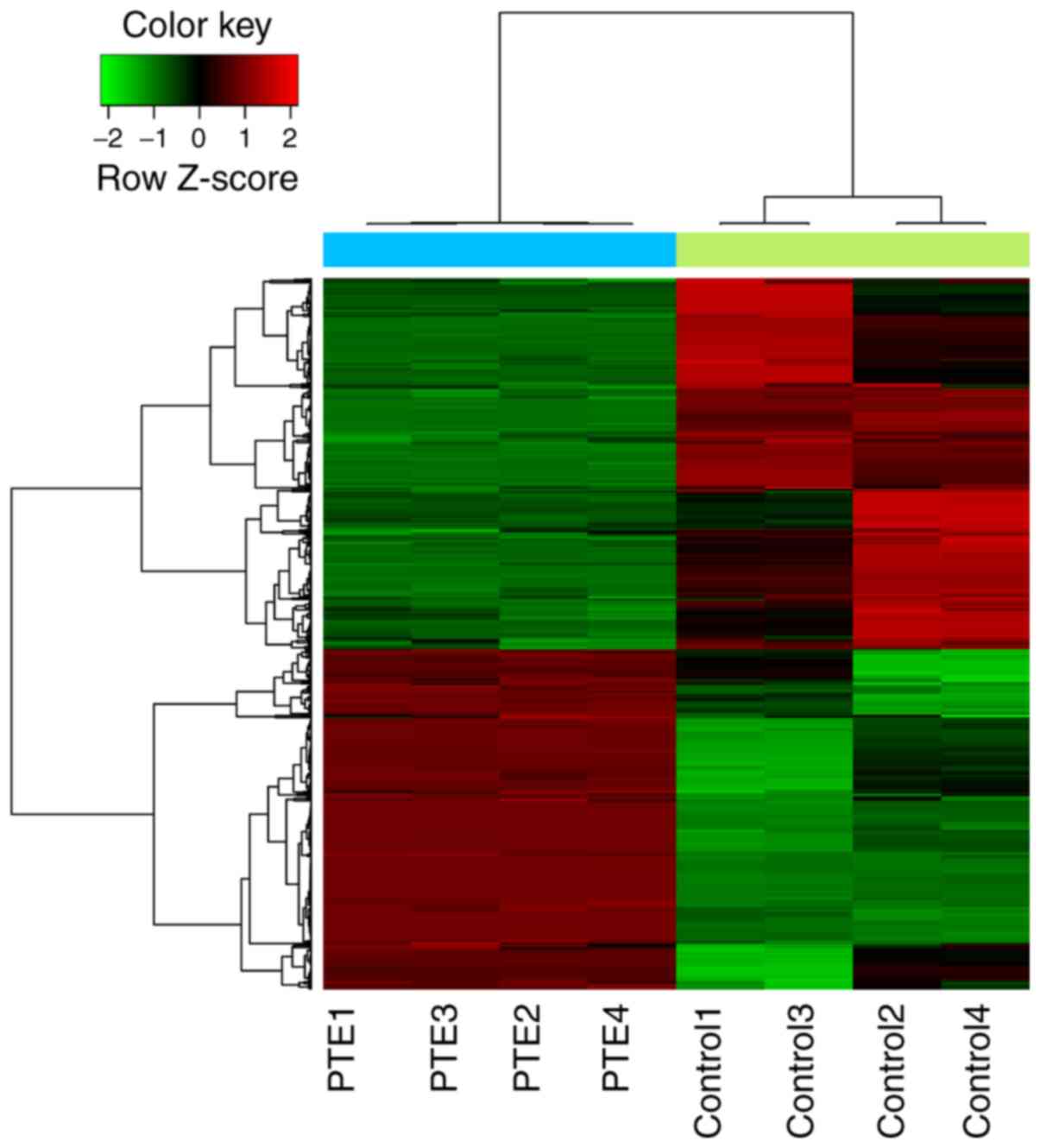
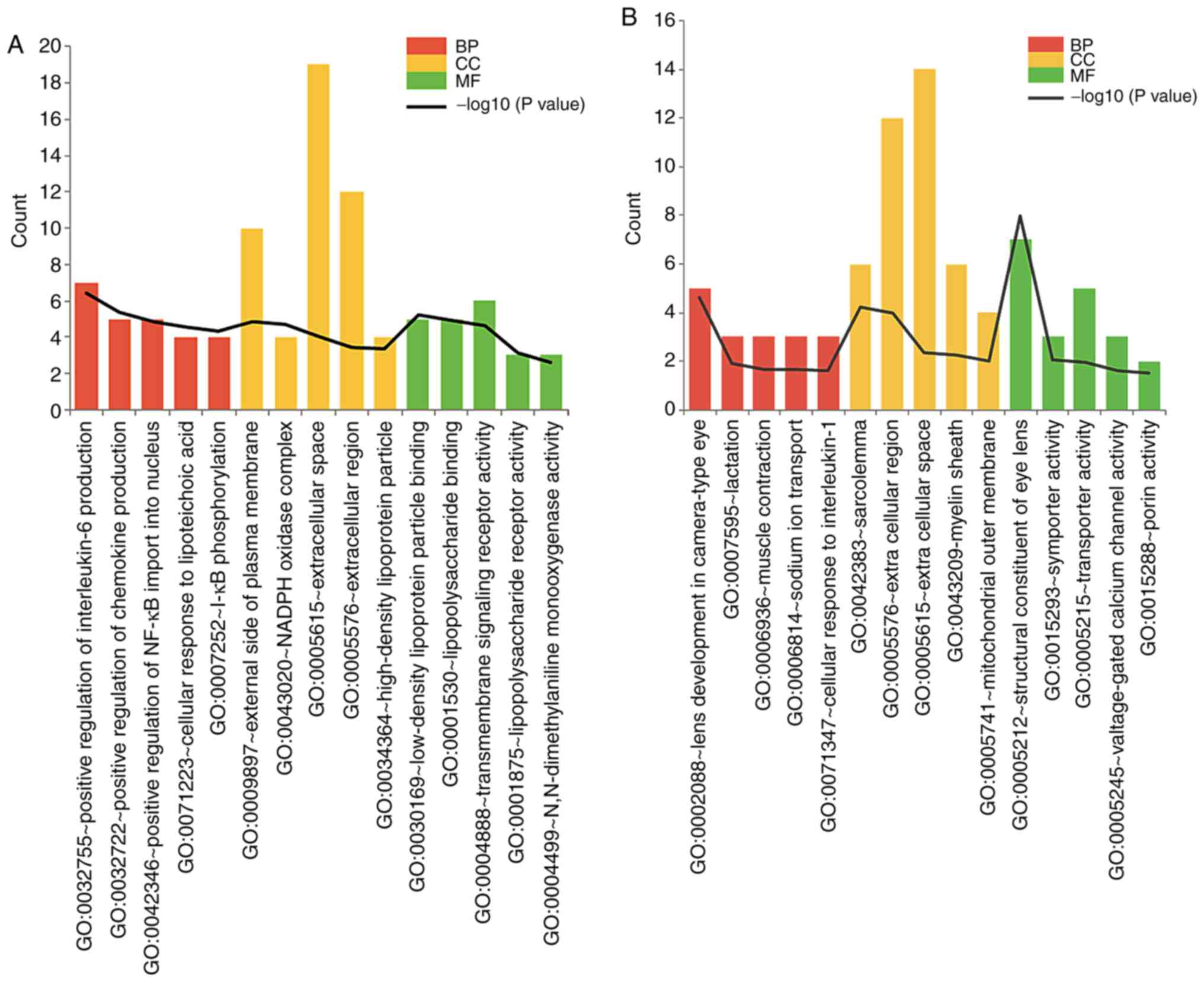
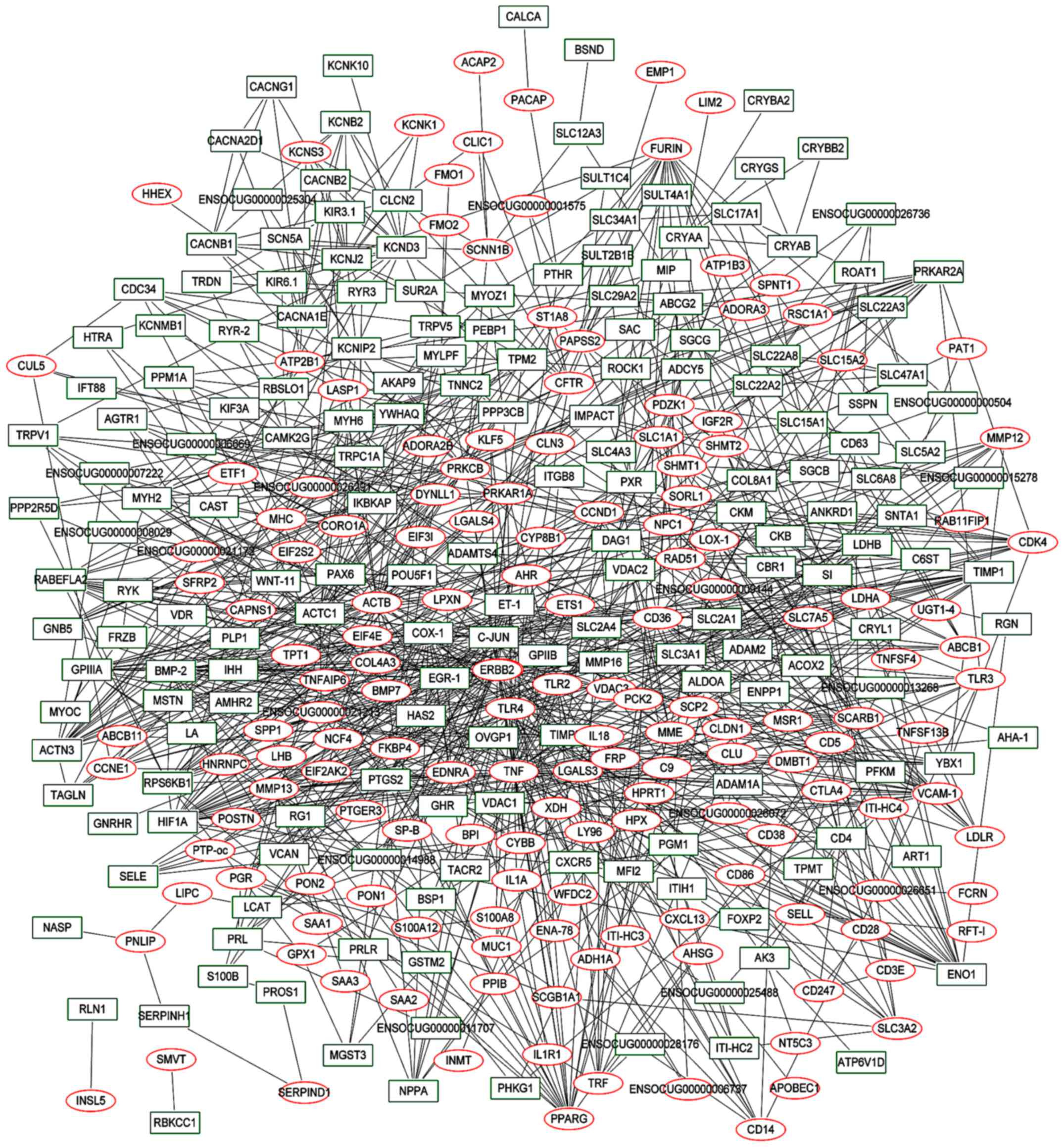
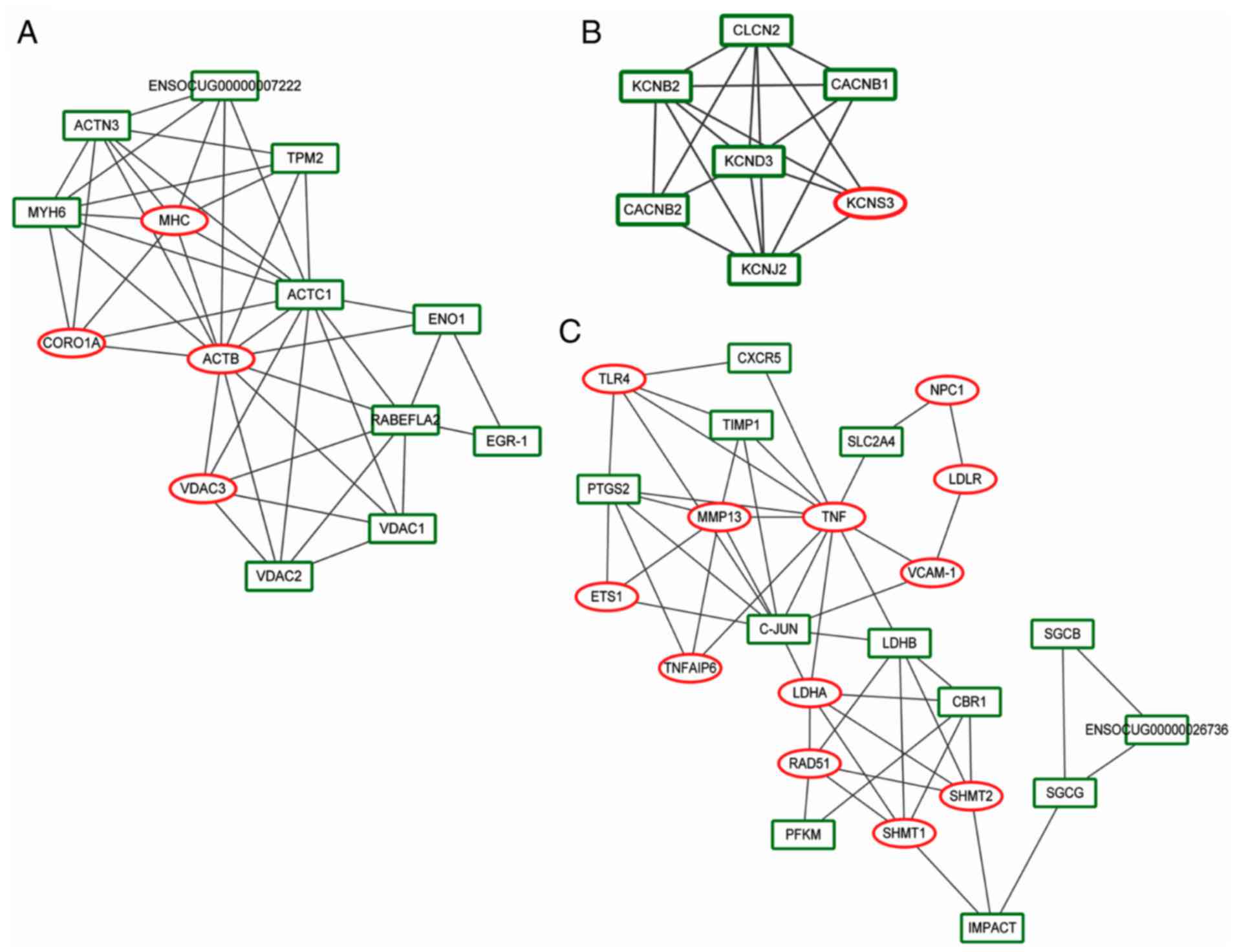
|
1
|
Santhakumar R, Ramalingam PK, Gayathri K,
Manjunath BV, Karuppusamy N, Vetriveeran B, Selvamani S, Vishnuram
P and Natarajan K: Pulmonary thromboembolism presenting as multiple
pulmonary cavities. J Assoc Physicians India. 64:85–86.
2016.PubMed/NCBI
|
|
2
|
Wood KE: Major pulmonary embolism: Review
of a pathophysiologic approach to the golden hour of
hemodynamically significant pulmonary embolism. Chest.
121:8779052002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kostadima E and Zakynthinos E: Pulmonary
embolism: Pathophysiology, diagnosis, treatment. Hellenic J
Cardiol. 48:94–107. 2007.PubMed/NCBI
|
|
4
|
Evans CE, Humphries J, Mattock K, Waltham
M, Wadoodi A, Saha P, Modarai B, Maxwell PH and Smith A: Hypoxia
and upregulation of hypoxia-inducible factor 1{alpha} stimulate
venous thrombus recanalization. Arterioscler Thromb Vasc Biol.
