Introduction
Neuropathic pain (NP) is a severe chronic condition
caused by injury or dysfunction affecting the somatosensory nervous
system (1). A total of >100
million Americans are thought to be affected by chronic pain
(2). NP is characterized by a
wide range of sensory, cognitive and affective symptoms, such as
tactile allodynia (burning pain resulting from noxious stimuli),
hyperalgesia, and spontaneous pain (3). Most NP patients may suffer from
depression, anxiety disorders, or other negative moods and various
therapies offer only partial relief to 40-60% of NP patients
(4). Despite great progress in
understanding NP's prognosis, a number of patients respond poorly
to current therapies. Therefore, it is necessary and urgent to
explore NP's molecular mechanisms, especially the association
between NP and anxiety, depression, and other mood disorders.
Chronic pain is associated with adaptations in
several brain networks involved in mood, motivation and reward.
Evidence indicates that the nucleus accumbens (NAc) is necessary
for expressing NP-like behavior (5), whereas the medial prefrontal cortex
(mPFC) is linked to a wide variety of cognitive functions critical
for social behavior, including personal traits (6). NP can lead to mPFC remodeling, which
associates it with emotional regulation of chronic pain (7). Periaqueductal gray (PAG) is the
primary center for descending pain modulation (8) that receive schronic pain and
temperature signals from the spinomesencephalic tract, hinting at
its key role in NP progression (9). Microarray profiling was previously
used to explore the altered gene expression of the murine nervous
system following nerve injury (10,11). This led to the identification of
IL-6, c-Jun and Plau as crucial genes
(12) and DNA binding, cell
cycle, and forkhead box protein as a major signaling pathway
involved in NP (13). Despite
these investigations profiling post nerve injury gene expression
using microarray analysis, NP's pathological mechanisms remain
poorly understood.
Recently, Descalzi et al (11) used RNA-sequencing technology to
explore NP; they found NP can affect the expression of multiple
genes in three distinct brain regions. However, they did not
explore differentially expressed mRNAs (DEMs) and microRNAs
(miRNAs/miR), especially the interactions between them. Based on
the microarray data deposited by Descalzi et al, DEMs and
differentially expressed miRNAs were screened from three distinct
brain regions (NAc, mPFC, and PAG) of the spared nerve injury (SNI)
and sham surgery murine model.
miRNA is a subset of non-coding, small RNAs, about
22nt long, that can combine with the 3′ untranslated region of
messenger RNA (mRNA) to regulate post-transcription gene expression
(14). miRNA binds to mRNA,
forming a complex regulatory network that plays a vital role in a
number of biological processes, such as cellular proliferation,
apoptosis, differentiation and metabolism (15). Biological networks have provided a
systems biology approach using data from DNA microarray, RNA-seq,
miRNA and signaling pathways (16). Gene Ontology (GO) annotation and
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment
analyses were used to analyze biological networks to reveal
potential synergism between gene and the organism (17). To achieve this, a miRNA-mRNA
regulatory network was constructed by integrating DEMs, related
miRNAs and major signaling pathways.
The present study results might provide further
understanding of molecular mechanisms involved in NP progression
and how differentially expressed mRNAs and miRNAs may serve as
potential targets for NP therapy.
Materials and methods
Microarray database
The microarray data under access number GSE91396
(11) were downloaded from the
National Center of Biotechnology Information (NCBI) Gene Expression
Omnibus (GEO; www.ncbi.nlm.nih.gov/) (18). In GSE91396, RNA-seq samples were
derived from three brain regions (NAc, mPFC and PAG) of animals two
and a half months after sham surgery or SNI on the sciatic nerve.
Bilateral punches were collected from the NAc, mPFC and PAG regions
from 12 adult male C57BL/6 mice in the sham or SNI surgery group.
The punches were pooled from two mice per sample. Thus, 36 samples
in GSE91396 were processed, including 18 (6 mPFC, 6 NAc and 6 PAG)
from sham surgery and 18 (6 mPFC, 6NAc and 6 PAG) from the SNI
surgery group. The SNI model of neuropathic pain was performed
under Avertine general anesthesia according to the study of
Descalzi et al (11).
Briefly, skin and muscle incisions were made on the left hind leg
at mid-thigh level, revealing the sciatic nerve and its three
branches. The common peroneal and sural nerves were carefully
ligated with 6.0 silk sutures, transected, and 1-2 mm sections of
each of these nerves were removed. The tibial nerve was left
intact. Skin was then sutured with silk 4.0 sutures. Sham surgery
mice underwent the same procedure, but all nerves were left
intact.
Gene expression profiles in TXT format were
downloaded from the NCBI database. The R3.4.1 preprocessCore
(19) version 1.40.0 (bioconductor.org/packages/2.4/bioc/html/preprocessCore.html)
was used to process the fragments per kilobase per million mapped
reads (FPKM) values. The limma package (20) version 3.32.5 (bioconductor.org/packages/release/bioc/html/limma.html)
was used to identify differentially expressed RNAs (mRNAs and
miRNAs) in the three brain regions that are significantly different
(P<0.05) and |log2 fold change (FC)| >0.585 were
considered as thresholds. The numbers of miRNAs and genes were
calculated. Subsequently, pheatmap (21) version 1.0.8 package (cran.r-project.org/web/packages/pheatmap/index.html)
in R3.4.1 software were used for unsupervised hierarchical
clustering analysis based on a correlation algorithm. Finally, the
Database for Annotation, Visualization and Integrated Discovery
(DAVID) (22,23) version 6.8 (david.ncifcrf. gov/)
was used to perform GO (Biology Process, Cellular Component, and
Molecular Function) and KEGG pathway enrichment analysis for the
identified differentially expressed genes (DEGs).
Analysis of the common differentially
expressed RNAs for all the three brain regions
Venn diagram (24)
(cran.r-project. org/web/packages/VennDiagram/index.html) version
1.6.17 in R3.4.1 software was used to visualize differentially
expressed RNAs. The common differentially expressed RNAs from all
the three brain regions were selected for further analysis.
The weighed gene co-expression network analysis
(WGCNA) algorithm provided topology properties for co-expression
network analysis (25). This
allows construction of scale-free networks, defines the
co-expression matrix and adjacency function, calculates different
node coefficients, and identifies functional modules associated
with disease from high through put data (26). The WGCNA package (27) version 1.61 in R3.4.1 software
(cran.r-project.org/web/packages/WGCNA/) was used to
analyze the correlation with disease status for common
differentially expressed RNAs. Genes in major modules were screened
by GO functional and KEGG pathway enrichment analysis; P<0.05
was considered to indicate a statistically significant
difference.
Construction and analysis of miRNA-mRNA
regulatory network
To construct an miRNA-mRNA regulatory network and
explore the correlations between miRNA and mRNA for mining for the
potential roles of these RNAs in NP, the TargetScan Release7.1
(28) (www.targetscan.org/vert_71/) database was used to
predict the target mRNAs of miRNAs. The miRNA-mRNA interactions
involving common DEMs were selected. According to co-expression
WGCNA results, the negative miRNA-mRNA interactions were used to
construct the miRNA-mRNA regulatory network that was visualized
using Cytoscape3.5.1 (29)
(www.cytoscape.org/). GO annotation and
KEGG pathway analysis was performed for target genes in the
regulatory network.
miRNA-mRNA-pathway network
construction
The Comparative Toxicogenomics Database (30) (CTD; ctd.mdibl. org/), a public
website and research tool launched by Mount Desert Island
Biological Laboratory, elucidates relationships between
genesproteins, diseases, phenotypes, GO annotations, pathways, and
interaction modules (31). CTD's
primary objective is to advance understanding of how
gene-environment interactions affect human health (32). 'Neuropathic pain' was used as the
keyword to search the KEGG pathways related to NP disease in CTD.
