Introduction
Cardiovascular diseases, particularly coronary heart
disease, remain the leading cause of mortality worldwide (1). Thus, the prevention and treatment of
cardiovascular disease is of utmost importance. Autophagy was
previously reported in human heart disease in the myocardial tissue
of patients with dilated cardiomyopathy (2). Autophagy plays an important role in
the development of a number of cardiovascular diseases, such as
myocardial ischemia and reperfusion injury, myocardial infarction,
cardiac hypertrophy and heart failure (3-6).
Autophagy removes old or excessive cell contents from
cardiomyocytes (7). Therefore,
autophagy plays an important role in the survival and functions of
cardiomyocytes.
Autophagy refers to the process of degrading protein
macromolecules and organelles in eukaryotic cells in autophagy
lysosomes, and is divided into macroautophagy, microautophagy and
chaperone-mediated autophagy (CMA). cmA refers to the translocation
of unfolded proteins into lysosomes through heat shock protein 70
(HSP70). Heat shock protein family A member 5 (Hspa5), also known
as binding immunoglobulin protein (BiP) or glucose-regulated
protein 78 (GRP-78), is a member of the HSP70 family (8), and is an endoplasmic reticulum (ER)
stress-associated protein which has an effect on cell protection
(9). Protein synthesis in the
center of ER is altered under starvation conditions so that
unfolded and misfolded proteins accumulate and result in ER stress
(10). This finally results in
cell autophagy (11). Hspa5 is
associated with autophagy and generally cardiac protection, which
is expressed as an ER stress chaperone synchronously with LC3II
(12,13).
MicroRNAs (miRNAs or miRs) are small non-coding RNAs
comprising 18-25 nucleotides and are the central regulatory factors
at the post-transcriptional level in animals and plants. miRNAs
bind to the 3'-untranslated region (3'-UTR) of their target mRNAs
and negatively regulate gene expression by accelerating mRNA
degradation or inhibiting mRNA translation (14). Several miRNAs have been shown to
affect autophagy and therefore control important processes that
contribute to cardiovascular diseases. For example, the knockdown
of miR-122 has been reported to protect H9c2 cardiomyocytes from
hypoxia-induced apoptosis and to promote autophagy (15). It has also been shown that miR-365
has the ability to accelerate cardiac hypertrophy by inhibiting
autophagy via the modulation of Skp2 expression (5). miR-181b-5p may also suppress
proliferation, migration and invasion and promote apoptosis in
astrocy-toma (16). miR-181b-5p
expression has also been shown to be associated with asthma
(17) and schizophrenia (18). However, the role of miR-181b-5p in
cardiomyocyte autophagy remains unknown.
In the present study, the role of miR-181b-5p in
cardiomyocyte autophagy was investigated. Hspa5 was predicted to be
a direct target of miR-181b-5p by bioinformatics analysis. The
present study further determined that the downregulation of
miR-181b-5p in starvation-induced cardiomyocyte autophagy may be
due to the targeting of Hspa5 via the phosphoinositide
3-kinase/Akt/mammalian target of rapamycin (mTOR) signaling
pathway, which plays a crucial role in cardiomyocyte
protection.
Materials and methods
Cell culture and treatment
The H9c2 cell line was obtained from the Cell Line
Bank of the Type Culture Collection of the Chinese Academy of
Sciences (Shanghai, China) and cultured in high glucose Dulbecco's
modified Eagle's medium (DMEM; Gibco/Thermo Fisher Scientific,
Inc., Waltham, MA, USA) supplemented with 10% fetal bovine serum
(FBS; Gibco/Thermo Fisher Scientific, Inc.), at 37°C in 5%
CO2.
Animal experiments
All animal protocols were approved by the
Experimental Animal Committee of the Second Affiliated Hospital of
Suchow University, Suchow, China. Neonatal rat ventricular myocytes
(NRVMs) were isolated from 2-day-old Sprague-Dawley rats (male,
n=5; Laboratory Animal Center of Soochow University, Suchow,
China). Briefly, the hearts from 2-day-old rats were aseptically
removed. Their ventricles were dissected, minced and trypsinized
overnight at 4°C. The following day, the cells were dissociated
with collagenase and plated for 2 h at 37°C. The non-adherent
cardiomyocytes were removed and plated in 24-well plates in
DMEM/F-12 medium containing 10% FBS and 0.1 mm bromodeoxyuridine
(Sigma-Aldrich/Merck KGaA, Darmstadt, Germany). A total of
1×105 cells/cm2 were seeded in a 24-well
plate for use in further experiments. This procedure yielded
cultures with a high proportion of cardiomyocytes; microscopic
observations determined that 90-95% of the cells were
cardiomyocytes, as assessed by the microscopic observation of cell
beating. To mimic starvation, cardiomyocytes were incubated in
Earle's Balanced Salt Solution (EBSS; Gibco/Thermo Fisher
Scientific, Inc.) for different periods of time (0, 2, 4, 6 and 8
h).
Cell transfection
The cells were plated into 6-well plates
(1×105 per well) and incubated at 37°C for 24 h. The
cells were either left untransfected or transiently transfected
with a miR-181b-5p mimic, miR-Con, miR-181b-5p inhibitor,
inhibitor-Con, siHspa5 or Con-siRNA (all from Guangzhou RiboBio Co.
Ltd., Guangzhou, China) using Lipofectamine 2000 (Invitrogen/Thermo
Fisher Scientific, Inc.) according to the manufacturer's
instructions. Once the transfection was successful, the following
experiments began.
Reverse transcription-quantitative PCR
(RT-qPCR)
The cells were plated in 6-well plates
(1×105 per well). After transfecting the cells for 48 h,
total RNA was extracted from the cells using TRIzol reagent
(Invitrogen/Thermo Fisher Scientific, Inc.). The concentration and
purity of the RNA was measured using a NanoDrop One Microvolume
UV-Vis Spectrophotometer (Thermo Fisher Scientific, Inc.) according
to the manufacturer's instructions. Complementary DNA (cDNA) was
synthesized using RevertAid First Strand cDNA (Fermentas, Waltham,
MA, USA) or the TaqMan MicroRNA Reverse Transcription kit (Applied
Biosystems, Foster City, CA, USA) according to the manufacturer's
instructions, then amplified using Power SYBR®-Green PCR
Master Mix or TaqMan MicroRNA Assay (Applied Biosystems). The
primers used were as follows: miR-181b-5p forward, 5'-ACA CTC CAG
CTG GGA CTT GGG CAC TGA AAC A-3' and reverse, 5'-TGG TGT CGT GGA
GTC G-3'; and U6 forward, 5'-CTC GCT TCG GCA GCA CA-3' and reverse,
5'-AAC GCT TCA CGA ATT TGC GT-3'. U6 was used for
normalization.
Western blot analysis
The H9c2 cardiomyocytes and NRVMs were plated in
6-well plates (1×105 per well). After transfecting the
cells for 48 h, total protein was extracted using the RIPA lysis
buffer (Beyotime Institute of Biotechnology, Shanghai, China). The
concentration and purity of the protein were measured using a
NanoDrop One Microvolume UV-Vis Spectrophotometer (Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions.
Protein samples were separated on SDS-PAGE gels (8%, 100-300 kDa;
10%, 30-100 kDa; 12%, 10-50 kDa) and transferred onto
polyvinylidene difluoride membranes (Millipore, Bedford, MA, USA).
The protein loaded was 35 µg per lane. The membranes were
blocked for 2 h in 5% skimmed milk at room temperature and then
incubated with primary antibodies (Beclin-1, sc-48341, 1:1,000;
Hspa5, sc-376768, 1:1,000; mTOR, sc-8319, 1:1,000; p-mTOR,
sc-293132, 1:1,000; Akt, sc-135829, 1:1,000; p-Akt, sc-271964,
1:1,000; PI3K, sc-293172, 1:1,000; all from Santa Cruz
Biotechnology; and p-PI3K, #4228, 1:1,000; cleaved caspase-3,
#9661, 1:1,000; Cell Signaling Technology, Danvers, MA, USA; Bcl-2,
sc-509, 1:1,000; Bax, sc-20067, 1:1,000; GAPDH, sc-47724, 1:1,000;
Santa Cruz Biotechnology) overnight at 4°C. The membranes were
washed with TBST 3 times, and incubated with horseradish
peroxidase-labeled secondary antibodies (7076; Cell Signaling
Technology) for 2 h at room temperature, and washed with TBST 3
times. Subsequently, the blots were detected using an enhanced
chemiluminescence kit and analyzed using ImageJ software.
Immunofluorescence
Cells on coverslips were fixed in 4%
paraformaldehyde for 20 min and permeabilized with 0.2% Triton
X-100 in PBS for 10 min, then blocked with PBS containing 2% bovine
serum albumin for 1 h at room temperature. Thereafter, the cells
were incubated with primary antibodies (LC3, #2775, 1:200, Cell
Signaling Technology) overnight at 4°C and incubated with secondary
antibodies (1647, 1:200, Invitrogen/Thermo Fisher Scientific, Inc.)
for 2 h at room temperature in the dark. Finally, the cells were
stained with 4',6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich/
Merck KGaA) for 5 min. The coverslips were washed with PBS after
each step. Images were captured using an Olympus IX50 inverted
fluorescence microscope (Olympus, Tokyo, Japan).
Cell apoptosis assay
The cells were plated in 6-well plates
(1×106 per well). After the cells were collected, they
were stained using an Annexin V-PE/7-amino-actinomycin D (7-AAD)
double staining kit (KeyGEN Biotech, Nanjing, China) according to
the manufacturer's instructions. Cell apoptosis was quantified
using FlowJo software (Tree Star, Inc., Ashland, OR, USA) on a
Beckman Coulter flow cytometer (Beckman Coulter, Indianapolis, IN,
USA).
Luciferase reporter assay
The potential miR-181b-5p-binding site in the 3'
untranslated region (3'-UTR) of the Hspa5 gene was predicted using
TargetScan (http://www.targetscan.org/cgi-bin/targetscan/vert_71/targetscan.cgi?mirg=hsa-miR-181b-5p).
