Introduction
Breast cancer is the most frequent type of cancer,
which is expected to account for 30% of all new cancer diagnoses in
women in the United States (1).
Ovarian cancer is also a significant contributor to morbidity and
mortality rates, ranking as the seventh most common type of cancer
and the eighth most common cause of cancer-associated mortality
among women (2). The majority of
breast cancer cases are a result of gene mutations, particularly
mutations in breast cancer 1 (BRCA1) and/or breast cancer 2
(BRCA2), which put individuals at high risk for developing a
secondary breast cancer and ovarian cancer (3). The BRCA1 and BRCA2
genes are tumor suppressors. BRCA1 is a large gene, which
comprises 24 exons located on chromosome 17 (17q21) and codes for a
1,863-amino acid protein with a zinc-binding Really Interesting New
Gene finger motif at the amino terminus and a conserved acidic
carboxyl terminal (BRCA1 C-terminal) motif (4,5).
The BRCA2 gene is located on chromosome 13 (13q12), which
codes for a 3,418-amino acid protein, and shares structural and
functional similarities with the BRCA1 protein. It is
currently understood that the normal protein products of
BRCA1 and BRCA2 genes are important in double-strand
DNA repairs by maintaining genomic integrity through RAD51
(6), and they are also involved
in pathways associated with homologous recombination (7). However, once either of these genes
is mutated or altered, DNA damage may not be repaired properly,
likely leading to the occurrence of cancer. In patients with
BRCA1 and/or BRCA2 mutations, the risk of breast
cancer is significantly higher, compared with that in the general
population, and the histological grade is also more aggressive
(8-10). In addition, men and women carrying
BRCA1 and/or BRCA2 mutations have a 50% chance of
passing the mutations on to their children, termed hereditary
breast and ovarian cancer syndrome, which is characterized by an
increased risk of breast cancer and ovarian cancer (11). Therefore, understanding the
expression and mutations of BRCA1/2 in breast cancer and
ovarian cancer is urgently required in clinical practice, as is
examining the mechanism of tumorigenesis.
Accumulated evidence has demonstrated that the
expression levels of BRCAs are altered in several types of human
cancer, and multiple mutations have been reported in breast cancer
and/or ovarian cancer (12,13). However, the comprehensive analysis
of the expression and mutation of BRCA, and its interaction
networks is required to provide valuable information for clinical
practice and evaluation of its mechanism. The present study mainly
investigated the expression levels, mutations and interaction
networks of BRCA1 and BRCA2 in breast cancer and
ovarian cancer using bioinformatics analyses, which aimed provide
insights to reveal the mechanism of BRCA genes ultimately
leading to breast or ovarian tumorigenesis.
Materials and methods
FIREHOSE analysis for BRCA1 and
BRCA2
The expression profile of BRCA1 and
BRCA2 across various human cancer types was examined through
the Broad Institute FireBrowse portal (http://firebrowse.org/). On the homepage,
‘BRCA1’ or ‘BRCA2’ were added to the search box and
‘View Expression Profile’ was selected. The boxplots produced
showed the expression level of the target gene, with red bars
representing tumor samples and blue bars representing normal
samples.
Oncomine database analysis
The mRNA levels of BRCA1 and BRCA2 in
breast cancer and ovarian cancer tissues were compared with their
matched normal tissues using The Cancer Genome Atlas (TCGA)
datasets in the Oncomine database (http://www.oncomine.org). The threshold used to obtain
the most significant probes of the queried gene for each microarray
data included a two-fold difference in expression between cancer
and normal tissues with a P-value of <1x10-4. For
each gene, the mRNA expression level in three independent datasets
was analyzed.
Kaplan-Meier Plotter analysis
The prognostic values of the BRCA1 and
BRCA2 genes in breast cancer and ovarian cancer were
analyzed using the Kaplan-Meier Plotter (http://kmplot.com/analysis/). Overall, the survival
rates of patients with high and low levels of BRCA1 or
BRCA2 were shown using a Kaplan-Meier survival plot.
Catalogue Of Somatic Mutations In Cancer
(COSMIC) analysis for BRCA1 and BRCA2 mutations
The COSMIC database (http://cancer.sanger.ac.uk/cosmic) was used for the
analysis of BRCA1 and BRCA2 mutations. Pie charts
were generated for a distribution overview and substitutions on the
coding strand in breast cancer and ovarian cancer.
cBioPortal analysis for alteration
frequency and interaction network of BRCA1 and BRCA2
The alteration frequencies of BRCA1 and
BRCA2 mRNA in breast cancer and ovarian cancer was
determined by using the cBioPortal for Cancer Genomics (http://www.cbioportal.org/). All searches and analyses
were performed according to the online instructions of cBioPortal
(14,15).