Pathways thus retrieved were compared with enriched pathways for
target genes in the regulatory network. The overlapping pathways
and related genes were hypothesized to play important roles in NP
pathogenesis.
Results
Data preprocessing and screening of
differentially expressed RNA
The transcriptional profiles downloaded from the GEO
database were preprocessed and, after data normalization, 21,193
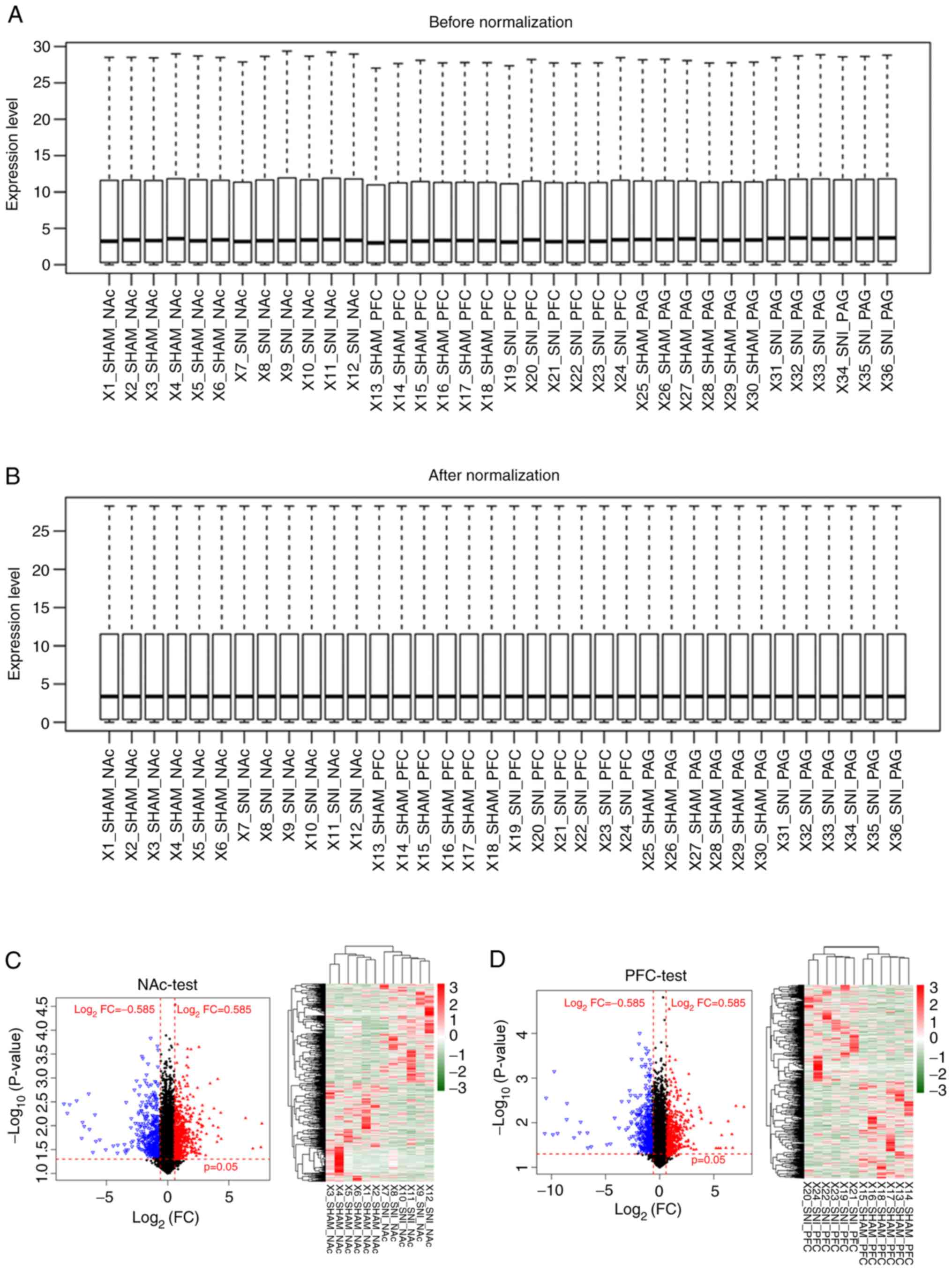
RNAs were identified, including 1,123 miRNAs and 20,070 mRNAs
(Fig. 1A and B). RNAs with a zero
median value were excluded, yielding 383 miRNAs and 17,654 mRNAs. A
total of 2,776 differentially expressed RNAs were identified (219
miRNAs and 2,557 mRNAs) in SNI samples compared with sham surgery
samples for further analysis (Table
I). The clustering heatmaps showed significant differences in
expression levels of differentially expressed RNAs between SNI and
sham surgery groups pertaining to the three distinct brain regions
(NAc, mPFC and PAG; Fig. 1C-E).
These results show that clustering heatmaps of differentially
expressed RNAs, identified from microarray datasets, may accurately
distinguish samples from SNI and sham surgeries.
 | Table IDifferentially expressed mRNAs and
miRNAs in three distinct brain regions (NAc, mPFC, PAG) related to
neuropathic pain. |
Table I
Differentially expressed mRNAs and
miRNAs in three distinct brain regions (NAc, mPFC, PAG) related to
neuropathic pain.
| Type | NAc
| mPFC
| PAG
|
|---|
| Down | Up | Down | Up | Down | Up |
|---|
| miRNA | 50 | 50 | 49 | 56 | 7 | 7 |
| mRNA | 489 | 491 | 437 | 422 | 95 | 623 |
| Total | 539 | 541 | 486 | 478 | 102 | 630 |
| 1,080 | | 964 | | 732 | |
GO functional and KEGG pathway enrichment
analysis for the DEGs
GO functional and KEGG pathway enrichment analyses
were performed for the DEGs identified above (Fig. 2). The DEGs in NAc were mainly
involved in DNA binding (GO:0003677, n=99), extracellular region
part (GO:0044421, n=82), transcription regulator activity
(GO:0030528, n=75), transcription factor activity (GO:0003700,
n=64) and extracellular space (GO:0005615, n=57; Fig. 2A). The DEGs in mPFC were mainly
involved in immune response (GO:0006955, n=41), transcription
factor activity (GO:0003700, n=41), extracellular region part
(GO:0044421, n=35), sequence-specific DNA binding (GO:0043565,
n=32) and positive regulation of the macromolecule metabolic
process (GO:0010604, n=32; Fig.
2B). The DEGs in PAG were mainly involved in DNA binding
(GO:0003677, n=83), transcription regulator activity (GO:0030528,
n=59), regulation of transcription from RNA polymerase II promoter
(GO:0006357, n=34), protein kinase activity (GO:0004672, n=31) and
structural molecule activity (GO:0005198, n=30; Fig. 2C).
