Introduction
Breast cancer is the most common form of female
cancer worldwide (1). It is a
phenotypically and genetically complex disease; the progression and
development of breast cancer has been associated with many factors
(2). According to the report of
the World Health Organization, there were 1.68 million cases of and
522,000 mortalities due to breast cancer in 2012 (3). Chemotherapy is the principal
therapeutic method for patients with breast cancer (4); doxorubicin (DOX) is one of the most
effective chemotherapeutic drugs for the treatment of breast
cancer, which may induce regression of metastatic breast cancer
(5). However, the effectiveness
of DOX is hindered by the capacity of tumor cells to develop
resistance to anticancer therapies (6). Therefore, understanding the
mechanism of cancer-specific drug resistance may help to inhibit or
overcome this resistance in breast cancer (7).
The causes of cancer-specific drug resistance have
been associated with drug-induced karyotypic alterations, random
drug-induced mutational events and non-mutational alterations of
gene function (8–10). Additionally, studies have reported
another mechanism of non-mutational regulation of gene function
mediated by non-protein-coding RNAs (ncRNAs) (11,12). Long (l)ncRNAs are RNA molecules
with >200 nucleotides (13).
Extensive studies have suggested that lncRNAs serve key regulatory
roles in numerous biological processes, which are increasingly
recognized as biomarkers of numerous types of cancer, including
breast cancer (14,15). lncRNA cancer susceptibility
candidate 9 (CASC9), located in the human chromosome 8q21.11, was
originally reported to be abnormally expressed in esophageal
squamous cell carcinoma (16).
Recent studies indicated that CASC9 is associated with a variety of
cancer types, including gastric cancer and nasopharyngeal carcinoma
(17,18). The role of CASC9 in breast cancer
and drug-resistant breast cancer remains to be examined.
To the best of our knowledge, the effect of CASC9 on
DOX-resistant breast cancer cells was investigated for the first
time in the present study. Additionally, the potential mechanisms
of CASC9 in breast cancer drug-resistant cells were evaluated by
investigating the interactions between CASC9 and multidrug
resistance 1 (MDR1). The present study aimed to provide some
theoretical basis for the underlying mechanism of DOX-resistant
breast cancer.
Materials and methods
Tissue collection
Paired breast cancer and adjacent normal breast
tissues were obtained from 48 female patients (age 50±11 years)
undergoing surgical breast cancer resection between January 2012
and December 2013 at The First Affiliated Hospital, University of
South China (Hengyang, China). The clinicopathological data of
patients are presented in Table
I. The patients did not receive local or systemic treatment
prior to surgery. All of the resected tissues were stored at −80°C
until total RNA extraction. The pathological stage, grade, nodal
status and estrogen receptor status of samples were appraised by an
experienced pathologist. American Joint Committee on Cancer stages
were used to characterize the stages of patient samples. The
experiments were approved by the Research Ethics Committee of the
First Affiliated Hospital, University of South China; all patients
provided written informed consent.
 | Table IClinicopathological data of
patients. |
Table I
Clinicopathological data of
patients.
| Clinicopathological
feature | No. patients | Expression of CASC9
| P-value |
|---|
| Lower | Higher |
|---|
| Age, years | | | | 0.246 |
| ≤50 | 20 | 4 | 16 | |
| >50 | 28 | 3 | 25 | |
| Histological
grade | | | | 0.023 |
| Grade 1 and 2 | 33 | 10 | 23 | |
| Grade 3 | 15 | 4 | 11 | |
| Tumor size, cm | | | | 0.342 |
| ≤2 | 26 | 4 | 22 | |
| >2 | 22 | 3 | 19 | |
| Lymph node
metastasis | | | | 0.017 |
| Absent | 18 | 15 | 3 | |
| Present | 30 | 8 | 22 | |
| AJCC stage | | | | 0.188 |
| I and II | 26 | 3 | 23 | |
| III and IV | 22 | 4 | 18 | |
| Lymphovascular
invasion | | | | 0.034 |
| Absent | 20 | 3 | 17 | |
| Present | 28 | 6 | 22 | |
| ER expression | | | | 0.026 |
| Negative | 14 | 3 | 11 | |
| Positive | 34 | 6 | 28 | |
Cell lines and cell culture
The human breast adenocarcinoma MCF-7 (HTB-22™) and
MCF-7/doxorubicin (DOX) cell lines were cultured using Iscove's
modified Dulbecco's medium (Sigma-Aldrich; Merck KGaA, Darmstadt,
Germany) containing 40 Ag/ml gentamicin and 10% newborn calf serum
(HyClone; GE Healthcare Life Sciences, Logan, UT, USA) at 37°C in
an atmosphere with 5% CO2. MDA-MB-231 (HTB-26™),
MDA-MB-157 (HTB-24™), and MDA-MB-468 (HTB-132™) human breast cancer
cell lines were maintained in Dulbecco's modified Eagle's medium
(DMEM; Sigma-Aldrich; Merck KGaA) supplemented with 10% fetal
bovine serum (FBS; Gibco; Thermo Fisher Scientific, Inc., Waltham,
MA, USA), 100 U/ml penicillin, and 100 μg/ml streptomycin.
The normal human mammary epithelial cell line (MCF10A; CRL-10317™)
was cultured in DMEM. All cell lines were purchased from the
American Type Culture Collection (Manassas, VA, USA). The
drug-resistant variant (MCF-7/DOX) of the MCF-7 cell line was
induced by stepwise selection following prolonged (>6 months)
treatment of MCF-7 cells with increasing concentrations of DOX at a
range of (0.5–25 nmol/l) in the medium (19). Following 6 months of culturing in
the presence of DOX, the half-maximal inhibitory concentrations
(IC50) for the MCF-7/DOX and parental MCF-7 cells were
24 and 1 Amol/l DOX, respectively. Cells were seeded at a density
of 0.5×106 viable cells per 100-mm plate, and the medium
was replaced every other day for 6 days. Trypsinized cells were
washed with PBS and frozen at −80°C immediately until subsequent
analyses.
Vector construction and transfection
The silencing vector pS-CASC9 and empty pSilencer
[negative control (NC), termed si-NC] were purchased from Guangzhou
FitGene Biotechnology Co., Ltd. (Guangdong, China), and MCF-7/DOX
cells at a density of 1×105 were transfected using
Lipofectamine® 3000 reagent (Invitrogen; Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocols, following 24 h of
culturing. The cell lines that expressed the vectors stably were
selected using 400 μg/ml Geneticin (Invitrogen; Thermo
Fisher Scientific, Inc.) for 2 weeks. Knockdown was confirmed and
measured by reverse transcription-quantitative polymerase chain
reaction (RT-qPCR). Enhancer of zeste homolog 2 (EZH2), MDR1 and
control siRNAs were synthe-sized by Shanghai GenePharma Co., Ltd.