Results
BRCA1 and BRCA2 are upregulated in breast
cancer and ovarian cancer
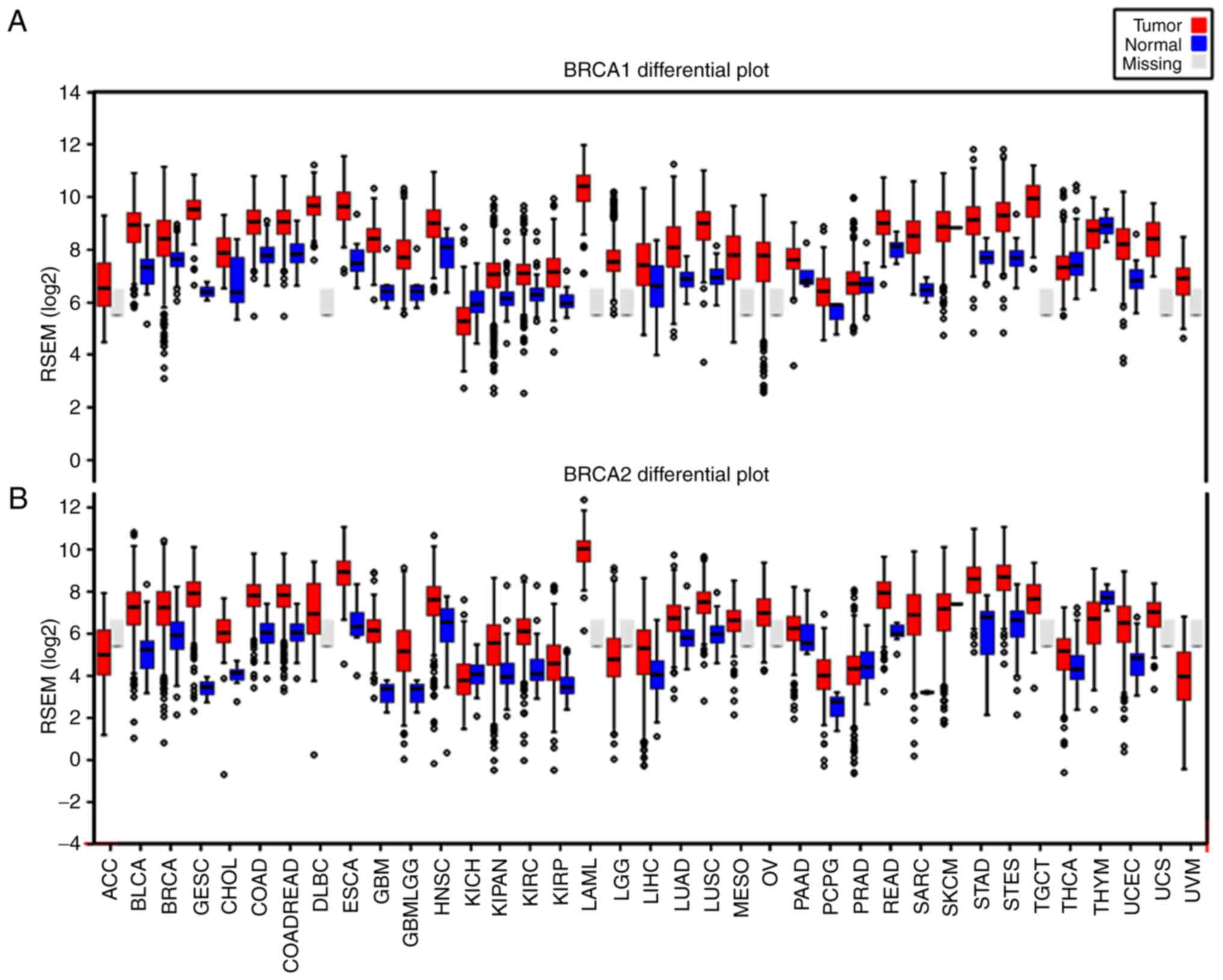
The gene expression levels of BRCA1 and
BRCA2 were surveyed in 38 cases of human cancer using the
TCGA database. The columns in Fig.
1 represent the accurate quantification of gene and isoform
expression from the RNA-Seq data. The results showed that higher
levels of BRCA1 (Fig. 1A)
and BRCA2 (Fig. 1B)
transcripts were observed in almost all cancer tissues, compared
with the levels in their matched normal tissues. Of note, the
BRCA1 and BRCA2 genes exhibited a similar expression
pattern in various cancer types, including breast cancer and