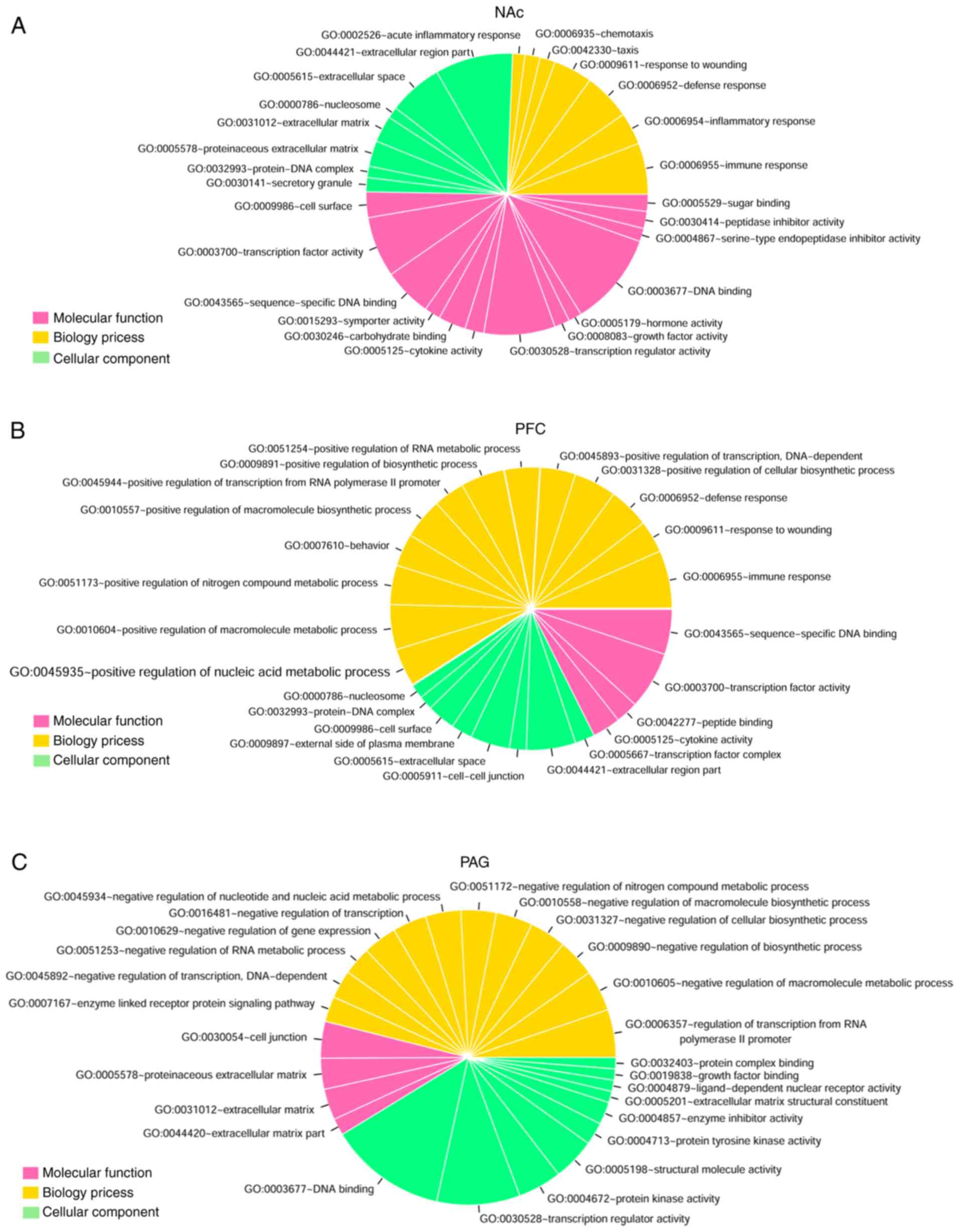 | Figure 2GO functional and KEGG pathway
enrichment analysis for the differentially expressed genes related
to neuropathic pain. The pie chart slice sizes correspond to the
number of annotated differentially expressed genes. GO functional
enrichment analysis results for the differentially expressed genes
in three distinct regions of brain (A) NAc, (B) mPFC and
periaqueductal gray, (C) PAG. The yellow, pink and green colors
represent Biological Process, Molecular Function and Cellular
Component, respectively. (D) KEGG pathway enrichment analysis
results for the differentially expressed genes in three distinct
regions of brain (NAc, mPFC and PAG). The pink, yellow and green
represent NAc, mPFC and PAG groups, respectively, and black dots
represent -log2 (P-value). GO, gene Ontology; KEGG,
Kyoto Encyclopedia of Genes and Genomes; NAc, nucleus accumbens;
mPFC, medial prefrontal cortex; PAG, periaqueductal gray. |
In addition, nine, six and four significant pathways
were identified for the DEGs identified in NAc, mPFC, and PAG,
respectively (Fig. 2D). There
were five overlapping pathways for NAc and mPFC, including
cytokine-cytokine receptor interaction (mmu04060), chemokine
signaling pathway (mmu04062), neuroactive ligand-receptor
interaction (mmu04080), JAK-signal transducer and activator of
transcription signaling pathway (mmu04630), and metabolism of
xenobiotics by cytochrome P450 (mmu00980). The DEGs in PAG were
mainly enriched in vascular endothelial growth factor (mmu04370)
and Notch (mmu04330) signaling pathways.
Analysis of critical gene modules related
to NP
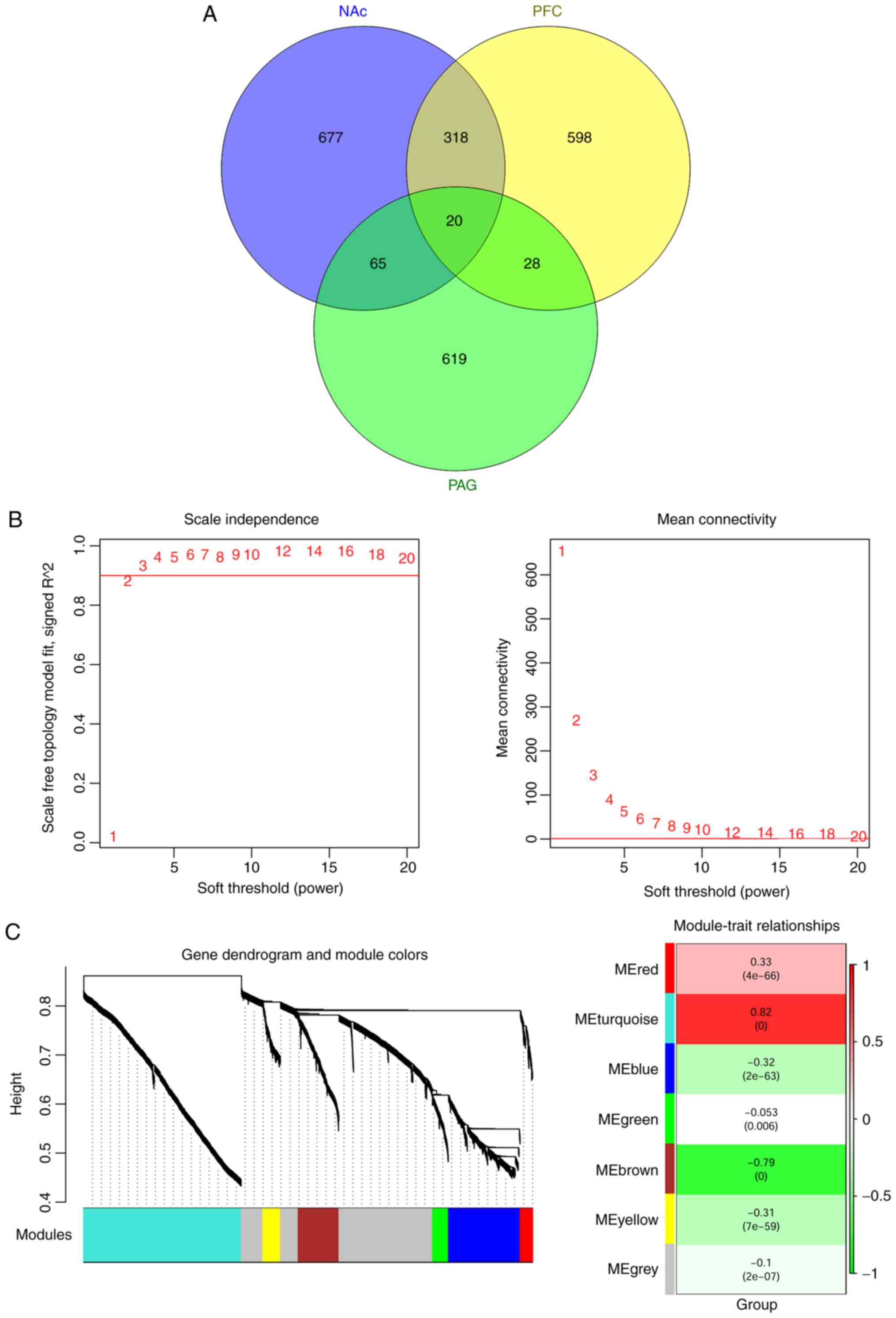
A total of 2,325 common differentially expressed
RNAs were identified in all the three brain regions (NAc, mPFC and
PAG), including 173 miRNAs and 2,152 mRNAs. These common
differentially expressed RNAs were analyzed using the WGCNA
algorithm to identify disease-related RNAs. The topological
matrix's scale-free distribution was calculated based on the
GSE91396 dataset. Once the square value of the correlation
coefficient reached 0.9 for the first time, the corresponding power
value (power=7) was selected to calculate community dissimilarity
of RNAs. After constructing the clustering dendrogram, the minimum
gene number was set as 50 and the cut Height= 0.99. This process
resulted in the identification of six modules-module-red,
module-turquoise, module-blue, module-green, module-brown and
module-yellow (Fig. 3).