30:2443–2451. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yılmaz M: Evaluation of Tp‑e interval,
Tp‑e/qt ratio and Tp‑e/Qtc ratio in patients with acute pulmonary
thromboembolism. Am J Cardiol. 121:e192018. View Article : Google Scholar
|
|
6
|
Battistini B: Modulation and roles of the
endothelins in the pathophysiology of pulmonary embolism. Canad J
Physiol Pharmacol. 81:555–569. 2003. View
Article : Google Scholar
|
|
7
|
Kimura H, Okada O, Tanabe N, Tanaka Y,
Terai M, Takiguchi Y, Masuda M, Nakajima N, Hiroshima K, Inadera H,
et al: Plasma monocyte chemoattractant protein-1 and pulmonary
vascular resistance in chronic thromboembolic pulmonary
hypertension. Am J Respir Crit Care Med. 164:319–324. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wan J, Lu LJ, Miao R, Liu J, Xu XX, Yang
T, Hu QH, Wang J and Wang C: Alterations of bone marrow‑derived
endothelial progenitor cells following acute pulmonary embolism in
mice. Exp Biol Med (Maywood). 235:989–998. 2010. View Article : Google Scholar
|
|
9
|
Raskob GE, Angchaisuksiri P, Blanco AN,
Buller H, Gallus A, Hunt BJ, Hylek EM, Kakkar A, Konstantinides SV,
McCumber M, et al: Thrombosis: A major contributor to global
disease burden. Arterioscler Thromb Vasc Biol. 34:2363–2371. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pieralli F, Olivotto I, Vanni S, Conti A,
Camaiti A, Targioni G, Grifoni S and Berni G: Usefulness of bedside
testing for brain natriuretic peptide to identify right ventricular
dysfunction and outcome in normotensive patients with acute
pulmonary embolism. Am J Cardiol. 97:1386–1390. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Kostrubiec M, Pruszczyk P, Bochowicz A,
Pacho R, Szulc M, Kaczynska A, Styczynski G, Kuch‑Wocial A,
Abramczyk P, Bartoszewicz Z, et al: Biomarker‑based risk assessment
model in acute pulmonary embolism. Eur Heart J. 26:2166–2172. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Babaoglu E, Hasanoglu HC, Senturk A,
Karalezli A, Kilic H, Aykun G and Oztuna D: Importance of
biomarkers in risk stratification of pulmonary thromboembolism
patients. J Investig Med. 62:328–331. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Tang Z, Wang X, Huang J, Zhou X, Xie H,
Zhu Q, Huang M and Ni S: Gene expression profiling of pulmonary
artery in a rabbit model of pulmonary thromboembolism. PLoS One.
11:e01645302016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
He X, Zhang Z, Zhang Q and Yuan G:
Selective recognition of G‑quadruplex in the vascular endothelial
growth factor gene with small‑molecule natural products by
electrospray ionization (ESI) mass spectrometry and circular
dichroism (CD) spectrometry. Canad J Chem. 90:55–59. 2012.
View Article : Google Scholar
|
|
15
|
An R, Hagiya Y, Tamura A, Li S, Saito H,
Tokushima D and Ishikawa T: Cellular phototoxicity evoked through
the inhibition of human ABC transporter ABCG2 by cyclin‑dependent
kinase inhibitors in vitro. Pharm Res. 26:449–458. 2009. View Article : Google Scholar
|
|
16
|
Wang L, Cao C, Ma Q, Zeng Q, Wang H, Cheng
Z, Zhu G, Qi J, Ma H, Nian H and Wang Y: RNA‑seq analyses of
multiple meristems of soybean: Novel and alternative transcripts,
evolutionary and functional implications. BMC Plant Biol.
14:1692014. View Article : Google Scholar
|
|
17
|
Szklarczyk D, Franceschini A, Kuhn M,
Simonovic M, Roth A, Minguez P, Doerks T, Stark M, Muller J, Bork
P, et al: The STRING database in 2011: Functional interaction
networks of proteins, globally integrated and scored. Nucleic Acids
Res. 39(Database Issue): D561–D568. 2011. View Article : Google Scholar :
|
|
18
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:249825042003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar
|
|
22
|
Davis AP, Grondin CJ, Lennon‑Hopkins K,
Saraceni‑Richards C, Sciaky D, King BL, Wiegers TC and Mattingly
CJ: The comparative toxicogenomics database’s 10th year
anniversary: Update 2015. Nucleic Acids Res. 43(Database Issue):
D914–D920. 2015. View Article : Google Scholar
|
|
23
|
Yan C, Wang X, Su H and Ying K: Recent
progress in research on the pathogenesis of pulmonary
thromboembolism: An old story with new perspectives. Biomed Res
Int. 2017.6516791:2017.
|
|
24
|
Wrobel JP, Thompson BR and Williams TJ:
Mechanisms of pulmonary hypertension in chronic obstructive
pulmonary disease: A pathophysiologic review. J Heart Lung
Transplant. 31:5572012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jo JY, Lee MY, Lee JW, Rho BH and Choi WI:
Leukocytes and systemic inflammatory response syndrome as
prognostic factors in pulmonary embolism patients. BMC Pulm Med.