ovarian cancer (Fig. 1).
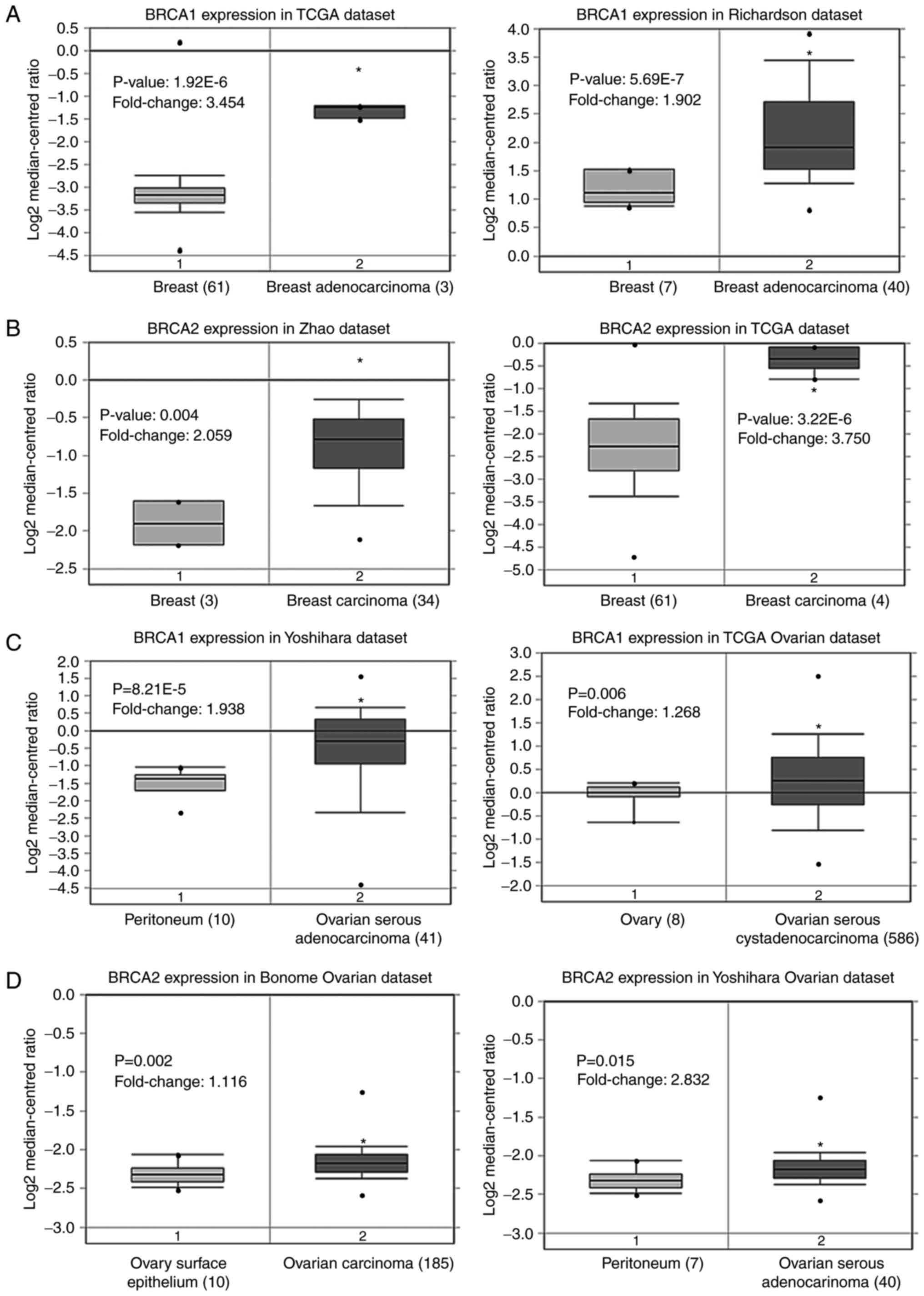
Oncomine analysis comparing cancer tissues with
normal tissues also showed that the BRCA1 and BRCA2
genes were significantly upregulated in breast cancer and ovarian
cancer tissues (Fig. 2A-D) in
three independent analyses, compared with corresponding normal
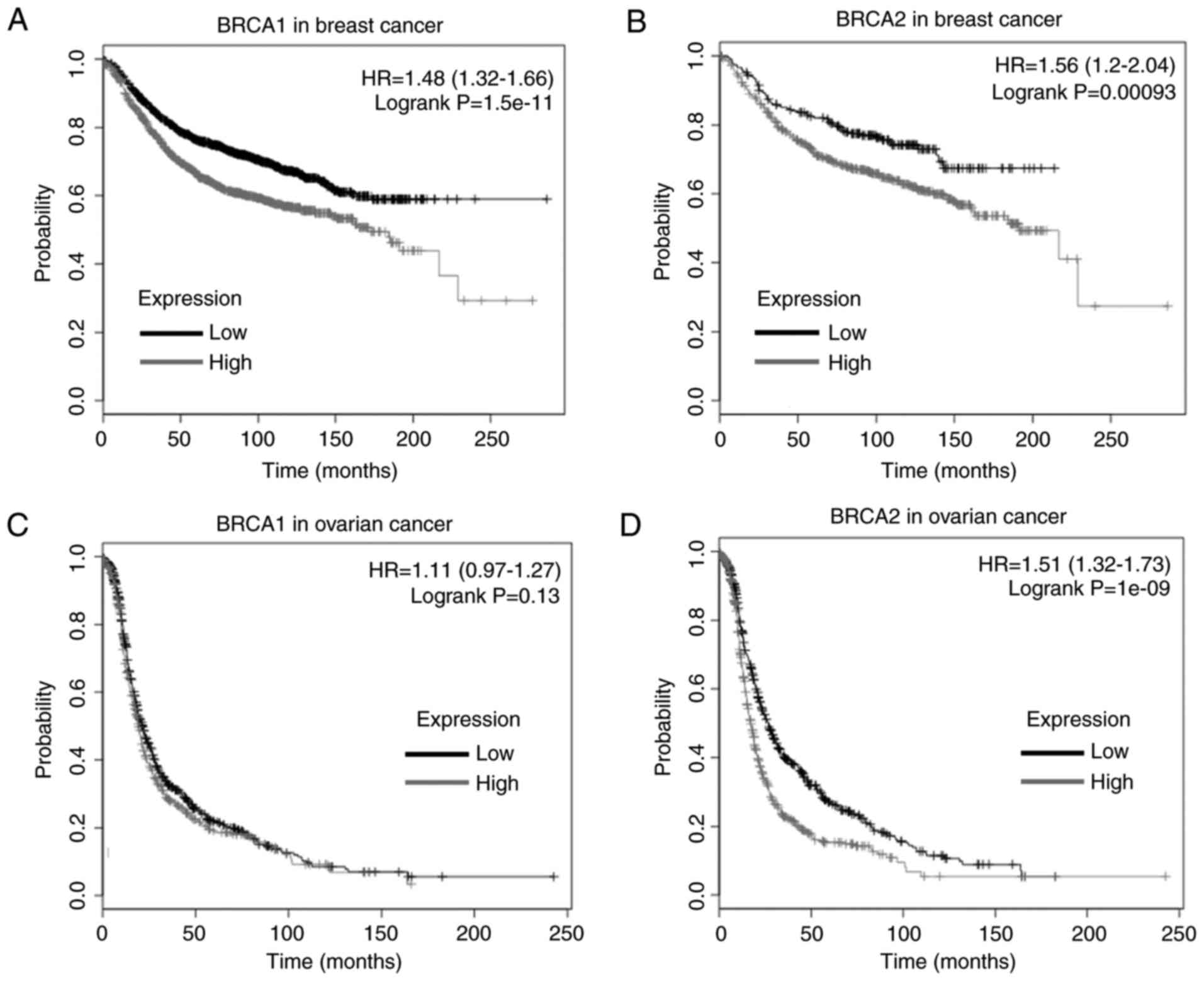
tissues (16,17). The results of the Kaplan-Meier
analysis revealed that a high expression level of BRCA1 was
correlated with a poor survival rate in breast cancer (P=1.51E-11;
Fig. 3A) and a high expression
level of BRCA2 was correlated with poor survival rates in
breast cancer (P=0.00093; Fig.