H9c2 cardiomyocytes were plated into 24-well plates
(1×105 per well), and co-transfected with a pmir-GLO
Dual-Luciferase miRNA Target Expression Vector (Promega Corp.,
Madison, WI, USA) (containing a wild-type or mutant Hspa5 3'UTR)
and the miR-181b-5p mimic or miR-Con using Lipofectamine 2000
(Invitrogen/Thermo Fisher Scientific, Inc.). After 48 h, the
luciferase activity was measured using a Dual Luciferase Reporter
Gene Assay kit (Beyotime, Shanghai, China) according to the
manufacturer's instructions.
Statistical analysis
All data are represented as the means ± standard
deviation. Significant differences were determined using one-way
ANOVA followed by the Tukey's Honestly Significant Difference test.
Data were analyzed using SPSS version 20.0 software (IBM Corp.,
Armonk, NY, USA). Statistical significance is indicated by values
of P<0.05 or P<0.01.
Results
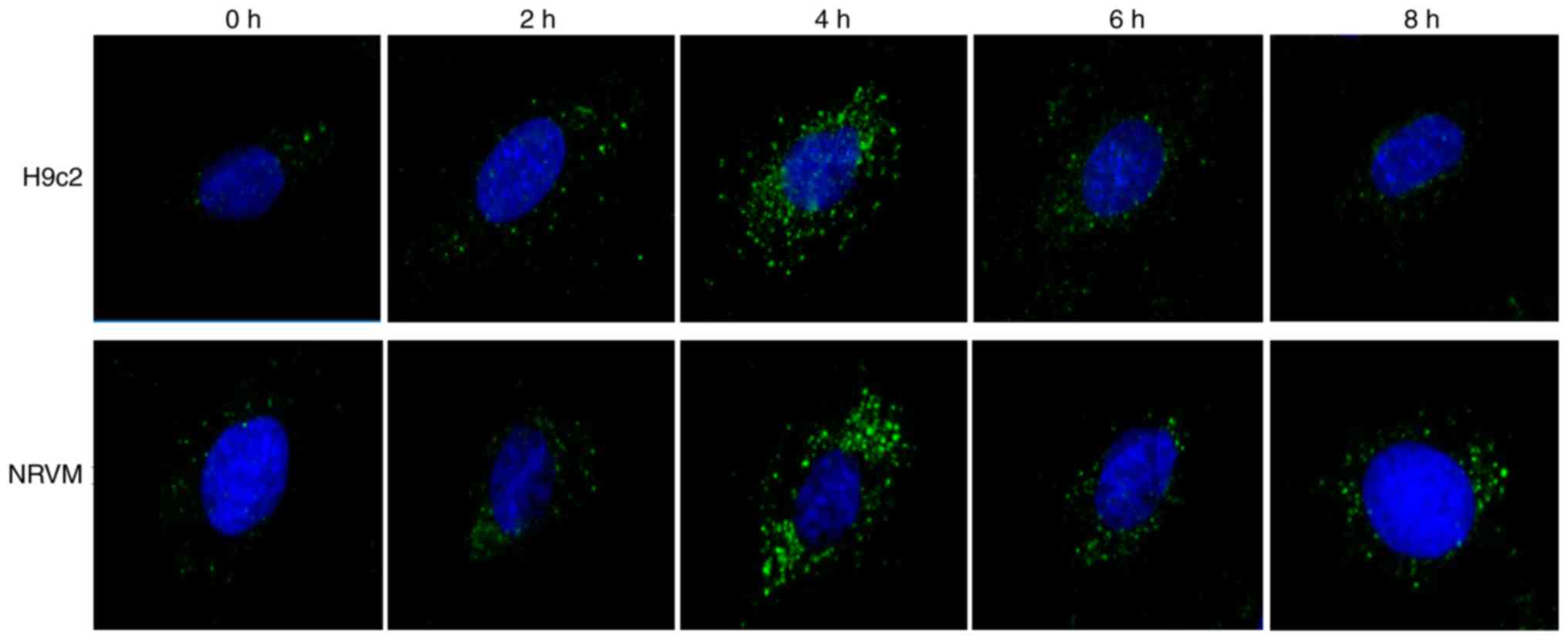
Starvation triggers autophagy, as well as
the suppression of miR-181b-5p expression in cardiomyocytes
To confirm that the autophagosomes were induced by
starvation, immunofluorescence was performed in H9c2 cardiomyocytes
and NRVMs cultured with EBSS for different time periods of time. As
shown in Fig. 1, the number of
LC3-GFP-positive vesicles increased in the starved cardiomyocytes,
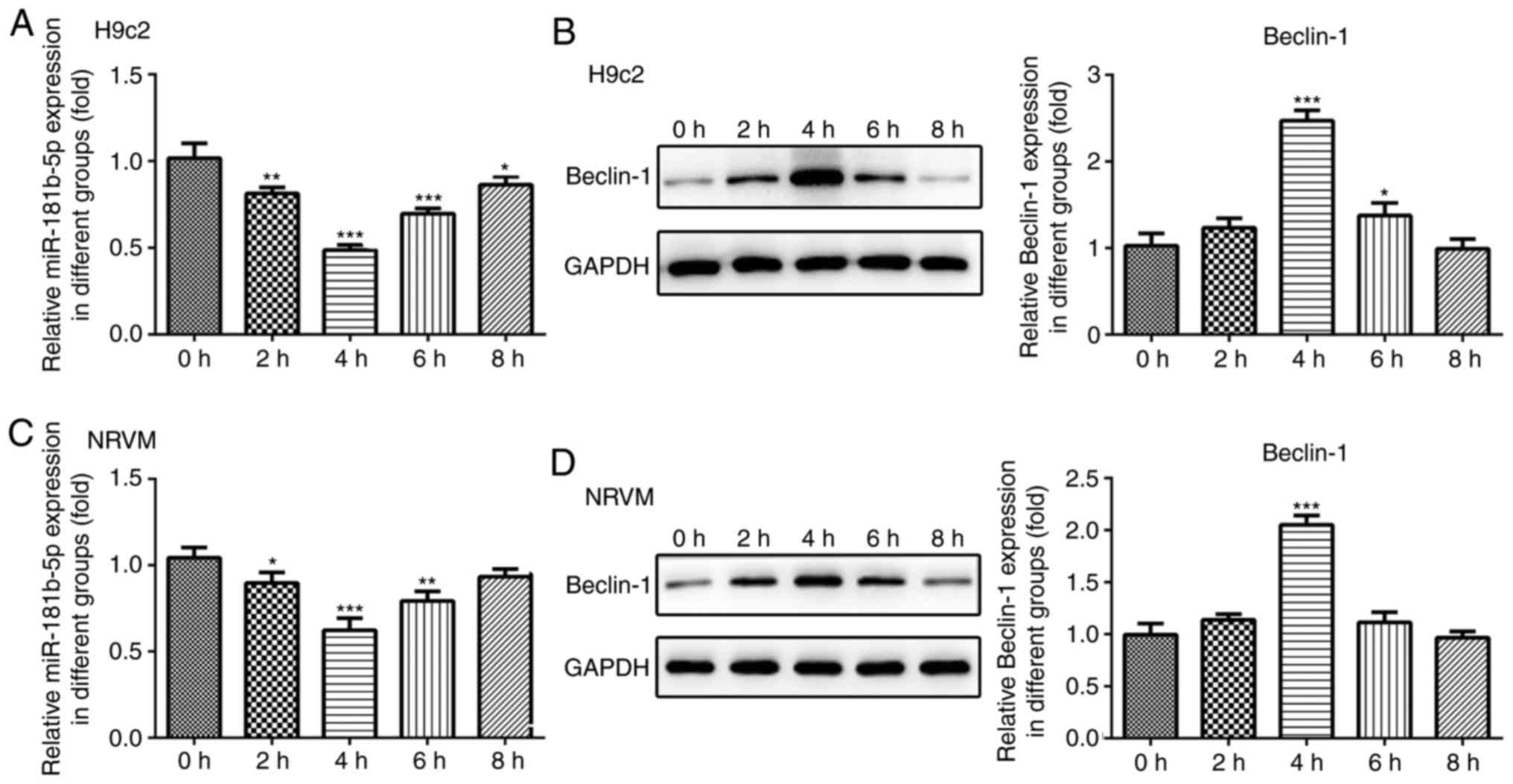
particularly after 4 h of starvation. Additionally, the protein
expression levels of Beclin-1 and Hspa5 were significantly
upregulated in the cardiomyocytes under starvation conditions after
4 h of starvation (Fig. 2B and
D). To assess the role of miR-181b-5p in the starved H9c2 and
NRVMs cardiomyocytes, RT-qPCR analysis was performed to measure the
expression of miR-181b-5p. As shown in Fig. 2A and C, the expression levels of
miR-181b-5p were decreased in the starved cardiomyocytes,
particularly after 4 h of starvation. Therefore, cell autophagy was
associated with miR-181b-5p. The cardiomyocytes starved for 4 h
were selected as the negative control (NC) for the subsequent
experiments.