13:742013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Halici B, Sarinc US, Günay E, Nural S, Sen
S, Akar O, Celik S and Unlu M: Assessment of inflammatory
biomarkers and oxidative stress in pulmonary thromboembolism:
Follow‑up results. Inflammation. 37:1186–1190. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Marchena Yglesias PJ, Nieto Rodríguez JA,
Serrano Martínez S, Belinchón Moya O, Cortés Carmona A, Díaz de
Tuesta A, Bruscas Alijarde MJ and Ruiz Ribó MD: Acute-phase
reactants and markers of inflammation in venous thromboembolic
disease: Correlation with clinical and evolution parameters. An Med
Interna. 23:105–110. 2006.In Spanish. PubMed/NCBI
|
|
28
|
Santosa F, Moerchel C, Berg C and Kröger
K: Disproportional increase of pulmonary embolism in young females
in Germany: Trends from 2005 to 2014. J Thromb Thrombolysis.
43:417–422. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Bird ST, Delaney JA, Etminan M, Brophy JM
and Hartzema AG: Drospirenone and non-fatal venous thromboembolism:
Is there a risk difference by dosage of ethinyl‑estradiol. J Thromb
Haemost. 11:1059–1068. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jiang X, Sung YK, Tian W, Qian J, Semenza
GL and Nicolls MR: Graft microvascular disease in solid organ
transplantation. J Mol Med (Berl). 92:797–810. 2014. View Article : Google Scholar
|
|
31
|
Rodriguesdiez R, Gonzálezguerrero C,
Ocañasalceda C, Rodriguesdiez RR, Egido J, Ortiz A, Ruizortega M
and Ramos AM: Calcineurin inhibitors cyclosporine A and tacrolimus
induce vascular inflammation and endothelial activation through
TLR4 signaling. Sci Rep. 6:279152016. View Article : Google Scholar
|
|
32
|
Rafiee P, Johnson CP, Li MS, Ogawa H,
Heidemann J, Fisher PJ, Lamirand TH, Otterson MF, Wilson KT and
Binion DG: Cyclosporine a enhances leukocyte binding by human
intestinal microvascular endothelial cells through inhibition of
p38 MAPK and iNOS. Paradoxical proinflammatory effect on the
micro-vascular endothelium. J Biol Chem. 277:35605–35615. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chavez‑Blanco A, Perez‑Sanchez V,
Gonzalez‑Fierro A, Vela‑Chavez T, Candelaria M, Cetina L, Vidal S
and Dueñas‑Gonzalez A: ER2 expression in cervical cancer as a
potential therapeutic target. BMC Cancer. 4:592004. View Article : Google Scholar
|
|
34
|
Balwierz AK: ERBB2 as a driver of an
invasive phenotype of cells grown in 3D culture and an important
regulator of oncogenic miRNAs’ expression in breast cancer.
2017.
|
|
35
|
Ahmed AR: HER2 expression is a strong
independent predictor of nodal metastasis in breast cancer. J Egypt
Natl Cancer Inst. 28:219–227. 2016. View Article : Google Scholar
|
|
36
|
Zhou X, De Luise C, Shen R, Bate A and
Gatto N: Incidence of pulmonary embolism among postmenopausal (pm)
women with Er+/HER2− breast cancer.
Pharmacoepidemiol Drug Safety. 24:372–373. 2015.
|
|
37
|
Massraro GD and Massaro D: Retinoic acid
treatment abrogates elastase-induced pulmonary emphysema in rats.
Nat Med. 3:675–677. 1997. View Article : Google Scholar
|
|
38
|
Jian‑Lin XU, Fan XY, Guo YL, Jin MA and
Wang WM: Effects of all-trans-retinoic acid on emphysema model in
rats. Chin J New Drugs Clin Remed. 29:109–114. 2010.In Chinese.
|
|
39
|
Faubert BL and Kaminski NE: AP‑1 activity
is negatively regulated by cannabinol through inhibition of its
protein components, c‑fos and c‑jun. J Leukoc Biol. 67(259):
2011–266. 2000. View Article : Google Scholar
|
|
40
|
Eferl R, Ricci R, Kenner L, Zenz R, David
JP, Rath M and Wagner EF: Liver tumor development. c‑Jun
antagonizes the proapoptotic activity of p53. Cell. 112:181–192.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Guo W, Pylayeva Y, Pepe A, Yoshioka T,
Muller WJ, Inghirami G and Giancotti FG: Beta 4 integrin amplifies
ErbB2 signaling to promote mammary tumorigenesis. Cell.
126:489–502. 2006. View Article : Google Scholar : PubMed/NCBI
|