3B). No significant correlation was found between the
expression level of BRCA1 and the survival rate of patients
for ovarian cancer (P=0.13; (Fig.
3C), whereas a high expression level of BRCA2 was
correlated with poor survival rates in ovarian cancer (P=1.0E-9;
Fig. 3D).
BRCA1 and BRCA2 mutations in breast
cancer and ovarian cancer
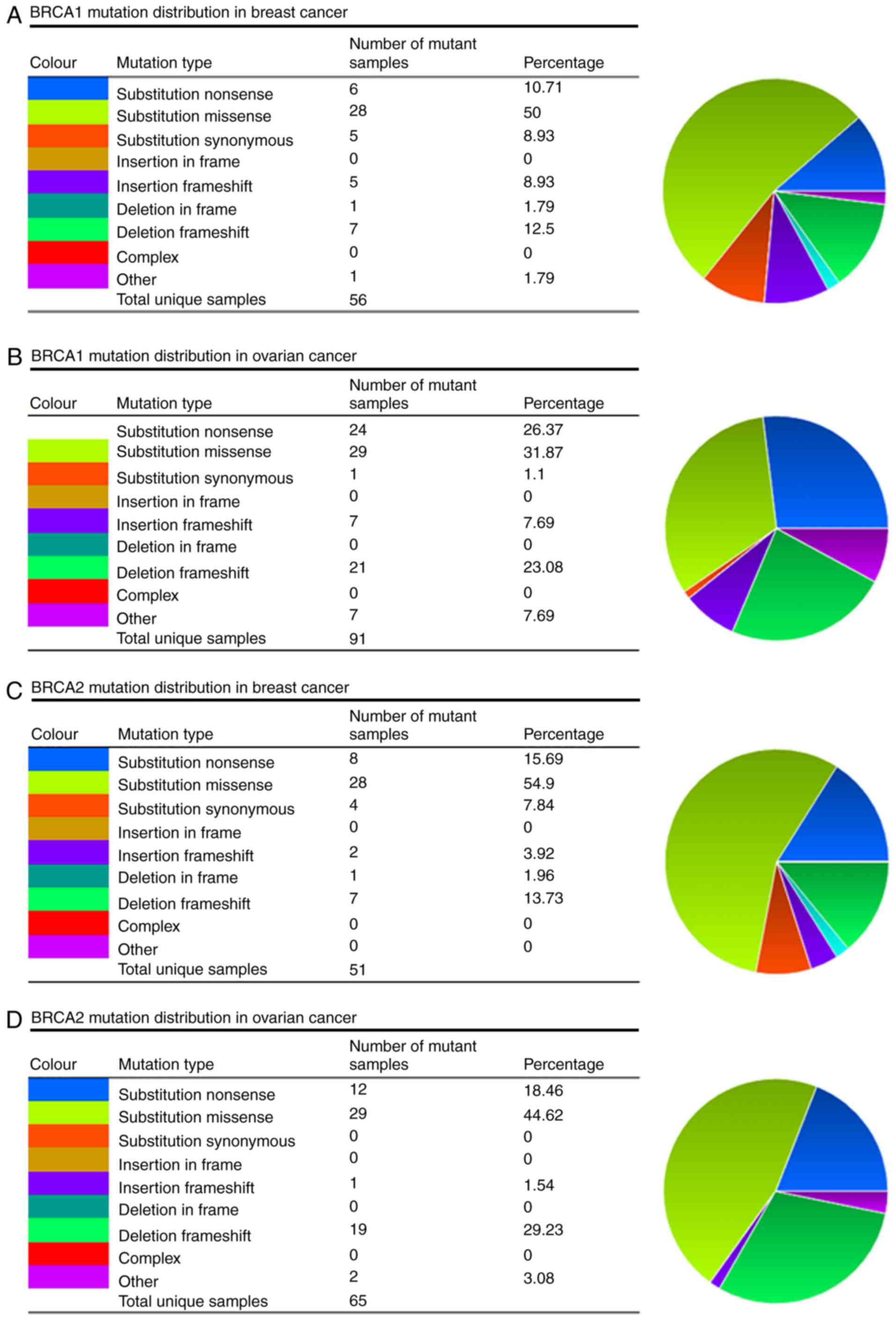
The present study evaluated the mutations of
BRCA1/2 using the COSMIC database, and the information
regarding the mutations of substitution missense, nonsense,
synonymous and insertion frame shift are presented in the pie-chart
shown in Fig. 4A-D. In breast
cancer and ovarian cancer, the most frequent mutation was
substitution missense in the BRCA1/2 genes. In the mutation
samples of breast cancer, 50% of the BRCA1 and 54.9% of the
BRCA2 mutations were substitution missense (Fig. 4A and C). In ovarian cancer, 31.87%
of the BRCA1 and 44.62% of the BRCA2 mutations were
substitution missense (Fig. 4B and
D). The mutation samples of breast cancer comprised 23.33%
G>A, 20.00% G>C and 20.00% G>T substitutions in the
BRCA1 coding strand (Fig.
4A), and 22.58% G>C and 19.35% G>A substitutions in the
BRCA2 coding strand (Fig.
