2018. View Article : Google Scholar
|
|
3
|
Cafiero C, Gigante M, Brunetti G, Simone
S, Chaoul N, Oranger A, Ranieri E, Colucci S, Pertosa GB, Grano M
and Gesualdo L: Inflammation induces osteoclast differentiation
from peripheral mononuclear cells in chronic kidney disease
patients: Crosstalk between the immune and bone systems. Nephrol
Dial Transplant. 33:65–75. 2018. View Article : Google Scholar
|
|
4
|
Tanaka S, Nakamura I, Inoue J, Oda H and
Nakamura K: Signal transduction pathways regulating osteoclast
differentiation and function. J Bone Miner Metab. 21:123–133. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Blair HC, Robinson LJ and Zaidi M:
Osteoclast signalling pathways. Biochem Biophys Res Commun.
328:728–738. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Del Fattore A, Teti A and Rucci N:
Osteoclast receptors and signaling. Arch Biochem Biophys.
473:147–160. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Seales EC, Micoli KJ and McDonald JM:
Calmodulin is a critical regulator of osteoclastic differentiation,
function, and survival. J Cell Biochem. 97:45–55. 2006. View Article : Google Scholar
|
|
8
|
Berridge MJ, Bootman MD and Roderick HL:
Calcium signalling: Dynamics, homeostasis and remodelling. Nat Rev
Mol Cell Biol. 4:517–529. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wagner E: Functions of AP1 (Fos/Jun) in
bone development. Ann Rheum Dis. 61:ii40–ii42. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Takayanagi H, Kim S, Koga T, Nishina H,
Isshiki M, Yoshida H, Saiura A, Isobe M, Yokochi T, Inoue J, et al:
Induction and activation of the transcription factor NFATc1 (NFAT2)
integrate RANKL signaling in terminal differentiation of
osteoclasts. Dev Cell. 3:889–901. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Linseman DA, Bartley CM, Le SS, Laessig
TA, Bouchard RJ, Meintzer MK, Li M and Heidenreich KA: Inactivation
of the myocyte enhancer factor-2 repressor histone deacetylase-5 by
endogenous Ca(2+)/calmodulin-dependent kinase II promotes
depolarization-mediated cerebellar granule neuron survival. J Biol
Chem. 278:41472–41481. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yao CH, Zhang P and Zhang L: Differential
protein and mRNA expression of CaMKs during osteoclastogenesis and
its functional implications. Biochem Cell Biol. 90:532–539. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sato K, Suematsu A, Nakashima T,
Takemoto-Kimura S, Aoki K, Morishita Y, Asahara H, Ohya K,
Yamaguchi A, Takai T, et al: Regulation of osteoclast
differentiation and function by the CaMK-CREB pathway. Nat Med.
12:1410–1416. 2006. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chang EJ, Ha J, Huang H, Kim HJ, Woo JH,
Lee Y, Lee ZH, Kim JH and Kim HH: The JNK-dependent CaMK pathway
restrains the reversion of committed cells during osteoclast
differentiation. J Cell Sci. 121:2555–2564. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bone HG, McClung MR, Roux C, Recker RR,
Eisman JA, Verbruggen N, Hustad CM, DaSilva C, Santora AC and Ince
BA: Odanacatib, a cathepsin-K inhibitor for osteoporosis: A
two-year study in postmenopausal women with low bone density. J
Bone Miner Res. 25:937–947. 2010.
|
|
16
|
Stoch S and Wagner J: Cathepsin K
inhibitors: A novel target for osteoporosis therapy. Clin Pharmacol
Ther. 83:172–176. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Missbach M, Jeschke M, Feyen J, Müller K,
Glatt M, Green J and Susa M: A novel inhibitor of the tyrosine
kinase Src suppresses phosphorylation of its major cellular
substrates and reduces bone resorption in vitro and in rodent
models in vivo. Bone. 24:437–449. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Fu YX, Gu JH, Wang Y, Yuan Y, Liu XZ, Bian
JC and Liu ZP: Involvement of the Ca2+ signaling pathway
in osteoprotegerin inhibition of osteoclast differentiation and
maturation. J Vet Sci. 16:151–156. 2015. View Article : Google Scholar :
|
|
19
|
Fu YX, Gu JH, Zhang YR, Tong XS, Zhao HY,
Yuan Y, Liu XZ, Bian JC and Liu ZP: Osteoprotegerin influences the
bone resorption activity of osteoclast. Int J Mol Med.