(Shanghai, China). The sequences were: Si-EZH2-1 sense,
GUGUAUGAGUUUAGAGUCATT-3′; si-MDR1 sense, CAGAAAGCUUAGUACCAAAdTdT;
and si-NC sense, UAACGACGCGACGACGUAAdTdT. Cells were seeded in a
6-well plate and cultured in antibiotic- and serum-free medium. At
60% confluence, cells were transfected using Oligofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.). Cells were refreshed
with regular medium after 4–6 h of transfection and subjected to
the measurement of knockdown efficiency using RT-qPCR.
To overexpress EZH2, the plasmid pcDNA-EZH2 was
constructed by introducing a BamHI-EcoRI fragment
containing the EZH2 cDNA into the same sites as pcDNA3.1. The
pcDNA-EZH2 plasmid was transfected into MCF-7/DOX and the cell line
stably expressing EZH2 was screened using G418.
RNA pull-down assay
The RNA pull-down assay was performed to identify
the CASC9-binding candidate using a Pierce Magnetic RNA-Protein
Pull-Down kit (Pierce; Thermo Fisher Scientific, Inc.). Briefly,
the target RNA and antisense control RNA were labeled with biotin
at the 3′ end and purified using a Pierce RNA 3′ End
Desthiobiotinylation kit (Thermo Fisher Scientific, Inc.). A
labeled RNA probe (50 pmol) was used to bind to streptavidin
magnetic beads following incubation in 1X RNA Capture buffer for 30
min at room temperature. Subsequently, 200 μg protein was
incubated in protein-RNA binding buffer for 150 min at 4°C with
agitation. The final RNA-magnetic bead-protein complexes were
washed three times with wash buffer. Subsequently, 12 μl
elution buffer was added to retrieve the pull-down protein
products. The retrieved proteins were resolved in gradient 4–12%
gel electrophoresis followed by mass spectrometry (MS)
identification. In detail, proteins precipitated by RNA pull-down
assays were subjected to NuPAGE 4–12% BisTris gel electrophoresis
and examined with silver stain using the Pierce Silver Stain kit
(cat. no. 24612; Pierce; Thermo Fisher Scientific, Inc.). Specific
bands only in the sense CASC9 lane were excised and analyzed by MS
(GeneScience Pharmaceuticals Co., Ltd., Beijing, China).
Cell survival assay
Cell survival was determined using an MTT assay.
Cells were plated in 96-well plates at 5×104 cells/well
and 20 μl of MTT solution (5 mg/ml; Sigma-Aldrich; Merck
KGaA) was added to each well for 4 h of incubation. The MTT
solution was then removed and 200 μl dimethyl sulfoxide
(Sigma-Aldrich; Merck KGaA) was added to dissolve the crystals.
Optical density was measured at a wavelength of 490 nm using a
microplate reader.
RT-Qpcr
Total RNA was isolated using the RNAiso™ Plus kit
(Takara Bio, Inc., Otsu, Japan), and 1 μg total RNA was
reverse-transcribed at 70°C into first-strand cDNA using a Takara
RNA PCR kit (AMV) v3.0 (Takara Biotechnology Co., Ltd., Dalian,
China). The primer sequences used were as follows: Human EZH2 gene
forward, 5′-GCCAGACTGGGAAGAAATCTG-3′ and reverse,
5′-TGTGCTGGAAAATCCAAGTCA-3′; MDR1 gene forward,
5′-CCCATCATTGCAATAGCAGG-3′ and reverse, 5′-GTTCAAACTTCTGCTCCTGA-3′;
and β-actin (internal control) forward, 5′-ACCCCCACTGAAAAAGATGA-3′
and reverse, 5′-ATCTTCAAACCTCATGATG-3′. Following heating to 94°C
for 2 min, the experimental reaction (50 μl) was subjected
to 32 cycles of 94°C for 30 sec, 61°C for 30 sec, and 72°C for 30
sec. The expression levels were calculated using the
2−ΔΔCq method (20).
Apoptosis analysis
Cells were trypsinized, and washed with cold PBS,
and subsequently suspended in PBS. The apoptotic cells were
detected by Annexin V and propidium iodide (PI) dual labeling using
an Annexin V-fluorescein isothiocyanate (FITC) kit (Beijing Biosea
Biotechnology Co., Ltd., Beijing, China), according to the
manufacturer's protocols. A total of 24 h following cell
transfection, breast cancer MCF-7 (HTB-22™) and MCF-7/DOX cells
were cultured in serum-free DMEM. The cells were harvested and
washed three times using PBS buffer (pH 7.4), and then resuspended
in staining buffer. Subsequently, 5 μl Annexin V-FITC and 5
μl PI was added into the cells and incubated at room
temperature for 10 min. The mixtures were analyzed using a FACScan
flow cytometer (BD Biosciences, Franklin Lakes, NJ, USA). Annexin
V-positive and PI-negative cells were considered to be apoptotic
cells. The apoptotic cells were analyzed using CellQuest software
(version 3.0; BD Biosciences).
Cell migration and invasion assays
For the migration assay, at 48 h post-transfection,
5×104 cells in serum-free medium were placed into the
upper chamber of an insert (8-mm pore size; EMD Millipore,
Billerica, MA, USA). For the invasion assay, 1×105 cells
in serum-free medium were placed into the upper chamber of an
insert coated with Matrigel. The lower chamber was filled with
medium containing 10% FBS. Following incubation at 37°C for 24 h,
the cells remaining on the upper membrane were removed with cotton
wool; cells migrating or invading through the membrane were stained
with methanol and 0.1% crystal violet at room temperature, and
imaged and counted using an IX71 inverted microscope at ×400
magnification (Olympus Corporation, Tokyo, Japan).
Western blot analysis
Cells were washed twice with ice-cold PBS, and lysed
using 2 ml lysis buffer (radioimmunoprecipitation assay buffer;
Sangon Biotech Co., Ltd., Shanghai, China). The supernatant was
collected following centrifugation at 6,000 x g for 15 min at 4°C
and cell lysates were matched for protein concentration using a
bicinchoninic acid protein assay kit (Pierce; Thermo Fisher
Scientific, Inc.). Protein samples (50 μg per lane) were
loaded onto a 10% SDS-PAGE gel and transferred onto nitrocellulose
membranes, and subsequently blocked in 5% non-fat milk at 4°C
overnight. The membrane was incubated with primary antibodies with
dilution 1:1,000 (Bcl2, cat. no. ab32124; Bax, cat. no. ab32503;
caspase-3, cat. no. ab32150; caspase-9, cat. no. ab138412; and
EZH2, cat. no. ab191250; all from Abcam, Cambridge, UK) for 2 h at
room temperature, and subsequently incubated with HRP-conjugated
secondary antibodies (cat. no. ab6721; dilution 1:5,000; Abcam) for
1 h at room temperature. The bands were visualized using enhanced
chemiluminescence substrates (Thermo Fisher Scientific, Inc.).
