Introduction
Pulmonary artery hypertension (PAH) is a
life-threatening disease, characterized by progressive remodeling
of the distal pulmonary arteries (PAs) due to increased cell
proliferation and resistance to apoptosis. PAH can lead to
right-sided heart failure and eventually death (1). At present, there is no cure for PAH,
and the goal of treatment is limited to delaying or preventing its
progression. Although significant advances have been made in
understanding the cellular and molecular events underlying the
pathogenesis of PAH, it remains an incurable disease. Therefore,
understanding the molecular mechanisms of vascular remodeling in
PAH may facilitate the development of more effective therapeutic
strategies that improve the prognosis and long-term survival of
patients with PAH.
Long non-coding RNAs (lncRNAs) are a sequence of
nucleotides >200 bp in length, which were once considered as
transcriptional 'noise' with no protein-encoding functions
(2). However, it is now evident
that they are involved in a variety of biological processes,
including cell differentiation, apoptosis and proliferation
(3). Metastasis-associated lung
adenocarcinoma transcript 1 (MALAT1) is a highly conserved lncRNA
that has been implicated in different cancer types, such as
non-small cell lung cancer (4),
prostate cancer (5) and gastric
cancer (6). Recently, MALAT1 has
received attention from researchers due to its observed role in the
progression of vascular endothelial cell dysfunction. One study
demonstrated that MALAT1 dysregulation significantly increased the
proliferation of vascular endothelial cells under hypoxic
conditions (7). In addition, it
was reported that MALAT1 regulates the proliferation of smooth
muscle cells and inhibits cardiac hypertrophy in PAH mice following
silencing of MALAT1 (8).
Together, these results suggest that MALAT1 serves a latent
biological role in the pathogenesis of hypoxia and PAH. Although
our understanding of the role of MALAT1 in PAH is increasing,
additional studies are required to further verify its role and
underlying mechanisms in PAH.
In addition to lncRNA, microRNAs (miRNAs/miRs) are a
class of non-coding single-stranded RNA molecules that are ~22
nucleotides in length. They function as key regulators of gene
expression in various biological processes at the transcriptional
or post-transcriptional level through targeting mRNAs for
degradation or suppressing translation (9). LncRNAs interact with miRNAs and
regulate each other's levels of expression and biological
activities. In addition, lncRNAs may function as competing
endogenous RNAs (ceRNAs) that inhibit miRNA expression and activity
(10). Of particular note, it has
been reported that MALAT1 harbors binding sites for several miRNAs,
such as miR-125b (11), miR-101b
(12) and miR-183 (13). However, there is limited research
regarding the interactions between MALAT1 and miRNAs in PAH.
The aim of the present study was to identify the
molecular mechanisms of MALAT1 in the pathogenesis of PAH. The
results demonstrated that the expression of MALAT1 was upregulated
in PA tissues and human pulmonary artery smooth muscle cells
(HPASMCs) of patients with PAH. Silencing of MALAT1 inhibited
HPASMC proliferation. The effect of MALAT1 overexpression exerted
the opposite effects. In addition, MALAT1 was demonstrated to
function as a ceRNA for hsa-miR-124-3p.1. MALAT1 participates in
pulmonary vascular remodeling by binding to hsa-miR-124-3p.1 to
form a molecular sponge. This promotes the expression of the
downstream target gene, Kruppel-like factor 5 (KLF5), which is also
involved in pulmonary vascular remodeling progression (14). The results of the current study
contribute to what is currently known regarding the role of MALAT1
in PAH and may provide new therapeutic targets and biomarkers for
the treatment of this disease.
Materials and methods
Patient criteria
Patients diagnosed with PAH that had undergone lung
transplantation surgery at Wuxi People's Hospital Affiliated to
Nanjing Medical University (Wuxi, China) between October 2014 and
October 2017 were included in the present study (Table I). Patients were 18-55 years of
age (mean ± standard deviation, 40±4 years) with a 6-min walking
distance of ≥100 and <500 m. PAH diagnosis was confirmed by
echocardiography and the diagnostic procedures conformed to the
2015 European Society of Cardiology and European Respiratory
Society guidelines for the diagnosis of pulmonary arterial
hypertension, pulmonary hypertension due to left heart disease,
pulmonary hypertension due to lung diseases and/or hypoxia, chronic
thromboembolic pulmonary hypertension, other PA obstructions and
pulmonary hypertension with unclear and/or multifactorial
mechanisms. Patients with severe obstructive pulmonary disease,
psychosis, a history of drug addiction or other diseases (chronic
liver disease, portal hypertension, chronic kidney disease,
amyloidosis and so on) and that had received prostacyclin,
endothelin receptor antagonist, L-arginine or sildenafil were
excluded.
 | Table ICharacteristics of PAH patients and
healthy donor. |
Table I
Characteristics of PAH patients and
healthy donor.
| Patient ID | Age, years | Sex | Ethnicity | Diagnosis/cause of
death | PAP, mm/Hg |
|---|
| PAH-01 | 55 | Female | Asian | HPAH | 90 |
| PAH-02 | 43 | Male | Asian | HPAH | 115 |
| PAH-03 | 50 | Male | Asian | HPAH | 94 |
| PAH-04 | 48 | Male | Asian | HPAH | 89 |
| PAH-05 | 47 | Female | Asian | HPAH | 112 |
| PAH-06 | 48 | Female | Asian | HPAH | 91 |
| PAH-07 | 52 | Female | Asian | HPAH | 90 |
| PAH-08 | 51 | Male | Asian | HPAH | 108 |
| Healthy
donor-01 | 50 | Female | Asian | Donor lung
tissue | N/A |
| Healthy
donor-02 | 43 | Female | Asian | Donor lung
tissue | N/A |
| Healthy
donor-03 | 48 | Female | Asian | Donor lung
tissue | N/A |
| Healthy
donor-04 | 47 | Male | Asian | Donor lung
tissue | N/A |
| Healthy
donor-05 | 44 | Male | Asian | Donor lung
tissue | N/A |
| Healthy
donor-06 | 52 | Male | Asian | Donor lung
tissue | N/A |
| Healthy
donor-07 | 55 | Female | Asian | Donor lung
tissue | N/A |
| Healthy
donor-08 | 53 | Male | Asian | Donor lung
tissue | N/A |
Preparation of human lung tissue
samples
Human PAH lung tissue samples were obtained from
lung transplantation patients at the Lung Transplant Group in Wuxi
People's Hospital Affiliated to Nanjing Medical University. This
study was approved by the Ethics Committee for Use of Human Samples
of the Nanjing Medical University, which was in accordance with The
Code of Ethics of the Helsinki Declaration of World Medical
Association for experiments involving humans. Each individual gave
written informed consent prior to their participation. Healthy lung
tissue samples were obtained from donors that were not suitable for
transplantation. The inclusion criteria for donors were as follows:
<50 years of age; a smoking history of <20 packs of
cigarettes per year; no chest trauma or sustained mechanical
ventilation for <1 week; FiO2, 1.0; positive
end-expiratory pressure, 5 cm; PaO2, >300 mmHg; chest
films indicating relatively clear lung fields; and bronchoscopy
results indicating a clear trachea. Lung tissue collection and
distal PA microscopic separation was performed according to the
protocol described previously (15,16).
Hematoxylin and eosin (H&E)
staining
Human lung tissue samples were sectioned into tissue
blocks and fixed in 4% paraformaldehyde overnight at 4°C. Tissues
were then dehydrated, cleared and embedded in paraffin. The
paraffin blocks were cut into 4-µm-thick sections and used
for H&E staining analysis (17). The sections were stained with
hematoxylin for 10 min at room temperature. The slices were rinsed
with water for 2 min and transferred to differentiation fluid (1%
hydrochloric acid) for 30 sec. Subsequently, the slices were place
in running water and washed for 30-60 min. Finally, the slices were
stained with eosin for 2-5 min at room temperature and then washed.
Images were captured using a light microscope (magnification,
×10).
Culture of HPASMCs
Primary HPASMCs were isolated from the explanted
lungs of patients undergoing transplantation for PAH, and from the
PAs of donor lungs. The primary culture of HPASMCs was performed
according to a previously published protocol (17). Using a light microscope, the
distal PAs were microdissected from lung explant tissues in a
dissection dish containing cold PBS. The separation steps were
performed according to a previously published study (18). The HPASMCs were subsequently
transferred to culture plates and incubated in SmGM-2 Smooth Muscle
Growth medium-2 BulletKit media (Lonza Group, Ltd.) containing 10%
(volume/volume) heat-inactivated FBS (Gibco; Thermo Fisher
Scientific, Inc.), 2 ng/ml human recombinant fibroblast growth
factor, 0.5 ng/ml human recombinant epidermal growth factor, 50
µg/ml gentamicin and 5 µg/ml insulin. Cells were
maintained at 37°C and 5% CO2 in an incubator for 1 week
(19). Passage 4-8 cells were
used for subsequent experiments.
Immunofluorescence analysis
To examine the purity of HPASMCs, the expression of
smooth muscle myosin heavy chain (SM-MHC) was analyzed by
immunofluorescence staining. The cells were fixed with 4%
polyformaldehyde for 15 min at 37°C, washed three times with cold
PBS for 5 min each time and then permeabilized with 0.5% Triton
X-100 (Sigma-Aldrich; Merck KGaA) for 10 min. The cells were
blocked in 5% bovine serum albumin (BSA) for 1 h at room
temperature and incubated overnight at 4°C with SM-MHC antibody
(cat. no. 21404-1-AP; 1:200; ProteinTech Group, Inc.), and
subsequently stained with a Fluorescein-conjugated Affinipure Goat
Anti-Rabbit IgG (H+L) secondary antibody (cat. no. SA00003-2;
1:100; ProteinTech Group, Inc.) for 1 h at room temperature. Slides
were then incubated for 5 min with DAPI. Finally, cell staining was
visualized by fluorescence microscopy.