4C). In the ovarian cancer mutation samples, there were 32.61%
G>T and 21.71% G>A substitutions in the BRCA1 coding
strand (Fig. 4B), and 23.68%
G>T and 21.05 G>A substitutions in the BRCA2 coding
strand (Fig. 4D).
The alteration frequencies of BRCA1 and
BRCA2 in breast cancer and ovarian cancer were also analyzed
using cBioportal. A total of eight studies on breast cancer and two
studies on ovarian cancer were included in the database. The
results showed that ~2-8% of breast cancer and 6-22% of ovarian
cancer clinical samples contained BRCA1 and/or BRCA2
mutations (Fig. 5A and B). In the
breast cancer clinical samples, there were 81 mutations observed in
BRCA1, 43 of which were missense mutations and 38 were
in-frame mutations and truncating; there were 88 mutations in
BRCA2, 42 of which were missense mutations and 46 were
in-frame mutations and truncating. In ovarian cancer, there were 48
mutations of the BRCA1 gene, only two of which were missense
mutations and 46 were in-frame mutations and truncating; there were
46 mutations in BRCA2, five of which were missense mutations
and 41 were in-frame mutations and truncating. Several common
mutations were observed in BRCA1 (E1346Kfs*20, E23Vfs*17 and
Q1756Pfs*74) and BRCA2 (V220Ifs*4, N1784Hfs*2 and
S1982Rfs*22) in breast cancer and ovarian cancer, determined by
pairwise-analysis (Tables I and
II).
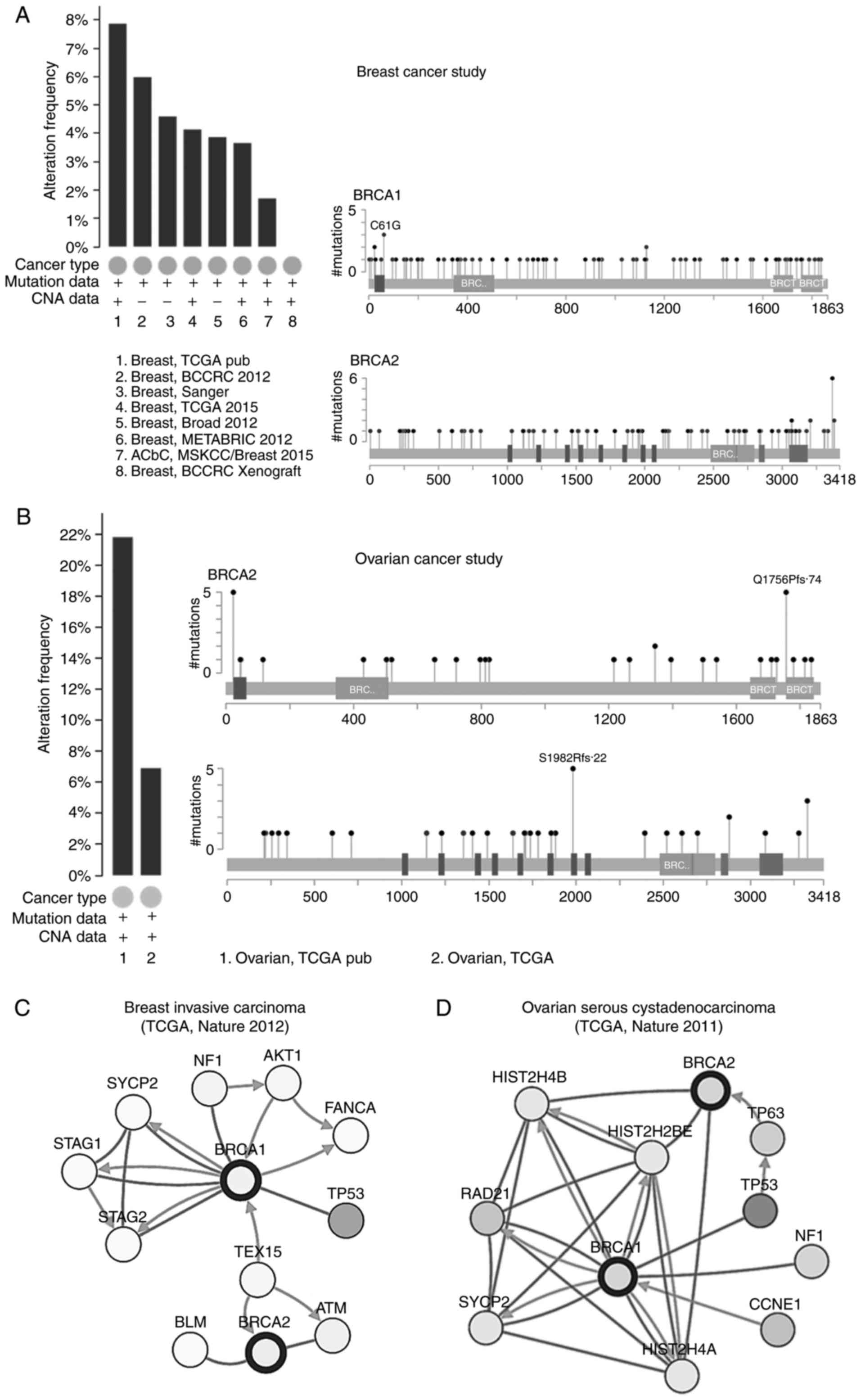 | Figure 5Alteration frequencies and
interaction networks of BRCA1 and BRCA2 in breast
cancer and ovarian cancer analyzed using cBioPortal. Mutation
analysis of BRCA1 and BRCA2 in (A) breast cancer and
(B) ovarian cancer. The results showed that ~2-8% of breast cancer
and 6-22% of ovarian cancer clinical samples contained the
BRCA1 and/or BRCA2 mutation. Interaction network
analysis for (C) breast cancer and (D) ovarian cancer using
cBioPortal. The spheres represent the genes in the interaction
networks, the green arrows mean ‘control expression of’, the brown
lines mean ‘complex with’ and the blue arrows mean the ‘control
state change of ’. TCGA, The Cancer Genome Atlas; BRCA1,
breast cancer 1; BRCA2, breast cancer 2; NF1, neurofibromin
1; TP53, tumor protein 53; TEX15, testis expressed 15; ATM, ataxia
telangiectasia mutated; BLM, bloom syndrome RecQ like helicase;
STAG1, stromal antigen 1; SYCP2, synaptonemal complex protein 2;
FANCA, fanconi anemia complementation group A; CCNE1, cyclin E1;
HIST2H, histone cluster 2; TP63, tumor protein 63. |
 | Table ICommon mutations in BRCA1 in
breast cancer and ovarian cancer. |
Table I
Common mutations in BRCA1 in
breast cancer and ovarian cancer.