Correlations between the RNA module and disease
characteristics (different regions and disease states) were
calculated and are shown in Table
II. As shown, genes in module-red (correlation=0.33,
P=4.0×10-66) and module-turquoise (correlation=0.82,
P<0.001) were positively correlated with disease
characteristics. Furthermore, the correlation values were
significantly increased compared with the module-gray (correlation
with disease=−0.10, P=2.0×10−07). Thus, the
differentially expressed RNAs in the red and turquoise modules were
used for further analysis.
 | Table IIDifferentially expressed RNAs in gene
modules related to neuropathic pain. |
Table II
Differentially expressed RNAs in gene
modules related to neuropathic pain.
| Module color | Correlation with
disease | P-value | #RNA | #miRNA | #mRNA |
|---|
| Blue | −0.32 |
2.0×10−63 | 412 | 11 | 401 |
| Brown | −0.79 | <0.0001 | 209 | 11 | 198 |
| Green | −0.05 |
6.0×10−3 | 87 | 0 | 87 |
| Grey | −0.10 |
2.0×10−7 | 665 | 118 | 547 |
| Red | 0.33 |
4.0×10−66 | 71 | 2 | 69 |
| Turquoise | 0.82 | <0.0001 | 781 | 31 | 750 |
| Yellow | −0.31 |
7.0×10−59 | 100 | 0 | 100 |
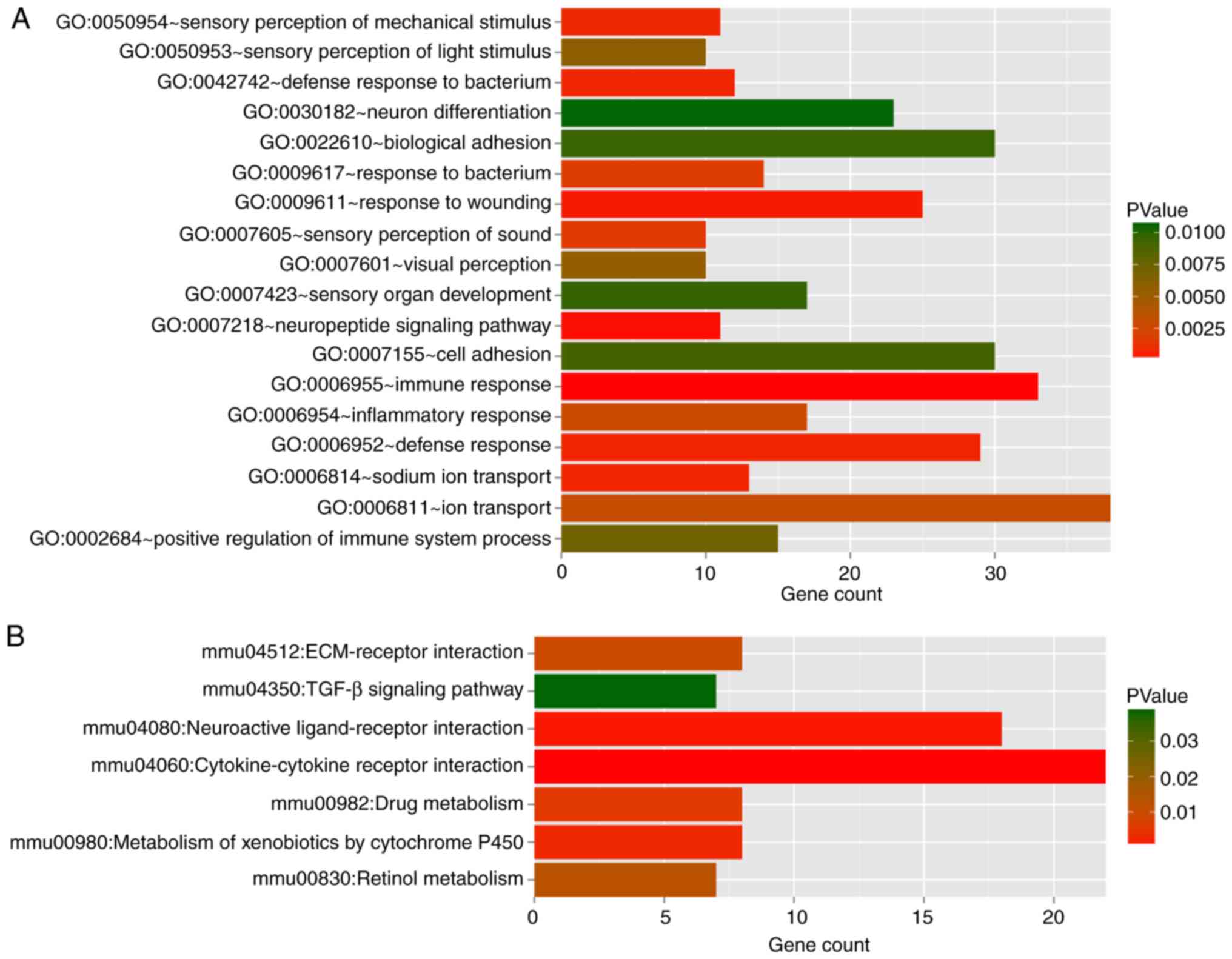
Pathway enrichment analyses by GO functional and
KEGG for DEGs in red and turquoise modules (Table III) showed 18 GO terms and seven
pathways (Fig. 4). The DEGs in
the two modules were mainly enriched in immune response
(GO:0006955), defense response (GO:0006952) and response to
wounding (GO:0009611). The major pathway categories for DEGs in the
two modules were cytokine-cytokine receptor interaction (mmu04060)
and neuroactive ligand-receptor interaction (mmu04080).
 | Table IIIGO terms and KEGG pathways for the
differential expressed mRNAs in module-red and
module-turquoise. |
Table III
GO terms and KEGG pathways for the
differential expressed mRNAs in module-red and
module-turquoise.
