Introduction
Similar to other human cancer types, breast cancer
(BRCA) is one of the most frequently diagnosed cancer types and is
the leading cause of cancer-associated mortality among females
worldwide. BRCA comprises a heterogeneous group of neoplasms; the
mechanisms underlying the development of this disease remain poorly
understood (1). The prognosis of
patients with BRCA varies and depends on the subtype, age and the
extent of the disease (2). BRCA
can be classified using several grading systems from the United
States National Cancer Institute Breast/Ovarian Cancer Family
Registry and systems based on histological type and grade (3,4),
which may affect the outcome and response to treatment. At present,
estrogen receptor (ER), progesterone receptor (PR) and human
epidermal-growth factor receptor 2 (HER2) are considered as three
biomarkers of cancer, and may be employed to aid the selection of
treatment for BRCA (5). Patients
with ER+ and/or PR+ BRCA, which are typical
of luminal tumors, tend to have better outcomes; however, patients
with BRCA of ER−, PR− and HER2−
status, typical of triple-negative breast cancer (TNBC), have
poorer outcomes (1). These two
subtypes of BRCA also exhibit distinct responses to chemotherapy
(6,7). Therefore, future studies focused on
BRCA subtypes should be conducted to improve the treatment and
prognosis of this disease.
Long non-coding RNAs (lncRNAs) are a form of RNA
that do not encode proteins; the discovery of numerous lncRNAs in
humans has notably improved the understanding of cancer and disease
(8,9). With increasing interest in human
lncRNAs and the availability of high-throughput technologies, the
number of BRCA-lncRNA associations identified has rapidly
increased. lncRNA metastasis associated lung adenocarcinoma
transcript 1 (MALAT1) could suppress the metastasis of BRCA
(10). lncRNA small nucleolar RNA
host gene 1 induced trastuzumab resistance in BRCA by regulating
polyadenylate-binding protein 1 expression via H3K27 acetylation
(11). A limited number of
previous studies have determined the expression and function of
lncRNAs in certain subtypes of BRCA. For example, lncRNA AWPPH
promoted the growth of TNBC by upregulating frizzled homolog 7
(12). Yang et al
(13) performed a comprehensive
analysis of the expression profiles of lncRNAs, mRNAs and microRNAs
(miRNAs) associated with competing endogenous RNA networks in TNBC;
however, the functions and mechanisms of the majority of lncRNAs
have not been characterized in BRCA, particularly in various
subtypes of this disease.
Transcription factors (TFs) strictly regulate gene
transcription by binding to genomic cis-regulatory elements present
in a specific sequence motif (14). Additionally, the efficacy of TFs
to regulate their target genes is affected by a variety of genetic
and epigenetic factors (15,16). The dysregulation of these global
regulatory mechanisms could contribute to the development of
diseases and cancer (17).
Accumulating evidence has demonstrated that lncRNAs mediate gene
expression and regulate transcription. For example, lncRNA MALAT1
controls the progression of the cell cycle by regulating the
expression of the oncogenic TF Myb-related protein B (18). lncRNA MALAT1 could promote
cellular proliferation by regulating the expression and pre-mRNA
processing of cell cycle-regulated TFs. In addition, lncRNA
colorectal cancer associated transcript 1-long isoform contributes
to regulating long-range interactions at the locus of the TF MYC
(19). These previous findings
suggested that lncRNAs may serve essential roles in regulating TFs;
however, only a few previous studies have determined the activity
of TFs mediated by lncRNAs in the subtypes of BRCA.
In the present study, a comprehensive and
computational approach was undertaken to systemically identify
lncRNA-mediated transcriptional dysregulation triplets (lncTDTs)
based on the expression profiles of TFs, genes and lncRNAs; their
previously experimentally identified interactions in
ER+/PR+, HER2− BRCA and TNBC were
further analyzed in the present study using bioinformatics methods.
A total of six regulatory profiles of lncTDTs were identified,
which exhibited distinct features in the two BRCA subtypes. The
diverse and common characteristics of the two subtypes were also
analyzed. Functional analysis revealed that the lncTDTs were
significantly associated with cancer-related Gene Ontology (GO)
terms and the PI3K/AKT signaling pathway. Additionally, certain
lncTDTs in ER+/PR+, HER2− BRCA and
TNBCs were detected. Collectively, the results of the present study
may provide further insight into the functions of lncRNAs and the
underlying mechanisms, and may serve as a basis for the
classification and treatment of BRCA.
Materials and methods
TF, gene and lncRNA expression profiles
of ER+/PR+, HER2− BRCA and
TNBC
Three same-sample expression profiles of TFs, genes
and lncRNAs were downloaded from The Cancer Genome Atlas (TCGA;
https://tcga-data.nci.nih.gov/;
https://portal.gdc.cancer.gov/exploration?filters=%7B''op''%3A''and''%2C''content''%3A%5B%7B''op''%3A''in''%2C''content''%3A%7B''field''%3A''cases.primary_site''%2C''value''%3A%5B''Breast''%5D%7D%7D%5D%7D).
The matched clinical information, including ER status, PR status,
HER2 status and survival data were also obtained. All BRCA samples
were divided into two sets: ER+/PR+,
HER2− BRCA and TNBC, based on clinical information. In
total, the ER+/PR+, HER2− BRCA
dataset included 422 cancer samples and 22 matched normal samples,
while the TNBC dataset contained 112 cancer samples and 3 matched
normal samples.
Collection of cancer-associated lncRNAs
and genes
To obtain more accurate information for analysis,
the data of BRCA-associated lncRNAs and genes were extracted.
Information regarding cancer-related lncRNAs was obtained from
lnc2cancer 2.0, which is an updated database that provides
comprehensive experimentally supported associations between lncRNAs
and human cancer (20).
Cancer-related genes were obtained from DisGeNET, which is the
largest publicly available database of genes and variants
associated with a variety of human diseases (21). A t-test was performed to identify
differentially expressed cancer-related genes and lncRNAs in
ER+/PR+, HER2− BRCA and TNBC.
P<0.05 was considered to indicate a statistically significant
difference. A total of 6,626 and 555 ER+/PR+,
HER2− BRCA-specific genes and lncRNAs, and 5,576 and 326
TNBC-specific genes and lncRNAs were respectively obtained. These
BRCA subtype-specific genes and lncRNAs were employed for
subsequent analysis.
Identification of
ER+/PR+, HER2− BRCA and
TNBC-specific TF-gene interactions
TF-gene interaction data were obtained from TRANSFAC
(22).
ER+/PR+, HER2− BRCA- and
TNBC-specific TF-gene interactions were extracted based on the
aforementioned conditions for the retrieval of BRCA
subtype-specific genes and co-expression data. Pearson correlation
coefficient (PCC) values were calculated for all cancer-associated
TF-gene interactions. Then, ER+/PR+,
HER2− BRCA and TNBC-specific TF-gene interactions were
considered, providing the absolute PCC values were >0.3.
Following filtering, 494 and 476 ER+/PR+,
HER2− BRCA- and TNBC-specific TF-gene interactions were
respectively identified and employed for subsequent analysis.
Identification of lncTDTs associated with
ER+/PR+, HER2− BRCA and TNBC
An integrated and computational approach was
developed to identify lncTDTs associated with patients with
ER+/PR+, HER2− BRCA and TNBC. For
each BRCA subtype-specific lncRNA, the BRCA samples were sorted
based on lncRNA expression; 25% of the samples exhibiting the
highest and lowest lncRNA expression levels were selected.