| Cancer study | Amino acid
change | Type | Copy number | Mutations in sample
(n) |
|---|
| Breast (TCGA) | E1346Kfs*20 | FS del | ShallowDel | 138 |
| Breast (TCGA) | E23Vfs*17 | FS del | Gain | 118 |
| Breast (TCGA) | Q1756Pfs*74 | FS ins | ShallowDel | 65 |
 | Table IICommon mutations in BRCA2 in
breast cancer and ovarian cancer. |
Table II
Common mutations in BRCA2 in
breast cancer and ovarian cancer.
| Cancer study | Amino acid
change | Type | Copy number | Mutations in sample
(n) |
|---|
| Breast (TCGA) | V220Ifs*4 | FS del | Diploid | 69 |
| Breast (TCGA) | N1784Hfs*2 | FS del | ShallowDel | 49 |
| Breast (TCGA) | S1982Rfs*22 | FS del | Diploid | 48 |
Interaction networks of BRCA1 and BRCA2
in breast cancer and ovarian cancer
The interaction networks of BRCA1 and
BRCA2 were analyzed using cBioportal (Fig. 5C and D). Among the genes involved
in the interaction network, >30% were able to form complexes
with BRCA1 and BRCA2 in breast cancer (30.6%) and
ovarian cancer (36.5%). In breast cancer, BRCA1 controlled
the expression of cyclin-dependent kinase inhibitor 1B (CDKN1B,
p27Kip1). The testis expressed 15 gene controlled
the state of BRCA1 and BRCA2, respectively. In
addition, it was found that the three genes, neurofibromin 1
(NF1), synaptonemal complex protein 2 (SYCP2) and
tumor protein 53 (TP53), were involved in breast cancer and
ovarian cancer. BRCA1 was able to form complex with the
proteins of these three genes, respectively, and also control the
state change of SYCP2 in breast cancer and ovarian cancer,
which is associated with cell cycle, mitotic and meiosis.
Discussion
In women, BRCA1 or BRCA2 mutations
result in a ~40-80% risk of developing breast cancer, and ~11-40%
risk of developing ovarian cancer, respectively (18-21). In the present study, the latest
evidence of the expression profiles and mutations of BRCA1
and BRCA2 were surveyed using bioinformatics analyses/TCGA
data portal and revealed that the BRCA genes were
significantly upregulated and mutated in various types of human
cancer, including breast cancer and ovarian cancer. Higher mRNA
levels of BRCA1 and BRCA2 were observed in tissues
samples of 23 types of cancer s, compared with their normal control
tissues, including breast cancer and ovarian cancer. This
upregulated expression pattern was further validated in three
independent RNA-seq datasets. Of note, a positive correlation was
identified between the mRNA expression level of the BRCA
genes and poor survival rates in breast cancer and ovarian cancer
by Kaplan-Meier analysis.
The gene expression levels of BRCA1 and
BRCA2 offer a potentially important tool for use in cancer
management. A study in lung cancer showed that the BRCA1
gene served as an indicator of chemoresistance, and the
reconstitution of wild-type BRCA1 function into lung cancer
cells resulted in thousands of fold increases in sensitivity to
paclitaxel and vinorelbine (22).
Another preclinical study in breast cancer demonstrated the
potential of using BRCA1 and BRCA2 dysfunction to
predict response to clinical treatment (23). However, another previous study
showed that the protein level of BRCA1 exhibited a
significant reduction in sporadic breast and ovarian cancer
(24,25), which may have been partially due
to the different splice variants or localization of the
BRCA1 protein. The expression levels of BRCA1 and
BRCA2 in human mammary epithelial and cancer cells vary with
cell cycle, which are expressed in a cell cycle-dependent manner,
peaking at the G1/S boundary (26).
In the present study, ~81 mutations in the
BRCA1 gene and 88 mutations in the BRCA2 gene were
found in breast cancer samples, and 48 mutations in BRCA1
and 46 mutations in BRCA2 were found in ovarian cancer
samples. Of note, three mutations in BRCA1 and three
mutations in BRCA2 were observed in both breast cancer and
ovarian cancer, which indicated that the effects of these mutations
may be common in breast cancer and ovarian cancer. In addition, the
common mutations in BRCA1 and BRCA2 were frame-shift
deletions, and are known to be oncogenic (27).
It is well known that BRCA1 and BRCA2
are involved in DNA repair, cell cycle checkpoint regulation and
transcription (28), and these
processes are dictated through crosstalk with a network of
proteins. It is now clear that the BRCA1 and BRCA2 proteins
co-localize with RAD51 complexes on mitotic and meiotic chromosomes
following exposure to ionizing radiation or hydroxyurea (7,29).