Statistical analysis
All experiments were independently repeated three
times. Values are presented as the mean ± standard deviation. SPSS
16.0 software (SPSS, Inc., Chicago, IL, USA) was used for
statistical analysis. The analysis of multiple groups was performed
with one-way analysis of variance with Dunnett's post hoc test.
P<0.05 was considered to indicate a statistically significant
difference.
Results
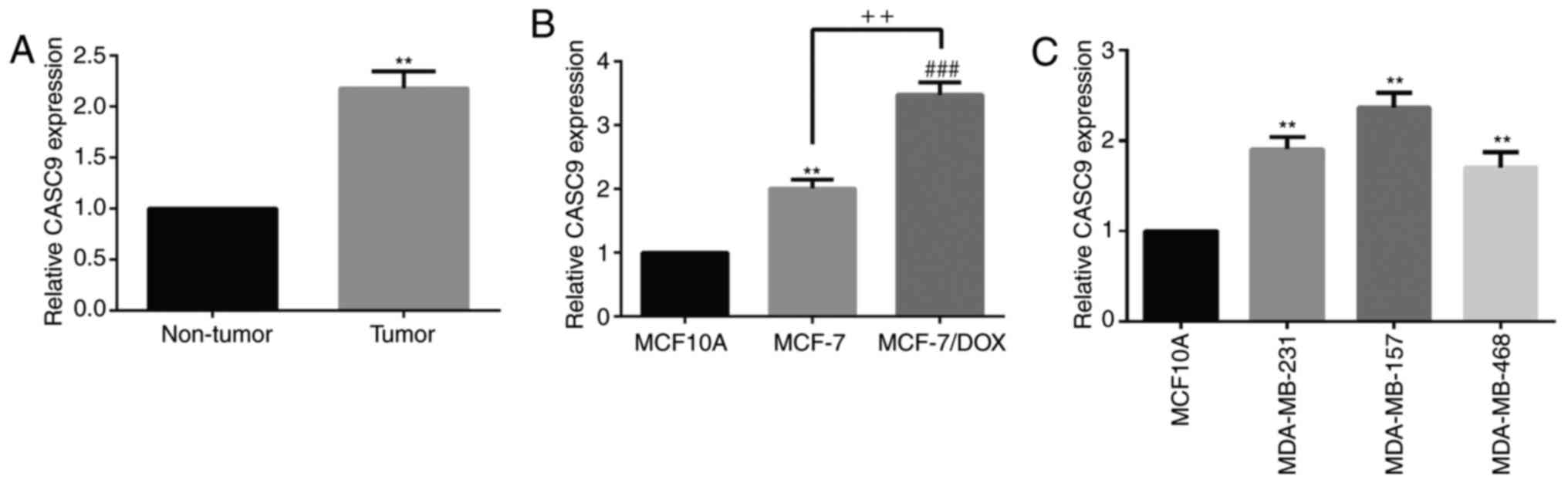
CASC9 is upregulated in breast cancer
tissues and breast cancer drug-resistant cell lines
To investigate the effects of CASC9 on breast
cancer, the expression of CASC9 in breast cancer tissues and cell
lines was detected. The results of the RT-qPCR analysis revealed
that CASC9 was significantly upregulated in breast cancer tissues
(Fig. 1A) and cell lines (MCF-7,
MCF-7/DOX, MDA-MB-231, MDA-MB-157 and MDA-MB-468) compared with in
adjacent normal tissues and normal human mammary epithelial cells
MCF10A (Fig. 1B and 1C). These results suggested that CASC9
may serve an important role in the pathogenesis and mechanism of
drug-resistance in breast cancer.
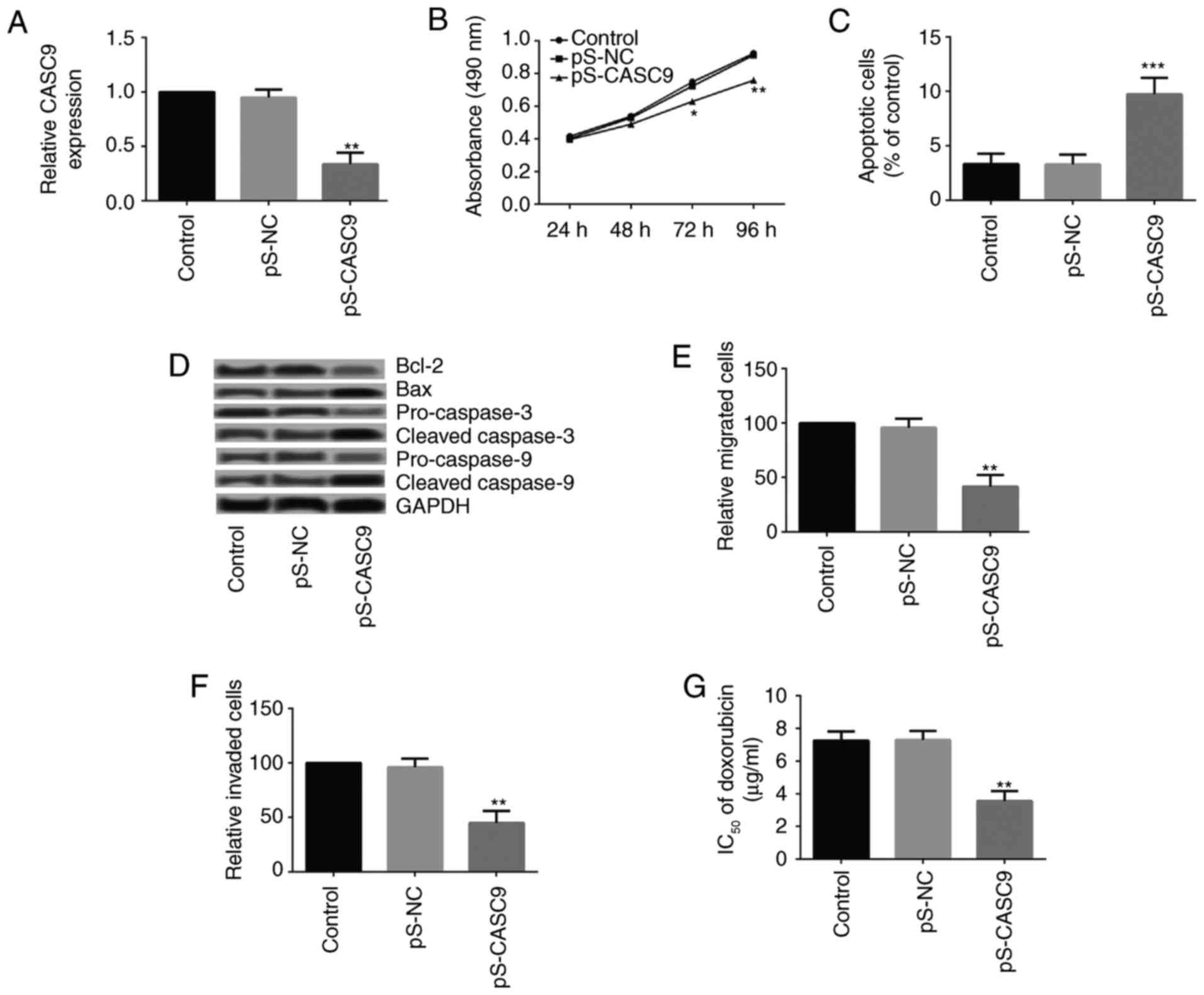
CASC9 knockdown inhibits the growth,
metastasis and chemoresistance of drug-resistant breast cancer
cells
To study whether abnormal expression of CASC9
affected the growth and metastasis of drug-resistant breast cancer
cells, MCF-7/DOX cells that stably expressed a CASC9 silencing
vector were established. The knockdown of CASC9 was confirmed by
RT-qPCR (Fig. 2A). Subsequently,
the cell viability, apoptosis, migration and invasion of MCF-7/DOX
cells following CASC9 knockdown was investigated. As presented in
Fig. 2B, CASC9 knockdown
significantly inhibited the cell viability of MCF-7/DOX cells from
72 h following cell culture. The results of the flow cytometry
demonstrated that CASC9 knockdown significantly increased the
apoptosis of MCF-7/DOX cells (Fig.