Additionally, interactions between TFs and genes were considered,
providing the interactions were affected by a certain lncRNA, based
on the independent analysis of each lncRNA-TF-gene triplet. The PPC
values between each TF and gene were respectively calculated in the
top and bottom 25% samples for a given lncRNA. A TF-gene
interaction was considered to be affected by an lncRNA providing
the absolute difference of the PCC values between the top and
bottom 25% samples was >0.4. This lncRNA-TF-gene interaction was
considered as a candidate lncTDT. For a particular lncRNA, all BRCA
subtype-specific TF-gene interactions were calculated. Furthermore,
1,000 random alterations of the sample labels of expression
profiles was performed to compare the absolute difference of PCC
values with changes in the absolute difference of these values to
determine significance. P<0.05 was selected as the threshold
value to obtain significant lncTDTs associated with
ER+/PR+, HER2− BRCA and TNBC.
Pattern classification of lncTDTs for
ER+/PR+, HER2− BRCA and TNBC
In order to further investigate the mechanism of
lncTDTs, the lncTDTs were separated into six profiles based on
their mode of regulation: i) Weaken inhibition, TFs can suppress
the expression of a gene, while an lncRNA could weaken this
inhibitory action; ii) strengthen inhibition, TFs inhibit the
expression of a gene, while an lncRNA could promote this inhibitory
action; iii) reverse inhibition, TFs induce the expression of gene,
while an lncRNA can invert these activating effects favoring
inhibition; iv) reverse activation, TFs inhibit the expression of a
gene, while an lncRNA can invert these inhibitory effects favoring
activation; v) weaken activation, TFs induce the expression of a
gene, but the effects of the TF are weakened by an lncRNA; and vi)
strengthen activation, TFs can induce the expression of a gene,
which is promoted by an lncRNA.
Construction and analysis of an lncTDT
network for ER+/PR+, HER2− BRCA
and TNBC
The networks of lncTDTs for
ER+/PR+, HER2− BRCA and TNBC were
constructed and the degree analysis was performed using Cytoscape
3.0 (http://www.cytoscape.org/).
Functional enrichment analysis for
lncTDTs in ER+/PR+, HER2− BRCA and
TNBC
For all TFs and genes associated with
ER+/PR+, HER2− BRCA and TNBC,
functional enrichment analyses were performed with the Enrichr tool
online web server using default parameters (23). GO terms (http://geneontology.org/; Accessed May 9, 2019;
P<0.01) and Kyoto Encyclopedia of Genes and Genomes (KEGG)
pathways (https://www.kegg.jp/; release 90.1;
P<0.05) associated with lncTDTs in
ER+/PR+, HER2− BRCA and TNBC were
determined.
Prognosis analysis for lncTDTs in
ER+/PR+, HER2− BRCA and TNBC
The regression coefficient was employed to verify
whether each lncTDT in ER+/PR+,
HER2− BRCA and TNBC was associated with survival based
on expression data. A multivariate cox regression model was
generated for each lncTDT and standardized Cox regression
coefficients were obtained. Then, a risk score was established for
each sample based on the expression profile of an lncTDT weighted
by the standardized Cox regression coefficient. Furthermore, the
BRCA samples were divided to high- and low-risk groups based on the
median risk score. Finally, Kaplan-Meier survival analysis of the
two groups was performed followed by a log-rank test. All analyses
were performed using R 3.3.2 software (https://www.r-project.org/).
Results
lncTDTs in TNBC and
ER+/PR+, HER2− BRCA
An integrated and computational approach was
performed to identify lncTDTs in TNBC and
ER+/PR+, HER2− BRCA based on
expression profiles and experimentally verified interactions. A
total of 2,645 and 1,417 lncTDTs were identified in TNBC and
ER+/PR+, HER2− BRCA; two lncTDT
networks were also constructed for TNBC and
ER+/PR+, HER2− BRCA (Fig. 1A and B; Table SI). There were 325 nodes and
1,417 edges in the TNBC lncTDT network; there were 583 nodes and
2,645 edges were identified in the ER+/PR+,
HER2− BRCA lncTDT network. The nodes included 218
lncRNAs, 107 TF-gene interactions, 33 TFs and 83 genes in the TNBC
lncTDT network (Fig. 1C). The
nodes included 410 lncRNAs, 173 TF-gene interactions, 69 TFs and
142 genes in the ER+/PR+, HER2−
BRCA lncTDT network (Fig. 1C).
The degrees of all nodes in the TNBC network (R2=0.61;
Fig. 1D) and the
ER+/PR+, HER2− BRCA network
(R2=0.85; Fig. 1E)
exhibited scale-free distribution, indicating that the networks
were biological regulatory networks. In addition, degree analysis
of the lncRNAs for each lncRNA in the networks for TNBC and
ER+/PR+, HER2− BRCA. The results
revealed that the lncRNA with the highest degree was RP11-82L18.2
in the TNBC lncTDT network. lncRNA RP11-82L18.2 was determined to
mediate 18 TF-gene interactions (Fig.
1F). The lncRNA with the highest degree was RAMP2 antisense RNA
1 (RAMP2-AS1) in the ER+/PR+,
HER2− BRCA lncTDT network; lncRNA RAMP2-AS1 mediated 30
TF-gene interactions (Fig. 1G).
These results indicated that lncRNAs serve essential and specific
roles in the lncTDT networks in TNBC and
ER+/PR+, HER2− BRCA.
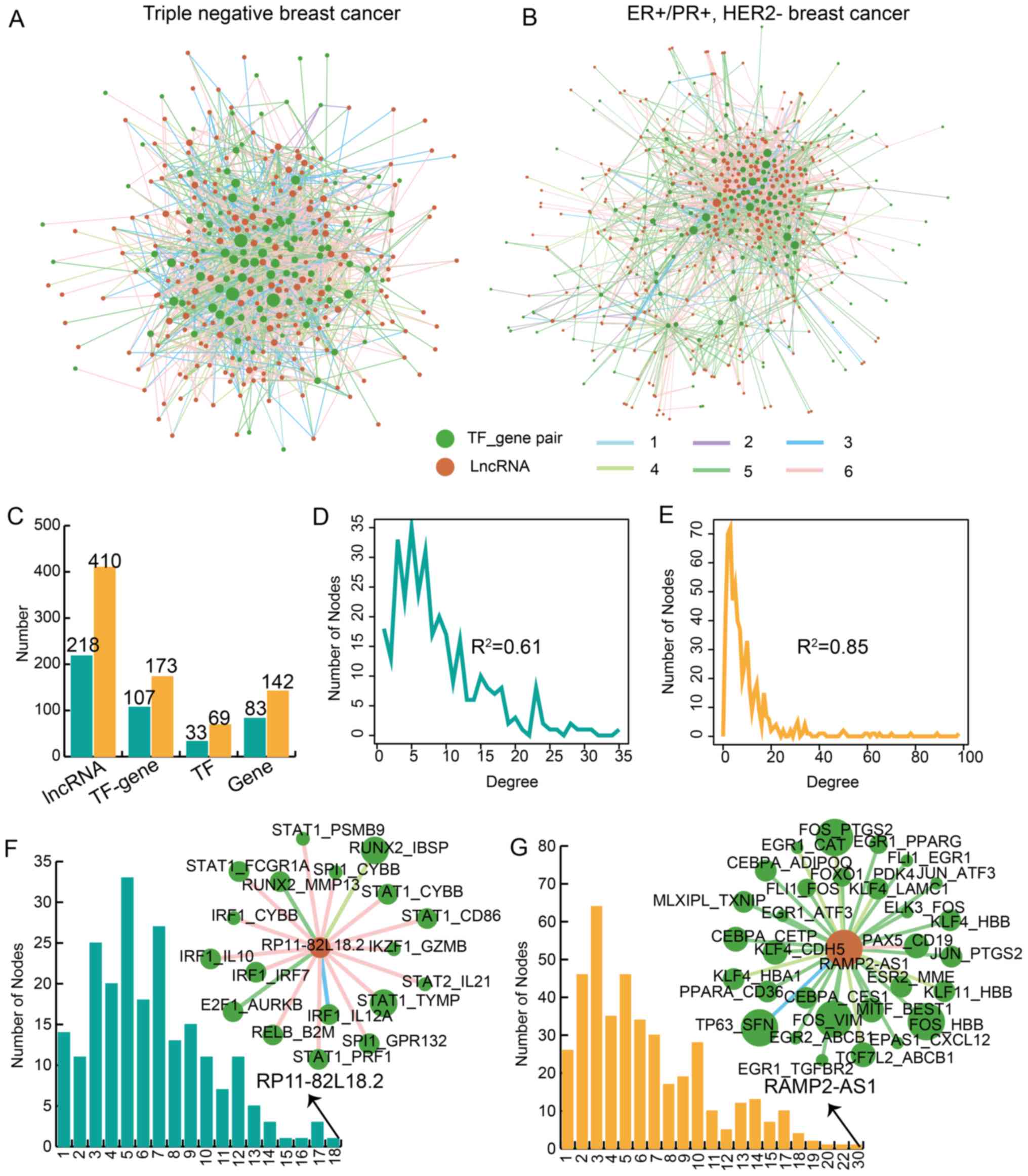 | Figure 1Identification of lncTDTs associated
with TNBC and ER+/PR+, HER2− BRCA.