A, Biology process
|
|---|
| Term | Count | P-value |
|---|
| GO:0006955 immune
response | 33 |
6.36×10−5 |
| GO:0007218
neuropeptide signaling pathway | 11 |
2.01×10−4 |
| GO:0009611 response
to wounding | 25 |
4.02×10−4 |
| GO:0006952 defense
response | 29 |
7.03×10−4 |
| GO:0006814 sodium
ion transport | 13 |
7.38×10−4 |
| GO:0042742 defense
response to bacterium | 12 |
7.42×10−4 |
| GO:0050954 sensory
perception of mechanical stimulus | 11 |
7.70×10−4 |
| GO:0007605 sensory
perception of sound | 10 |
1.55×10−3 |
| GO:0009617 response
to bacterium | 14 |
1.78×10−3 |
| GO:0006954
inflammatory response | 17 |
2.72×10−3 |
| GO:0006811 ion
transport | 38 |
3.05×10−3 |
| GO:0007601 visual
perception | 10 |
5.42×10−3 |
| GO:0050953 sensory
perception of light stimulus | 10 |
5.79×10−3 |
| GO:0002684 positive
regulation of immune system process | 15 |
7.24×10−3 |
| GO:0007155 cell
adhesion | 30 |
9.03×10−3 |
| GO:0022610
biological adhesion | 30 |
9.42×10−3 |
| GO:0007423 sensory
organ development | 17 |
9.56×10−3 |
| GO:0030182 neuron
differentiation | 23 |
1.06×10−2 |
B, KEGG pathway
|
|---|
| Term | Count | P-value |
|---|
| mmu04060:
Cytokine-cytokine receptor interaction | 22 |
5.36×10−6 |
| mmu04080:
Neuroactive ligand-receptor interaction | 18 |
1.20×10−3 |
| mmu00980:
Metabolism of xenobiotics by cytochrome P450 | 8 |
2.68×10−3 |
| mmu00982: Drug
metabolism | 8 |
5.50×10−3 |
| mmu04512:
ECM-receptor interaction | 8 |
9.50×10−3 |
| mmu00830: Retinol
metabolism | 7 |
1.30×10−2 |
| mmu04350: TGF-β
signaling pathway | 7 |
3.85×10−2 |
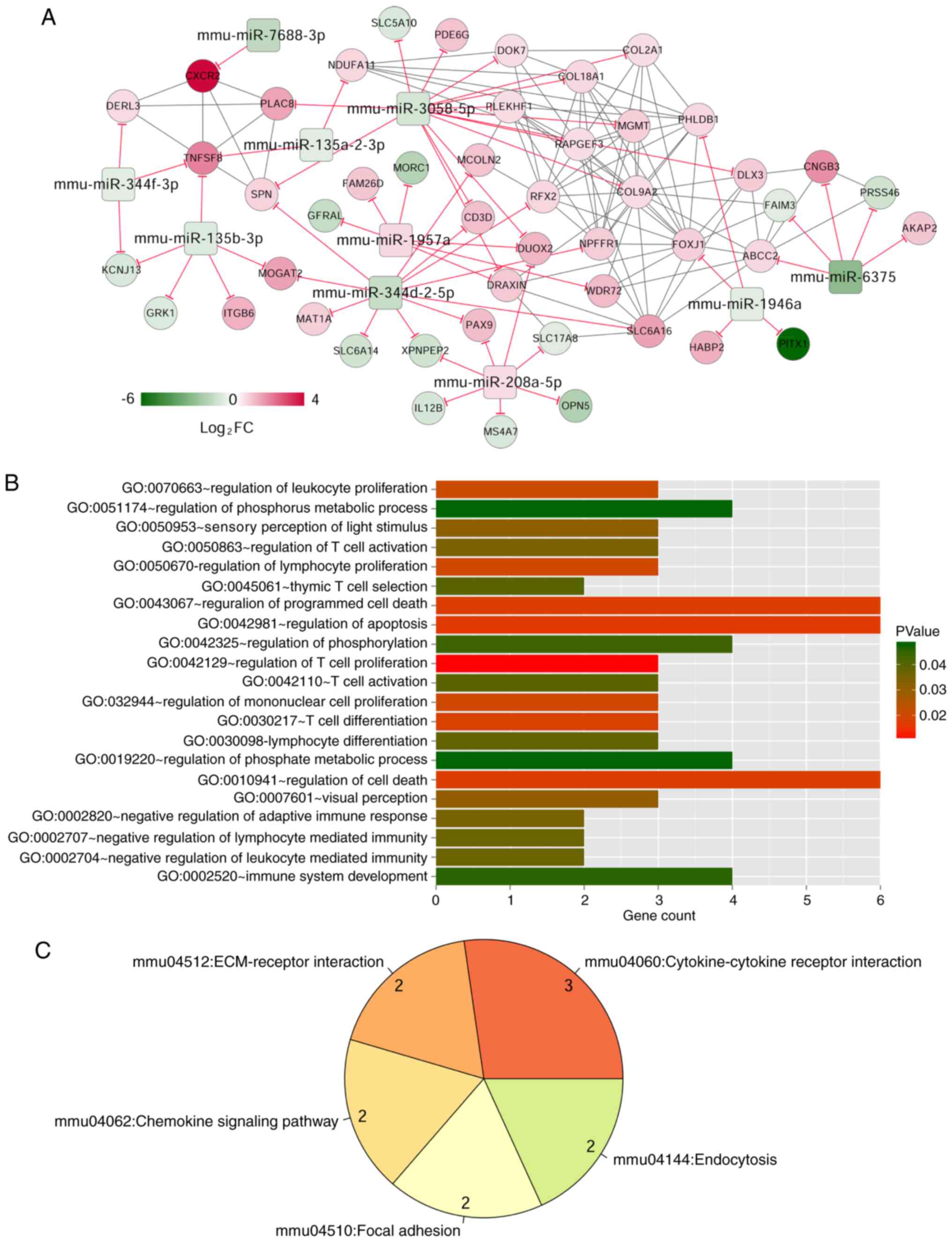
The mRNA-miRNA regulatory network
construction
Target gene prediction of 33 miRNAs in red (2
miRNAs) and turquoise (31 miRNAs) modules using TargetScan Release
7.1 database resulted in overlapping target genes, which were
matched with 819 mRNAs in red (69 mRNAs) and turquoise (750 mRNAs)
modules, involving 558 pairs of negative miRNA-mRNA regulatory
interactions. The correlation coefficients >0.6 for these
relationships, based on the WGCNA algorithm, yielded 59 miRNA-mRNA
interactions and 172 mRNA-mRNA interactions that facilitated
construction of a miRNA-mRNA regulatory network (Fig. 5), which consisted of 58 nodes,
including 10 miRNAs and 48 mRNAs.
The GO functional and KEGG pathway enrichment
analysis showed that DEGs in the miRNA-mRNA regulatory network were
enriched in 21 GO terms and five pathways. The GO terms included
regulating T cell proliferation (GO:0042129) and T cell
differentiation (GO:0030217). The major pathway was
cytokine-cytokine receptor interaction (mmu04060; Table IV).
 | Table IVGO terms and KEGG pathways for the
critical genes in mRNA-microRNA regulatory network. |
Table IV
GO terms and KEGG pathways for the
critical genes in mRNA-microRNA regulatory network.