2C). The expression levels of apoptosis-associated proteins
[apoptosis regulator Bcl-2 (Bcl-2), apoptosis regulator BAX (Bax),
caspase-3 and caspase-9] are presented in Fig. 2D. The expression levels of Bcl-2,
pro-caspase-3 and pro-caspase-9 decreased significantly following
CASC9 knockdown, whereas Bax, cleaved-caspase-3 and
cleaved-caspase-9 increased markedly. A Transwell assay revealed
that CASC9 knockdown in MCF-7/DOX cells significantly impeded cell
migration and invasion (Fig. 2E
and 2F).
In addition, the IC50 of DOX in MCF-7/DOX
cells following CASC9 knockdown was detected. CASC9 knockdown in
MCF-7/DOX cells significantly decreased the IC50 of DOX
(Fig. 2G).
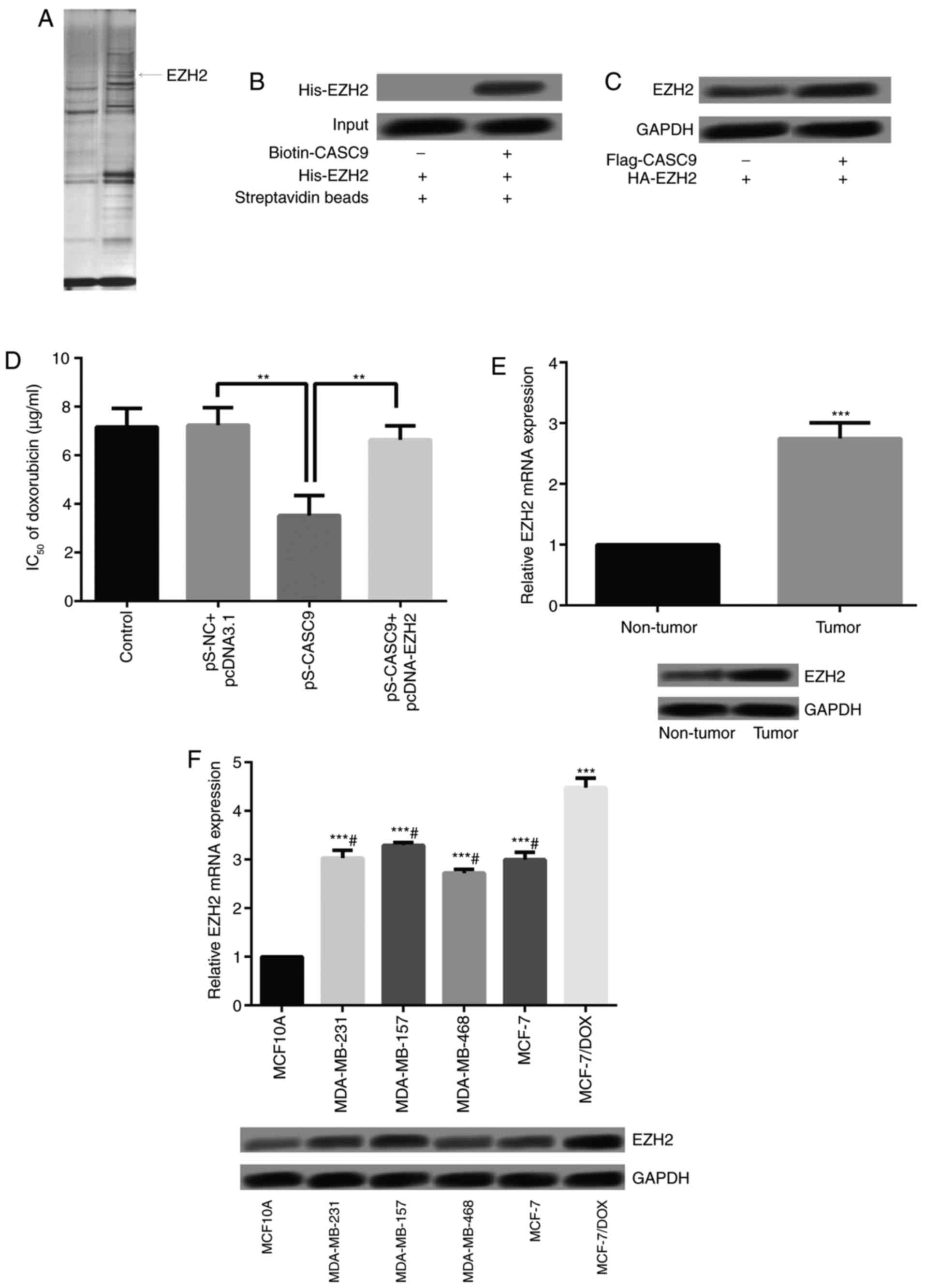
Identification of EZH2 as a CASC9 binding
protein
To further understand the underlying mechanism of
CASC9 in breast cancer cells, a protein that potentially interacts
with CASC9 was investigated using RNA pull-down assays followed by
MS (Fig. 3A). EZH2 is a
transcriptional repressor, which has been reported to be a marker
of aggressive breast cancer (21); EZH2 may interact with CASC9.