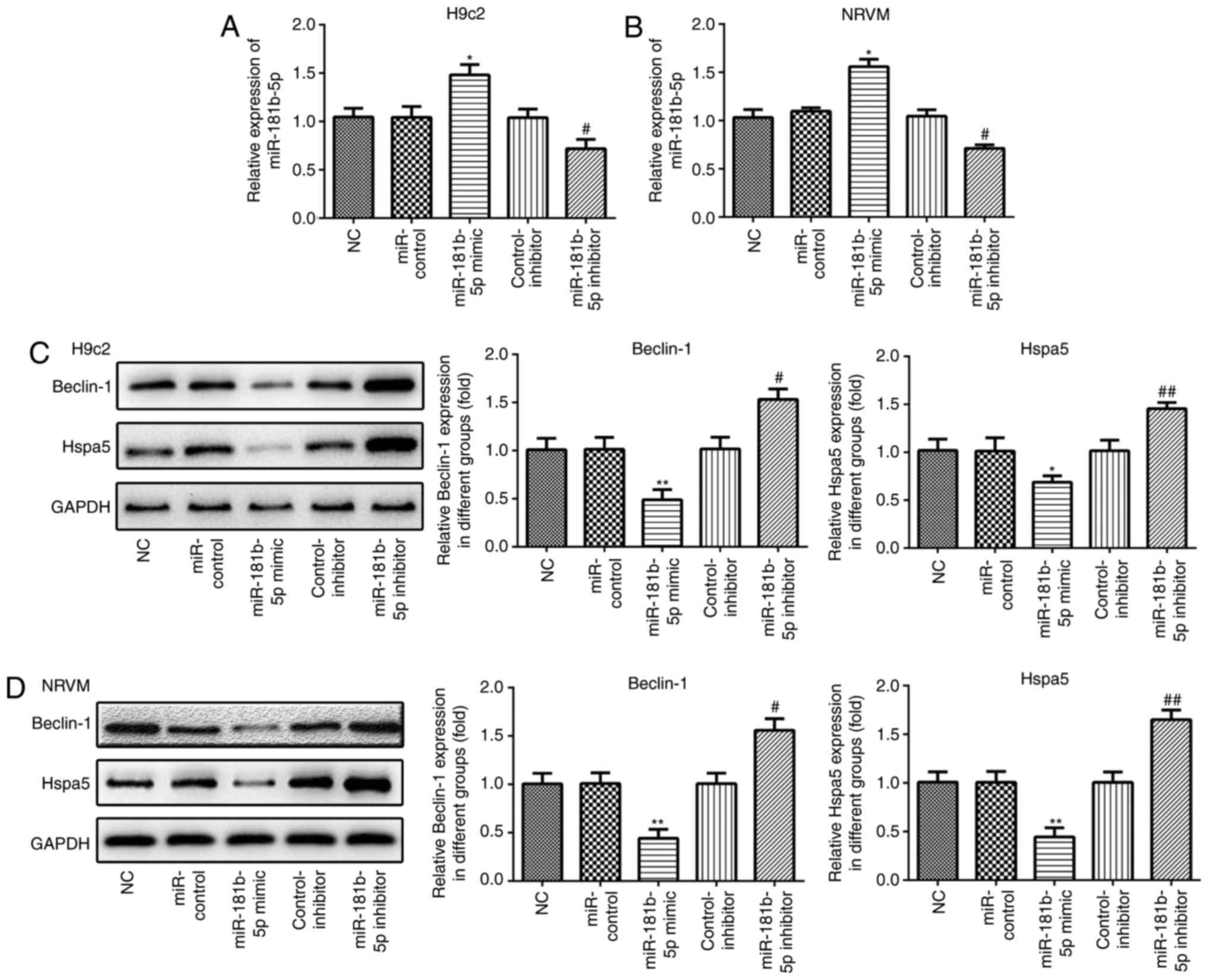
miR-181b-5p regulates Beclin-1 and Hspa5
expression in the cardiomyocytes under starvation conditions
RT-qPCR was performed to measure the expression of
miR-181b-5p in the transfected cardiomyocytes. As shown in Fig. 3A and B, the expression of
miR-181b-5p was markedly upregulated in the miR-181b-5p mimic group
and downregulated in the miR-181b-5p inhibitor group, compared with
their respective control groups. In accordance with the results of
western blot analysis, the overexpression of miR-181b-5p inhibited
the expression of Beclin-1 and Hspa5, while the inhibition of
miR-181b-5p promoted the expression of these proteins in the
starved H9c2 cardiomyocytes (Fig.
3C) and NRVMs (Fig. 3D).
These data suggest that miR-181b-5p regulates Beclin-1 and Hspa5
expression in starved H9c2 cardiomyocytes and NRVMs.
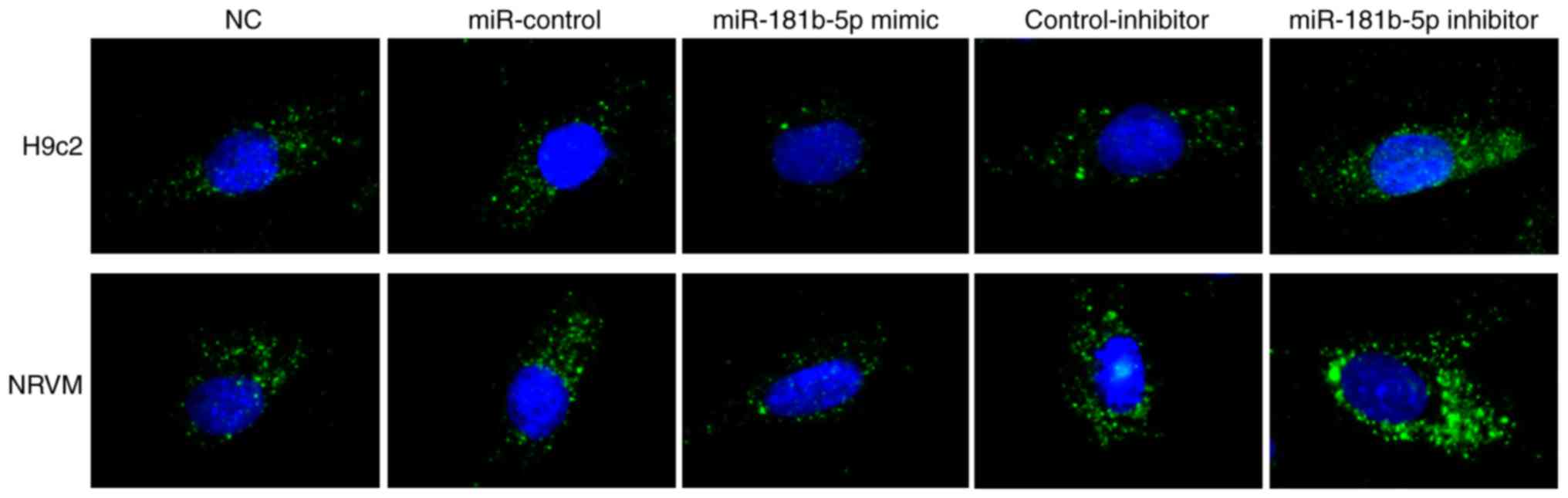
miR-181b-5p regulates starvation-induced
cardiomyocyte autophagy
To determine the role of miR-181b-5p in autophagy,
immunofluorescence was performed to observe the formation of
autophagosomes by detecting LC3B in transfected cardiomyocytes. As
shown in Fig. 4, no marked
differences were observed between the NC, miR-Con and inhibitor-Con
groups. The miR-181b-5p mimic group exhibited markedly reduced
autophagy compared with the miR-Con group. However, the inhibition
of miR-181b-5p in the starved H9c2 cardiomyocytes and NRVMs
markedly promoted autophagy compared with the inhibitor-Con group.
These data indicated that the downregulation of miR-181b-5p
promoted autophagy in the starved H9c2 cardiomyocytes and
NRVMs.
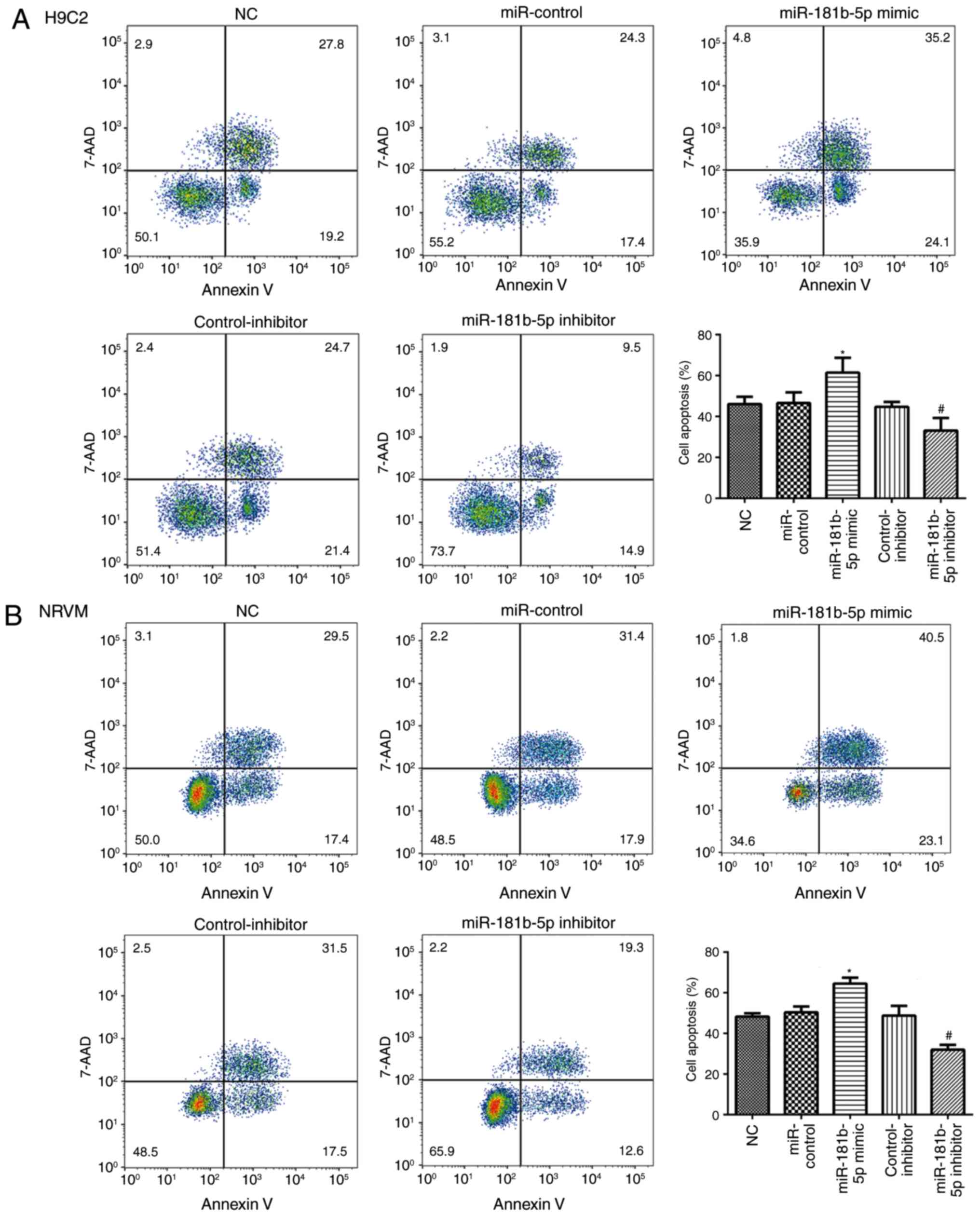
miR-181b-5p regulated cell apoptosis in
starved cardiomyocytes
To explore the role of miR-181b-5p in cell
apoptosis, flow cytometry and western blot analysis were performed
to measure cell apoptosis in the transfected cardiomyocytes. As
shown in Fig. 5, no significant
differences were observed in the apoptosis of the cells in the NC,
miR-control and control-inhibitor. However, the overexpres-sion of
miR-181b-5p in the starved H9c2 cardiomyocytes and NRVMs
significantly promoted cell apoptosis compared with the miR-Con
group. However, the inhibition of miR-181b-5p in the starved H9c2
cardiomyocytes and NRVMs significantly inhibited cell apoptosis
compared with the inhibitor-Con group. The results of western blot
analysis also revealed that the overexpression of miR-181b-5p
inhibited the protein expression levels of Bcl-2, while it
increased the protein expression levels of Bax and cleaved
caspase-3. Additionally, the inhibition of miR-181b-5p enhanced the
protein expression levels of Bcl-2, while it decreased the protein
expression levels of Bax and cleaved caspase-3 (Figs. 6A and B, and 7A and B). The
Bcl-2/Bax ratio was similar to the trend observed with the
expression of Bcl-2 (Figs. 6C and
7C). These data indicated that
miR-181b-5p regulated cell apoptosis in the starved H9c2
cardiomyocytes and NRVMs.