Transfection
A short-hairpin RNA (shRNA) sequence targeting
MALAT1 and a negative control sequence were obtained from Shanghai
GenePharma Co., Ltd. The hsa-miR124-3p.1 mimic, mimic negative
control, inhibitor and inhibitor negative control were also
purchased from Shanghai GenePharma Co., Ltd. The sequences for each
were as follows: hsa-miR124-3p.1 mimic, sense, 5′-UAA GGC ACG CGG
UGA AUG CCA-3′; miR-124 antisense, 3′-UAA UUC CGU GCG CCA CUU
ACG-5′; hsa-miR124-3p.1 inhibitor, 5′-GGC AUU CAC CGC GUG CCU
UA-3′; sh-MALAT1, 5′-GAG TTG TGC TGC TAT CTT A-3′. hsa-miR124-3p.1
mimic and inhibitor were transfected at a concentration of 50
µM. The mimic negative control and inhibitor negative
control were transfected at a concentration of 50 µM. The
overexpression plasmid containing the MALAT1 sequence
(pcDNA-MALAT1) and empty vector (pcDNA) were synthesized by
Shanghai GenePharma Co., Ltd. HPASMCs were transfected with
shMALAT1 and the shRNA negative control at a final concentration of
20 nmol/l using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.), according to the manufacturer's protocol.
Furthermore, HPASMCs were transfected with pcDNA-MALAT1 and pcDNA
at a final concentration of 5 mg/l using Lipofectamine
2000®, according to the manufacturer's protocol. After
6-8 h, the transfection reagents were removed and the cells were
further cultured in DMEM containing 5% FBS for 24 h prior to
subsequent experiments.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from PAs and HPASMCs using
TRIzol reagent (Thermo Fisher Scientific, Inc.). First strand cDNA
synthesis was achieved using a high capacity cDNA reverse
transcription kit (Applied Biosystems; Thermo Fisher Scientific,
Inc.). Reactions were performed for 30 min at 60°C, followed by
heat inactivation for 2 min at 94°C. miRNA was extracted using a
miRcute Plus miRNA First-Strand cDNA Synthesis kit (Tiangen
Biotech, Co., Ltd.). qPCR analysis was performed using FastFire
qPCR PreMix (SYBR-Green; Tiangen Biotech, Co., Ltd.) and the ABI
StepOne Real-Time PCR System (Applied Biosystems; Thermo Fisher
Scientific, Inc). Reactions were performed using the following
thermocycling conditions: Pre-denaturation at 95°C for 1 min; 40
cycles of 95°C for 5 sec, 60°C for 15 sec, and a final step at 72°C
for 15 sec. GAPDH, β-actin and U6 were used as endogenous controls
for MALAT1, KLF5 and hsa-miR-124-3p.1 expression analysis. The
primers used were as follows: MALAT1 forward, 5′-GCA GGC GTT GTG
CGT AGA G-3′, and reverse, 5′-TTG CCG ACC TCA CGG ATT-3′; GAPDH
forward, 5′-CGC ATT GCC AGA CAT ATC AGC-3′, and reverse, 5′-AGG TGA
AGC AGG CTC AAT CAA-3′; hsa-miR-124-3p.1 forward, 5′-ACA CTC CAG
CTG GGT AAG GCA CGC GGT G-3′, and reverse, 5′-TGG TGT CGT GGA GTC
G-3′; U6 forward, 5′-CTC GCT TCG GCA GCA CA-3′, and reverse, 5′-TGG
TGT CGT GGA GTC G-3′; KLF5 (212 bp) forward, 5′-AGC TCA CCT GAG GAC
TCA TA-3′, and reverse, 5′-GTG CGC AGT GCT CAG T TC T-3′; and
β-actin, forward, 5′-TGA GAG GGA AAT CGT GCG TGA C-3′, and reverse,
5′-AAG AAG GAA GGC TGG AAA AGA G-3′. Relative target expression
levels were calculated using the 2−ΔΔCt method (20).
Western blotting
Protein samples were extracted from PAs and HPASMCs
by lysing them in RIPA buffer for 30 min on ice. Samples were then
sonicated with 20 KHz frequency on ice for 1 min and centrifuged at
14,000 × g for 15 min at 4°C. The protein concentration was
quantified using a BCA kit (Nanjing KeyGen Biotech Co., Ltd.).
Total protein (20 µg) was separated by 10% SDS-PAGE,
transferred to nitrocellulose membranes and subjected to western
blotting according to the previously published protocols (15,16). Nitrocellulose membranes were
incubated at 4°C overnight with the following specific antibodies:
proliferating cell nuclear antigen (PCNA; cat. no. 10205-2-AP;
1:1,000; ProteinTech Group, Inc.), cyclin A1 (cat. no. 13295-1-AP;
1:1,000; ProteinTech Group, Inc.), KLF5 (cat. no. ab137676; 1:500;
Abcam), cyclin D1 (cat. no. 60186-1-lg; 1:1,000; ProteinTech Group,
Inc.), cyclin E1 (cat. no. 11554-1-AP; 1:1,000; ProteinTech Group,
Inc.) and β-actin (cat. no. 60008-1-lg; 1:5,000; ProteinTech Group,
Inc.). Membranes were subsequently incubated with goat-anti-rabbit
IgG secondary antibodies (cat. no. SA00001-2; 1:10,000; Nanjing
KeyGen Biotech Co., Ltd.) or goat-anti-mouse IgG secondary antibody
(cat. no. SA00001-1; 1:10,000; Nanjing KeyGen Biotech Co., Ltd),
shaken and incubated at room temperature for 1 h. Imaging was then
performed using the ECL plus detection reagent (Thermo Fisher
Scientific, Inc.). Blots were imaged by VersaDoc™ MP 4000 (Bio-Rad
Laboratories, Inc.) and quantified using PDQuest Advanced 2D
analysis software (version 8.0; Bio-Rad Laboratories, Inc.).
Cell Counting kit-8 (CCK-8) assay
To examine the rate of cell proliferation, HPASMCs
were seeded at a density of 0.5×104 cells in 96-well
microtiter plates and cultured without serum for 24 h. The cells
were then transfected with the aforementioned sequences. Following
incubation at 37°C for 48 h, 20 µl CCK-8 reagent was added
and the cells were incubated for a further 4 h at 37°C. The
absorbance was read at 450 nm using a Synergy™ 2 multifunctional
enzyme standard instrument.
Cell cycle analysis
To investigate the role of MALAT1 in cell cycle
progression, the cell cycle distribution of HPASMCs trans-fected
with sh-MALAT1 or pcDNA-MALAT1 was analyzed by flow cytometry.
Following transfection, cells were collected by trypsinization,
centrifuged at 425 × g for 5 min at room temperature, and then
resuspended in 1 ml cold PBS. The cells were then washed twice with
PBS and fixed in 70% ethanol at 4°C overnight. Cell fragments were
resuspended in 200 µl PBS and 200 µl RNase A for 10
min at room temperature. At this point, the cells were stained with
200 µl propidium iodide at 4°C for 10 min in the dark.
HPASMCs were then filtered once through 400-mesh sieves before they
were analyzed using a FACSCalibur Flow Cytometer (BD Biosciences).
The data were analyzed using ModFit software (version 4.1; Verity
Software House, Inc., Topsham, ME, USA).
Target prediction and luciferase reporter
gene activity assay
TargetScan 7.2 (http://www.targetscan.org/) and DIANA-LncBase
(http://omictools.com/diana-lncbase-tool) were used for
target prediction. The wild-type or mutant 3′-untranslated region
(3′-UTR) of MALAT1 containing the predicted hsa-miR-124-3p.1
binding site was purchased from Shanghai GenePharma Co., Ltd. and
inserted into the pmirGLO Dual-Luciferase miRNA Target Expression
Vector (Promega Corporation). Similarly, the 3′-UTR of KLF5
containing the predicted target sites for hsa-miR-124-3p.1 was
synthesized by PCR amplification. This fragment was inserted into
the pmirGLO dual luciferase miRNA target expression vector (Promega
Corporation) to generate the KLF5-wild type reporter vector. This
vector and hsa-miR-124-3p.1 were subsequently co-transfected into
HPASMCs using Lipofectamine 2000® transfection reagent
(Thermo Fisher Scientific, Inc.). After 6-8 h, the transfection
reagents were removed and the HPASMCs were further cultured in DMEM
containing 5% FBS for 48 h. Luciferase activity was measured using
a dual luciferase reporter assay (Promega Corporation).
Renilla luciferase activity was used for normalization.
RNA immunoprecipitation (RIP) assay
An RIP assay was used to investigate the interaction
between MALAT1 and hsa-miR-124-3p.1 using the EZ-Magna RIP™ RNA
Binding Protein Immunoprecipitation kit (Merck KGaA). The cells
were first lysed using the lysis buffer (catalog no. 17-701; Merck
KGaA) before they were incubated with an anti-human argonaute
RNA-induced silencing complex (RISC) catalytic component 2 (AGO2)
antibody (Merck KGaA) coated on magnetic beads in RIP buffer. Input
and normal IgG were selected as controls for the experiment. RNA
was isolated and reverse transcribed into cDNA before MALAT1 and
hsa-miR-124-3p.1 levels were analyzed by RT-qPCR.
Scratch wound healing assay
To determine cell migration, HPASMCs were
transferred into 6-well plates and cultured to 90% confluence. A
sterile pipette tip was then used to generate a vertical 'wound' ~1
mm in diameter. Cell migration in the 6-well plates was measured as
described previously (15).
HPASMCs were washed with cold PBS and images were captured under a
light microscope to record the wound width at 0 h. The culture
medium was then replaced with medium containing 5% FBS. Following
incubation for 24 h, the cell images were again captured under a
microscope and the degree of migration was marked to quantify the
cell migration ability.
Statistical analysis
Experiments were repeated at least three times and
all data were presented as the mean ± standard error of mean.