Therefore, CASC9 was conjugated to D-Biotin, and the associations
between CASC9 and EZH2 were analyzed using recombinant EZH2 via the
in vitro streptavidin-binding assay. As a result, EZH2 was
demonstrated to be a binding protein of CASC9 (Fig. 3B). To investigate whether CASC9
affected the stability of EZH2, MCF-7 cells were transfected with
Flag-CASC9 and the expression of EZH2 was detected. As presented in
Fig. 3C, overexpressed CASC9 may
significantly increase the protein expression of EZH2. Furthermore,
the IC50 of DOX in MCF-7/DOX cells following CASC9
knockdown and EZH2 overexpression was determined. CASC9 knockdown
significantly decreased the IC50 of DOX (Fig. 3D). Conversely, EZH2 overexpression
reversed the inhibitory effect of CASC9 knockdown on the
IC50 of DOX, which suggested that CASC9 promoted
DOX-resistance by binding with EZH2.
EZH2 expression is increased in breast
cancer tissues and breast cancer drug-resistant cell lines
The expression levels of EZH2 in breast cancer
tissues and cells was detected. As presented in Fig. 3E, EZH2 expression levels were
significantly higher in breast cancer tissues compared with in the
adjacent normal tissues. Additionally, EZH2 expression within the
breast cancer cells (MDA-MB-231, MDA-MB-157 and MDA-MB-468) and
MCF-7/DOX cells was significantly higher compared with in MCF10A
cells. Additionally, compared with MCF-7, MDA-MB-231, MDA-MB-157
and MDA-MB-468, EZH2 expression in MCF-7/DOX was significantly
increased (Fig. 3F).
Effects of EZH2 siRNA on the cell growth,
metastasis and chemoresistance of drug-resistant breast cancer
cells
To investigate whether the abnormal expression of
EZH2 affected the growth and metastasis of drug-resistant breast
cancer cells, MCF-7/DOX cells were transfected with EZH2 siRNA. The
expression levels of EZH2 decreased significantly post-transfection
(Fig. 4A and B). Cell viability,
apoptosis, migration and invasion following EZH2 silencing were
detected. As Presented in Fig.
4C, EZH2 silencing significantly inhibited the viability of
MCF-7/DOX cells from 72 h following cell culture. Flow cytometry
revealed that EZH2 silencing significantly increased the apoptotic
rate of MCF-7/DOX cells (Fig.
4D). Additionally, the expression levels of Bcl-2,
pro-caspase-3 and pro-caspase-9 decreased markedly, while those of
Bax, cleaved-caspase-3 and cleaved-caspase-9 increased markedly
following EZH2 knockdown (Fig.
4E). The Transwell assay revealed that EZH2 silencing
significantly inhibited cell migration and invasion (Fig. 4F and G). The IC50 of
MCF-7/DOX cells to DOX following EZH2 knockdown were investigated;
EZH2 knockdown in MCF-7/DOX cells decreased the IC50 of
DOX significantly (Fig. 4H).
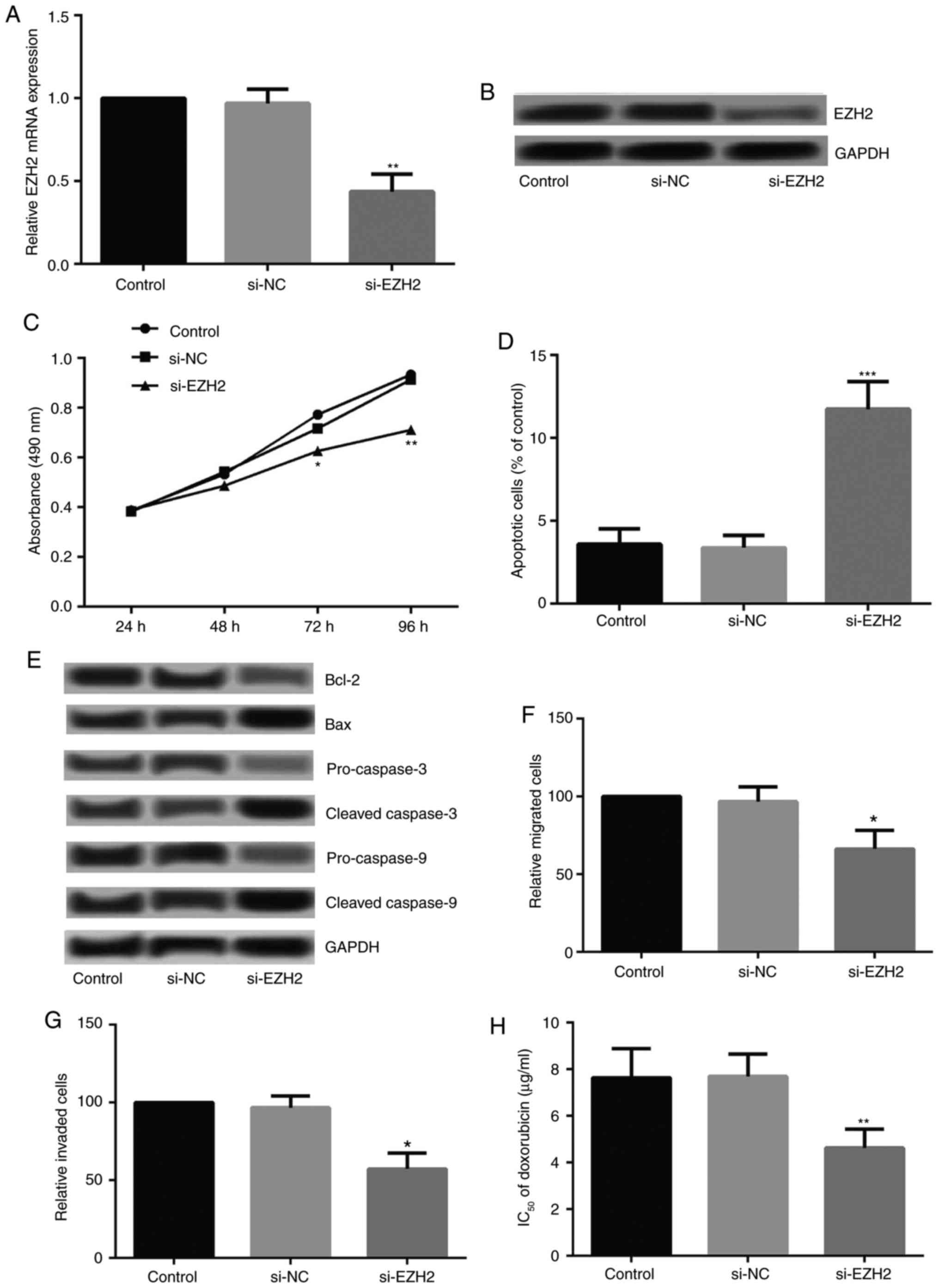 | Figure 4Effects of EZH2 on the growth and
metastasis of drug-resistant breast cancer cells MCF-7/DOX.