A, Biology process
|
|---|
| Term | Count | P-value | Genes |
|---|
| GO:0042129
regulation of T cell proliferation | 3 | 0.011 | FOXJ1, IL12B,
SPN |
| GO:0042981
regulation of apoptosis | 6 | 0.016 | COL18A1, PLEKHF1,
GFRAL, MGMT, |
| | | COL2A1, SPN |
| GO:0043067
regulation of programmed cell death | 6 | 0.017 | COL18A1, PLEKHF1,
GFRAL, MGMT, COL2A1, SPN |
| GO:0010941
regulation of cell death | 6 | 0.017 | COL18A1, PLEKHF1,
GFRAL, MGMT, COL2A1, SPN |
| GO:0030217 T cell
differentiation | 3 | 0.018 | CD3D, IL12B,
SPN |
| GO:0032944
regulation of mononuclear cell proliferation | 3 | 0.020 | FOXJ1, IL12B,
SPN |
| GO:0050670
regulation of lymphocyte proliferation | 3 | 0.020 | FOXJ1, IL12B,
SPN |
| GO:0070663
regulation of leukocyte proliferation | 3 | 0.021 | FOXJ1, IL12B,
SPN |
| GO:0007601 visual
perception | 3 | 0.031 | OPN5, PDE6G,
CNGB3 |
| GO:0050953 sensory
perception of light stimulus | 3 | 0.031 | OPN5, PDE6G,
CNGB3 |
| GO:0050863
regulation of T cell activation | 3 | 0.035 | FOXJ1, IL12B,
SPN |
| GO:0002820 negative
regulation of adaptive immune response | 2 | 0.035 | FOXJ1, SPN |
| GO:0002704 negative
regulation of leukocyte mediated immunity | 2 | 0.037 | FOXJ1, SPN |
| GO:0002707 negative
regulation of lymphocyte mediated immunity | 2 | 0.037 | FOXJ1, SPN |
| GO:0030098
lymphocyte differentiation | 3 | 0.038 | CD3D, IL12B,
SPN |
| GO:0042110 T cell
activation | 3 | 0.040 | CD3D, IL12B,
SPN |
| GO:0045061 thymic T
cell selection | 2 | 0.040 | CD3D, SPN |
| GO:0042325
regulation of phosphorylation | 4 | 0.044 | DOK7, RAPGEF3,
PDE6G, GRK1 |
| GO:0002520 immune
system development | 4 | 0.046 | CD3D, FOXJ1, IL12B,
SPN |
| GO:0051174
regulation of phosphorus metabolic process | 4 | 0.048 | DOK7, RAPGEF3,
PDE6G, GRK1 |
|
GO:0019220-regulation of phosphate
metabolic process | 4 | 0.048 | DOK7, RAPGEF3,
PDE6G, GRK1 |
B, KEGG pathway
|
|---|
| Term | Count | P-value | Genes |
|---|
| mmu04060:
Cytokine-cytokine receptor interactiona | 3 | 0.010 | CXCR2, IL12B,
TNFSF8 |
| mmu04512:
ECM-receptor interaction | 2 | 0.017 | ITGB6, COL2A1 |
| mmu04062: Chemokine
signaling pathwaya | 2 | 0.034 | CXCR2, GRK1 |
| mmu04510: Focal
adhesion | 2 | 0.037 | ITGB6, COL2A1 |
| mmu04144:
Endocytosisa | 2 | 0.037 | CXCR2, GRK1 |
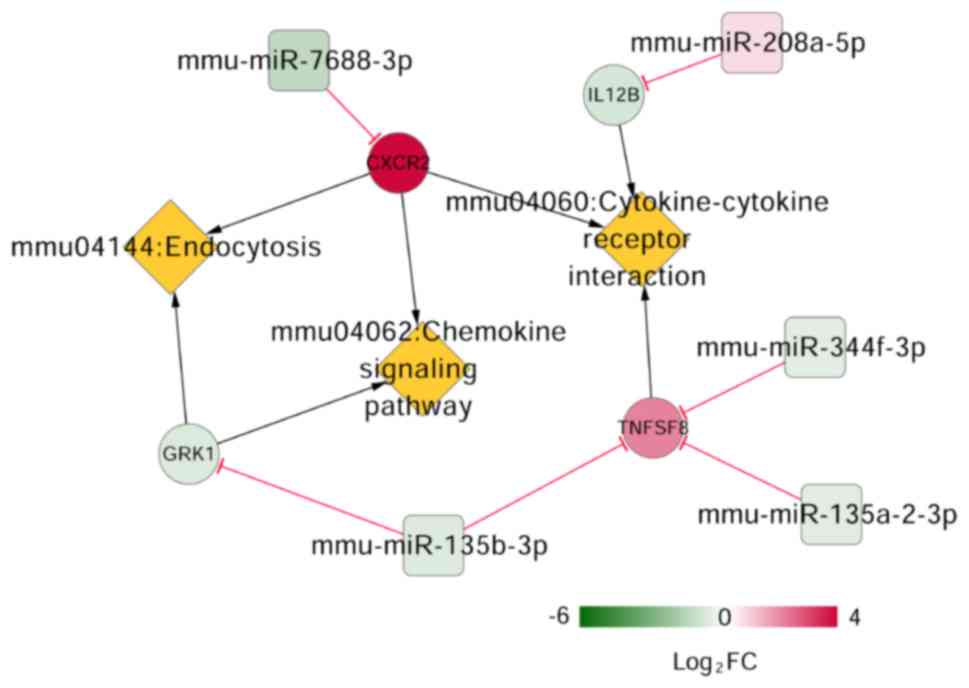
Crucial miRNAs, mRNAs and pathways
related to NP
Keyword ('neuropathic pain') search of the CTD
resulted in 88 pathways. Three pathways overlapped when the 88
pathways were compared to previously-identified enriched pathways
for the DEGs in the mRNA-miRNA regulatory network. These pathways,
cytokine-cytokine receptor interaction, chemokine signaling pathway
and endocytosis were involved in four DEGs, including interleukin
(IL)-8 receptor β (CXCR2), subunit β of interleukin (IL)-12
(IL12B), CD153 (also known as TNFSF8), and rhodopsin
kinase (GRK1). The CXCR2 can be targeted by
miR-7688-3p, IL12B by miR-208a-5p, GRK1
by miR-135b-3p and TNFSF8 by miR-344f-3p,
miR-135b-3p, and miR-135a-2-3p, respectively
(Fig. 6).
Discussion
In this study, 2,776 differentially expressed RNAs
(219 miRNAs and 2,557 mRNAs) were identified in SNI compared with
the sham surgery samples. In the three brain regions (NAc, mPFC and
PAG), there were 2,325 common differentially expressed RNAs (173
miRNAs and 2,152 mRNAs). Two important modules (red and turquoise
module) were identified as related to NP using WGCNA for the common
differentially expressed RNAs. The miRNA-mRNA regulatory network
was constructed based on the differentially expressed RNAs in the
red and turquoise modules. The DEGs in miRNA-mRNA regulatory
network were enriched in 21 GO terms and 5 pathways. Three pathways
in the miRNA-target gene-pathway regulatory network, including
cytokine-cytokine receptor interaction, chemokine signaling pathway
and endocytosis, comprised four important DEGs (CXCR2,
IL12B, TNFSF8, and GRK1) that were related to
NP.
Cytokines are a group of small proteins (5-20 kDa)
produced by a range of cells, including immune cells-macrophages, B
lymphocytes, T lymphocytes, mast cells, as well as endothelial
cells and various stromal cells (33). Cytokines have important roles in
the immune system (34).
Cytokines, along with chemokines, interferons (IFN), ILs, colony
stimulation factor and tumor necrosis factor (TNF) are involved in
responsiveness to trauma, pain, and infection (35). In the present study, three DEGs
(CXCR2, IL12B and TNFSF8) participated in
cytokine-cytokine receptor interaction. IL12B located on
chromosome 5q31-33 encodes the p40 subunit of IL-12, an
immunomodulatory cytokine (36).
IL12B polymorphisms are associated with asthma and psoriasis
(37). A recent study showed that
significant decreases in systemic concentrations of chemokines,
IL-12 and IFNγ, were observed in nerve-injured Foxp3+
regulatory T cell-depleted transgenic mice; decrease in IL-12
promoted pain hypersensitivity in this model (38). The immune system, particularly T
cells, plays a key role in mediating NP. IL-12p70, also referred to
as IL-12, is a pro-inflammatory cytokine secreted by activated
hematopoietic phagocytic cells; Chen et al (39) investigated pain response following
systemic administration of IL-12p70 and IL-12p40 homodimer and
found that IL-12p40 exhibited significant anti-nociceptive effects
in a rat model of chronic NP. The present study found that
IL12B was dysregulated in brain tissues of NP mice, which is
consistent with previous studies (37-39). This dysregulation may be
effectuated by miRNA, one of which, miR-208a-5p, was found
in the miRNA-target gene-pathway regulatory network, to target
IL12B. Findings in the literature show miR-208a to
regulate cardiac hypertrophy and conduction in mice (40), whereas the expression level of
miR-208b progressively declined after spinal cord injury in
humans (41). The results of the
present study suggest miR-208a-5p may play a vital role in
NP pathogenesis by regulating IL12B expression levels.