(A) lncTDT network for TNBC. (B) lncTDT network for
ER+/PR+, HER2− BRCA. Green and
dark orange nodes represent the TF-gene pairs and lncRNAs,
respectively. The different colored edges represent various
regulatory patterns. Light blue (1), purple (2), dark blue (3), light green (4), dark green (5) and pink (6) represent weaken inhibition,
strengthen inhibition, reverse inhibition, reverse activation,
weaken activation and strengthen activation, respectively. The size
of the nodes indicates the degree of nodes. (C) Bar plots indicate
the number of lncRNAs, TF-gene pairs, TFs and genes. Plots indicate
the degree distribution for (D) TNBC and (E)
ER+/PR+, HER2− BRCA. Bar plots
demonstrate the degree of lncRNAs in (F) TNBC and (G)
ER+/PR+, HER2− BRCA. Teal and
orange represent TNBC and ER+/PR+,
HER2− BRCA, respectively. lncTDT, lncRNA-mediated
transcriptional dysregulation triplet; TNBC, triple-negative breast
cancer; ER, estrogen receptor; PR, progesterone receptor; HER2,
human epidermal-growth factor receptor 2; BRCA, breast cancer; TF,
transcription factor; lncRNA, long non-coding RNA; RAMP2-AS1, RAMP2
antisense RNA 1. |
Complex patterns of lncTDTs in TNBC and
ER+/PR+, HER2− BRCA
TFs are known to activate or inhibit gene
expression; however, this is a complex process in diseases, such as
cancer. In addition, lncRNAs can influence the effects of TFs on
their target genes; thus, these lncRNA-mediated TFs-gene
interactions were defined as lncTDTs. The regulatory patterns of
lncTDTs in TNBC and ER+/PR+, HER2−
BRCA are complex. The lncTDTs were divided into six patterns based
on the regulatory action of lncRNAs and TFs (Fig. 2A). In total, there were, 1.27,
0.42, 18.77, 13.20, 24.49 and 41.85% lncTDTs involving lncRNAs that
could weaken inhibition, strengthen inhibition, reverse inhibition,
reverse activation, weaken activation and strengthen activation of
TF-regulated gene expression, respectively for TNBC. In addition,
there were 0.076, 0.87, 7.18, 4.23, 33.50 and 54.14 lncTDTs
involving lncRNAs that could weaken inhibition, strengthen
inhibition, reverse inhibition, reverse activation, weaken
activation and strengthen activation of the effects of TF on gene
expression for ER+/PR+, HER2− BRCA
(Fig. 2B). In TNBC and
ER+/PR+, HER2− BRCA, the most
common regulatory pattern was strengthened activation, while the
least common were weakened inhibition and strengthened inhibition.
Additionally, it was determined that an lncRNA can exhibit various
regulatory patterns; for example, a particular lncRNA could weaken,
strengthen or reverse the effects of different TFs on gene
expression. The cancer-associated lncRNA HOX transcript antisense
RNA (HOTAIR), which is an important cancer-related lncRNA (24), could weaken activation and reverse
inhibition of gene expression in TNBC (Fig. 2D). Furthermore, HOTAIR was
proposed to reverse inhibition, and strengthen or weaken activation
in ER+/PR+, HER2− BRCA (Fig. 2E); only the most significant
HOTAIR-related lncTDTs are shown. The most common patterns of
HOTAIR in TNBC and ER+/PR+, HER2−
BRCA were weaken activation and strengthen activation,
respectively. These results indicated the complexity of lncTDTs in
TNBC and ER+/PR+, HER2− BRCA.
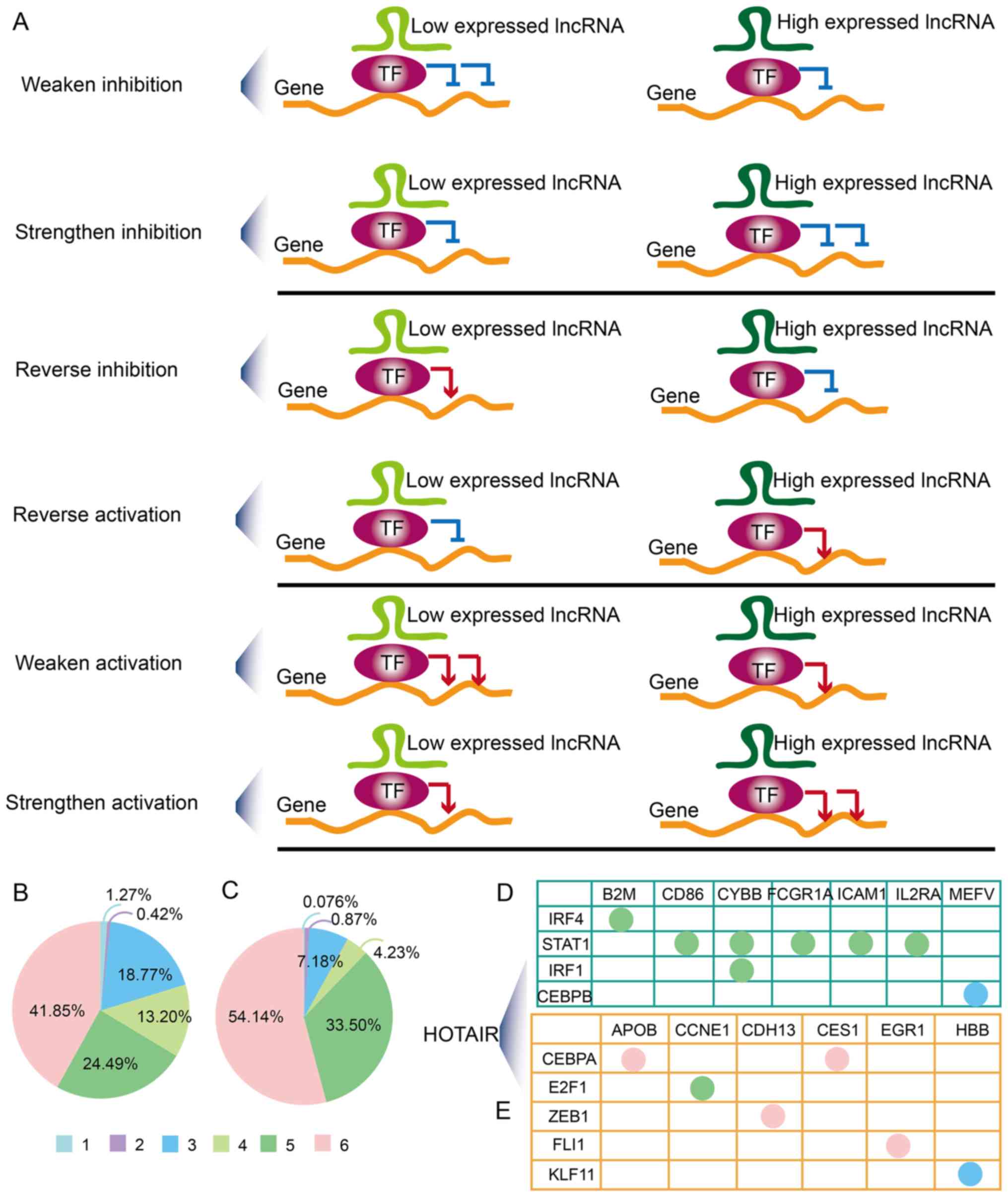 | Figure 2Complex regulatory patterns. (A) A
total of six regulatory patterns were identified. Green, burgundy
and orange represent lncRNAs, TFs and genes, respectively. The pie
charts demonstrate the distribution of the regulatory patterns in
(B) TNBC and (C) ER+/PR+, HER2−
BRCA. Light blue (1), purple
(2), dark blue (3), light green (4), dark green (5) and pink (6) represent weaken inhibition,
strengthen inhibition, reverse inhibition, reverse activation,
weaken activation and strengthen activation, respectively.