Certain other proteins have been reported to interact with BRCA1,
including ataxia telangiectasia mutated (ATM)/ATM-related kinase,
checkpoint kinase 2, and aurora A protein kinase, to regulate cell
cycle progression (30). In the
present study, three genes (NF1, SYCP2 and
TP53) were found to be associated with BRCA1 in
breast cancer and ovarian cancer. NF1 is a tumor suppressor
gene, which comprises 60 exons coding for the protein
neurofibromin, which is associated with neurofibromatosis-noonan
syndrome and neurofibromatosis, type 1. Among its associated
pathways are the mitogen-activated protein kinase signaling pathway
and Ras signaling pathway (31).
The NF1 and BRCA1 genes are located in the long arm
of chromosome 17, and the involvement of NF1 in breast
cancer has been suggested in previous publications (32,33). SYCP2 is a testis-specific
human gene with aberrant expression in human
papillomavirus-positive cancer (34), and head and neck squamous cell
carcinoma (35). In the present
study, it was found that BRCA1 was able to form a complex
with NF1 and SYCP2, respectively, in breast cancer
and ovarian cancer, which suggested that the BRCA1 gene
interacts with NF1 and SYCP2 directly or indirectly
in cell cycle regulation. However, further investigations are
required to discern the complex mechanisms underlying these
observations in the future.
In conclusion, the findings of the present study
revealed not only the increased expression pattern of the
BRCA1 and BRCA2 genes in breast cancer and ovarian
cancer, but also provided an understanding on the mutations and
interaction networks of these two genes in the types of cancers
mentioned. The results also provide significant insight into
certain mutations and proteins involved in the interaction network,
the roles of which may be common in breast cancer and ovarian
cancer.
Acknowledgements
Not applicable.
Funding
This study was financially supported by the Shenzhen
Science and Technology Program (Basic Research Project; grant no.
JCYJ20170307141840188) and the Innovation and Technology fund of
Longhua Shenzhen (grant no. 20150925A0410013).
Availability of data and materials
The data that support the findings of this study are
available from The Cancer Genome Atlas: http://cancergenome.nih.gov/.
Authors' contributions
ZW performed the experiments and drafted the
manuscript. YZ and QD performed the Oncomine analysis and
cBioportal analysis. JZ and HL conceived the study design, obtained
funding for the study, and drafted and revised the manuscript. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2017. CA Cancer J Clin. 67:7–30. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shi T, Wang P, Xie C, Yin S, Shi D, Wei C,
Tang W, Jiang R, Cheng X, Wei Q, et al: BRCA1 and BRCA2 mutations
in ovarian cancer patients from China: Ethnic-related mutations in
BRCA1 associated with an increased risk of ovarian cancer. Int J
Cancer. 140:2051–2059. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Eeles RA: Screening for hereditary cancer
and genetic testing, epitomized by breast cancer. Eur J Cancer.
35:1954–1962. 1999. View Article : Google Scholar
|
|
4
|
Miki Y, Swensen J, Shattuck-Eidens D,
Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM,
Ding W, et al: A strong candidate for the breast and ovarian cancer
susceptibility gene BRCA1. Science. 266:66–71. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chapman MS and Verma IM: Transcriptional
activation by BRCA1. Nature. 382:678–679. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Kolinjivadi AM, Sannino V, de Antoni A,
Técher H, Baldi G and Costanzo V: Moonlighting at replication forks
- a new life for homologous recombination proteins BRCA1, BRCA2 and
RAD51. FEBS Lett. 591:1083–1100. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Chen J, Silver DP, Walpita D, Cantor SB,
Gazdar AF, Tomlinson G, Couch FJ, Weber BL, Ashley T, Livingston DM
and Scully R: Stable interaction between the products of the BRCA1
and BRCA2 tumor suppressor genes in mitotic and meiotic cells. Mol
Cell. 2:317–328. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Narod SA, Brunet JS, Ghadirian P, Robson
M, Heimdal K, Neuhausen SL, Stoppa-Lyonnet D, Lerman C, Pasini B,
de los Rios P, et al: Tamoxifen and risk of contralateral breast
cancer in BRCA1 and BRCA2 mutation carriers: A case-control study.
Hereditary breast cancer clinical study group Lancet.
356:1876–1881. 2000.
|
|
9
|
Sakorafas GH and Tsiotou AG: Genetic
predisposition to breast cancer: A surgical perspective. Br J Surg.
87:149–162. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nicoletto MO, Donach M, De Nicolo A,
Artioli G, Banna G and Monfardini S: BRCA-1 and BRCA-2 mutations as
prognostic factors in clinical practice and genetic counselling.
Cancer Treat Rev. 27:295–304. 2001. View Article : Google Scholar
|
|
11
|
Cicero G, De Luca R, Dorangricchia P, Lo
Coco G, Guarnaccia C, Fanale D, Calò V and Russo A: Risk perception
and psychological distress in genetic counselling for hereditary
breast and/or ovarian cancer. J Genet Couns. 26:999–1007. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Alvarez C, Aravena A, Tapia T, Rozenblum
E, Solís L, Corvalán A, Camus M, Alvarez M, Munroe D, Maass A and
Carvallo P: Different Array CGH profiles within hereditary breast
cancer tumors associated to BRCA1 expression and overall survival.