TNFSF8, a ligand of cluster of
differentiation (CD)30 (or CD153), exhibits polymorphisms that
showed significant associations with spondylarthritis in a French
cohort (42). In addition,
TNFSF8 is a susceptibility gene in excessive inflammatory
responses (43). Since
inflammation is a key pathophysiological process in NP (44), TNFSF8 might be a major
regulator in NP inflammatory responses. The miRNA-target
gene-pathway regulatory network revealed differentially expressed
TNFSF8 could be targeted by three miRNAs
(miR-344f-3p, miR-135b-3p and miR-135a-2-3p).
It has been reported that miR-135a modulates inflammatory
molecules, IL-6, IL-1β and TNF-α, which enhances inflammatory
responses of vascular smooth muscle cells involved in vascular
disease complications (45,46). Another study showed that
overexpressing the miR-344b-1-3p inhibitor in alveolar macrophages
significantly increased the expression of TNF-α, IL-1β and
macrophage inflammatory protein (MIP)-2 (47). Together, the results of the
present study indicate that differentially expressed IL12B
targeted by miR-208a-5p and TNFSF8 targeted by
miR-344f-3p, miR-135b-3p and miR-135a-2-3p,
might play critical roles in NP through cytokine-cytokine receptor
interaction.
CXCR2 and GRK1 are chemokines that
were identified in this study to be involved in NP progression.
CXCR2 (or IL 8 receptor β) is a chemokine receptor whose
interaction with MIP elicits chronic neuroinflammation through
neutrophil accumulation and hyperacetylation of histone H3 leading
to NP (48).
CXCL1/CXCR2 signaling plays a vital
role in pathological pain, including peripheral and central
sensitization (49). CXCL1
sensitizes primary peripheral neurons by directly acting on
CXCR2 (50). In the
central nervous system, CXCL1/CXCR2 signaling increases
N-methyl-D-aspartate receptor currents in neurons and promotes
expression of genes related to neuroplasticity, contributing to
prolonged chronic pain (51).
The enzyme phosphorylating rhodopsin receptor,
rhodopsin kinase (GRK1), was identified in the late 1970s as
controlling vision (52). Nerve
injury, high norepinephrine concentration and abnormal
β2-adrenoreceptor functions indicate high sympathetic nerve
activity and dysfunction (53).
Phosphorylation by GRK1/2, under high norepinephrine concentrations
and high sympathetic nerve activity, induced β2-adrenoreceptor
internalization by recruiting β-arrestin-1 to the receptor, leading
adrenoreceptors to either recycle to the membrane or traffic to
lysosomes for degradation (54).
Taken together, it was speculated that CXCR2 targeted by
miR-7688-3p and GRK1 targeted by miR-135b-3p
might be involved in NP progression through chemokine signaling
pathway and endocytosis.
The present study has limitations. The small sample
size available for analysis may have obscured rare interactions.
Additional validation of the roles of DEGs and miRNAs in NP
progression is warranted.
In conclusion, 2,776 differentially expressed RNAs
(219 miRNAs and 2,557 mRNAs) were identified in the NP SNI model.
Differentially expressed IL12B, targeted by
miR-208a-5p, as well as TNFSF8 dysregulated by
miR-344f-3p, miR-135b-3p and miR-135a-2-3p,
might play critical roles in NP through cytokine-cytokine receptor
interaction. CXCR2 targeted by miR-7688-3p and
GRK1 targeted by miR-135b-3p might be involved in NP
progression through chemokine signaling pathway and endocytosis.
The present study has provided new insights into the regulatory
mechanisms of NP, which merits their potential as candidates for
therapies for this debilitating condition.
Acknowledgments
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
requests.
Authors' contributions
SXL and CGZ made substantial intellectual
contributions to the study design. HL, HQW, SXL and HJZ searched
and downloaded microarray data from the Gene Expression Omnibus
database. HL, HQW, SXL and HJZ made substantial contributions to
the analysis and interpretation of microarray dataset. SXL and CGZ
were involved in revising the manuscript critically for important
intellectual content. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bennett GJ and Xie YK: A peripheral
mononeuropathy in rat that produces disorders of pain sensation
like those seen in man. Pain. 33:87–107. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Institute of Medicine (US) Committee on
Advancing Pain Research Care, and Education: Relieving pain in
America: A blueprint for transforming prevention, care, education,
and research. Institute of Medicine (US); 2011
|
|
3
|
Baron R: Mechanisms of disease:
Neuropathic pain-a clinical perspective. Nat Clin Pract Neurol.
2:95–106. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dworkin RH, O'Connor AB, Backonja M,
Farrar JT, Finnerup NB, Jensen TS, Kalso EA, Loeser JD, Miaskowski
C, Nurmikko TJ, et al: Pharmacologic management of neuropathic
pain: Evidence-based recommendations. Pain. 132:237–251. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chang PC, Pollema-Mays SL, Centeno MV,
Procissi D, Contini M, Baria AT, Martina M and Apkarian AV: Role of
nucleus accumbens in neuropathic pain: Linked multi-scale evidence
in the rat transitioning to neuropathic pain. Pain. 155:1128–1139.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Weaver KE and Richardson AG: Medial
prefrontal cortex, secondary hyperalgesia, and the default mode
network. J Neurosci. 29:11424–11425. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Seifert F, Bschorer K, De Col R, Filitz J,
Peltz E, Koppert W and Maihöfner C: Medial prefrontal cortex
activity is predictive for hyperalgesia and pharmacological
antihyperalgesia. J Neurosci. 29:6167–6175. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Oh MY, Abosch A, Kim SH, Lang AE and
Lozano AM: Long-term hardware-related complications of deep brain
stimulation. Neurosurgery. 50:1268–1274. 2002.PubMed/NCBI
|
|
9
|
Du L, Wang SJ, Cui J, He WJ and Ruan HZ:
The role of HCN channels within the periaqueductal gray in
neuropathic pain. Brain Res. 1500:36–44. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Griffin RS, Costigan M, Brenner GJ, Ma CH,
Scholz J, Moss A, Allchorne AJ, Stahl GL and Woolf CJ: Complement
induction in spinal cord microglia results in anaphylatoxin
C5a-mediated pain hypersensitivity. J Neurosci. 27:8699–8708. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Descalzi G, Mitsi V, Purushothaman I,
Gaspari S, Avrampou K, Loh YE, Shen L and Zachariou V: Neuropathic
pain promotes adaptive changes in gene expression in brain networks
involved in stress and depression. Sci Signal. 10:2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu H, Xia T, Xu F, Ma Z and Gu X:
Identification of the key genes associated with neuropathic pain.
Mol Med Rep. 17:6371–6378. 2018.PubMed/NCBI
|
|
13
|
Chen CJ, Liu DZ, Yao WF, Gu Y, Huang F,
Hei ZQ and Li X: Identification of key genes and pathways
associated with neuropathic pain in uninjured dorsal root ganglion
by using bioinformatic analysis. J Pain Res. 10:2665–2674. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cline MS, Smoot M, Cerami E, Kuchinsky A,
Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross
B, et al: Integration of biological networks and gene expression
data using Cytoscape. Nat Protoc. 2:2366–2382. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Habibi I, Emamian ES and Abdi A:
Quantitative analysis of intracellular communication and signaling
errors in signaling networks. BMC Syst Biol. 8:892014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 39:D991–D995.
2013.
|
|
19
|
Yan M, Song M, Bai R, Cheng S and Yan W:
Identification of potential therapeutic targets for colorectal
cancer by bioinformatics analysis. Oncol Lett. 12:5092–5098. 2016.
View Article : Google Scholar
|
|
20
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Diao C, Xi Y and Xiao T: Identification
and analysis of key genes in osteosarcoma using bioinformatics.