Regulatory patterns of the most significant HOTAIR-mediated
lncRNA-mediated transcriptional dysregulation triplets in (D) TNBC
and (E) ER+/PR+, HER2− BRCA. TNBC,
triple-negative breast cancer; ER, estrogen receptor; PR,
progesterone receptor; HER2, human epidermal-growth factor receptor
2; BRCA, breast cancer; TF, transcription factor; lncRNA, long
non-coding RNA; HOTAIR, HOX transcript antisense RNA. |
Diverse and common characteristics of
lncTDTs in TNBC and ER+/PR+, HER2−
BRCA
TNBC and ER+/PR+,
HER2− BRCA are two important subtypes of BRCA. The
mechanisms underlying the development and progression of these
subtypes differ. Therefore, investigating the common
characteristics of lncTDTs in TNBC and
ER+/PR+, HER2− BRCA may improve
the understanding of this disease. In the present study, the
lncRNAs, TFs, genes and TF-gene interactions in TNBC and
ER+/PR+, HER2− BRCA were analyzed.
The majority of these molecules differed between TNBC and
ER+/PR+, HER2− BRCA; however, 142
lncRNAs, 18 TFs, 23 genes and 23 TF-gene interactions were reported
in the two BRCA subtypes (Fig.
3A-D). In addition, five common lncTDTs were identified between
TNBC and ER+/PR+, HER2− BRCA
(Fig. 3E). In the present study,
80% of the common lncTDTs exhibited the same regulation pattern
(Fig. 3F); these lncTDTs
included: NCK1-antisense RNA 1 (NCK1-AS1)/nuclear factor of
activated T cells 2 (NFATC2)/CD3g molecule,
NCK1-AS1/NFATC2/cytotoxic T-lymphocyte associated protein 4, small
nucleolar RNA host gene 6 (SNHG6)/interferon regulatory factor 1
(IRF1)/interleukin 12B (IL12B) and SNHG6.1/IRF1/IL12B, which
exhibited weakened activation (Fig.
3G). Of all the common lncTDTs, only PRC1 antisense RNA 1
(PRC1-AS1)/NK2 homeobox 5 (NKX2-5)/procollagen-lysine,
2-oxoglutarate 5-dioxygenase 1 (PLOD1) revealed a different
regulatory pattern in TNBC and ER+/PR+,
HER2− BRCA. In addition, PRC1-AS1/NKX2-5/PLOD1 exhibited
reversed inhibition and strengthened inhibition patterns (Fig. 3H). It was identified that lncRNA
NCK1-AS1 serves an important role in the two BRCA subtypes; lncRNA
NCK1-AS1 could promote proliferation and induce cell cycle
progression in cervical cancer (25). Downregulation of lncRNA NCK1-AS1
was reported to increase chemosensitivity to cisplatin in cervical
cancer (26). In the present
study, lncRNA NCK1-AS1 was determined to regulate 12 and 14 TF-gene
interactions in TNBC and ER+/PR+,
HER2− BRCA, respectively (Fig. 3I). The regulatory patterns in
ER+/PR+, HER2− BRCA are varied and
complex. The present results indicated that the lncTDTs possessed
common and specific characteristics in TNBC and
ER+/PR+, HER2− BRCA.
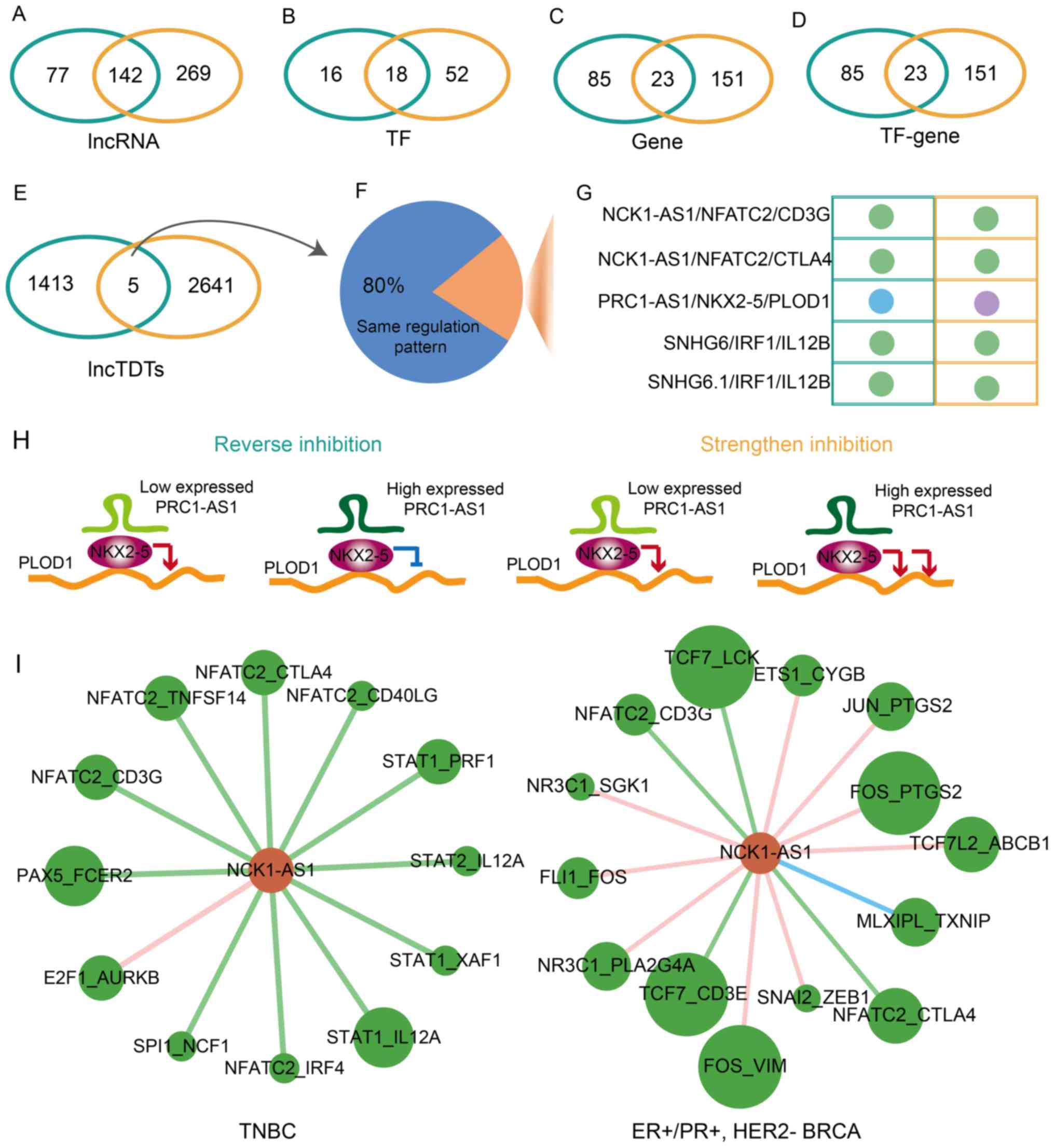 | Figure 3Common and specific lncTDTs for TNBC
and ER+/PR+, HER2− BRCA. Venn
diagrams demonstrating the association between (A) lncRNAs, (B)
TFs, (C) genes, (D) TF-gene pairs and (E) lncTDTs. Teal indicates
TNBC and orange indicates ER+/PR+, HER2- BRCA. (F) Pie chart
indicates the percentage of the same regulatory pattern among
common lncTDTs. (G) Regulatory patterns of common lncTDTs. The
nodes represent various regulatory patterns. Light blue, purple and
dark green represent weaken inhibition, strengthen inhibition and
weaken activation, respectively. (H) Regulatory pattern of lncTDT
PRC1-AS1/NKX2-5/PLOD1. Green, burgundy and orange represent
lncRNAs, TFs and genes, respectively. (I) Subnetworks of lncRNA