Bmc Cancer. 16:2192016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ewald IP, Vargas FR, Moreira MA, Filho CM,
da Cunha DR, Ramos JP, Ribeiro PL, Caleffi M, Giuglani R and
Ashton-Prolla P: Prevalence of BRCA1 and BRCA2 founder mutations in
Brazilian hereditary breast and ovarian cancer families. J Clin
Oncol. 26:2008. View Article : Google Scholar
|
|
14
|
Gao J, Aksoy BA, Dogrusoz U, Dresdner G,
Gross B, Sumer SO, Sun Y, Jacobsen A, Sinha R, Larsson E, et al:
Integrative analysis of complex cancer genomics and clinical
profiles using the cBio-Portal. Sci Signal. 6:2013. View Article : Google Scholar
|
|
15
|
Cerami E, Gao J, Dogrusoz U, Gross BE,
Sumer SO, Aksoy BA, Jacobsen A, Byrne CJ, Heuer ML, Larsson E, et
al: The cBio cancer genomics portal: An open platform for exploring
multidimensional cancer genomics data. Cancer Discov. 2:401–404.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhao H, Langerød A, Ji Y, Nowels KW,
Nesland JM, Tibshirani R, Bukholm IK, Kåresen R, Botstein D,
Børresen-Dale AL and Jeffrey SS: Different gene expression patterns
in invasive lobular and ductal carcinomas of the breast. Mol Biol
Cell. 15:2523–2536. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Richardson AL, Wang ZC, De Nicolo A, Lu X,
Brown M, Miron A, Liao X, Iglehart JD, Livingston DM and Ganesan S:
X chromosomal abnormalities in basal-like human breast cancer.
Cancer Cell. 9:121–132. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mavaddat N, Peock S, Frost D, Ellis S,
Platte R, Fineberg E, Evans DG, Izatt L, Eeles RA, Adlard J, et al:
Cancer risks for BRCA1 and BRCA2 mutation carriers: Results from
prospective analysis of EMBRACE. J Natl Cancer Inst. 105:812–822.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chen S and Parmigiani G: Meta-analysis of
BRCA1 and BRCA2 penetrance. J Clin Oncol. 25:1329–1333. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Blay P, Santamaria I, Pitiot AS, Luque M,
Alvarado MG, Lastra A, Fernández Y, Paredes A, Freije JM and Balbín
M: Mutational analysis of BRCA1 and BRCA2 in hereditary breast and
ovarian cancer families from Asturias (Northern Spain). BMC Cancer.
13:2432013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Petrucelli N, Daly MB and Feldman GL:
Hereditary breast and ovarian cancer due to mutations in BRCA1 and
BRCA2. Genet Mod. 12:245–259. 2010. View Article : Google Scholar
|
|
22
|
Taron M, Rosell R, Felip E, Mendez P,
Souglakos J, Ronco MS, Queralt C, Majo J, Sanchez JM, Sanchez JJ
and Maestre J: BRCA1 mRNA expression levels as an indicator of
chemoresistance in lung cancer. Hum Mol Genet. 13:2443–2449. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Farmer H, McCabe N, Lord CJ, Tutt AN,
Johnson DA, Richardson TB, Santarosa M, Dillon KJ, Hickson I,
Knights C, et al: Targeting the DNA repair defect in BRCA mutant
cells as a therapeutic strategy. Nature. 434:917–921. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kim MC, Choi JE, Lee SJ and Bae YK:
Coexistent loss of the expressions of BRCA1 and p53 predicts poor
prognosis in triple-negative breast cancer. Ann Surg Oncol.
23:3524–3530. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ju LL, Zhao CY, Ye KF, Yang H and Zhang J:
Expression and clinical implication of Beclin1, HMGB1, p62,
survivin, BRCA1 and ERCC1 in epithelial ovarian tumor tissues. Eur
Rev Med Pharmacol Sci. 20:1993–2003. 2016.PubMed/NCBI
|
|
26
|
Rajan JV, Wang M, Marquis ST and Chodosh
LA: Brca2 is coordinately regulated with Brca1 during proliferation
and differentiation in mammary epithelial cells. Proc Natl Acad Sci
USA. 93:13078–13083. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Cancer Genome Atlas Network: Comprehensive
molecular portraits of human breast tumours. Nature. 490:61–70.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Welcsh PL and King MC: BRCA1 and BRCA2 and
the genetics of breast and ovarian cancer. Hum Mol Genet.
10:705–713. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Parvin S, Islam MS, Al-Mamun MM, Islam MS,
Ahmed MU, Kabir ER and Hasnat A: Association of BRCA1, BRCA2,
RAD51, and HER2 gene polymorphisms with the breast cancer risk in
the Bangladeshi population. Breast Cancer. 24:229–237. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Christou CM and Kyriacou K: BRCA1 and its
network of interacting partners. Biology. 2:40–63. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Brems H, Beert E, de Ravel T and Legius E:
Mechanisms in the pathogenesis of malignant tumours in
neurofibromatosis type 1. Lancet Oncol. 10:508–515. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Campos B, Balmaña J, Gardenyes J,
Valenzuela I, Abad O, Fàbregas P, Volpini V and Díez O: Germline
mutations in NF1 and BRCA1 in a family with neurofibromatosis type
1 and early-onset breast cancer. Breast Cancer Res Treat.
139:597–602. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Madanikia SA, Bergner A, Ye X and Blakeley
JO: Increased risk of breast cancer in women with NF1. Am J Med
Genet A. 158A:3056–3060. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Masterson L, Sorgeloos F, Winder D,
Lechner M, Marker A, Malhotra S, Sudhoff H, Jani P, Goon P and
Sterling J: Deregulation of SYCP2 predicts early stage human
papillomavirus-positive oropharyngeal carcinoma: A prospective
whole transcriptome analysis. Cancer Sci. 106:1568–1575. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Pannone G, Santoro A, Papagerakis S, Lo
Muzio L, De Rosa G and Bufo P: The role of human papillomavirus in
the pathogenesis of head & neck squamous cell carcinoma: An
overview. Infect Agent Cancer. 6:42011. View Article : Google Scholar
|