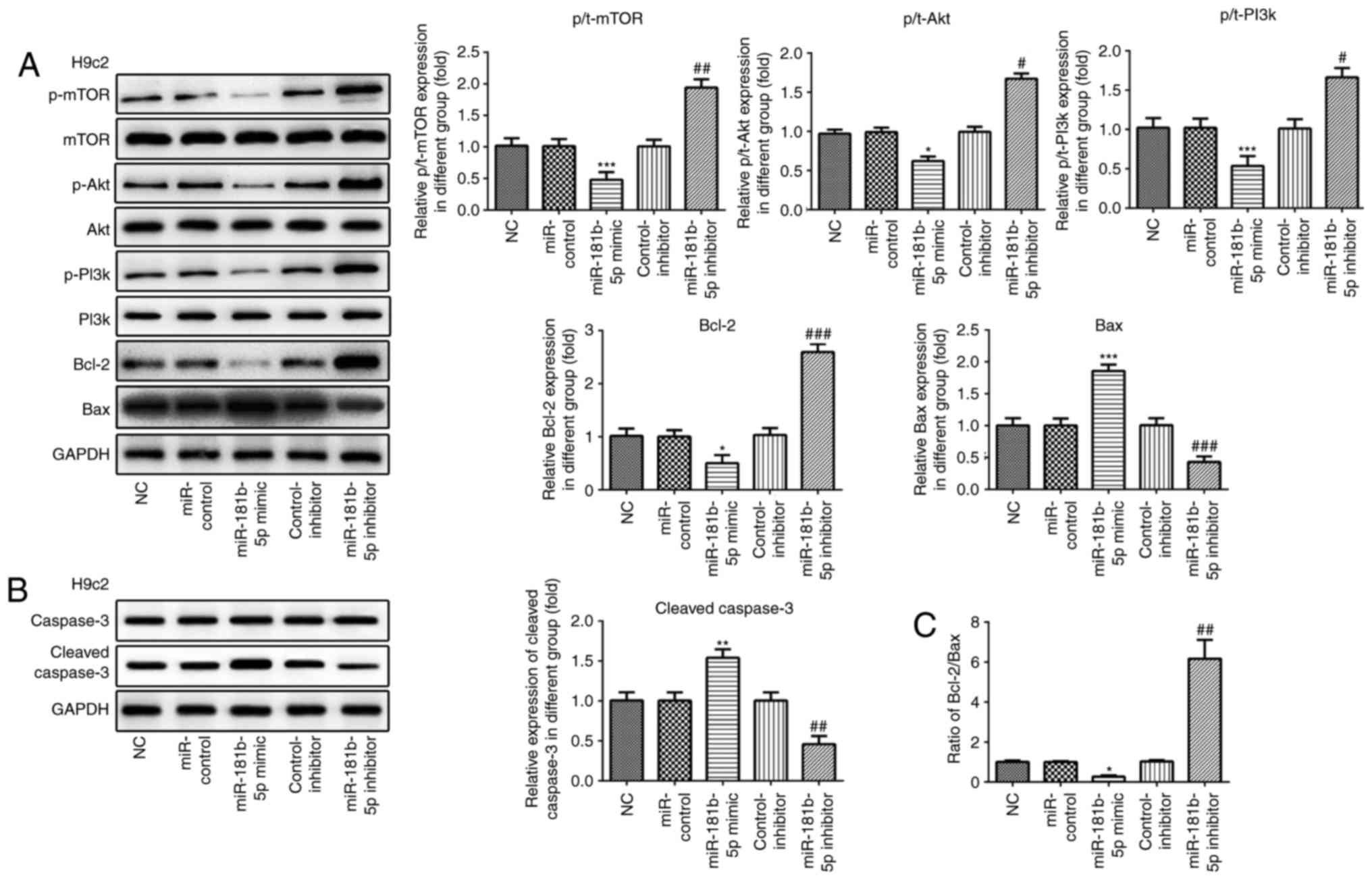 | Figure 6The PI3K/Akt/mTOR signaling pathway
and apoptosis-related proteins are regulated by miR-181b-5p in H9c2
cardiomyocytes. (A and B) The protein expression of p-mTOR, mTOR,
p-AKT, AKT, p-PI3K, PI3K, Bcl-2, Bax, caspase-3, cleaved caspase-3
and their grayscale scanning analysis. (C) The relative expression
ratio of Bcl-2/Bax. NC, untransfected cells; miR-control, cells
transfected with miR-181b-5p mimic control; miR-181b-5p mimic,
cells overexpressing miR-181b-5p; control-inhibitor, cells
transfected with miR-181b-5p inhibitor control; miR-181b-5p
inhibitor, cells in which miR-181b-5p was knocked down.
*P<0.05, **P<0.01 and
***P<0.001 vs. NC; #P<0.05,
##P<0.01 and ###P<0.001 vs. inhibitor
control. |
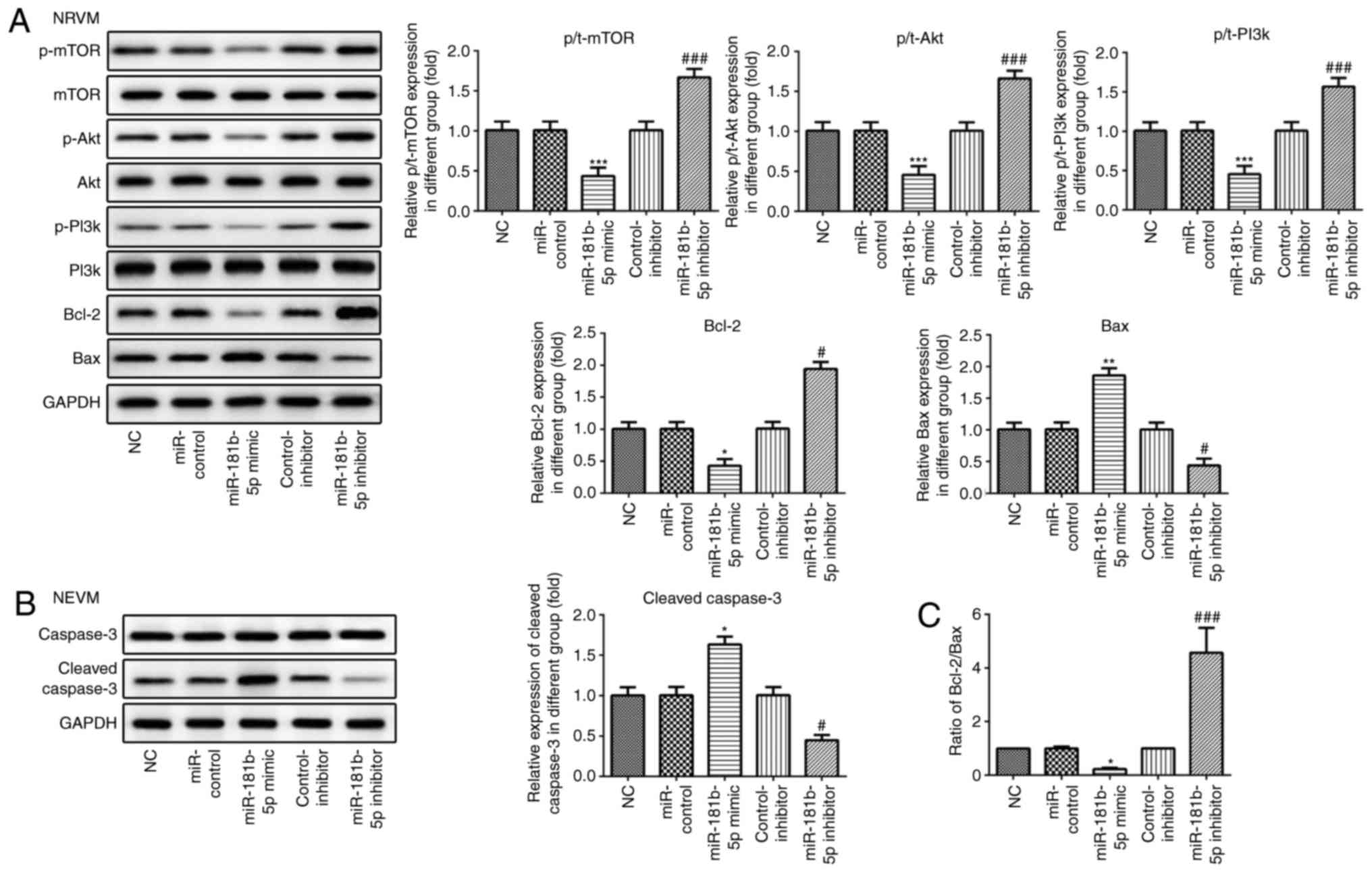 | Figure 7The PI3K/Akt/mTOR signaling pathway
and apoptosis-related proteins are regulated by miR-181b-5p in
NRVMs. (A and B) The protein expression of p-mTOR, mTOR, p-AKT,
AKT, p-PI3K, PI3K, Bcl-2, Bax, caspase-3, cleaved caspase-3 and
their grayscale scanning analysis. (C) The relative expression
ratio of Bcl-2/Bax. NC, untransfected cells; miR-control, cells
transfected with miR-181b-5p mimic control; miR-181b-5p mimic,
cells overexpressing miR-181b-5p; control-inhibitor, cells
transfected with miR-181b-5p inhibitor control; miR-181b-5p
inhibitor, cells in which miR-181b-5p was knocked down.