NCK1-AS1 in TNBC and ER+/PR+,
HER2− BRCA. Green and dark orange nodes represent the
TF-gene pairs and lncRNAs, respectively. The different colored
edges represent various regulatory patterns. Dark blue, dark green
and pink represent reverse inhibition, weaken activation and
strengthen activation, respectively. lncTDTs, long non-coding
RNA-mediated transcriptional dysregulation triplets; TNBC,
triple-negative breast cancer; ER, estrogen receptor; PR,
progesterone receptor; HER2, human epidermal-growth factor receptor
2; BRCA, breast cancer; lncRNAs, long non-coding RNAs; TF,
transcription factor; PRC1-AS1, PRC1 antisense RNA 1; NKX2-5, NK2
homeobox 5; PLOD1, procollagen-lysine,2-oxoglutarate 5-dioxygenase
1. |
lncTDTs exhibit cancer-associated
functions in TNBC and ER+/PR+,
HER2− BRCA
To further understand the functions and mechanisms
of lncTDTs in TNBC and ER+/PR+,
HER2− BRCA, GO and KEGG pathway functional enrichment
analyses were performed for genes and TFs associated with lncTDTs.
The lncTDTs in TNBC or ER+/PR+,
HER2− BRCA were enriched for certain cancer-associated
functions, including 'positive regulation of transcription,
DNA-templated', 'negative regulation of transcription,
DNA-templated', 'positive regulation of gene expression' and
'cellular response to cytokine stimulus' (Fig. 4A and B). Additionally, the lncTDTs
in TNBC and ER+/PR+, HER2− BRCA
were both enriched in the PI3K/AKT signaling pathway which is a key
pathway for cancer development and treatment (data not shown) and
this pathway is key in the development and treatment of cancer
(27-29); alterations in the PI3K/AKT
signaling pathway also contribute to drug resistance in BRCA
(30,31). In the present study, numerous
genes in lncTDTs were enriched in this pathway for TNBC and
ER+/PR+, HER2− BRCA (Fig. 4C). Of note, lncTDTs were
associated with the PI3K/AKT signaling pathway in the two BRCA
subtypes; however, the genes differed. Furthermore, a common gene,
CD19, was determined to be associated with the PI3K/AKT signaling
pathway in TNBC and ER+/PR+, HER2−
BRCA. These results indicated that lncTDTs may serve essential
roles in TNBC and ER+/PR+, HER2−
BRCA by affecting certain important biological processes and
pathways.
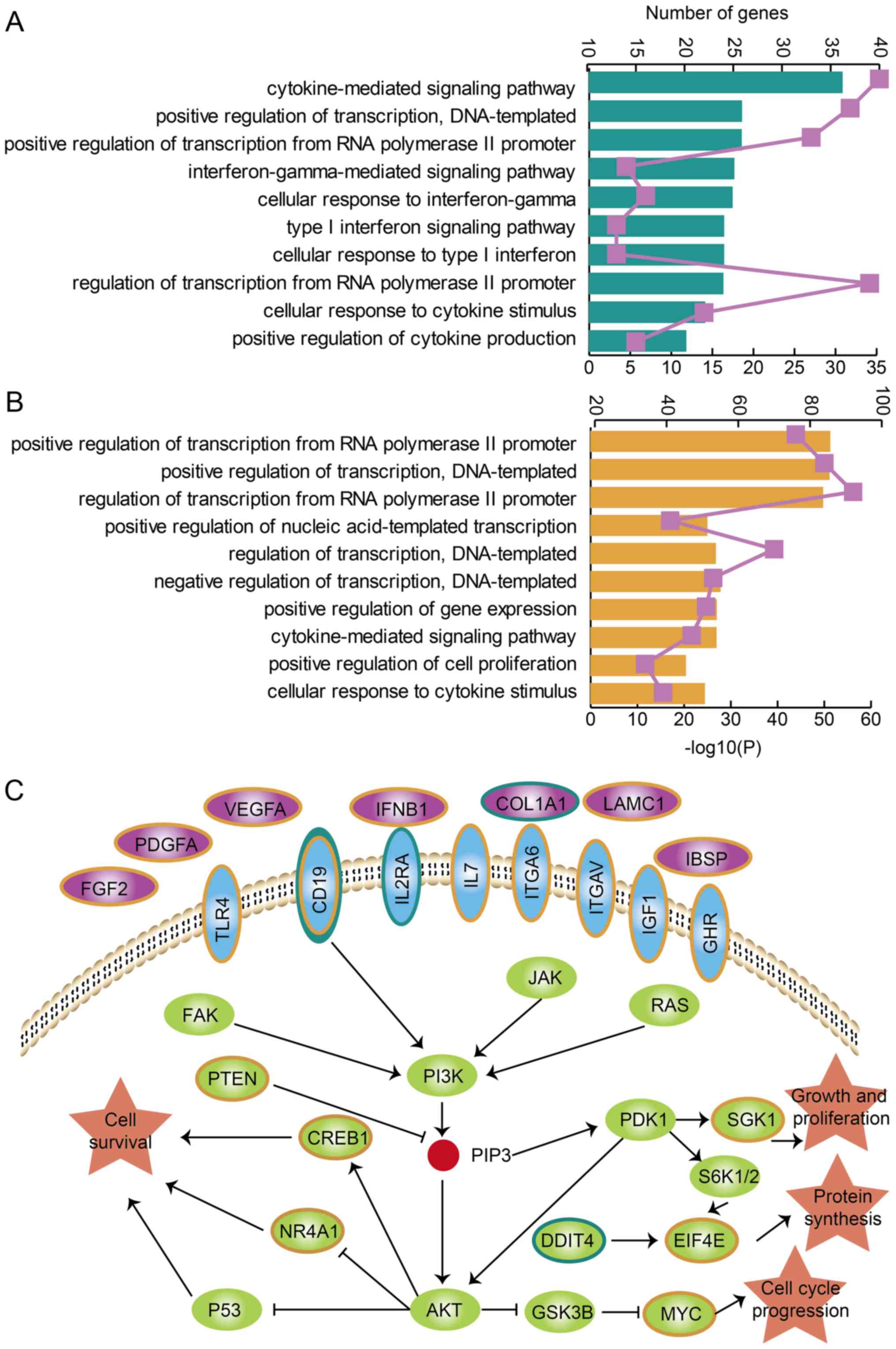 | Figure 4Functional analysis for lncTDTs in
TNBC and ER+/PR+, HER2− BRCA. Gene
Ontology terms enriched for genes and transcription factors in
lncTDTs for (A) TNBC and (B) ER+/PR+,
HER2− BRCA are presented and ranked by
-log10(P) values. The purple lines represent the number
of enriched genes. (C) PI3K/AKT signaling pathway and the genes
associated with TNBC and ER+/PR+,
HER2− BRCA, which are presented as teal and orange,
respectively. The purple, blue, green and orange shapes represent
the genes in the cell membrane, transmembrane, intracellular
membrane and biology process, repectively. lncTDTs, long non-coding
RNA-mediated transcriptional dysregulation triplets; TNBC,
triple-negative breast cancer; ER, estrogen receptor; PR,
progesterone receptor; HER2, human epidermal-growth factor receptor
2; BRCA, breast cancer. |
lncTDTs may serve as candidate prognostic
biomarkers in TNBC and ER+/PR+,
HER2− BRCA
A risk score-based analysis on the expression of
the lncRNA, TF and gene of an lncTDT was performed for the
prediction of survival to evaluate the potential of lncTDTs as