Relative expression levels of EZH2 following cell transfection,
confirmed by (A) reverse transcription-quantitative polymerase
chain reaction and (B) western blotting. Alterations in the (C)
cell viability and (D) apoptosis of MCF-7/DOX following EZH2
silencing. (E) Expression levels of apoptosis-associated proteins
(Bcl-2, Bax, caspase-3 and caspase-9) in MCF-7/DOX following EZH2
silencing. Alterations in the (F) cell migration and (G) invasion
abilities of MCF-7/DOX cells following EZH2 silencing. (H)
Alterations in the IC50 of DOX in MCF-7/DOX cells
following EZH2 silencing. *P<0.05,
**P<0.01 and ***P<0.001 vs. the control
group. Bcl-2, apoptosis regulator Bcl-2; Bax, apoptosis regulator
BAX; DOX, doxorubicin; EZH2, enhancer of zeste homolog; NC,
negative control; si, small interfering RNA. |
Effects of EZH2 siRNA on the expression
of MDR1/P-glycoprotein (P-gp)
Previously, it was suggested that the multidrug
resistance of tumors is associated with the MDR1 gene (22). Therefore, the expression levels of
the MDR1 gene and its encoded protein P-gp in drug-resistant breast
cancer cells were detected. The expression levels of MDR1/P-gp in
MCF-7/DOX cells increased significantly compared with in MCF-7,
MDA-MB-231, MDA-MB-157 and MDA-MB-468 cells (Fig. 5A).
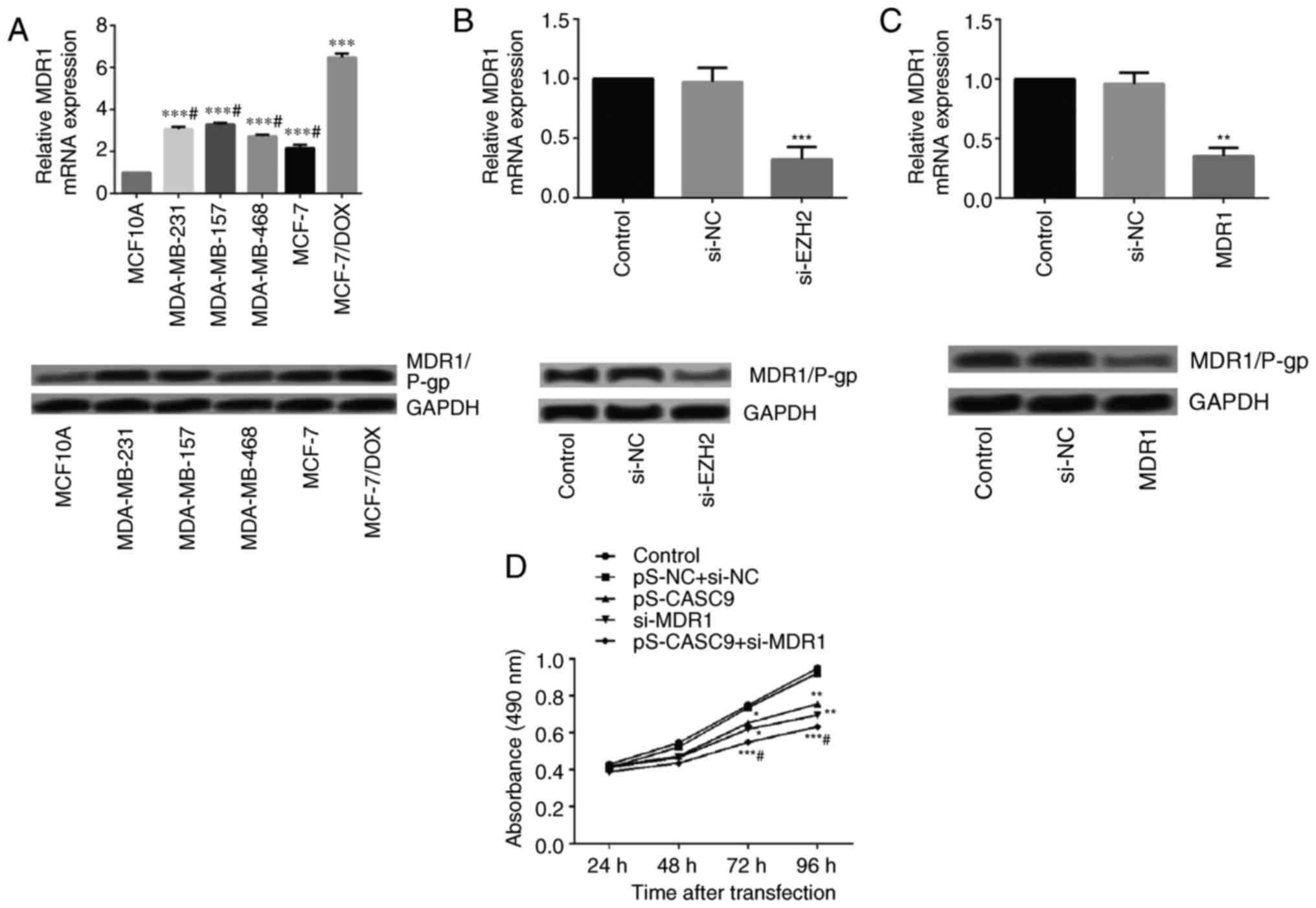 | Figure 5Effects of EZH2 on the growth and
metastasis of drug-resistant breast cancer cells MCF-7/DOX. (A)
Relative expression levels of MDR1/P-gp in MCF-7, MDA-MB-231,
MDA-MB-157, and MDA-MB-468 and MCF-7/DOX cells detected by RT-qPCR
and western blotting. ***P<0.001 vs. MCF10A group;
#P<0.05 vs. MCF-7/DOX group. (B) Relative expression
levels of MDR1/P-gp following EZH2 silencing as detected by RT-qPCR
and western blotting. ***P<0.001 vs. control group.
(C) Relative expression levels of MDR1 following cell transfection
as confirmed by RT-qPCR and western blotting.
**P<0.01 vs. control group. Alterations in the (D)
cell viability of MCF-7/DOX following CASC9 and MDR1 knockdown.
*P<0.05, **P<0.01 and
***P<0.001 vs. pS-NC + si-NC group;
#P<0.05 vs. pS-CASC9 group. (E) Apoptosis of
MCF-7/DOX following CASC9 and MDR1 knockdown.
**P<0.01 and ***P<0.001 vs. pS-NC +
si-NC group; #P<0.05 vs. pS-CASC9 group. (F)
Expression levels of apoptosis-associated proteins (Bcl-2, Bax,
caspase-3 and caspase-9) in MCF-7/DOX following CASC9 and MDR1
knockdown. Alterations in the (G) cell migration and (H) invasion
abilities of MCF-7/DOX cells following CASC9 and MDR1 knockdown.