Oncol Lett. 15:2789–2794. 2018.PubMed/NCBI
|
|
22
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar
|
|
24
|
Chen H and Boutros PC: VennDiagram: A
package for the generation of highly-customizable Venn and Euler
diagrams in R. BMC Bioinformatics. 12:352011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Oldham MC, Konopka G, Iwamoto K,
Langfelder P, Kato T, Horvath S and Geschwind DH: Functional
organization of the transcriptome in human brain. Nat Neurosci.
11:1271–1282. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liao Q, Liu C, Yuan X, Kang S, Miao R,
Xiao H, Zhao G, Luo H, Bu D, Zhao H, et al: Large-scale prediction
of long non-coding RNA functions in a coding-non-coding gene
co-expression network. Nucleic Acids Res. 39:3864–3878. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fromm B, Billipp T, Peck LE, Johansen M,
Tarver JE, King BL, Newcomb JM, Sempere LF, Flatmark K, Hovig E and
Peterson KJ: A uniform system for the annotation of vertebrate
microRNA genes and the evolution of the human microRNAome. Annu Rev
Genet. 49:213–242. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Davis AP, King BL, Mockus S, Murphy CG,
Saraceni-Richards C, Rosenstein M, Wiegers T and Mattingly CJ: The
comparative toxicogenomics database: Update 2011. Nucleic Acids
Res. 39:D1067–D1072. 2011. View Article : Google Scholar :
|
|
31
|
Mattingly CJ, Rosenstein MC, Davis AP,
Colby GT, Forrest JN Jr and Boyer JL: The comparative
toxicogenomics database: A cross-species resource for building
chemical-gene interaction networks. Toxicol Sci. 92:587–595. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mattingly CJ, Colby GT, Rosenstein MC,
Forrest JN Jr and Boyer JL: Promoting comparative molecular studies
in environmental health research: An overview of the comparative
toxicogenomics database (CTD). Pharmacogenomics J. 4:5–8. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lackie J: A Dictionary of Biomedicine. 1st
ed. Oxford University Press Inc; New York, NY: 2010
|
|
34
|
Rittner HL, Brack A and Stein C: Pain and
the immune system. Br J Anaesth. 101:40–44. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Okamoto K, Martin DP, Schmelzer JD, Mitsui
Y and Low PA: Pro- and anti-inflammatory cytokine gene expression
in rat sciatic nerve chronic constriction injury model of
neuropathic pain. Exp Neurol. 169:386–391. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Huang D, Cancilla MR and Morahan G:
Complete primary structure, chromosomal localisation, and
definition of polymorphisms of the gene encoding the human
interleukin-12 p40 subunit. Genes Immun. 1:515–520. 2000.
View Article : Google Scholar
|
|
37
|
Randolph AG, Lange C, Silverman EK,
Lazarus R, Silverman ES, Raby B, Brown A, Ozonoff A, Richter B and
Weiss ST: The IL12B gene is associated with asthma. Am J Hum Genet.
75:709–715. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lees JG, Duffy SS, Perera CJ and
Moalem-Taylor G: Depletion of Foxp3+ regulatory T cells
increases severity of mechanical allodynia and significantly alters
systemic cytokine levels following peripheral nerve injury.
Cytokine. 71:207–214. 2015. View Article : Google Scholar
|
|
39
|
Chen IF, Khan J, Noma N, Hadlaq E, Teich
S, Benoliel R and Eliav E: Anti-nociceptive effect of IL-12 p40 in
a rat model of neuropathic pain. Cytokine. 62:401–406. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Callis TE, Pandya K, Seok HY, Tang RH,
Tatsuguchi M, Huang ZP, Chen JF, Deng Z, Gunn B, Shumate J, et al:
MicroRNA-208a is a regulator of cardiac hypertrophy and conduction
in mice. J Clin Invest. 119:2772–2786. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Boon H, Sjögren RJ, Massart J, Egan B,
Kostovski E, Iversen PO, Hjeltnes N, Chibalin AV, Widegren U and
Zierath JR: MicroRNA-208b progressively declines after spinal cord
injury in humans and is inversely related to myostatin expression.
Physiol Rep. 3:2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zinovieva E, Kadi A, Letourneur F, Cagnard
N, Izac B, Vigier A, Said-Nahal R, Elewaut D, de Vlam K,
Pimentel-Santos F, et al: Systematic candidate gene investigations
in the SPA2 locus (9q32) show an association between TNFSF8 and
susceptibility to spondylarthritis. Arthritis Rheum. 63:1853–1859.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fava VM, Cobat A, Van Thuc N, Latini AC,
Stefani MM, Belone AF, Ba NN, Orlova M, Manry J, Mira MT, et al:
Association of TNFSF8 regulatory variants with excessive
inflammatory responses but not leprosy per se. J Infect Dis.
211:968–977. 2015. View Article : Google Scholar
|
|
44
|
Shi J, Jiang K and Li Z: MiR-145
ameliorates neuropathic pain via inhibiting inflammatory responses
and mTOR signaling pathway by targeting Akt3 in a rat model.
Neurosci Res. 134:10–17. 2018. View Article : Google Scholar
|
|
45
|
Lu X, Yin D, Zhou B and Li T: MiR-135a
promotes inflammatory responses of vascular smooth muscle cells
from db/db mice via downregulation of FOXO1. Int Heart J.
59:170–179. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Du XJ and Lu JM: MiR-135a represses
oxidative stress and vascular inflammatory events via targeting
toll-like receptor 4 in atherogenesis. J Cell Biochem.
119:6154–6161. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Xu H, Wu Y, Li L, Yuan W, Zhang D, Yan Q,
Guo Z and Huang W: MiR-344b-1-3p targets TLR2 and negatively
regulates TLR2 signaling pathway. Int J Chron Obstruct Pulmon Dis.
12:627–638. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kiguchi N, Kobayashi Y, Maeda T, Fukazawa
Y, Tohya K, Kimura M and Kishioka S: Epigenetic augmentation of the
macrophage inflammatory protein 2/C-X-C chemokine receptor type 2
axis through histone H3 acetylation in injured peripheral nerves
elicits neuropathic pain. J Pharmacol Exp Ther. 340:577–587. 2012.
View Article : Google Scholar
|
|
49
|
Silva RL, Lopes AH, Guimarães RM and Cunha
TM: CXCL1/CXCR2 signaling in pathological pain: Role in peripheral
and central sensitization. Neurobiol Dis. 105:109–116. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Manjavachi MN, Costa R, Quintão NL and
Calixto JB: The role of keratinocyte-derived chemokine (KC) on
hyperalgesia caused by peripheral nerve injury in mice.
Neuropharmacology. 79:17–27. 2014. View Article : Google Scholar
|
|
51
|
Chen G, Park CK, Xie RG, Berta T,
Nedergaard M and Ji RR: Connexin-43 induces chemokine release from
spinal cord astrocytes to maintain late-phase neuropathic pain in
mice. Brain. 137:2193–2209. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Bayburt TH, Vishnivetskiy SA, McLean MA,
Morizumi T, Huang CC, Tesmer JJ, Ernst OP, Sligar SG and Gurevich
VV: Monomeric rhodopsin is sufficient for normal rhodopsin kinase
(GRK1) phosphorylation and arrestin-1 binding. J Biol Chem.
286:1420–1428. 2011. View Article : Google Scholar :
|
|
53
|
Elenkov IJ, Iezzoni DG, Daly A, Harris AG
and Chrousos GP: Cytokine dysregulation, inflammation and
well-being. Neuroimmunomodulation. 12:255–269. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Bellinger DL and Lorton D: Sympathetic
nerve hyperactivity in the spleen: Causal for nonpathogenic-driven
chronic immune-mediated inflammatory diseases (IMIDs)? Int J Mol
Sci. 19:2018. View Article : Google Scholar : PubMed/NCBI
|