Statistical differences between or among groups were analyzed using
a Student's t-test or one-way ANOVA followed by Bonferroni's test
using the GraphPad Prism software package (version 5.0; GraphPad
Software, Inc.). P<0.05 was considered to indicate a
statistically significant difference.
Results
MALAT1 is highly expressed in PA tissues
and HPASMCs derived from patients with PAH
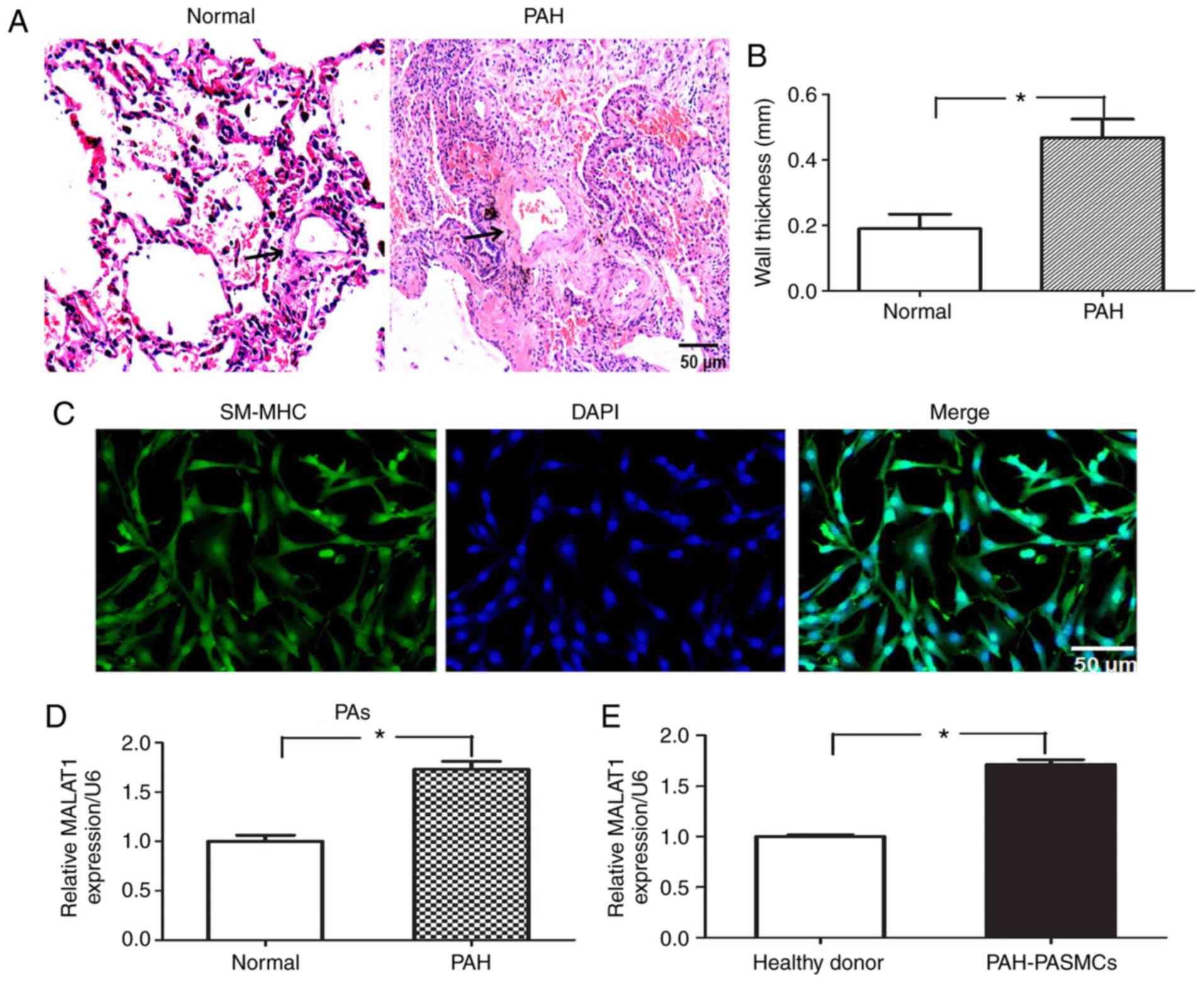
The vascular wall morphology of the lung tissue
samples was analyzed by H&E staining. Compared with the healthy
volunteers, the vascular wall thickness of patients with PAH was
significantly increased in medium-sized PAs (P<0.05; Fig. 1A and B). The purity of HPASMCs was
verified using smooth muscle myosin heavy chain antibody (Fig. 1C). To determine whether MALAT1 may
be involved in the pathologic process of PAH, a total of eight
paired PAH and normal PA tissue samples from patients with PAH were
used to determine the expression of MALAT1 by RT-qPCR analysis. As
demonstrated in Fig. 1D, the
average expression level of MALAT1 in PAs from PAH tissues was
significantly increased compared with the normal PAs tissues
(P<0.05). In addition, RT-qPCR analysis revealed that the
expression of MALAT1 was significantly upregulated in HPASMCs from
patients with PAH compared with those from healthy donors
(P<0.05; Fig. 1E). These
results suggest that MALAT1 is highly expressed in PA tissues and
HPASMCs derived from patients with PAH.
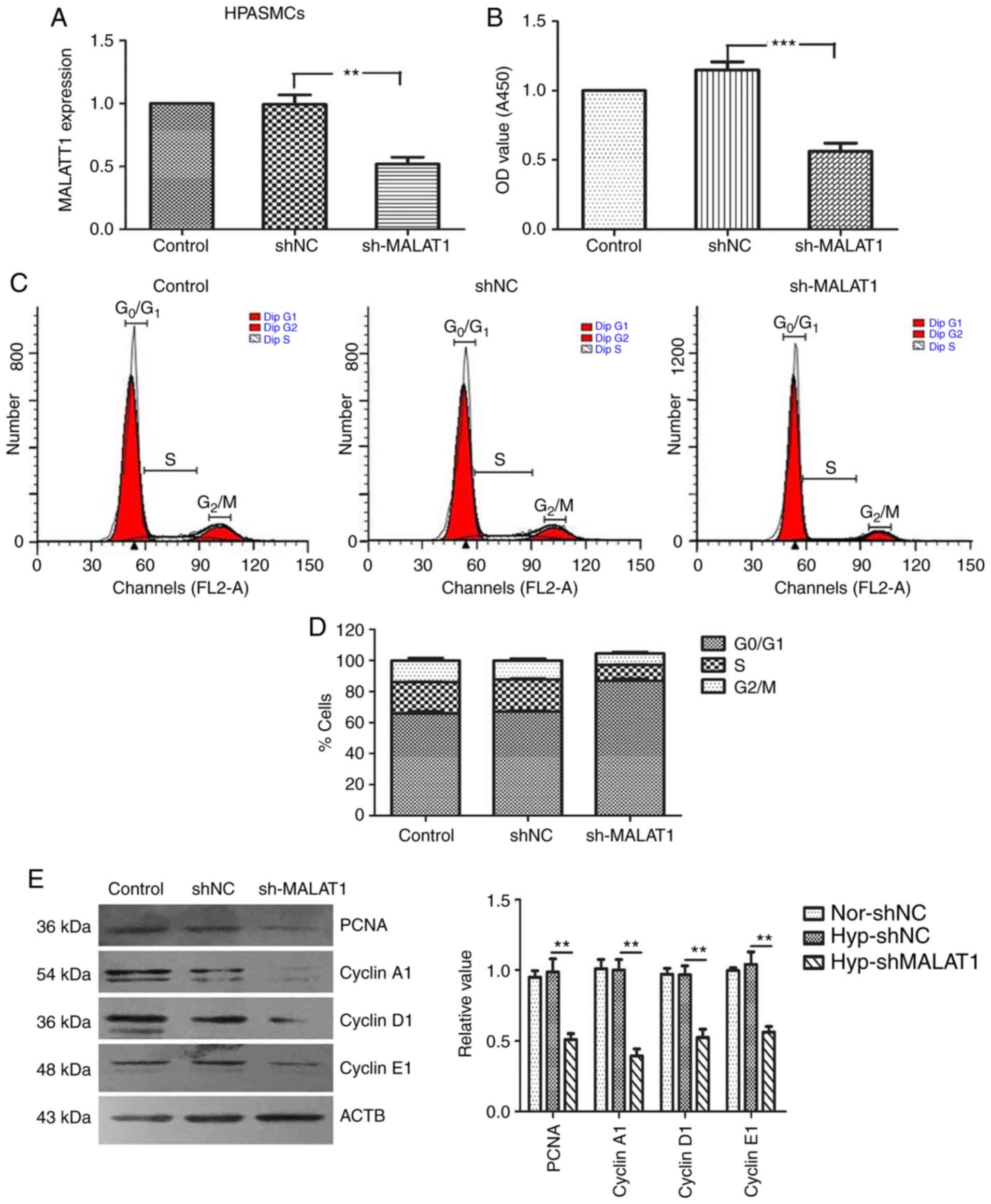
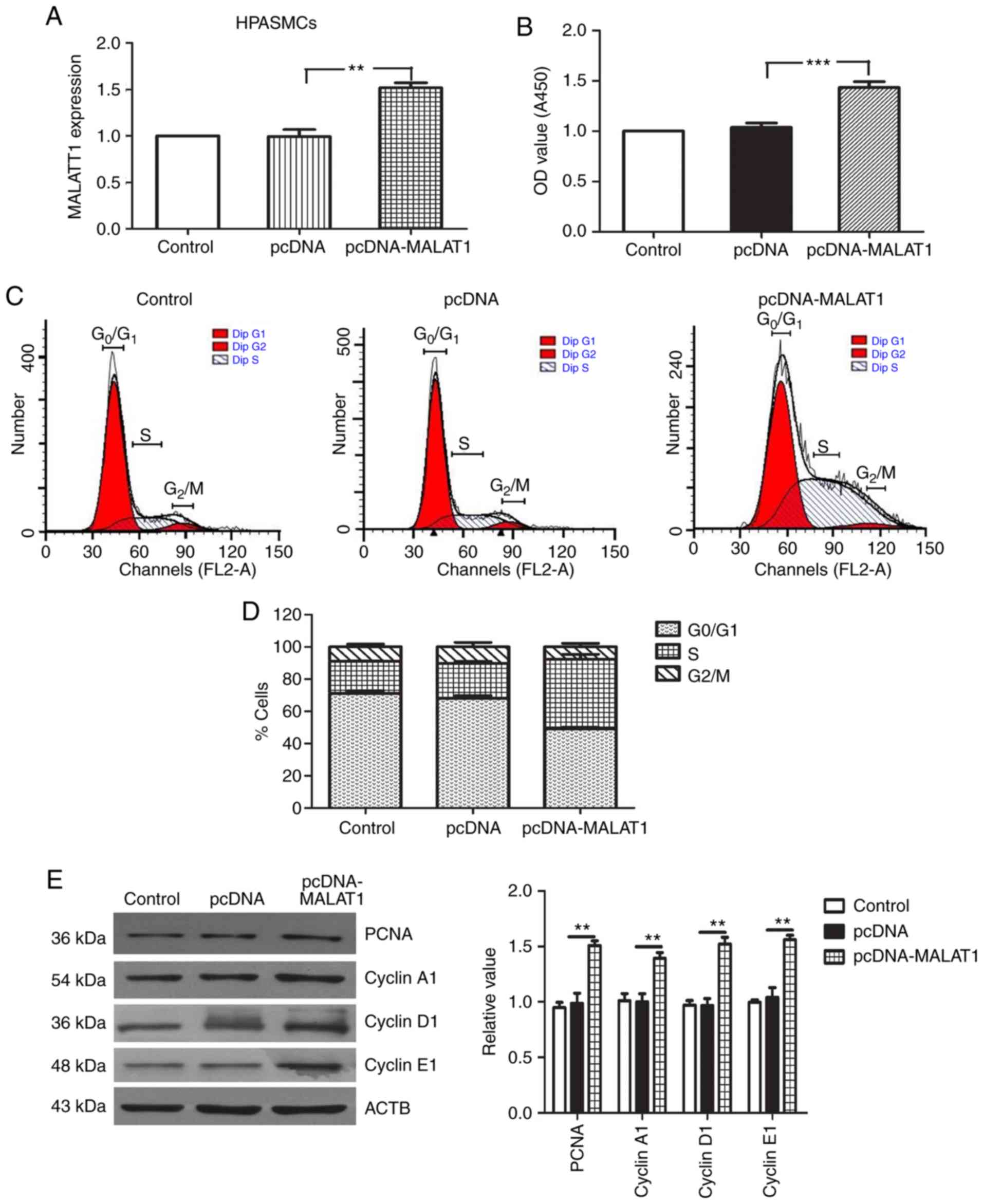
MALAT1 affects the proliferation and
migration of HPASMCs
To verify the role of MALAT1 in the cellular
processes of HPASMCs, the proliferation and migration of HPASMCs
transfected with sh-MALAT1 and pcDNA-MALAT1 were analyzed. As
demonstrated in Figs. 2 and
3, the transfection efficiency of
sh-MALAT1 and pcDNA-MALAT1 was confirmed by RT-qPCR analysis. A
CCK-8 assay was then used to determine the growth rate of HPASMCs.
The results demonstrated that knockdown of MALAT1 significantly
inhibited the growth of HPASMCs when compared with the negative
control group (P<0.001; Fig.
2B). By contrast, MALAT1 overexpression promoted cell