*P<0.05, **P<0.01 and
***P<0.001 vs. NC; #P<0.05 and
###P<0.001 vs. inhibitor control. NRVMs, neonatal rat
ventricular myocytes. |
miR-181b-5p regulates the PI3K/Akt/mTOR
signaling pathway in the starved cardiomyocytes
Western blot analysis was carried out to examine the
effects of miR-181b-5p on the PI3K/Akt/mTOR signaling pathway. As
shown in Figs. 6 and 7, the overexpression of miR-181b-5p in
the starved H9c2 cardiomyocytes and NRVMs significantly decreased
the protein expression levels of p-mTOR, p-Akt and p-PI3K, compared
with the miR-Con group. However, the inhibition of miR-181b-5p in
the starved H9c2 cardiomyocytes and NRVMs significantly increased
the protein expression levels of p-mTOR, p-Akt and p-PI3K, compared
with the inhibitor-Con group. These results indicated that
miR-181b-5p regulated the PI3K/Akt/mTOR signaling pathway in the
starved H9c2 cardiomyocytes and NRVMs.
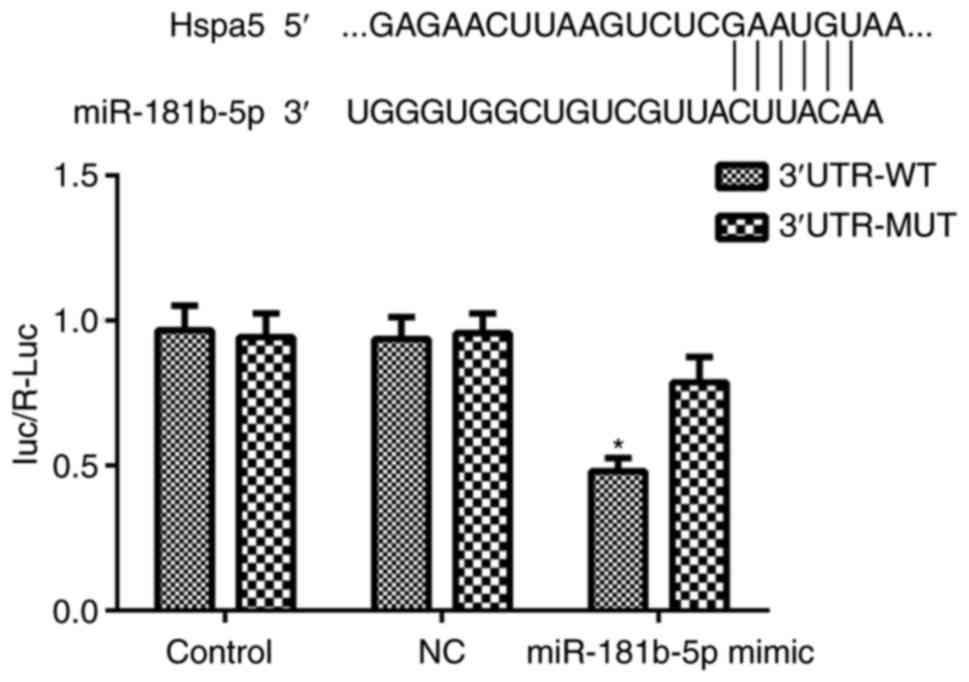
Hspa5 is a direct target of
miR-181b-5p
To elucidate the mechanisms through which
miR-181b-5p regulates autophagy and apoptosis in starved H9c2
cardiomyocytes and NRVMs, the potential targets of miR-181b-5p were
investigated using TargetScan, which identified the 3'-UTR region
of Hspa5 mRNA as a match to miR-181b-5p. According to a previous
study, Hspa5 participates in cardiac protection via autophagy
(19). Thus, in this study, a
dual-luciferase reporter assay was performed to examine whether
Hspa5 is a direct target of miR-181b-5p. As shown in Fig. 8, the miR-181b-5p mimic
significantly inhibited the dual-luciferase activity of the
3'UTR-WT of Hspa5 compared with the NC group. These data suggest
that Hspa5 is a direct target of miR-181b-5p and that miR-181b-5p
negatively regulates Hspa5.
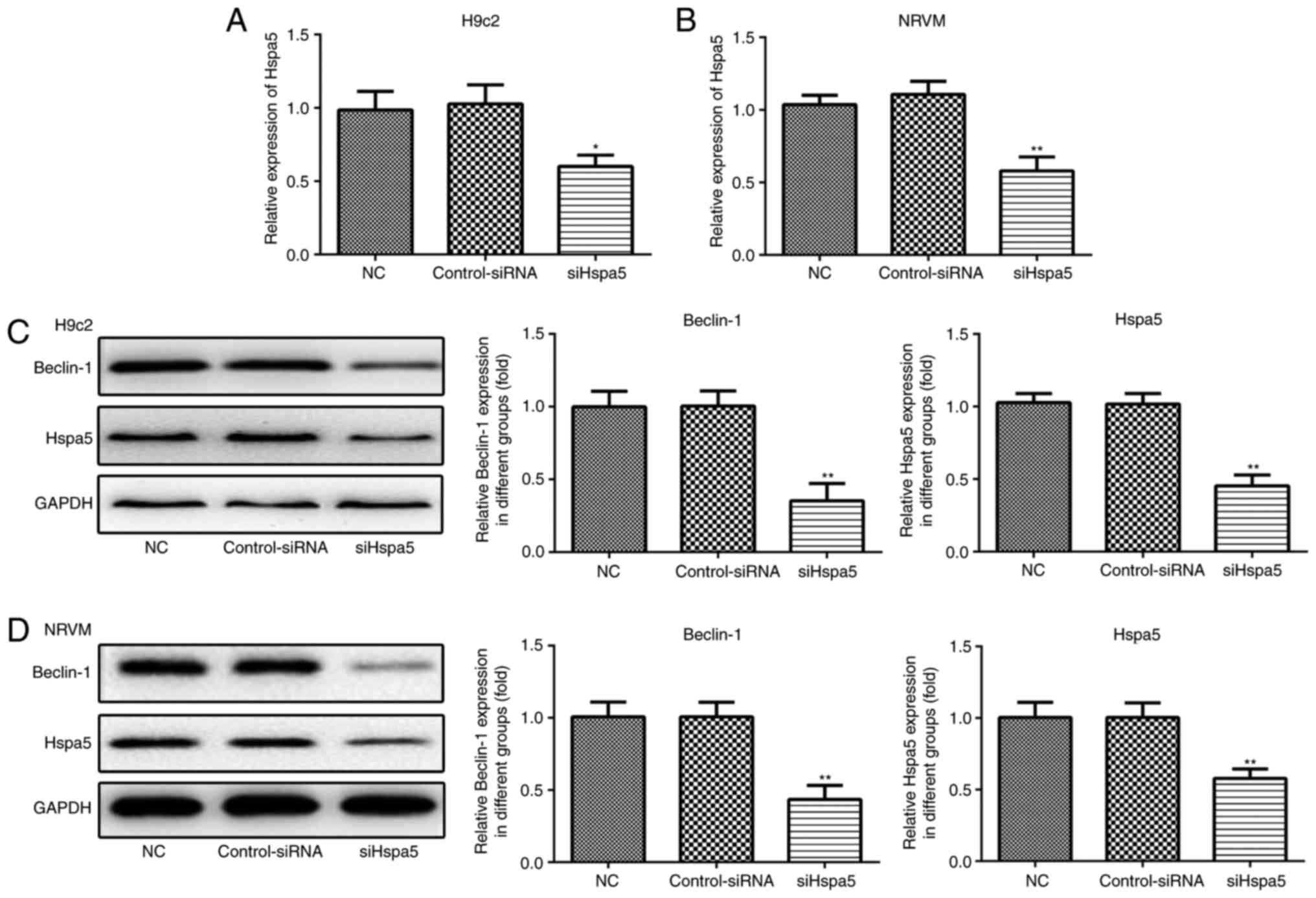
siHspa5 inhibits autophagy in starved
cardiomyocytes
RT-qPCR and western blot analysis were performed to
measure the expression levels of Hspa5 in the transfected
cardiomyocytes. As shown in Fig. 9A
and B, no significant difference was observed in the expression
of Hspa5 between the NC and siRNA-Con group; however, the mRNA and
protein expression levels of Hspa5 were downregulated in the
siHspa5 group, which indicated the successful transfection of
siHspa5. Western blot analysis was then performed to measure
Beclin-1 expression. siHspa5 inhibited the protein expression of
Beclin-1 in the H9c2 cardiomyocytes (Fig. 9C) and NRVMs (Fig. 9D). These data suggest that siHspa5
regulates Beclin-1 expression in starved H9c2 cardiomyocytes and
NRVMs.