*P<0.05, **P<0.01 and
***P<0.001 vs. pS-NC + si-NC group;
#P<0.05 vs. pS-CASC9 group. Bcl-2, apoptosis
regulator Bcl-2; Bax, apoptosis regulator BAX; DOX, doxorubicin;
EZH2, enhancer of zeste homolog; NC, negative control; P-gp,
P-glycoprotein; pS, vector; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; si, small
interfering RNA; CASC9, cancer susceptibility candidate 9; MDR1,
multidrug resistance protein 1. |
It has been reported that RNA interference-mediated
EZH2 depletion may reduce MDR1 expression and sensitize
multidrug-resistant tumor cells to chemotherapy (23–25). Considering the well-established
role of EZH2 in regulating MDR1/P-gp expression in certain cancer
types, the effects of EZH2 depletion on MDR1/P-gp levels were
detected in drug-resistant breast cancer cells. As presented in
Fig. 5B, EZH2 silencing resulted
in suppressed MDR1/P-gp expression compared with in cells
transfected with control or siRNA-NC.
Effects of CASC9 on the cell growth,
metastasis and chemoresistance of drug-resistant breast cancer
cells via regulation of MDR1 expression
To further explore the mechanisms underlying the
influence of CASC9 on the cell growth, metastasis and
chemoresistance of MCF-7/DOX cells, MDR1 was silenced by
transfecting MCF-7/DOX cells with MDR1 siRNA. The knockdown effect
was confirmed by measuring the mRNA and protein expression levels
(Fig. 5C). Subsequent analysis
demonstrated that compared with the CASC9 knockdown and control
groups, cell viability in the CASC9 knockdown + si-MDR1 group
decreased significantly (Fig.
5D). Additionally, CASC9 knockdown + si-MDR1 further promoted
apoptosis in MCF-7/DOX cells (Fig.
5E), and affected the expression of apoptosis-associated
proteins (Fig. 5F). Furthermore,
significant reductions in the migration and invasion of MCF-7/DOX
cells were detected in the CASC9 knockdown + si-MDR1 group compared
with the CASC9 knockdown and control groups (Fig. 5G and 5H).
Discussion
The results of the present study revealed that CASC9
was upregulated in breast cancer tissues and cell lines, in
addition to drug-resistant breast cancer cells. CASC9 knockdown
inhibited the growth and metastasis of drug-resistant breast cancer
cells, and decreased the IC50 of DOX in MCF-7/DOX cells.
In addition, further study indicated that CASC9 bound to EZH2 which
regulated the MDR1 gene, a crucial factor in drug resistance. These
interactions may serve an important role in the development of
breast cancer cell resistance to chemotherapeutic drugs.
CASC9 was originally detected in esophageal squamous
cell carcinoma, and a higher expression level of CASC9 was
correlated with poor differentiation in esophageal squamous cell
carcinoma (16,26). A recent study demonstrated that
CASC9 was frequently overexpressed in gastric cancer. Furthermore,
it may promote cell growth and chemoresistance to adriamycin in
gastric cancer (27). To the best
of our knowledge, the present was the first to suggest the
upregulation of CASC9 in breast cancer. Furthermore, CASC9
knockdown inhibited cell growth and metastasis, and reduced the
chemoresistance of drug-resistant breast cancer cells to DOX. These
findings were consistent with the previously mentioned studies, and
may indicate the important role of CASC9 in human cancer.
EZH2, a transcriptional repressor, has been
suggested to serve a critical role in the tumorigenic process as it
has been revealed to be overexpressed in a number of malignancies,
including lymphoma (28),
prostate cancer (29) and bladder
cancer (30). Importantly,
numerous studies have confirmed that increased expression of EZH2
is associated with a high histological grade and worse survival in
breast cancer, suggesting its promising role as a prognostic
biomarker in aggressive breast cancer (21,31). In the present study, RNA pull-down
assays revealed that EZH2 potentially interacted with CASC9.
Overexpressed CASC9 significantly increased the protein expression
of EZH2. In addition, EZH2 silencing inhibited cell growth and
metastasis, and reduced the IC50 of DOX in MCF-7/DOX
cells. Therefore, CASC9 may serve roles in breast cancer
progression and chemoresistance to DOX by binding to EZH2.
Studies have reported that the multidrug resistance
of a tumor may be caused by MDR1/P-gp, which is encoded by the
human MDR1 gene. MDR1/P-gp is an integral membrane protein, whose
function is the energy-dependent export of substances from the
inside of cells and from membranes to the outside (32,33). MDR1/P-gp is considered to render
tumor cells resistant to chemotherapy via the effective elimination
of these agents from cancer cells (34). Notably, numerous studies have
reported that the silencing of EZH2 may lead to decreases in MDR1
expression (23,24). The results of the present study
revealed that EZH2 silencing suppressed the expression of MDR1/P-gp
in MCF-7/DOX cells, suggesting that EZH2 may be involved in the
transcriptional regulation of MDR1, consistent with the
aforementioned studies.
In conclusion, the present study demonstrated the
oncogenic role of CASC9 in breast cancer and drug-resistant breast
cancer cells by binding to EZH2 and regulating the MDR1 gene. These
findings indicated that the modulation of CASC9 expression may be a
promising target in therapy of breast cancer and drug-resistant
breast cancer.
Funding
The present study was supported by the Natural
Science Foundation of Hunan Province (grant no. 2015JJ2119).
Availability of data and materials
The data that support the findings of this study are
available from the First Affiliated Hospital of University of South
China (Hengyang, China) but restrictions apply to the availability
of these data, which were used under license for the current study,
and so are not publicly available. Data are however available from
the authors upon reasonable request and with permission of the
First Affiliated Hospital of University of South China.
Authors' contributions
BJ wrote the manuscript and conducted the MTT assay,
RNA pull-down assay, western blotting and RT-qPCR. YL designed the
study and provided the foundation of the study. XQ, HZ and YT
performed the cellular apoptosis analysis. QF and YJ contributed to
the data analysis. ML and XW helped to collect data.
Ethics approval and consent to
participate
The experiments were approved by the Research Ethics
Committee of the First Affiliated Hospital, University of South
China (Hengyang, China); all patients provided written informed
consent.
Patient consent for publication
All patients provided written informed consent.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Ferlay J, Héry C, Autier P and
Sankaranarayanan R: Global burden of breast cancer. Breast cancer
epidemiology. Springer; pp. 1–19. 2010
|
|
2
|
Riaz M, van Jaarsveld MT, Hollestelle A,
Prager-van der Smissen WJ, Heine AA, Boersma AW, Liu J, Helmijr J,
Ozturk B, Smid M, et al: miRNA expression profiling of 51 human
breast cancer cell lines reveals subtype and driver
mutation-specific miRNAs. Breast Cancer Res. 15:R332013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
World Health Organization: World Cancer
Report. 2014
|
|
4
|
Eifel P, Axelson JA, Costa J, Crowley J,
Curran WJ Jr, Deshler A, Fulton S, Hendricks CB, Kemeny M,
Kornblith AB, et al: National institutes of health consensus
development conference statement: Adjuvant therapy for breast
cancer, november 1-3, 2000. J Natl Cancer Inst. 93:979–989. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Blum RH and Carter SK: Adriamycin. A new
anticancer drug with significant clinical activity. Ann Intern Med.