proliferation when compared with the negative control group
(Fig. 3B). To examine whether
MALAT1 may regulate cell cycle progression, flow cytometry was used
to analyze the cell cycle distribution. The results indicated that
MALAT1 silencing reduced the percentage of cells (Fig. 2C and D), while MALAT1
overexpression increased the percentage of cells (Fig. 3C and D), in the G2/M
and S phases. It has been reported that cell cycle proteins (such
as: Cyclin A1/D1/E1) serve key roles in S and G2/M
phases, and have been widely recognized as markers of cell
proliferation (21,22). To understand the effect of MALAT1
on the regulation of cyclin proteins, western blotting was
performed to analyze the expression levels of PCNA, cyclin A1,
cyclin D1 and cyclin E1. The results demonstrated that transfection
of HPASMCs with sh-MALAT1 significantly inhibited the expression of
cell cycle proteins, PCNA, cyclin A1, cyclin D1 and cyclin E1 when
compared with the controls (P<0.01; Fig. 2E). By contrast, MALAT1
overexpression signifi-cantly increased the expression levels of
these proteins when compared with the controls (P<0.01; Fig. 3E).
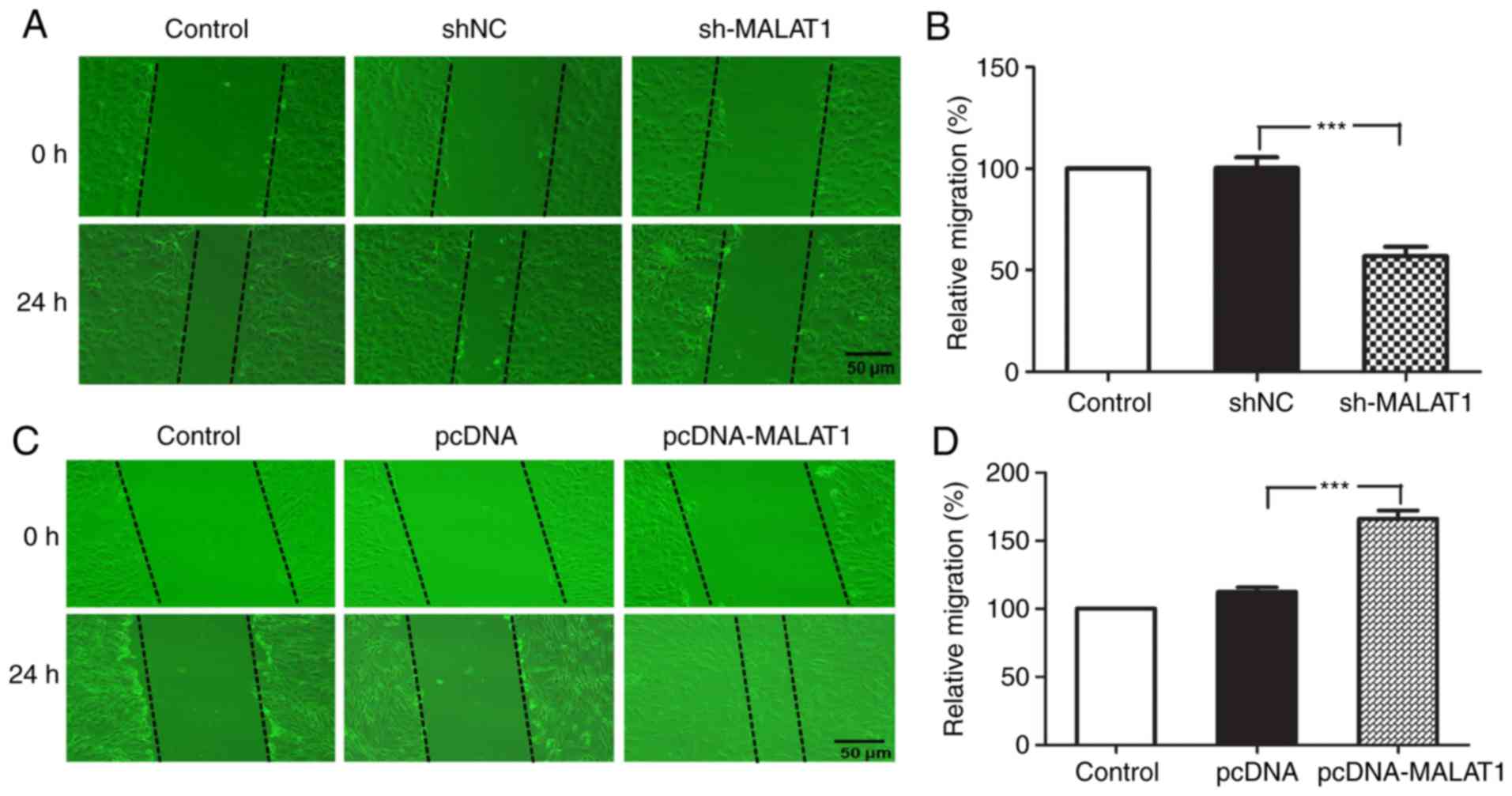
To examine whether MALAT1 may affect the migratory
ability of HPASMCs in vitro, the role of MALAT1 in HPASMC
cell migration was analyzed using a scratch wound healing assay.
The results demonstrated that MALAT1 silencing significantly
inhibited cell migration in vitro when compared with the
negative control group (P<0.001; Fig. 4A and B). By contrast, MALAT1
overexpression significantly promoted the migration of HPASMCs
(P<0.001; Fig. 4C and D).
These results suggest that increased expression of MALAT1 may be a
lethal risk factor for PAH and that a reduction in MALAT1
expression may exert a protective effect on the occurrence and
development of the disease.
MALAT1 functions as a ceRNA that binds
directly to hsa-miR-124-3p
LncRNAs may function as ceRNAs in different types of
carcinomas (23). Previous
studies have also indicated that MALAT1 may promote bladder cancer
(11), liver fibrosis (12) and melanoma (13) progression by functioning as a
ceRNA that targets miR-125b, miR-101b and miR-183, respectively.