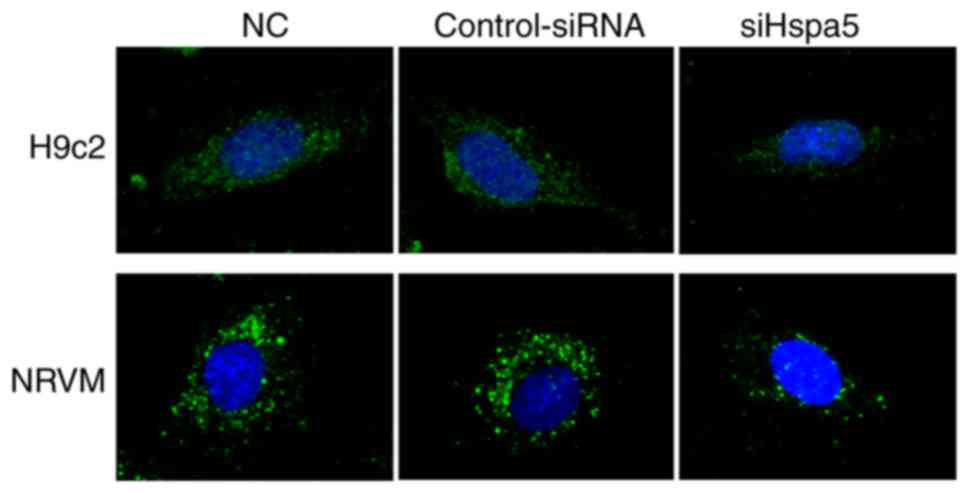
To determine the role of Hspa5 in autophagy,
immunofluorescence was performed to observe the autophagosomes in
the transfected cardiomyocytes. As shown in Fig. 10, siHspa5 markedly reduced
autophagy in the H9c2 and NRVMs compared with the Con-siRNA group.
These data indicated that Hspa5 regulated autophagy in the starved
H9c2 cardiomyocytes and NRVMs.
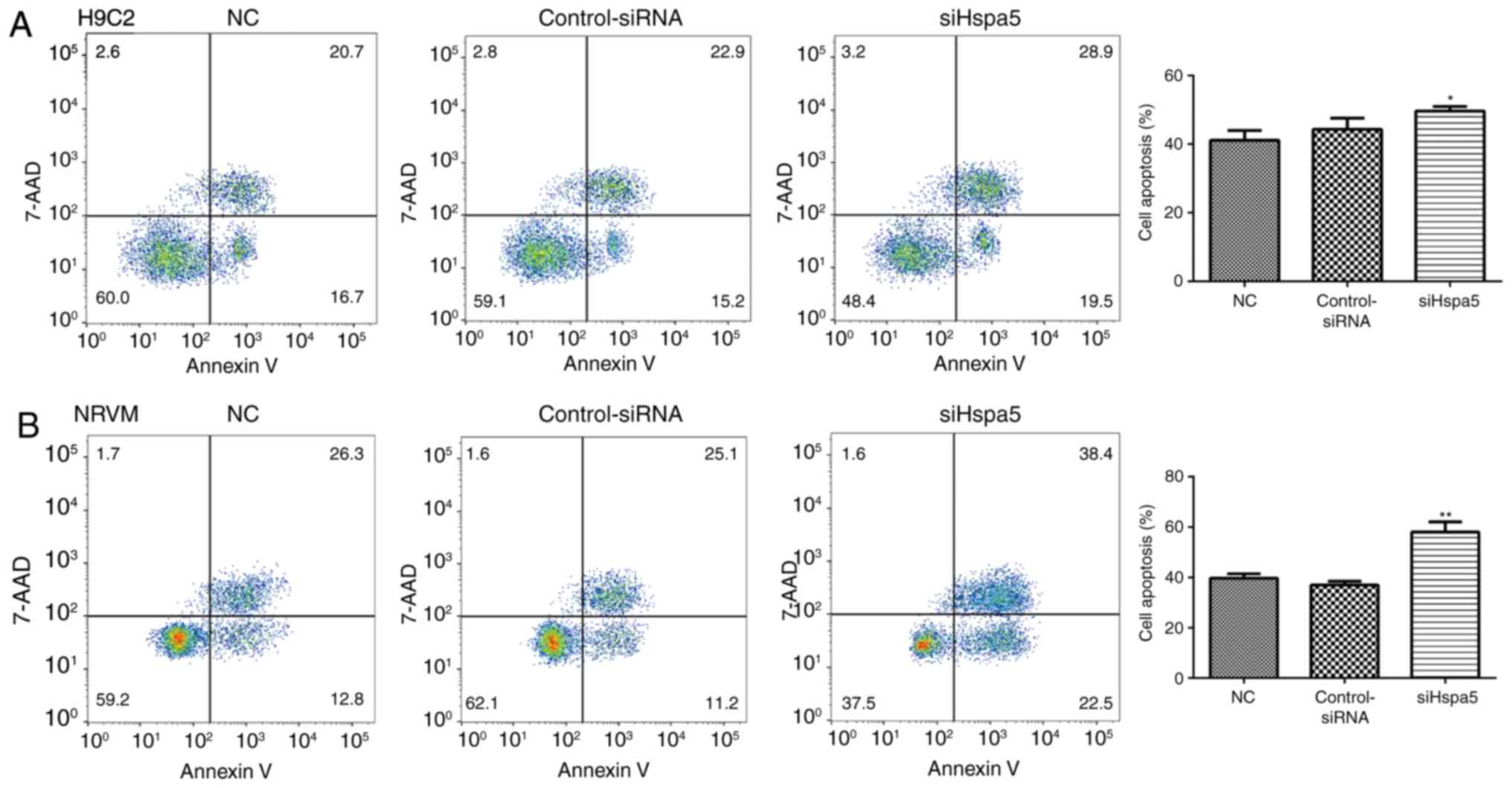
siHspa5 promotes the apoptosis of starved
cardiomyocytes
To explore the role of Hspa5 in cell apoptosis, flow
cytometry and western blot analysis were performed to measure the
apoptosis of the transfected cardiomyocytes. As shown in Fig. 11, transfection with siHspa5
significantly promoted the apoptosis of H9c2 and NRVMs compared
with the Con-siRNA group. Western blot analysis also revealed that
siHspa5 inhibited the protein expression levels of Bcl-2, while it
increased the protein expression levels of Bax and cleaved
caspase-3 (Figs. 12A and B, and
13A and B). The Bcl-2/Bax ratio
also exhibited a similar to Bcl-2 expression (Figs. 12C and 13C). These data indicated that siHspa5
induced the apoptosis of starved H9c2 and NRVMs.