80:249–259. 1974. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen JS, Agarwal N and Mehta K:
Multidrug-resistant MCF-7 breast cancer cells contain deficient
intracellular calcium pools. Breast Cancer Res Treat. 71:237–247.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Herman J, Mangala L and Mehta K:
Implications of increased tissue transglutaminase (TG2) expression
in drug-resistant breast cancer (MCF-7) cells. Oncogene.
25:3049–3058. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Roberti A, La Sala DL and Cinti C:
Multiple genetic and epigenetic interacting mechanisms contribute
to clonally selection of drug-resistant tumors: Current views and
new therapeutic prospective. J Cell Physiol. 207:571–581. 2006.
View Article : Google Scholar
|
|
9
|
Duesberg P, Li R, Sachs R, Fabarius A,
Upender MB and Hehlmann R: Cancer drug resistance: The central role
of the karyotype. Drug Resist Updat. 10:51–58. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Fojo T: Multiple paths to a drug
resistance phenotype: Mutations, translocations, deletions and
amplification of coding genes or promoter regions, epigenetic
changes and microRNAs. Drug Resist Updat. 10:59–67. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sevignani C, Calin GA, Siracusa LD and
Croce CM: Mammalian microRNAs: A small world for fine-tuning gene
expression. Mamm Genome. 17:189–202. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bushati N and Cohen SM: microRNA
functions. Annu Rev Cell Dev Biol. 23:175–205. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Spizzo R, Almeida MIE, Colombatti A and
Calin GA: Long non-coding RNAs and cancer: A new frontier of
translational research? Oncogene. 31:4577–4587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:382011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shore AN, Herschkowitz JI and Rosen JM:
Noncoding RNAs involved in mammary gland development and
tumorigenesis: There's a long way to go. J Mammary Gland Biol
Neoplasia. 17:43–58. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Pan Z, Mao W, Bao Y, Zhang M, Su X and Xu
X: The long noncoding RNA CASC9 regulates migration and invasion in
esophageal cancer. Cancer Med. 5:2442–2447. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Song P, Jiang B, Liu Z, Ding J, Liu S and
Guan W: A three-lncRNA expression signature associated with the
prognosis of gastric cancer patients. Cancer Med. 6:1154–1164.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Su X, Li G and Liu W: The long noncoding
RNA cancer susceptibility candidate 9 promotes nasopharyngeal
carcinogenesis via stabilizing HIF1α. DNA Cell Biol. 36:394–400.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chekhun VF, Lukyanova NY, Kovalchuk O,
Tryndyak VP and Pogribny IP: Epigenetic profiling of
multidrug-resistant human MCF-7 breast adenocarcinoma cells reveals
novel hyper- and hypomethylated targets. Mol Cancer Ther.
6:1089–1098. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
21
|
Kleer CG, Cao Q, Varambally S, Shen R, Ota
I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, et al: EZH2
is a marker of aggressive breast cancer and promotes neoplastic
transformation of breast epithelial cells. Proc Natl Acad Sci USA.
100:11606–11611. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hoffmeyer S, Burk O, Von Richter O, Arnold
HP, Brockmöller J, Johne A, Cascorbi I, Gerloff T, Roots I,
Eichelbaum M, et al: Functional polymorphisms of the human
multidrug-resistance gene: Multiple sequence variations and
correlation of one allele with P-glycoprotein expression and
activity in vivo. Proc Natl Acad Sci USA. 97:3473–3478. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tang B, Zhang Y, Liang R, Gao Z, Sun D and
Wang L: RNAi-mediated EZH2 depletion decreases MDR1 expression and
sensitizes multidrug-resistant hepatocellular carcinoma cells to
chemotherapy. Oncol Rep. 29:1037–1042. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhou W, Wang J, Man WY, Zhang QW and Xu
WG: siRNA silencing EZH2 reverses cisplatin-resistance of human
non-small cell lung and gastric cancer cells. Asian Pac J Cancer
Prev. 16:2425–2430. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang Y, Liu G, Lin C, Liao G and Tang B:
Silencing the EZH2 gene by RNA interference reverses the drug
resistance of human hepatic multidrug-resistant cancer cells to
5-Fu. Life Sci. 92:896–902. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Cao W, Wu W, Shi F, Chen X, Wu L, Yang K,
Tian F, Zhu M, Chen G, Wang W, et al: Integrated analysis of long
noncoding RNA and coding RNA expression in esophageal squamous cell
carcinoma. Int J Genomics. 2013:4805342013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shang C, Sun L, Zhang J, Zhao B, Chen X,
Xu H and Huang B: Silence of cancer susceptibility candidate 9
inhibits gastric cancer and reverses chemoresistance. Oncotarget.
8:15393–15398. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Visser HP, Gunster MJ, Kluin-Nelemans HC,
Manders EM, Raaphorst FM, Meijer CJ, Willemze R and Otte AP: The
Polycomb group protein EZH2 is upregulated in proliferating,
cultured human mantle cell lymphoma. Br J Haematol. 112:950–958.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Varambally S, Dhanasekaran SM, Zhou M,
Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt
RG, Otte AP, et al: The polycomb group protein EZH2 is involved in
progression of prostate cancer. Nature. 419:624–629. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Raman JD, Mongan NP, Tickoo SK, Boorjian
SA, Scherr DS and Gudas LJ: Increased expression of the polycomb
group gene, EZH2, in transitional cell carcinoma of the bladder.
Clin Cancer Res. 11:8570–8576. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Collett K, Eide GE, Arnes J, Stefansson
IM, Eide J, Braaten A, Aas T, Otte AP and Akslen LA: Expression of
enhancer of zeste homologue 2 is significantly associated with
increased tumor cell proliferation and is a marker of aggressive
breast cancer. Clin Cancer Res. 12:1168–1174. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Callaghan R, Luk F and Bebawy M:
Inhibition of the multidrug resistance P-glycoprotein: Time for a
change of strategy? Drug Metab Dispos. 42:623–631. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Rahbari NN, Mehrabi A, Mollberg NM, Müller
SA, Koch M, Büchler MW and Weitz J: Hepatocellular carcinoma:
Current management and perspectives for the future. Ann Surg.
253:453–469. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Takara K, Sakaeda T and Okumura K: An
update on overcoming MDR1-mediated multidrug resistance in cancer
chemotherapy. Curr Pharm Des. 12:273–286. 2006. View Article : Google Scholar : PubMed/NCBI
|