PAH and cancer share the common features, such as abnormal cell
proliferation and apoptosis resistance (24). Therefore, the authors of the
present study hypothesized that MALAT1 may exert similar effects in
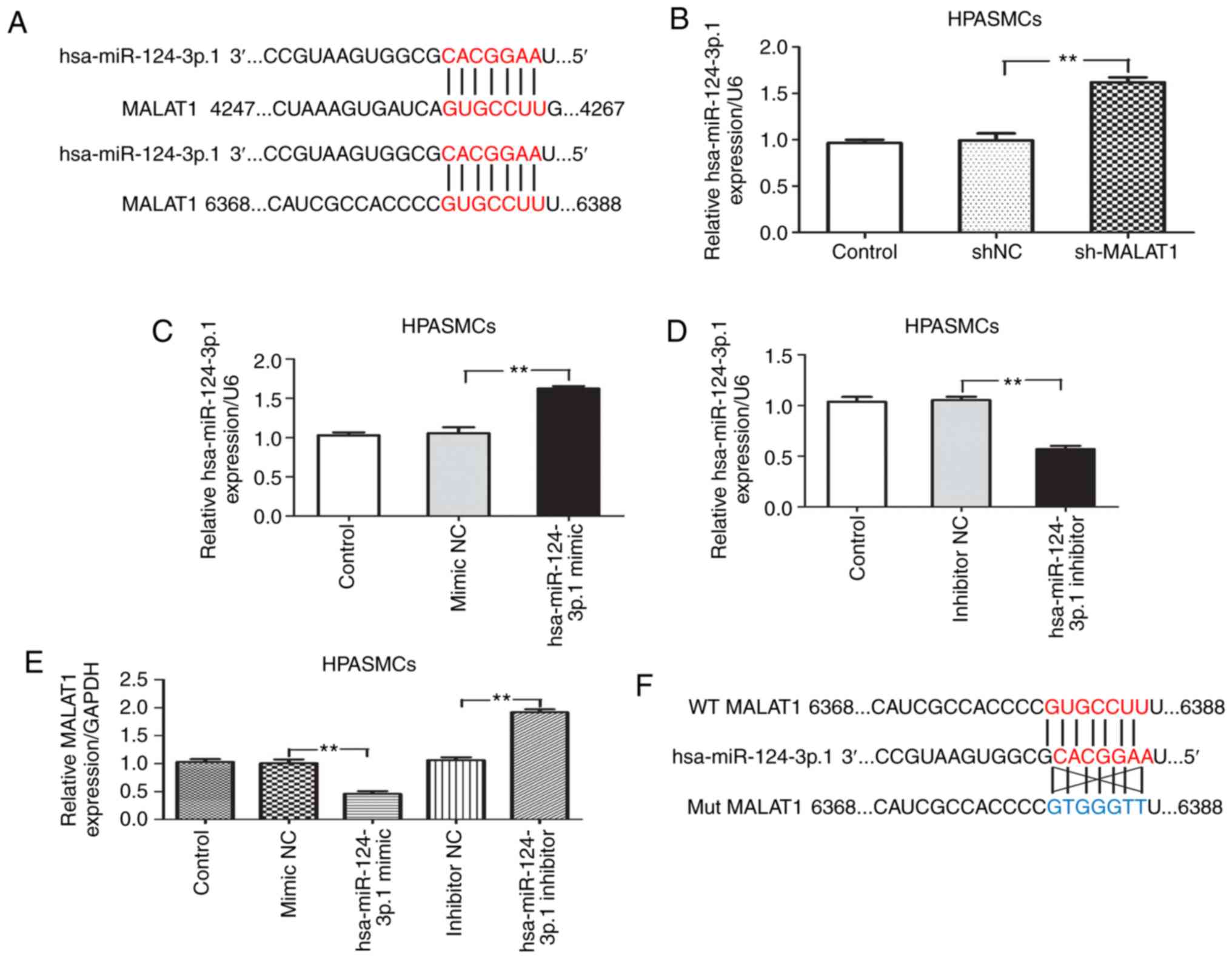
PAH. The presence of a putative binding site for hsa-miR-124-3p.1
in the MALAT1 transcript was identified using LncBase Predicted v.2
(Fig. 5A). RT-qPCR experiments
revealed that the expression of hsa-miR-124-3p.1 was significantly
upregulated when MALAT1 was inhibited (P<0.01; Fig. 5B). The hsa-miR-124-3p.1 mimic or
inhibitor was transfected into HPASMCs to examine the transfection
efficiency (Fig. 5C and D). It
was found that endogenous MALAT1 expression was significantly
decreased in hsa-miR-124-3p.1 mimic-transfected cells (P<0.01),
while transfection with the hsa-miR-124-3p.1 inhibitor increased
MALAT1 expression (P<0.01; Fig.
5E). These results indicate a negative correlation between
MALAT1 and hsa-miR-124-3p.1. Luciferase reporter plasmids harboring
wild-type MALAT1 and mutant MALAT1 3′-UTR sequences containing the
predicted hsa-miR-124-3p.1 binding sites were generated (Fig. 5F). Dual luciferase reporter assays
demonstrated that co-transfection of HPASMCs with the
hsa-miR-124-3p.1 mimic resulted in a significant decrease in
luciferase activity in cells transfected with wild-type MALAT1
(P<0.01), whereas the luciferase activity of cells transfected
with the mutant form remained unaffected (Fig. 5G). The RIP assay results
demonstrated that MALAT1 and hsa-miR-124-3p.1 were significantly
enriched in AGO2 immunoprecipitates when compared with the
IgG-pellet (P<0.01), indicating that MALAT1 and hsa-miR-124-3p.1
were located in the same RISC (Fig.
5H). This suggests that MALAT1 interacts with hsa-miR-124-3p.1
and functions as a ceRNA. Together, these findings prompted an
investigation of the role of hsa-miR-124-3p.1 in PAH. Therefore,
the expression of hsa-miR-124-3p.1 in 8 paired PAH and normal
tissues were analyzed by RT-qPCR. The expression of
hsa-miR-124-3p.1 was significantly reduced in PAH tissues when
compared with normal PA tissues (P<0.01; Fig. 5I). Subsequently, hsa-miR-124-3p.1
expression in HPASMCs derived from patients with PAH and healthy
donors were compared. The results demonstrated that
hsa-miR-124-3p.1 expression was significantly reduced in HPASMCs
from patients with PAH when compared with healthy donors
(P<0.05). This effect was significantly reversed when HPASMCs
were transfected with shMALAT1 (P<0.05; Fig. 5J).
KLF5 is a direct target of
hsa-miR-124-3p.1 and is aberrantly expressed in PA tissues from
patients with PAH and hypoxic HPASMCs
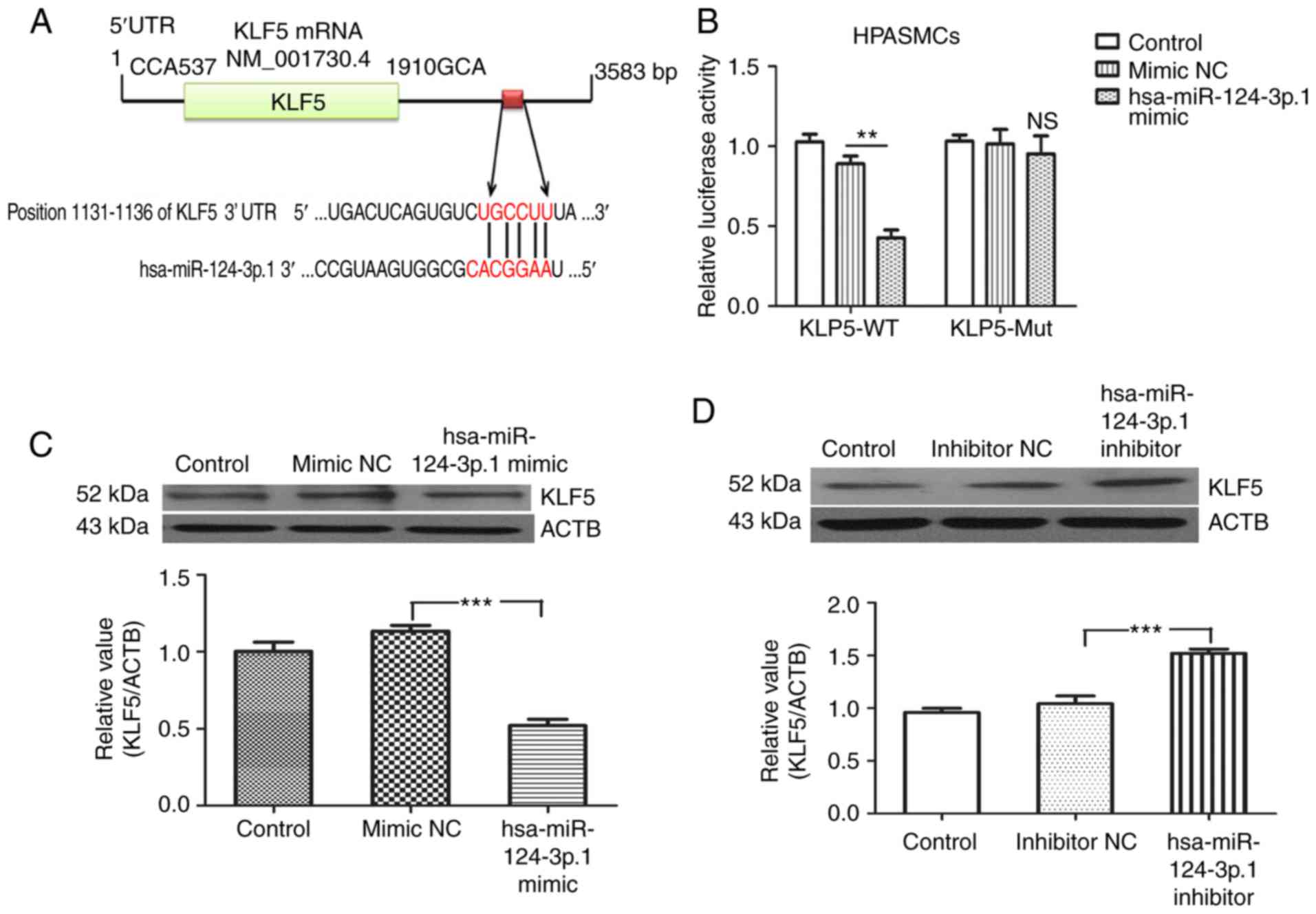
To investigate the mechanism of action of
hsa-miR-124-3p.1 in HPASMCs, bioinformatics analysis (http://www.targetscan.org/) was used to identify a
complementary binding site between hsa-miR-124-3p.1 and the 3′-UTR
of KLF5 (Fig. 6A). This suggested
that KLF5 may be a target of hsa-miR-124-3p.1. Luciferase reporter
plasmids containing the wild-type and mutant hsa-miR-124-3p.1
binding site sequences within the KLF5 3′-UTR were generated. Dual
luciferase reporter assays demonstrated that co-transfection of
HPASMCs with the hsa-miR-124-3p.1 mimic and wild-type sequences was
associated with a significant decrease in luciferase activity
(P<0.01), whereas no obvious changes in luciferase activity was
observed in cells co-transfected with the mutant sequence (Fig. 6B). Western blotting analysis
indicated that the expression of KLF5 was significantly reduced in
hsa-miR-124-3p.1 mimic-transfected cells (P<0.01; Fig. 6C). By contrast, KLF5 was observed
to be significantly upregulated in the hsa-miR-124-3p.1
inhibitor-transfected group (P<0.01; Fig. 6D).