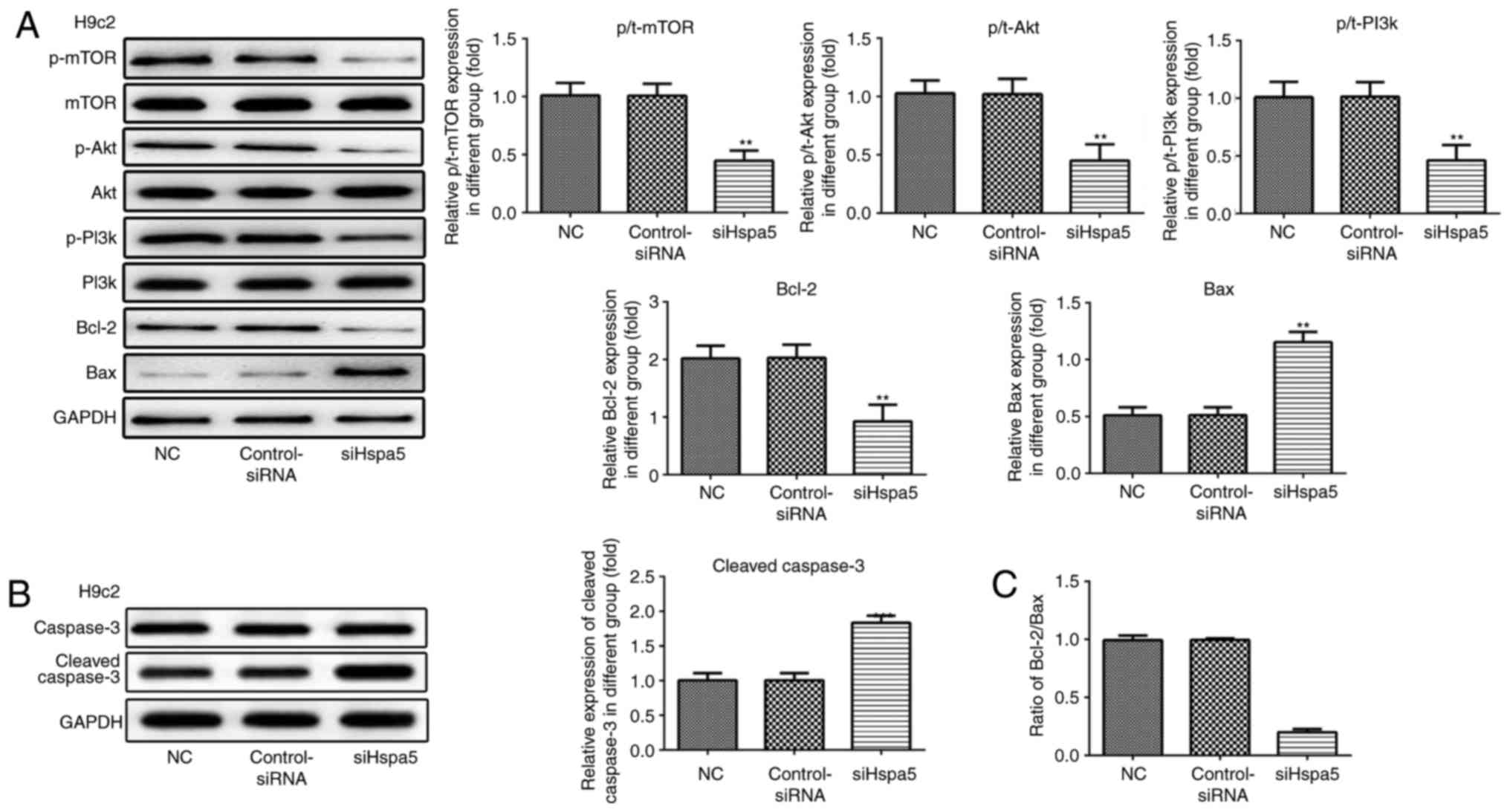
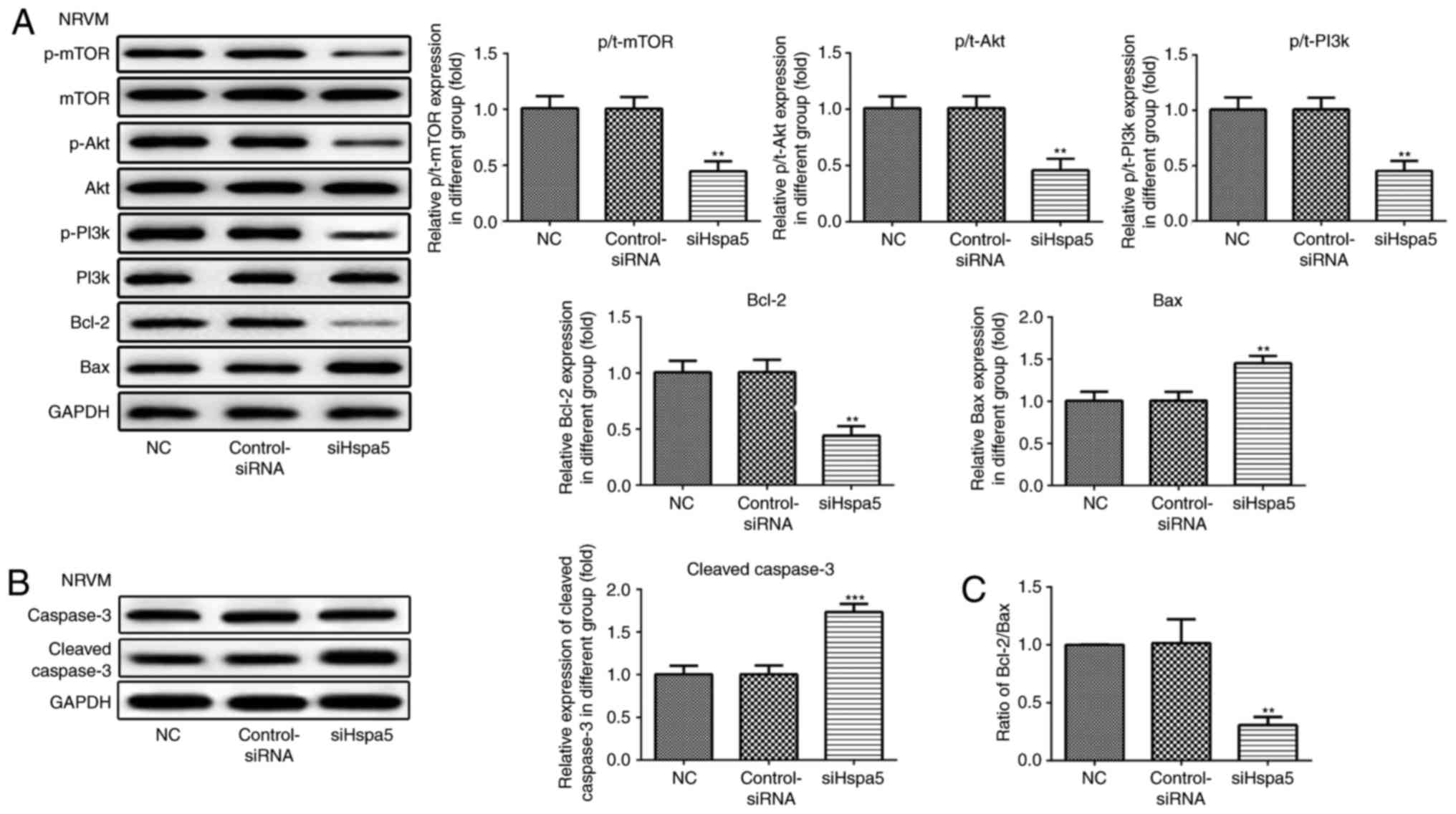
siHspa5 regulates the PI3K/Akt/mTOR
signaling pathway in starved cardiomyocytes
Western blot analysis was carried out to investigate
the effect of Hspa5 on the PI3K/Akt/mTOR signaling pathway. As
shown in Figs. 12 and 13, siHspa5 significantly decreased the
protein expression of p-mTOR, p-Akt and p-PI3K in the H9c2 and
NRVMs, compared with the Con-siRNA group. These results indicated
that Hspa5 regulates the PI3K/Akt/mTOR pathway in starved H9c2
cardiomyocytes and NRVMs.
Discussion
Previous studies have demonstrated that miRNAs play
an essential role in cardiovascular disease. To the best of our
knowledge, he current study is the first study to show the effects
of miR-181b-5p on cardiomyocytes, and that the downregulation of
miR-181b-5p can protect cardiomyocytes by promoting autophagy and
inhibiting apoptosis. Further investigations into the underlying
mechanisms indicated that autophagy and apoptosis were affected in
cardiomyo-cytes, as miR-181b-5p directly targeted Hspa5 and that
the PI3K/Akt/mTOR signaling pathway was downstream of Hspa5. Thus,
the Hspa5/PI3K/Akt/mTOR signaling pathway is activated by the
downregulation of miR-181b-5p in starved cardiomyocytes.
Autophagy is a biological process regulated by
various factors (20). Autophagy
helps cells respond to various stress factors inside and outside
the cell, including hunger, insulin deficiency, growth factors
deficiency and endoplasmic reticulum stress (21). In this study, H9c2 cardiomyocytes
and NRVMs were cultured in EBSS to establish a starvation model.
The results indicated that starvation triggered autophagosome
formation, and upregulated the protein expression of Beclin-1 in
H9c2 cardiomyocytes and NRVMs, particularly after 4 h of
starvation. In addition, miR-181b-5p expression was downregulated
in the starved cardiomyocytes, which revealed the possible
association between miR-181b-5p and autophagy.
miR-181b-5p has been reported in several diseases,
including astrocytoma, schizophrenia, pancreatic ductal
adenocarcinoma and non-small lung cancer (16,18,22,23). However, the effects of miR-181b-5p
have not yet been evaluated to the same extent in myocardial
diseases. A recent study found that miR-181b-5p expression was
markedly decreased in cardiac dysfunction progression, such as
diabetic cardiomyopathy (24). In
addition, miR-181b-5p expression has been shown to be upregulated
in heart failure resulting from cardiomyopathy (25). In this study, the effects of
miR-181b-5p on cardiomyocyte autophagy and apoptosis under
starvation conditions were elucidated in vitro. The results
revealed that miR-181b-5p downregulation promoted cell survival by
promoting autophagy and inhibiting apoptosis. Beclin-1 plays a
critical role in the regulation of both autophagy and cell death
(26). Hamacher-Brady et
al demonstrated that Beclin-1 overexpression promoted autophagy
and inhibited the apoptosis-related protein, Bax, to protect the
cardiomyocytes (27). In this
study, miR-181b-5p negatively regulated Beclin-1 expression and
positively regulated Bax expression. It has previously been
demonstrated that the increased expression levels of cleaved
caspase-3 and cleaved caspase-9 are affected by the release of
cytochrome c, which is related to mitochondrial apoptosis.
Bcl-2 inhibits apoptosis by binding to Bax; this binding plays a
role in the formation of mitochondrial outer membrane pores so that
mitochondria cannot release cytochrome c (28-30). In this study, cleaved caspase-3
and Bax were downregulated, while Bcl-2 was upregulated in
cardiomyocytes transfected with miR-181b-5p inhibitors. These
results indicate that the miR-181b-5p-mediated apoptosis of starved
cardiomyocytes may be as a result of mitochondrial apoptosis. Thus,
miR-181b-5p downregulation may promote autophagy via
Beclin-1-dependent autophagy and inhibit mitochondrial apoptosis;
however, further studies are required to validate this hypothesis.
Cytochrome c expression and mitochondrial membrane potential
warrant further investigation in the future.
miRNAs act by targeting multiple genes. Hspa5, also
known as GRP78, is an endoplasmic reticulum stress-related protein
that plays an important role in cell protection by preventing
protein aggregation (9). Hspa5
has been reported to play a role in starvation-induced
cardiomyocyte autophagy (31).
Since Hspa5 upregulation has been reported to be beneficial to the
treatment of cardiovascular diseases, its association with
miR-181b-5p was evaluated in this study. The results indicated that
miR-181b-5p negatively and directly regulated Hspa5 in
cardiomyocytes. Furthermore, cardiomyocytes transfected with
Hspa5-siRNA exhibited a decreased autophagy and increased
apoptosis. Hspa5 inhibition also downregulated the expression of
Beclin-1. Based on the role of Beclin-1 in autophagy and apoptosis,
it may be downstream of Hspa5, but the association between Hspa5
and Beclin-1 requires further investigation. It may also be
hypothesized that Hspa5 is a direct target of miR-181b-5p in the
autophagy of starved cardiomyocytes.
In this study, the PI3K/Akt/mTOR signaling pathway
was downregulated when miR-181b-5p was overexpressed or Hspa5 was
inhibited. The PI3K/Akt/mTOR signaling pathway is a recognized
signaling pathway that regulates numerous biological functions in
cardiomyocytes, including cell viability, apoptosis and autophagy
(32,33). This may be a downstream of Hspa5;
however, further investigations are required.