prognostic biomarkers in the two BRCA subtypes. A total of 16.08
and 7.86% lncTDTs were significantly associated with survival in
TNBC and ER+/PR+, HER2− BRCA,
respectively (Fig. 5A and B); few
lncTDTs (0.28 and 1.36%) exhibited a highly significant association
with survival in the two respective BRCA subtypes. In addition, the
prognostic value of HOTAIR-mediated lncTDTs in the two BRCA
subtypes was investigated. Certain HOTAIR-mediated lncTDTs in TNBC
were associated with survival (data not shown). A total of seven
HOTAIR-mediated lncTDTs in ER+/PR+,
HER2− BRCA were significantly related to survival
(Fig. 5C), including HOTAIR/E2F
transcription factor 1 (E2F1)/cyclin E1 (CCNE1), HOTAIR/CCAAT
enhancer binding protein α (CEBPA)/apolipoprotein B,
HOTAIR/CEBPA/carboxylesterase 1, HOTAIR/signal transducer and
activator of transcription 5A/insulin like growth factor 1,
HOTAIR/estrogen receptor 2/membrane metalloendopeptidase,
HOTAIR/Kruppel like factor 11/hemoglobin subunit β and HOTAIR/early
growth response 2/ATP binding cassette subfamily B member 1. Of
these HOTAIR-mediated lncTDTs, HOTAIR/E2F1/CCNE1 was determined to
be related to survival with the highest degree of significance
(P=0.02); the lncTDTs that were associated with survival in TNBC
and ER+/PR+, HER2− BRCA with a
high degree of significance are presented in Fig. 5D and E. The patients in the
high-risk group exhibited shorter survival than those in the
low-risk group. The results of the present study indicated that
lncTDTs may collectively influence the survival of patients with
TNBC or ER+/PR+, HER2− BRCA; thus,
lncTDTs could be considered as potential biomarkers for these
subtypes of BRCA.
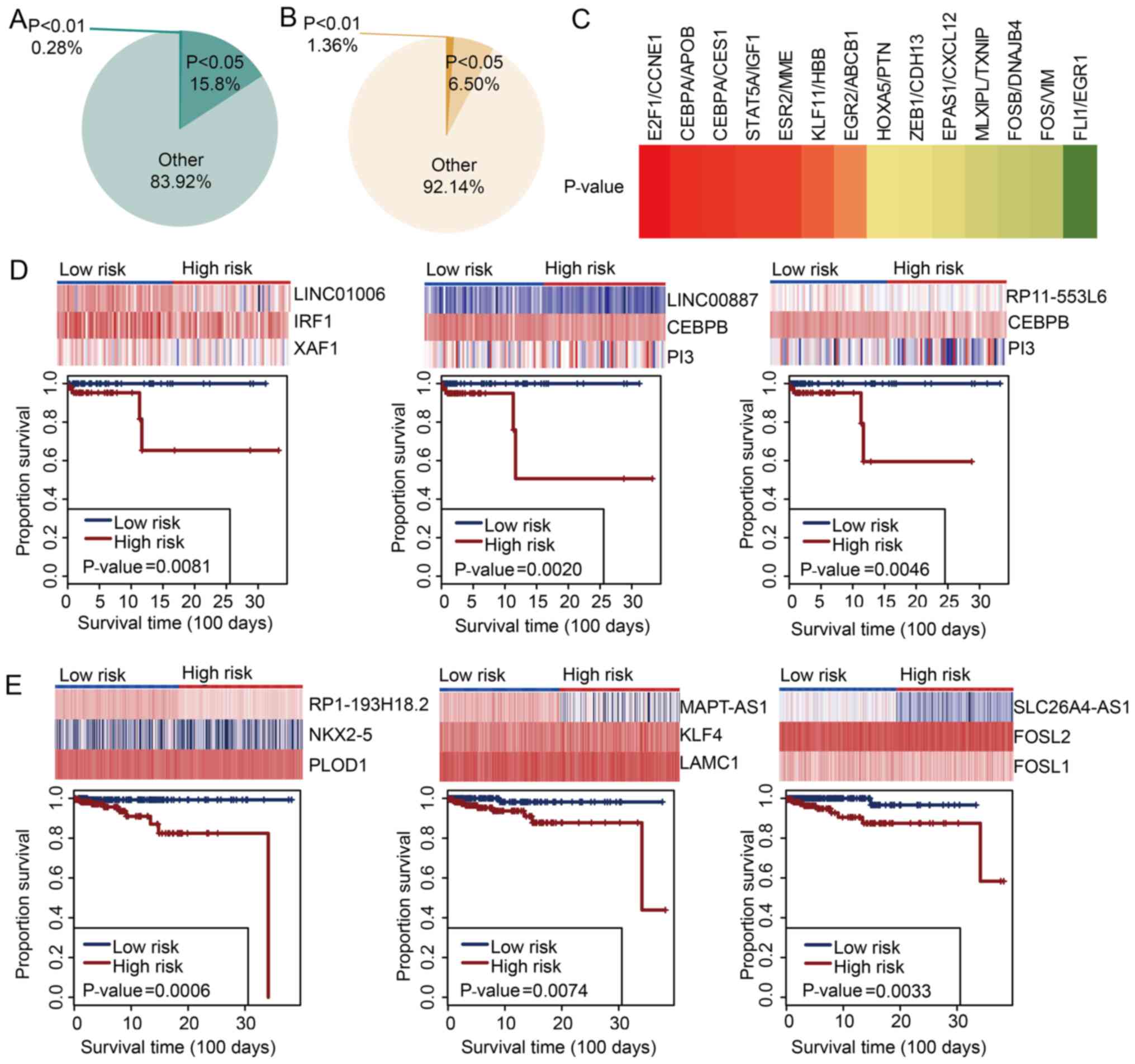 | Figure 5lncTDTs are associated with survival
in TNBC and ER+/PR+, HER2− BRCA.
Pie charts indicate the percentage of survival-related lncTDTs in
(A) TNBC and (B) ER+/PR+, HER2−
BRCA. (C) Survival analyses for HOX transcript antisense
RNA-mediated lncTDTs in ER+/PR+,
HER2− BRCA; highly significant associations are
presented in red. Orange/red indicate significant associations and
yellow/green indicate non-significant associations. Examples of
survival-related lncTDTs in (D) TNBC and (E)
ER+/PR+, HER2− BRCA. lncTDTs, long
non-coding RNA-mediated transcriptional dysregulation triplets;
TNBC, triple-negative breast cancer; ER, estrogen receptor; PR,
progesterone receptor; HER2, human epidermal-growth factor receptor
2; BRCA, breast cancer. |
Discussion
In present study, an integrated and computational
approach was developed to identify lncTDTs in
ER+/PR+, HER2− BRCA and TNBC based
on expression profiles and experimentally verified TF-gene
interactions. The regulatory patterns of these lncTDTs were complex
and could be divided into six types, including weaken inhibition,
strengthen inhibition, reverse inhibition, reverse activation,
weaken activation and strengthen activation. In addition, it was
proposed that an lncRNA may regulate different TF-gene
interactions, while an interaction could also be regulated by
various lncRNAs. In addition, the common and diverse
characteristics of ER+/PR+, HER2−
BRCA and TNBC were investigated. The majority of lncTDTs were BRCA
subtype-specific and only five common lncTDTs were reported; four
of these five common lncTDTs exhibited a common regulatory pattern.