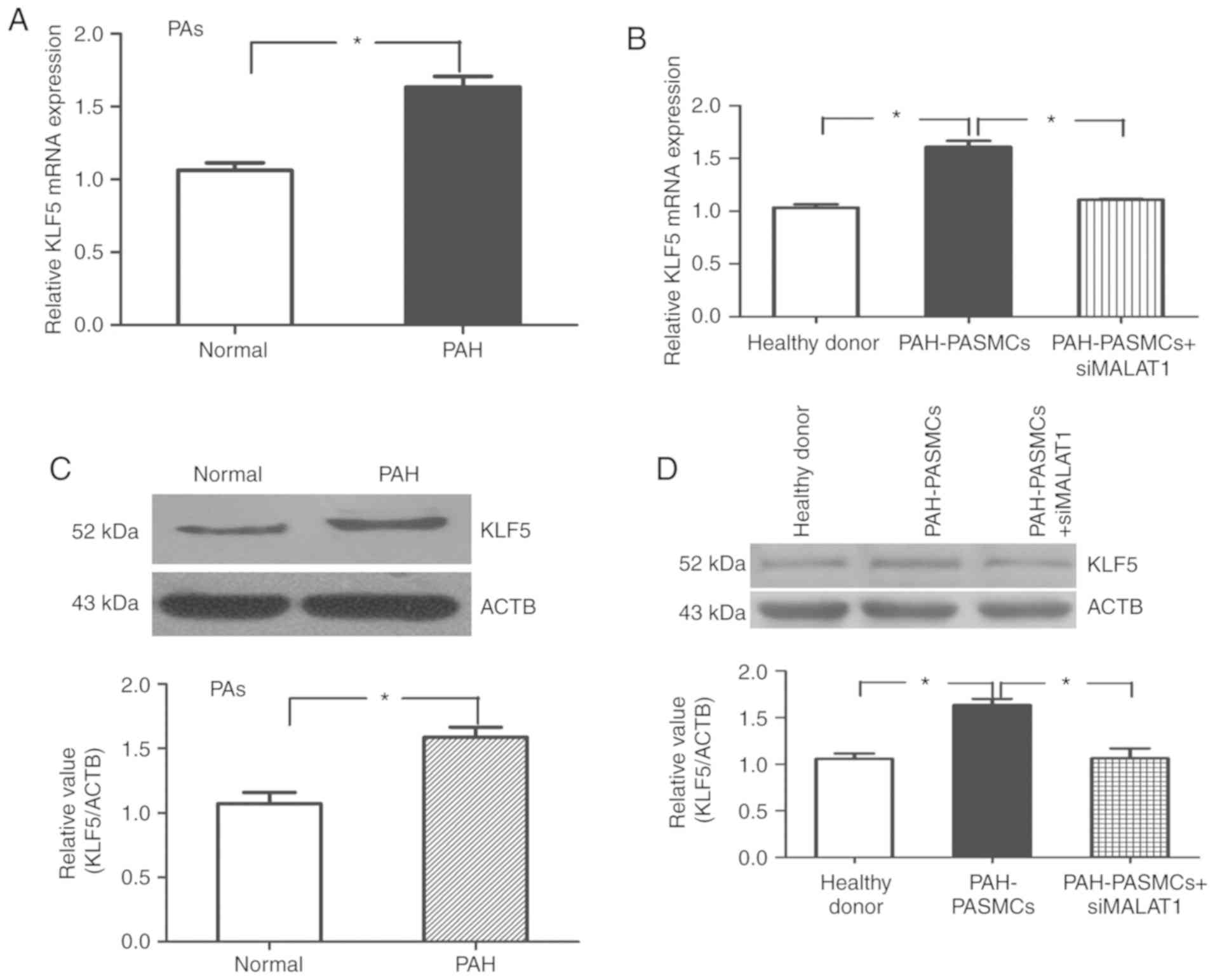
KLF5 has been reported to serve a crucial role in
hypoxia-induced vascular remodeling (14). Therefore, to further examine the
role of KLF5 in PAH, the expression of KLF5 in the PAs derived from
patients with PAH and healthy donors was examined. Western blotting
and RT-qPCR results demonstrated significantly increased levels of
KLF5 expression in the PAs from patients with PAH compared with
healthy donors (P<0.05; Fig. 7A
and C). In parallel PA tissue experiments, KLF5 mRNA and
protein expression levels were also significantly increased in
HPASMCs derived from patients with PAH, and this effect was
inhibited by transfection with shMALAT1 (P<0.01; Fig. 7B and D).
MALAT1 regulates the growth of HPASMCs
via hsa-miR-124-3p.1 and KLF5
To examine the interaction between MALAT1,
hsa-miR-124-3p.1 and KLF5, the expression of KLF5 protein in
HPASMCs transfected with sh-MALAT1, hsa-miR-124-3p.1 mimic or
inhibitors were analyzed. MALAT1 silencing reduced the expression
of KLF5 at the mRNA and protein levels (Fig. 8A and B). The same effect on KLF5
expression was observed in cells transfected with hsa-miR-124-3p.1
mimics, whereas the hsa-miR-124-3p.1 inhibitor group showed the
opposite effect. Of particular note, transfection with the
hsa-miR-124-3p.1 inhibitor counteracted the downregulation of KLF5
expression induced by MALAT1 knockdown (Fig. 8C).
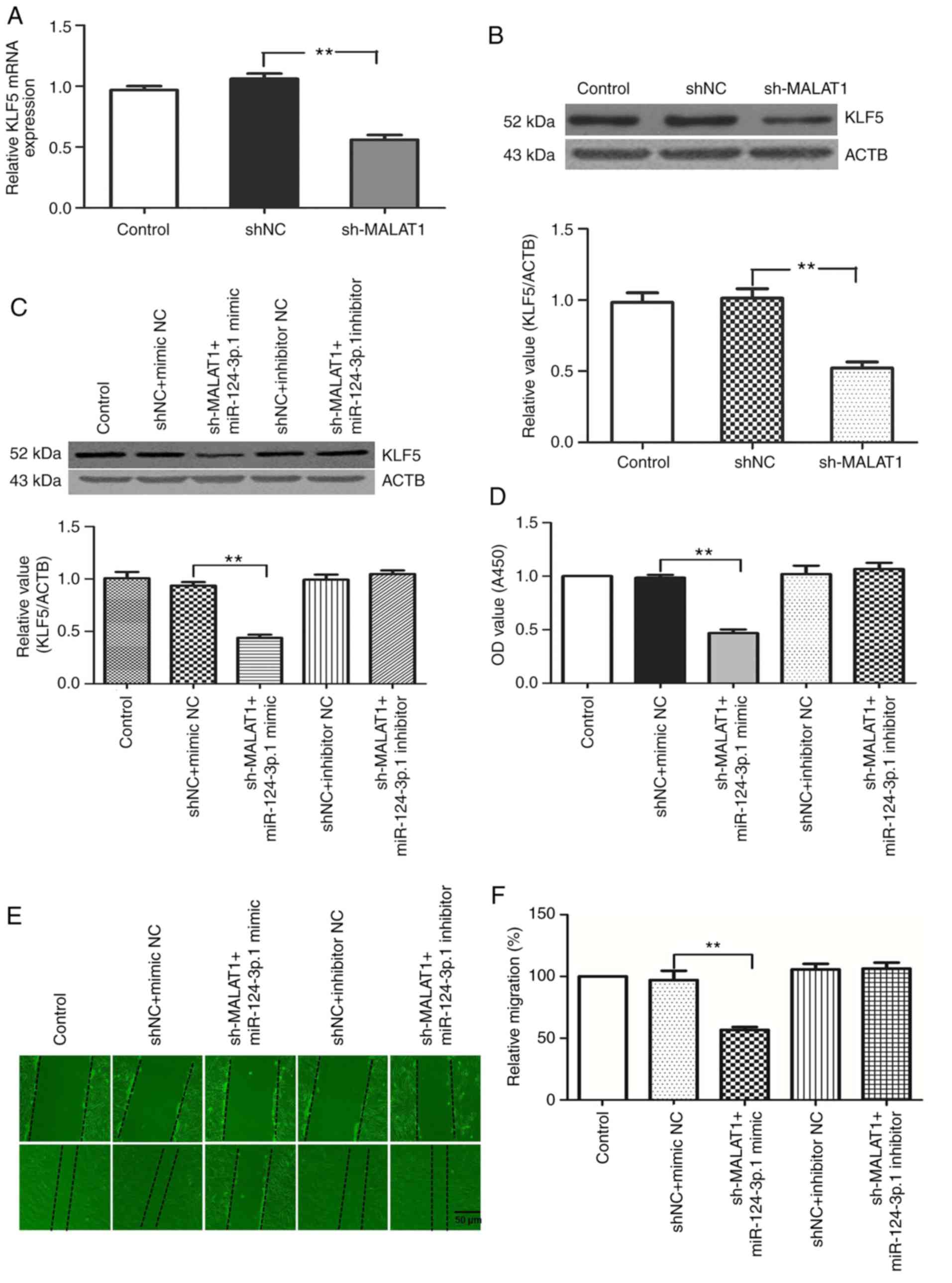 | Figure 8MALAT1 knockdown suppresses the
expression of KLF5 via upregulating hsa-miR-124-3p.1. KLF5 (A) mRNA
and (B) protein expression levels in HPASMCs transfected with
sh-MALAT1. (C) Protein expression levels of KLF5 in HPASMCs
co-transfected with sh-MALAT1 and hsa-miR-124-3p.1 mimic or
inhibitor. HPASMCs were co-transfected with (D) sh-MALAT1 mimic or
inhibitor. HPASMCs were co-transfected with (E) hsa-miR-124-3p.1
mimic or inhibitor. Changes in (D) cell viability and (E) migration
were assessed and (F) analyzed. (G) Cell cycle analysis of HPASMCs
co-transfected with sh-MALAT1 and hsa-miR-124-3p.1 mimic or
inhibitor. (H) Histogram presenting the percentage of cells in the
G0/G1, S and G2/M phases. (I)
Protein expression levels of PCNA, cyclin A1, cyclin D1 and cyclin
E1 were analyzed by western blotting following co-transfection of
HPASMCs with sh-MALAT1 and hsa-miR-124-3p.1 mimic or inhibitor.