In conclusion, in this study, the effects of
miR-181b-5p and its targets on starved cardiomyocyte injury were
assessed and the underlying mechanisms were revealed. miR-181b-5p
mainly mediated autophagy and apoptosis. In addition, the effects
of miR-181b-5p were mediated via Hspa5 and PI3K/Akt/mTOR may be the
downstream signaling pathway. The PI3K/Akt/mTOR pathway signaling
pathway is a well-known autophagy pathway. Animal studies of the
miR-181b-5p-mediated effects may reveal the association among
miR-181b-5p, autophagy and apoptosis in more detail and therefore
may be performed in the future. This study demonstrates that
miR-181b-5p may be a potential target in the treatment and/or
prevention of cardiomyocyte injury.
Acknowledgments
Not applicable.
Funding
This study was funded by the Key Technology
Application Research of Suzhou grants (SS201639 to JZ, SS201638 to
JC, SS201763 to HX), the Basic Research of Medical Science of
Suzhou grant (SYSD2017090 to PC) and the National Natural Science
Foundation of China grant (81372024 to JZ).
Availability of data and materials
The analyzed data sets generated during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
LC and XC equal contributed to this article,
including in the study design, data analysis and data
interpretation. LC drafted the article. PC and JC collected data
and analyzed the data. HX and JZ were involed in the conception and
design of the study, and edited the language of the manuscript. All
authors have read and approved the final manuscript.
Ethics approval and consent to
participate
All animal protocols were approved by the
Experimental Animal Committee of the Second Affiliated Hospital of
Suchow University, Suchow, China.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Benjamin EJ, Virani SS, Callaway CW,
Chamberlain AM, Chang AR, Cheng S, Chiuve SE, Cushman M, Delling
FN, Deo R, et al: Heart disease and stroke statistics-2018 update:
A report from the american heart Association. Circulation.
137:e67–e492. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shimomura H, Terasaki F, Hayashi T,
Kitaura Y, Isomura T and Suma H: Autophagic degeneration as a
possible mechanism of myocardial cell death in dilated
cardiomyopathy. Jpn Circ J. 65:965–968. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Ma S, Wang Y, Chen Y and Cao F: The role
of the autophagy in myocardial ischemia/reperfusion injury. Biochim
Biophys Acta. 1852.271–276. 2015.
|
|
4
|
Wang K, Liu CY, Zhou LY, Wang JX, Wang M,
Zhao B, Zhao WK, Xu SJ, Fan LH, Zhang XJ, et al: APF lncRNA
regulates autophagy and myocardial infarction by targeting
miR-188-3p. Nat Commun. 6:67792015. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wu H, Wang Y, Wang X, Li R and Yin D:
MicroRNA-365 accelerates cardiac hypertrophy by inhibiting
autophagy via the modulation of Skp2 expression. Biochem Biophys
Res Commun. 484:304–310. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Takemura G, Kanamori H, Okada H, Miyazaki
N, Watanabe T, Tsujimoto A, Goto K, Maruyama R, Fujiwara T and
Fujiwara H: Anti-apoptosis in nonmyocytes and pro-autophagy in
cardiomyocytes: Two strategies against postinfarction heart failure
through regulation of cell death/degeneration. Heart Fail Rev.
23:759–772. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Palikaras K, Lionaki E and Tavernarakis N:
Mitophagy: In sickness and in health. Mol Cell Oncol.
3:e10563322015. View Article : Google Scholar
|
|
8
|
Ortiz C and Cardemil L: Heat-shock
responses in two leguminous plants: A comparative study. J Exp Bot.
52:1711–1719. 2001.PubMed/NCBI
|
|
9
|
Petrovski G, Das S, Juhasz B, Kertesz A,
Tosaki A and Das DK: Cardioprotection by endoplasmic reticulum
stress-induced autophagy. Antioxid Redox Signal. 14:2191–2200.
2011. View Article : Google Scholar
|
|
10
|
Eizirik DL, Cardozo AK and Cnop M: The
role for endoplasmic reticulum stress in diabetes mellitus. Endocr
Rev. 29:42–61. 2008. View Article : Google Scholar
|
|
11
|
Yorimitsu T, Nair U, Yang Z and Klionsky
DJ: Endoplasmic reticulum stress triggers autophagy. J Biol Chem.
281:30299–30304. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang L, Hong Q, Lv Y, Feng Z, Zhang X, Wu
L, Cui S, Hou K, Su H, Huang Z, et al: Autophagy can repair
endoplasmic reticulum stress damage of the passive Heymann
nephritis model as revealed by proteomics analysis. J Proteomics.
75:3866–3876. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang PL, Lun M, Teng J, Huang J, Blasick
TM, Yin L, Herrera GA and Cheung JY: Preinduced molecular
chaperones in the endoplasmic reticulum protect cardiomyocytes from
lethal injury. Ann Clin Lab Sci. 34:449–457. 2004.
|
|
14
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhang Z, Li H, Chen S, Li Y, Cui Z and Ma
J: Knockdown of MicroRNA-122 protects H9c2 cardiomyocytes from
hypoxia-induced apoptosis and promotes autophagy. Med Sci Monit.
23:4284–4290. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zhi F, Wang Q, Deng D, Shao N, Wang R, Xue
L, Wang S, Xia X and Yang Y: MiR-181b-5p downregulates NOVA1 to
suppress proliferation, migration and invasion and promote
apoptosis in astrocytoma. PLoS One. 9:e1091242014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huo X, Zhang K, Yi L, Mo Y, Liang Y, Zhao
J, Zhang Z, Xu Y and Zhen G: Decreased epithelial and plasma
miR-181b-5p expression associates with airway eosinophilic
inflammation in asthma. Clin Exp Allergy. 46:1281–1290. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Alacam H, Akgun S, Akca H, Ozturk O,
Kabukcu BB and Herken H: miR-181b-5p miR-195-5p and miR-301a-3p are
related with treatment resistance in schizophrenia. Psychiatry Res.
245:200–206. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lin R, Su Z, Tan X, Su Y, Chen Y, Shu X,
Liang S, Wang J and Xie S: Effect of endoplasmic reticulum stress
and autophagy in the regulation of post-infarct cardiac repair.
Arch Med Res. 2018. View Article : Google Scholar
|
|
20
|
Yang Z and Klionsky DJ: An overview of the
molecular mechanism of autophagy. Curr Top Microbiol Immunol.
335:1–32. 2009.PubMed/NCBI
|
|
21
|
He C and Klionsky DJ: Regulation
mechanisms and signaling pathways of autophagy. Annu Rev Genet.
43:67–93. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tomihara H, Yamada D, Eguchi H, Iwagami Y,
Noda T, Asaoka T, Wada H, Kawamoto K, Gotoh K, Takeda Y, et al:
MicroRNA-181b-5p ETS1, and the c-Met pathway exacerbate the
prognosis of pancreatic ductal adenocarcinoma after radiation
therapy. Cancer Sci. 108:398–407. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tian F, Shen Y, Chen Z, Li R, Lu J and Ge
Q: Aberrant miR-181b-5p and miR-486-5p expression in serum and
tissue of non-small cell lung cancer. Gene. 591:338–343. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Copier CU, Leon L, Fernandez M, Contador D
and Calligaris SD: Circulating miR-19b and miR-181b are potential
biomarkers for diabetic cardiomyopathy. Sci Rep. 7:135142017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Marques FZ, Vizi D, Khammy O, Mariani JA
and Kaye DM: The transcardiac gradient of cardio-microRNAs in the
failing heart. Eur J Heart Fail. 18:1000–1008. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhong Y, Wang QJ, Li X, Yan Y, Backer JM,
Chait BT, Heintz N and Yue Z: Distinct regulation of autophagic
activity by Atg14L and Rubicon associated with Beclin 1-
phosphatidylinositol-3-kin ase complex. Nat Cell Biol. 11:468–476.
2009. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hamacher-Brady A, Brady NR and Gottlieb
RA: Enhancing macroautophagy protects against ischemia/reperfusion
injury in cardiac myocytes. J Biol Chem. 281:29776–29787. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Lin M, Tang S, Zhang C, Chen H, Huang W,
Liu Y and Zhang J: Euphorbia factor L2 induces apoptosis in A549
cells through the mitochondrial pathway. Acta Pharm Sin B. 7:59–64.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Tao YW, Lin YC, She ZG, Lin MT, Chen PX
and Zhang JY: Anticancer activity and mechanism investigation of
beauvericin isolated from secondary metabolites of the mangrove
endophytic fungi. Anticancer Agents Med Chem. 15:258–266. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang JY, Lin MT, Tung HY, Tang SL, Yi T,
Zhang YZ, Tang YN, Zhao ZZ and Chen HB: Bruceine D induces
apoptosis in human chronic myeloid leukemia K562 cells via
mitochondrial pathway. Am J Cancer Res. 6:819–826. 2016.PubMed/NCBI
|
|
31
|
Chen L, Wang FY, Zeng ZY, Cui L, Shen J,
Song XW, Li P, Zhao XX and Qin YW: MicroRNA-199a acts as a
potential suppressor of cardiomyocyte autophagy through targeting
Hspa5. Oncotarget. 8:63825–63834. 2017.PubMed/NCBI
|
|
32
|
Hou X, Hu Z, Xu H, Xu J, Zhang S, Zhong Y,
He X and Wang N: Advanced glycation endproducts trigger autophagy
in cadiomyocyte via RAGE/PI3K/AKT/mTOR pathway. Cardiovasc
Diabetol. 13:782014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Yu W, Sun H, Zha W, Cui W, Xu L, Min Q and
Wu J: Apigenin attenuates adriamycin-Induced cardiomyocyte
apoptosis via the PI3K/AKT/mTOR pathway. Evid Based Complement
Alternat Med. 2017.2590676:2017.
|