However, lncTDT PRC1-AS1/NKX2-5/PLOD1 revealed a reverse inhibition
pattern in TNBC and a strengthen inhibition pattern in
ER+/PR+, HER2− BRCA. Therefore,
the regulatory pattern for PRC1-AS1/NKX2-5/PLOD1 was reversed in
ER+/PR+, HER2− BRCA and TNBC. The
functional analysis demonstrated that the lncTDTs were associated
with the PI3K/AKT signaling pathway in
ER+/PR+, HER2− BRCA and TNBC.
Survival analysis revealed that lncTDTs could serve as candidate
prognostic biomarkers in these two subtypes of BRCA.
At present, the status of ER, PR and HER2 serves as
a major reference for the administration of targeted adjuvant
therapy for BRCA (32,33). TNBC is a distinct subclass of BRCA
with a high degree of aggressiveness (34). Classifying the two BRCA subtypes
may improve the understanding of the mechanism underlying the
formation of BRCA and aid the development of targeted treatment. Li
et al (35) revealed the
vascular features of triple negative breast carcinomas using
dynamic magnetic resonance imaging. Li et al (36) reported that patients with TNBC had
unique clinicopathological characteristics and poorer prognosis.
Previous studies have employed high-throughput molecular profiles
or other genetic risk factors to classify BRCA subtypes (37,38). Jiang et al (39) analyzed the subtypes and treatment
strategies of TNBC. These previous findings indicated that TNBC and
other subtypes exhibit specific molecular and therapeutic features.
In addition, the results of the present study may provide novel
insight for the classification of ER+/PR+,
HER2− BRCA and TNBC. It was identified that the majority
of lncTDTs were specific to BRCA subtypes, and only five common
lncTDTs were observed in ER+/PR+,
HER2− BRCA and TNBC. The results indicated the high
degree of variation between ER+/PR+,
HER2− BRCA and TNBC at the lncTDT level.
The regulatory patterns of lncTDTs were complex and
indicated that lncRNAs can serve a variety of functions. Previous
studies have employed several approaches, such as co-expression
analyses and investigations into interactions with miRNAs to
determine the functions of lncRNAs in TNBC (40-42). In the present study, the roles of
lncRNAs in ER+/PR+, HER2− BRCA and
TNBC were further examined based on lncTDTs. Approximately one-half
of the lncTDTs exhibited strengthened activation regulation, in
which lncRNAs could promote the ability of TFs to activate gene
expression. Reverse inhibition and reverse activation were two
major regulatory patterns by which lncRNAs reversed the regulatory
effects of TF-gene interactions. The percentage of lncTDTs in
ER+/PR+, HER2− BRCA and TNBC
notably differed. The lncRNA HOTAIR, a cancer-related lncRNA, was
investigated, which had been verified to be associated with BRCA in
36 previous studies retrieved via lnc2cancer 2.0 (20). In TNBC, HOTAIR regulated eight
TF-gene interactions and the major regulation pattern was weakened
activation. In ER+/PR+, HER2−
BRCA, HOTAIR regulated 14 TF-gene interactions and the major
regulation pattern was strengthened activation. These results
indicated the different functions of HOTAIR in
ER+/PR+, HER2− BRCA and TNBC.
However, the number of control samples in the present study was
small and experiments are required to verify the present results.
Future studies should aim to conduct analysis using data from more
control samples to validate the present computational approach and
findings.
Collectively, the present study performed an
integrated and computational approach to identify lncTDTs in
ER+/PR+, HER2− BRCA and TNBC. A
total of six regulatory patterns for lncTDTs were proposed; the
mechanisms underlying the modes of regulation were notably complex.
The common and specific characteristics of lncTDTs between
ER+/PR+, HER2− BRCA and TNBC were
also determined. Functional and survival analyses revealed lncTDTs
as potential biomarkers for the prognosis of
ER+/PR+, HER2− BRCA and TNBC.
Supplementary Materials
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the
current study are available from the corresponding author on
reasonable request.
Authors' contributions
ZD, WG and SG conceived and designed the present
study. ZD, WG, YL and JS performed the experiments and analyzed the
data. YS and TC validated and improved the computational approach
in the present study. ZD wrote the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Acknowledgments
Not applicable.
References
|
1
|
Onitilo AA, Engel JM, Greenlee RT and
Mukesh BN: Breast cancer subtypes based on ER/PR and Her2
expression: Comparison of clinicopathologic features and survival.
Clin Med Res. 7:4–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Partridge AH, Hughes ME, Warner ET,
Ottesen RA, Wong YN, Edge SB, Theriault RL, Blayney DW, Niland JC,
Winer EP, et al: Subtype-dependent relationship between young age
at diagnosis and breast cancer survival. J Clin Oncol.
34:3308–3314. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Elston CW: Classification and grading of
invasive breast carcinoma. Verh Dtsch Ges Pathol. 89:35–44.
2005.
|
|
4
|
Longacre TA, Ennis M, Quenneville LA, Bane
AL, Bleiweiss IJ, Carter BA, Catelano E, Hendrickson MR, Hibshoosh
H, Layfield LJ, et al: Interobserver agreement and reproducibility
in classification of invasive breast carcinoma: An NCI breast
cancer family registry study. Mod Pathol. 19:195–207. 2006.
View Article : Google Scholar
|
|
5
|
Zhang L, Yu Q, Wu XC, Hsieh MC, Loch M,
Chen VW, Fontham E and Ferguson T: Impact of chemotherapy relative
dose intensity on cause-specific and overall survival for stage
I-III breast cancer: ER+/PR+,
HER2− vs. triple-negative. Breast Cancer Res Treat.
169:pp. 175–187. 2018, View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lee HJ, Song IH, Seo AN, Lim B, Kim JY,
Lee JJ, Park IA, Shin J, Yu JH, Ahn JH and Gong G: Correlations
between molecular subtypes and pathologic response patterns of
breast cancers after neoadjuvant chemotherapy. Ann Surg Oncol.
22:392–400. 2015. View Article : Google Scholar
|
|
7
|
Kuerer HM, Newman LA, Smith TL, Ames FC,
Hunt KK, Dhingra K, Theriault RL, Singh G, Binkley SM, Sneige N, et
al: Clinical course of breast cancer patients with complete
pathologic primary tumor and axillary lymph node response to
doxorubicin-based neoadjuvant chemotherapy. J Clin Oncol.
17:460–469. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Huarte M: The emerging role of lncRNAs in
cancer. Nat Med. 21:1253–1261. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Prensner JR and Chinnaiyan AM: The
emergence of lncRNAs in cancer biology. Cancer Discov. 1:391–407.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kim J, Piao HL, Kim BJ, Yao F, Han Z, Wang
Y, Xiao Z, Siverly AN, Lawhon SE, Ton BN, et al: Long noncoding RNA
MALAT1 suppresses breast cancer metastasis. Nat Genet.
50:1705–1715. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Dong H, Wang W, Mo S, Liu Q, Chen X, Chen
R, Zhang Y, Zou K, Ye M, He X, et al: Long non-coding RNA SNHG14
induces trastuzumab resistance of breast cancer via regulating
PABPC1 expression through H3K27 acetylation. J Cell Mol Med.
22:4935–4947. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang K, Li X, Song C and Li M: LncRNA
AWPPH promotes the growth of triple-negative breast cancer by
up-regulating frizzled homolog 7 (FZD7). Biosci Rep. 38:2018.