**P<0.01. MALAT1, metastasis-associated lung
adenocarcinoma transcript 1; KLF5, Kruppel-like factor 5; miR,
microRNA; HPASMCs, human pulmonary artery smooth muscle cells;
shRNA, short-hairpin RNA; PCNA, proliferating cell nuclear antigen;
ACTB, β-actin; NC, negative control. |
The effect of MALAT1 on the proliferation and
migration of HPASMCs was largely offset by the miR-124-3p.1 mimic
in MALAT1-silenced cells, while there was no obvious change in
these characteristics in the control group and the MALAT1 plus
miR-124-3p.1-silenced group. This suggests that MALAT1 knockdown
was unable to suppress HPASMC proliferation and migration when
hsa-miR-124-3p.1 was inhibited (Fig.
8D-I). The results indicate that MALAT1 silencing may inhibit
the growth and migration of HPASMCs by sponging hsa-miR-124-3p.1.
Ultimately, these results demonstrate that MALAT1 may regulate KLF5
expression by binding competitively to hsa-miR-124-3p, thus
promoting the growth of HASMCs.
Discussion
Pulmonary vascular remodeling is a multifactorial
pathological process characterized by the abnormal proliferation
and migration of HPASMCs, as well as the resistance of these cells
to apoptosis. The identification of suitable therapeutic targets
that suppress vascular remodeling is important. Emerging evidence
has increased understanding of the multifaceted role of lncRNAs as
modifying factors of vascular remodeling. For instance, MALAT1
(7) and transforming growth
factor-β2-overlapping transcript 1 (25) have been found to promote vascular
endothelial cell dysfunction, while long intergenic RNA-p21
(26), lnc-Ang362 (27) and maternally expressed 3 (19) contribute to vascular smooth muscle
cell proliferation. MALAT1 was originally discovered in non-small
cell lung cancer and is used as a prognostic marker for lung cancer
metastasis (4,28). Previous studies have reported that
MALAT1 is aberrantly expressed in numerous solid tumors and is
involved in the proliferation and metastasis of several cancers
(29,30). However, accumulating evidence
suggests that MALAT1 may also regulate multiple pathological
vascular remodeling processes (7,8).
However, the role and precise mechanisms of MALAT1 in pulmonary
vascular remodeling is not fully understood. The present study
reports that MALAT1 is upregulated in the PA tissues and HPASMCs
derived from patients with PAH. In addition, knockdown of MALAT1
reduced HPASMC viability and proliferation, with a greater number
of cells in the G0/G1 phase. Bioinformatics
analysis predicted an interaction between MALAT1 and
hsa-miR-124-3p.1. A negative regulatory feedback loop between
MALAT1 and hsa-miR-124-3p.1 was also subsequently identified, and
the function of MALAT1 as a ceRNA by sponging hsa-miR-124-3p.1 in
HPASMCs was demonstrated.
A number of previous studies have reported that
increased expression of MALAT1 may serve an important role in the
development of different tumors (4,5,31).
In addition, MALAT1 was reported to be an important regulator of
angiogenesis (7). The present
study primarily focused on the effects of MALAT1 on pulmonary
vascular remodeling and found that MALAT1 expression was increased
in PA tissues and HPASMCs derived from patients with PAH. These
results are consistent with the results of a previous study
(8). In the present study,
knockdown of MALAT1 in HPASMCs suppressed viability and inhibited
cell cycle progression through the G0/G1
phase. The protein expression levels of PCNA, cyclin A1, cyclin D1
and cyclin E1 were also downregulated, which is consistent with the
cell cycle analysis results. By contrast, these effects were
reversed following overexpression of MALAT1.
Previous studies have demonstrated that numerous
lncRNAs function as ceRNAs, which inhibit normal miRNA activity and
modulate miRNA signaling pathways (32-34). Therefore, the authors of the
current study proposed that an association between MALAT1 and miRNA
in PAH may exist. Bioinformatics analysis revealed multiple miRNA
binding sites in the MALAT1 sequence. However, RT-qPCR results
indicated that only hsa-miR-124-3p.1 was able to repress the
expression of MALAT1 RNA in HPASMCs. These results were supported
by luciferase reporter and RIP assay analyses. Therefore, the
present study provides strong evidence to suggest that MALAT1 may
function as a ceRNA for hsa-miR-124-3p.1. In addition, a negative
correlation between MALAT1 and hsa-miR-124-3p.1 in PAH progression
was observed.
Previous studies have demonstrated that miR-124
affects the inflammatory phenotype, proliferation and migration of
pulmonary vascular fibroblasts (35,36). Consistently, the results of the
current study demonstrated that hsa-miR-124-3p.1 was down-regulated
in PA tissues and HPASMCs derived from patients with PAH, implying
that the expression of hsa-miR-124-3p.1 may be involved in the
development PAH. KLF5 was also identified as a direct functional
effector of hsa-miR-124-3p.1. A series of reports have demonstrated
that KLF5 is elevated in human lung biopsies and HPASMCs isolated
from the distal PAs of patients with PAH when compared with those
obtained from normal patients (14,37,38). The results of the current study
provide evidence to suggest that MALAT1 may function as a ceRNA for
hsa-miR-124-3p.1. In addition, the expression levels of MALAT1 were
observed to be downregulated in HPASMCs transfected with
hsa-miR-124-3p.1 mimics, while its expression was upregulated in
the hsa-miR-124-3p.1 inhibitor-transfected group of cells.
Furthermore, luciferase assays demonstrated that hsa-miR-124-3p.1
reduced the activity of the wild-type MALAT1 3′-UTR but had no
effect on the mutant sequence. The results of the RIP assay
indicated that MALAT1 and hsa-miR-124-3p.1 formed part of the RISC.
Together, these results suggest that MALAT1 functions as a ceRNA
and interacts with hsa-miR-124-3p.1. As such, MALAT may promote
pulmonary vascular remodeling progression in vivo by
operating as a ceRNA.
It was found that MALAT1, a ceRNA, shared the
complementary sequence of hsa-miR-124-3p.1 with its target KLF5 by
bioinformatics software analysis. In addition, the expression of
KLF5 was observed to be influenced by MALAT1 in HPASMCs. MALAT1
silencing reduced the mRNA and protein levels of KLF5. The effect
of MALAT1 on KLF5 expression was reversed by transfection of cells
with the hsa-miR-124-3p.1 inhibitor. In addition, KLF5 expression
was downregulated when HPASMCs were co-transfected with sh-MALAT1
and hsa-miR-124-3p.1 mimics, while hsa-miR-124-3p.1 suppression
exhibited the opposite effects. Furthermore, silencing of MALAT1
expression and overexpression of hsa-miR-124-3p.1 inhibited the
proliferation and migration of HPASMCs. Taken together, the results
suggest that the MALAT1/hsa-miR-124-3p.1/KLF5 axis may serve an
important role in the development of PAH.
In conclusion, the expression of MALAT1,
hsa-miR-124-3p.1 and KLF5 in HPASMCs and PA tissues were examined
in the present study. MALAT1 promoted the growth and migration of
HPASMCs potentially via sponging hsa-miR-124-3p.1 and KLF5.
Understanding the molecular mechanisms of MALAT1 in PAH may provide
a novel approach for developing effective therapeutic interventions
for patients with PAH.
Funding
This study was supported by National Natural Science
Foundation of China (grant no. 81500039).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
DW performed H&E staining, the Cell Counting
Kit-8 assay, luciferase reporter gene activity assay and RNA
immunoprecipitation assay, and wrote the manuscript. HX conducted
western blotting and reverse transcription-quantitative polymerase
chain reaction experiments. BW performed flow cytometry assays. SJ
performed cell culture, cell transfection and the wound healing
assay. HP was responsible for patient sample collection and
acquisition of clinical data. RW designed the present study and the
experiments. JC interpreted and analyzed the data. All authors read
and approved the final version of the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee for the use of human samples of Wuxi People's Hospital
Affiliated to Nanjing Medical University (Wuxi, China), which was
in accordance with the code of ethics of the Declaration of
Helsinki developed by the World Medical Association. Each
individual provided written informed consent prior to their
participation.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no conflict of
interest.
Acknowledgments
Not applicable.
References
|
1
|
Voelkel NF, Gomez-Arroyo J, Abbate A,
Bogaard HJ and Nicolls MR: Pathobiology of pulmonary arterial
hypertension and right ventricular failure. The Euro Respir J.