31:1411–1417. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(−delta delta C(t)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
21
|
Ono T and Nakashima T: Recent advances in
osteoclast biology. Histochem Cell Biol. 149:325–341. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pixley FJ and Stanley ER: CSF-1 regulation
of the wandering macrophage: Complexity in action. Trends Cell
Biol. 14:628–638. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ross FP and Teitelbaum SL: avb3 and
macrophage colony-stimulating factor: Partners in osteoclast
biology. Immunol Rev. 208:88–105. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Arai F, Miyamoto T, Ohneda O, Inada T,
Sudo T, Brasel K, Miyata T, Anderson DM and Suda T: Commitment and
differentiation of osteoclast precursor cells by the sequential
expression of c-Fms and receptor activator of nuclear factor kappaB
(RANK) receptors. J Exp Med. 190:1741–1754. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lacey DL, Timms E, Tan HL, Kelley MJ,
Dunstan CR, Burgess T, Elliott R, Colombero A, Elliott G, Scully S,
et al: Osteoprotegerin ligand is a cytokine that regulates
osteoclast differentiation and activation. Cell. 93:165–176. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Negishi-Koga T and Takayanagi H:
Ca2+-NFATc1 signaling is an essential axis of osteoclast
differentiation. Immunol Rev. 231:241–256. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lorenzo J, Horowitz M and Choi Y:
Osteoimmunology: Interactions of the bone and immune system. Endocr
Rev. 29:403–440. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zeng X, Zhang Y, Wang S, Wang K, Tao L,
Zou M, Chen N, Xu J, Liu S and Li X: Artesunate suppresses
RANKL-induced osteoclastogenesis through inhibition of
PLCγ1-Ca2+-NFATc1 signaling pathway and prevents
ovariectomy-induced bone loss. Biochem Pharmacol. 124:57–68. 2017.
View Article : Google Scholar
|
|
29
|
Soysa NS, Alles N, Aoki K and Ohya K:
Osteoclast formation and differentiation: An overview. J Med Dent
Sci. 59:65–74. 2012.PubMed/NCBI
|
|
30
|
Saltel F, Chabadel A, Bonnelye E and
Jurdic P: Actin cytoskeletal organisation in osteoclasts: A model
to decipher transmigration and matrix degradation. Eur J Cell Biol.
87:459–468. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Dolmetsch RE, Lewis RS, Goodnow CC and
Healy JI: Differential activation of transcription factors induced
by Ca2+ response amplitude and duration. Nature.
386:855–858. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Dolmetsch RE, Xu K and Lewis RS: Calcium
oscillations increase the efficiency and specificity of gene
expression. Nature. 392:933–936. 1998. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tomida T, Hirose K, Takizawa A, Shibasaki
F and Iino M: NFAT functions as a working memory of Ca2+
signals in decoding Ca2+ oscillation. EMBO J.
22:3825–3832. 2008. View Article : Google Scholar
|
|
34
|
Koga T, Matsui Y, Asagiri M, Kodama T, De
Crombrugghe B, Nakashima K and Takayanagi H: NFAT and Osterix
cooperatively regulate bone formation. Nat Med. 11:880–885. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Hofbauer LC, Neubauer A and Heufelder AE:
Receptor activator of nuclear factor-kappaB ligand and
osteoprotegerin: Potential implications for the pathogenesis and
treatment of malignant bone diseases. Cancer. 92:460–470. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Price CP, Kirwan A and Vader C:
Tartrate-resistant acid phosphatase as a marker of bone resorption.
Clin Chem. 41:641–643. 1995.PubMed/NCBI
|
|
37
|
Vaananen HK, Zhao H, Mulari M and Halleen
JM: The cell biology of osteoclast function. J Cell Sci.
113:377–381. 2000.PubMed/NCBI
|
|
38
|
Lehenkari P, Hentunen TA, Laitala-Leinonen
T, Tuukkanen J and Väänänen HK: Carbonic anhydrase II plays a major
role in osteoclast differentiation and bone resorption by effecting
the steady state intracellular pH and Ca2+. Exp Cell
Res. 242:128–137. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Teitelbaum SL, Tondravi MM and Ross FP:
Osteoclasts, macrophages, and the molecular mechanisms of bone
resorption. J Leuk Biol. 61:381–388. 1997. View Article : Google Scholar
|
|
40
|
Kusano K, Miyaura C, Inada M, Tamura T,
Ito A, Nagase H, Kamoi K and Suda T: Regulation of matrix
metalloproteinases (MMP-2, -3, -9, and -13) by interleukin-1 and
interleukin-6 in mouse calvaria: Association of MMP induction with
bone resorption. Endocrinology. 139:1338–1345. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Wagner EF and Eferl R: Fos/AP-1 proteins
in bone and the immune system. Immunol Rev. 208:126–140. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ikeda F, Nishimura R, Matsubara T, Tanaka
S, Inoue J, Reddy SV, Hata K, Yamashita K, Hiraga T, Watanabe T, et
al: Critical roles of c-Jun signaling in regulation of NFAT family
and RANKL-regulated osteoclast differentiation. J Clin Invest.
114:475–484. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Takayanagi H: The role of NFAT in
osteoclast formation. Ann N Y Acad Sci. 1116:227–237. 2007.
View Article : Google Scholar : PubMed/NCBI
|