View Article : Google Scholar
|
|
13
|
Yang R, Xing L, Wang M, Chi H, Zhang L and
Chen J: Comprehensive analysis of differentially expressed profiles
of lncRNAs/mRNAs and miRNAs with associated ceRNA networks in
triple-negative breast cancer. Cell Physiol Biochem. 50:473–488.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Spitz F and Furlong EE: Transcription
factors: From enhancer binding to developmental control. Nat Rev
Genet. 13:613–626. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Flores M, Hsiao TH, Chiu YC, Chuang EY,
Huang Y and Chen Y: Gene regulation, modulation, and their
applications in gene expression data analysis. Adv Bioinformatics.
2013:3606782013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang K, Saito M, Bisikirska BC, Alvarez
MJ, Lim WK, Rajbhandari P, Shen Q, Nemenman I, Basso K, Margolin
AA, et al: Genome-wide identification of post-translational
modulators of transcription factor activity in human B cells. Nat
Biotechnol. 27:829–839. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Riley T, Sontag E, Chen P and Levine A:
Transcriptional control of human p53-regulated genes. Nat Rev Mol
Cell Biol. 9:402–412. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tripathi V, Shen Z, Chakraborty A, Giri S,
Freier SM, Wu X, Zhang Y, Gorospe M, Prasanth SG, Lal A and
Prasanth KV: Long noncoding RNA MALAT1 controls cell cycle
progression by regulating the expression of oncogenic transcription
factor B-MYB. PLoS Genet. 9:pp. e10033682013, View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Xiang JF, Yin QF, Chen T, Zhang Y, Zhang
XO, Wu Z, Zhang S, Wang HB, Ge J, Lu X, et al: Human colorectal
cancer-specific CCAT1-L lncRNA regulates long-range chromatin
interactions at the MYC locus. Cell Res. 24:513–531. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Gao Y, Wang P, Wang Y, Ma X, Zhi H, Zhou
D, Li X, Fang Y, Shen W, Xu Y, et al: Lnc2Cancer v2.0: Updated
database of experimentally supported long non-coding RNAs in human
cancers. Nucleic Acids Res. 47:D1028–D1033. 2019. View Article : Google Scholar :
|
|
21
|
Piñero J, Bravo À, Queralt-Rosinach N,
Gutiérrez-Sacristán A, Deu-Pons J, Centeno E, García-García J, Sanz
F and Furlong LI: DisGeNET: A comprehensive platform integrating
information on human disease-associated genes and variants. Nucleic
Acids Res. 45:D833–D839. 2017. View Article : Google Scholar :
|
|
22
|
Wingender E, Dietze P, Karas H and Knüppel
R: TRANSFAC: A database on transcription factors and their DNA
binding sites. Nucleic Acids Res. 24:238–241. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kuleshov MV, Jones MR, Rouillard AD,
Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM,
Lachmann A, et al: Enrichr: A comprehensive gene set enrichment
analysis web server 2016 update. Nucleic Acids Res. 44:W90–W97.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li H, Jia Y, Cheng J, Liu G and Song F:
LncRNA NCK1-AS1 promotes proliferation and induces cell cycle
progression by crosstalk NCK1-AS1/miR-6857/CDK-1 pathway. Cell
Death Dis. 9:1982018. View Article : Google Scholar
|
|
26
|
Zhang WY, Liu YJ, He Y and Chen P:
Suppression of long noncoding RNA NCK1-AS1 increases
chemosensitivity to cisplatin in cervical cancer. J Cell Physiol.
234:4302–4313. 2019. View Article : Google Scholar
|
|
27
|
Hennessy BT, Smith DL, Ram PT, Lu Y and
Mills GB: Exploiting the PI3K/AKT pathway for cancer drug
discovery. Nat Rev Drug Discov. 4:988–1004. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Osaki M, Oshimura M and Ito H: PI3K-Akt
pathway: Its functions and alterations in human cancer. Apoptosis.
9:667–676. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Luo J, Manning BD and Cantley LC:
Targeting the PI3K-Akt pathway in human cancer: Rationale and
promise. Cancer Cell. 4:257–262. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chandarlapaty S, Sakr RA, Giri D, Patil S,
Heguy A, Morrow M, Modi S, Norton L, Rosen N, Hudis C and King TA:
Frequent mutational activation of the PI3K-AKT pathway in
trastuzumab-resistant breast cancer. Clin Cancer Res. 18:6784–6791.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Berns K, Horlings HM, Hennessy BT,
Madiredjo M, Hijmans EM, Beelen K, Linn SC, Gonzalez-Angulo AM,
Stemke-Hale K, Hauptmann M, et al: A functional genetic approach
identifies the PI3K pathway as a major determinant of trastuzumab
resistance in breast cancer. Cancer Cell. 12:395–402. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Anderson WF and Matsuno R: Breast cancer
heterogeneity: A mixture of at least two main types? J Natl Cancer
Inst. 98:948–951. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Perou CM, Sørlie T, Eisen MB, van de Rijn
M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA,
et al: Molecular portraits of human breast tumours. Nature.
406:747–752. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Haffty BG, Yang Q, Reiss M, Kearney T,
Higgins SA, Weidhaas J, Harris L, Hait W and Toppmeyer D:
Locoregional relapse and distant metastasis in conservatively
managed triple negative early-stage breast cancer. J Clin Oncol.
24:5652–5657. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li SP, Padhani AR, Taylor NJ, Beresford
MJ, Ah-See ML, Stirling JJ, d'Arcy JA, Collins DJ and Makris A:
Vascular characterisation of triple negative breast carcinomas
using dynamic MRI. Eur Radiol. 21:1364–1373. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li Y, Zhang N, Zhang H and Yang Q:
Comparative prognostic analysis for triple-negative breast cancer
with metaplastic and invasive ductal carcinoma. J Clin Pathol.
72:418–424. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Häberle L, Hein A, Rübner M, Schneider M,
Ekici AB, Gass P, Hartmann A, Schulz-Wendtland R, Beckmann MW, Lo
WY, et al: Predicting triple-negative breast cancer subtype using
multiple single nucleotide polymorphisms for breast cancer risk and
several variable selection methods. Geburtshilfe Frauenheilkd.
77:667–678. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sorlie T, Perou CM, Tibshirani R, Aas T,
Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey
SS, et al: Gene expression patterns of breast carcinomas
distinguish tumor subclasses with clinical implications. Proc Natl
Acad Sci USA. 98:10869–10874. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jiang YZ, Ma D, Suo C, Shi J, Xue M, Hu X,
Xiao Y, Yu KD, Liu YR, Yu Y, et al: Genomic and transcriptomic
landscape of triple-negative breast cancers: Subtypes and treatment
strategies. Cancer Cell. 35:428–440.e5. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Shin VY, Chen J, Cheuk IW, Siu MT, Ho CW,
Wang X, Jin H and Kwong A: Long non-coding RNA NEAT1 confers
oncogenic role in triple-negative breast cancer through modulating
chemo-resistance and cancer stemness. Cell Death Dis. 10:2702019.
View Article : Google Scholar
|
|
41
|
Fan CN, Ma L and Liu N: Comprehensive
analysis of novel three-long noncoding RNA signatures as a
diagnostic and prognostic biomarkers of human triple-negative
breast cancer. J Cell Biochem. 120:3185–3196. 2019. View Article : Google Scholar
|
|
42
|
Yang F, Shen Y, Zhang W, Jin J, Huang D,
Fang H, Ji W, Shi Y, Tang L, Chen W, et al: An androgen receptor
negatively induced long non-coding RNA ARNILA binding to miR-204
promotes the invasion and metastasis of triple-negative breast
cancer. Cell Death Differ. 25:2209–2220. 2018. View Article : Google Scholar : PubMed/NCBI
|



