40:1555–1565. 2012. View Article : Google Scholar
|
|
2
|
Uchida S and Dimmeler S: Long noncoding
RNAs in cardiovascular diseases. Circ Res. 116:737–750. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhao XY and Lin JD: Long noncoding RNAs: A
new regulatory code in metabolic control. Trends Biochem Sci.
40:586–596. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA, and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ren S, Liu Y, Xu W, Sun Y, Lu J, Wang F,
Wei M, Shen J, Hou J, Gao X, et al: Long noncoding RNA MALAT-1 is a
new potential therapeutic target for castration resistant prostate
cancer. J Urol. 190:2278–2287. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Qi Y, Ooi HS, Wu J, Chen J, Zhang X, Tan
S, Yu Q, Li YY, Kang Y, Li H, et al: MALAT1 long ncRNA promotes
gastric cancer metastasis by suppressing PCDH10. Oncotarget.
7:12693–12703. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Michalik KM, You X, Manavski Y,
Doddaballapur A, Zörnig M, Braun T, John D, Ponomareva Y, Chen W,
Uchida S, et al: Long noncoding RNA MALAT1 regulates endothelial
cell function and vessel growth. Circ Res. 114:1389–1397. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Brock M, Schuoler C, Leuenberger C,
Bühlmann C, Haider TJ, Vogel J, Ulrich S, Gassmann M, Kohler M and
Huber LC: Analysis of hypoxia-induced noncoding RNAs reveals
metastasis-associated lung adenocarcinoma transcript 1 as an
important regulator of vascular smooth muscle cell proliferation.
Exp Biol Med (Maywood). 242:487–496. 2017. View Article : Google Scholar
|
|
9
|
Lund E, Guttinger S, Calado A, Dahlberg JE
and Kutay U: Nuclear export of microRNA precursors. Science.
303:95–98. 2004. View Article : Google Scholar
|
|
10
|
Wang X, Li M, Wang Z, Han S, Tang X, Ge Y,
Zhou L, Zhou C, Yuan Q and Yang M: Silencing of long noncoding RNA
MALAT1 by miR-101 and miR-217 inhibits proliferation, migration,
and invasion of esophageal squamous cell carcinoma cells. J Biol
Chem. 290:3925–3935. 2015. View Article : Google Scholar :
|
|
11
|
Han Y, Liu Y, Zhang H, Wang T, Diao R,
Jiang Z, Gui Y and Cai Z: Hsa-miR-125b suppresses bladder cancer
development by down-regulating oncogene SIRT7 and oncogenic long
non-coding RNA MALAT1. FEBS Lett. 587:3875–3882. 2013. View Article : Google Scholar
|
|
12
|
Yu F, Lu Z, Cai J, Huang K, Chen B, Li G,
Dong P and Zheng J: MALAT1 functions as a competing endogenous RNA
to mediate Rac1 expression by sequestering miR-101b in liver
fibrosis. Cell Cycle. 14:3885–3896. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sun Y, Cheng H, Wang G, Yu G, Zhang D,
Wang Y, Fan W and Yang W: Deregulation of miR-183 promotes melanoma
development via lncRNA MALAT1 regulation and ITGB1 signal
activation. Oncotarget. 8:3509–3518. 2017.
|
|
14
|
Li X, He Y, Xu Y, Huang X, Liu J, Xie M
and Liu X: KLF5 mediates vascular remodeling via HIF-1α in hypoxic
pulmonary hypertension. Am J Physiol Lung Cell Mol Physiol.
310:L299–L310. 2016. View Article : Google Scholar
|
|
15
|
Nie X, Dai Y, Tan J, Chen Y, Qin G, Mao W,
Zou J, Chang Y, Wang Q and Chen J: α-Solanine reverses pulmonary
vascular remodeling and vascular angiogenesis in experimental
pulmonary artery hypertension. J Hypertens. 35:2419–2435. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Mao W, Xia W and Chen J: Interobserver
variability in grading acute rejection after lung transplantation.
Chest. 145:416–417. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nie X, Qin G, Mao W, Wang W, Chang Y, Wei
D, Zhou M, Wu B and Chen J: Axis inhibition protein 2 deficiency
leads to hypoxic pulmonary hypertension through beta-catenin
signaling pathway. J Hypertens. 34:877–892. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Peng G, Xu J, Liu R, Fu Z, Li S, Hong W,
Chen J, Li B and Ran P: Isolation, culture and identification of
pulmonary arterial smooth muscle cells from rat distal pulmonary
arteries. Cytotechnology. 69:831–840. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sun Z, Nie X, Sun S, Dong S, Yuan C, Li Y,
Xiao B, Jie D and Liu Y: Long non-coding RNA MEG3 downregulation
triggers human pulmonary artery smooth muscle cell proliferation
and migration via the p53 signaling pathway. Cell Physiol Biochem.
42:2569–2581. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
21
|
Liu Y, Ma C, Zhang Q, Yu L, Ma J, Zhang L,
Hao X, Cao F, Wang L and Zhu D: The key role of transforming growth
factor-beta receptor I and 15-lipoxygenase in hypoxia-induced
proliferation of pulmonary artery smooth muscle cells. Int J
Biochem Cell Biol. 44:1184–1202. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Guppy A, Jamal-Hanjani M and Pickering L:
Anticancer effects of metformin and its potential use as a
therapeutic agent for breast cancer. Future Oncol. 7:727–736. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Qu J, Li M, Zhong W and Hu C: Competing
endogenous RNA in cancer: A new pattern of gene expression
regulation. Int J Clin Exp Med. 8:17110–17116. 2015.
|
|
24
|
Voelkel NF, Cool C, Lee SD, Wright L,
Geraci MW and Tuder RM: Primary pulmonary hypertension between
inflammation and cancer. Chest. 114(Suppl 3): pp. 225S–230S. 1998,
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Huang S, Lu W, Ge D, Meng N, Li Y, Su L,
Zhang S, Zhang Y, Zhao B and Miao J: A new microRNA signal pathway
regulated by long noncoding RNA TGFB2-OT1 in autophagy and
inflammation of vascular endothelial cells. Autophagy.
11:2172–2183. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wu G, Cai J, Han Y, Chen J, Huang ZP, Chen
C, Cai Y, Huang H, Yang Y, Liu Y, et al: LincRNA-p21 regulates
neointima formation, vascular smooth muscle cell proliferation,
apoptosis, and atherosclerosis by enhancing p53 activity.
Circulation. 130:1452–1465. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Leung A, Trac C, Jin W, Lanting L and
Akbany A: Novel long noncoding RNAs are regulated by angiotensin II
in vascular smooth muscle cells. Circ Res. 113:266–278. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Sun Y and Ma L: New insights into long
non-coding RNA MALAT1 in cancer and metastasis. Cancers (Basel).
11:E2162019. View Article : Google Scholar
|
|
29
|
Wang SH, Zhang WJ, Wu XC, Weng MZ, Zhang
MD, Cai Q, Zhou D, Wang JD and Quan ZW: The lncRNA MALAT1 functions
as a competing endogenous RNA to regulate MCL-1 expression by
sponging miR-363-3p in gallbladder cancer. J Cell Mol Med.
20:2299–2308. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li L, Chen H, Gao Y, Wang YW, Zhang GQ,
Pan SH, Ji L, Kong R, Wang G, Jia YH, et al: Long noncoding RNA
MALAT-1 promotes aggressive pancreatic cancer proliferation and
metastasis via the stimulation of autophagy. Mol Cancer Ther.
15:2232–2243. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wu XS, Wang XA, Wu WG, Hu YP, Li ML, Ding
Q, Weng H, Shu YJ, Liu TY, Jiang L, et al: MALAT 1 p romotes the
proliferation and metastasis of gallbladder cancer cells by
activating the ERK/MAPK pathway. Cancer Biol Ther. 15:806–814.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Poliseno L, Salmena L, Zhang J, Carver B,
Haveman WJ and Pandolfi PP: A coding-independent function of gene
and pseudo-gene mRNAs regulates tumour biology. Nature.
465:1033–1038. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Cesana M, Cacchiarelli D, Legnini I,
Santini T, Sthandier O, Chinappi M, Tramontano A and Bozzoni I: A
long noncoding RNA controls muscle differentiation by functioning
as a competing endogenous RNA. Cell. 147:358–369. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ebert MS and Sharp PA: MicroRNA sponges:
Progress and possibilities. RNA. 16:2043–2050. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang D, Zhang H, Li M, Frid MG, Flockton
AR, McKeon BA, Yeager ME, Fini MA, Morrell NW, Pullamsetti SS, et
al: MicroRNA-124 controls the proliferative, migratory, and
inflammatory phenotype of pulmonary vascular fibroblasts. Circ Res.
114:67–78. 2014. View Article : Google Scholar :
|
|
36
|
Kang K, Peng X, Zhang X, Wang Y, Zhang L,
Gao L, Weng T, Zhang H, Ramchandran R, Raj JU, et al: MicroRNA-124
suppresses the transactivation of nuclear factor of activated T
cells by targeting multiple genes and inhibits the proliferation of
pulmonary artery smooth muscle cells. J Biol Chem. 288:25414–25427.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Courboulin A, Tremblay VL, Barrier M,
Meloche J, Jacob MH, Chapolard M, Bisserier M, Paulin R, Lambert C,
Provencher S and Bonnet S: Krüppel-like factor 5 contributes to
pulmonary artery smooth muscle proliferation and resistance to
apoptosis in human pulmonary arterial hypertension. Respir Res.
12:1282011. View Article : Google Scholar
|
|
38
|
Abe K, Sugiura H, Hashimoto Y, Ichikawa T,
Koarai A, Yamada M, Numakura T, Onodera K, Tanaka R, Sato K, et al:
Possible role of Krüppel-like factor 5 in the remodeling of small
airways and pulmonary vessels in chronic obstructive pulmonary
disease. Respir Res. 17:72016. View Article : Google Scholar
|