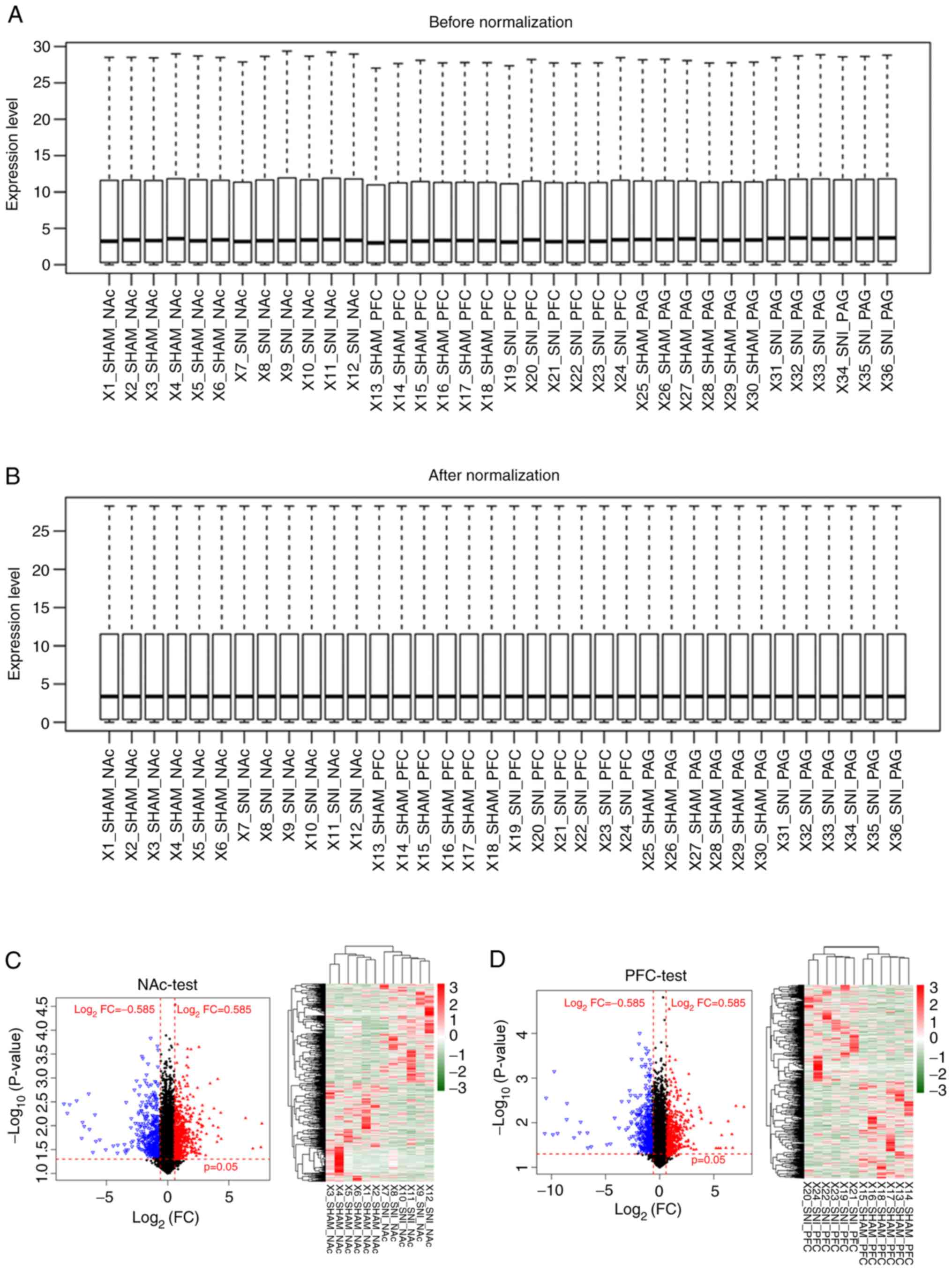
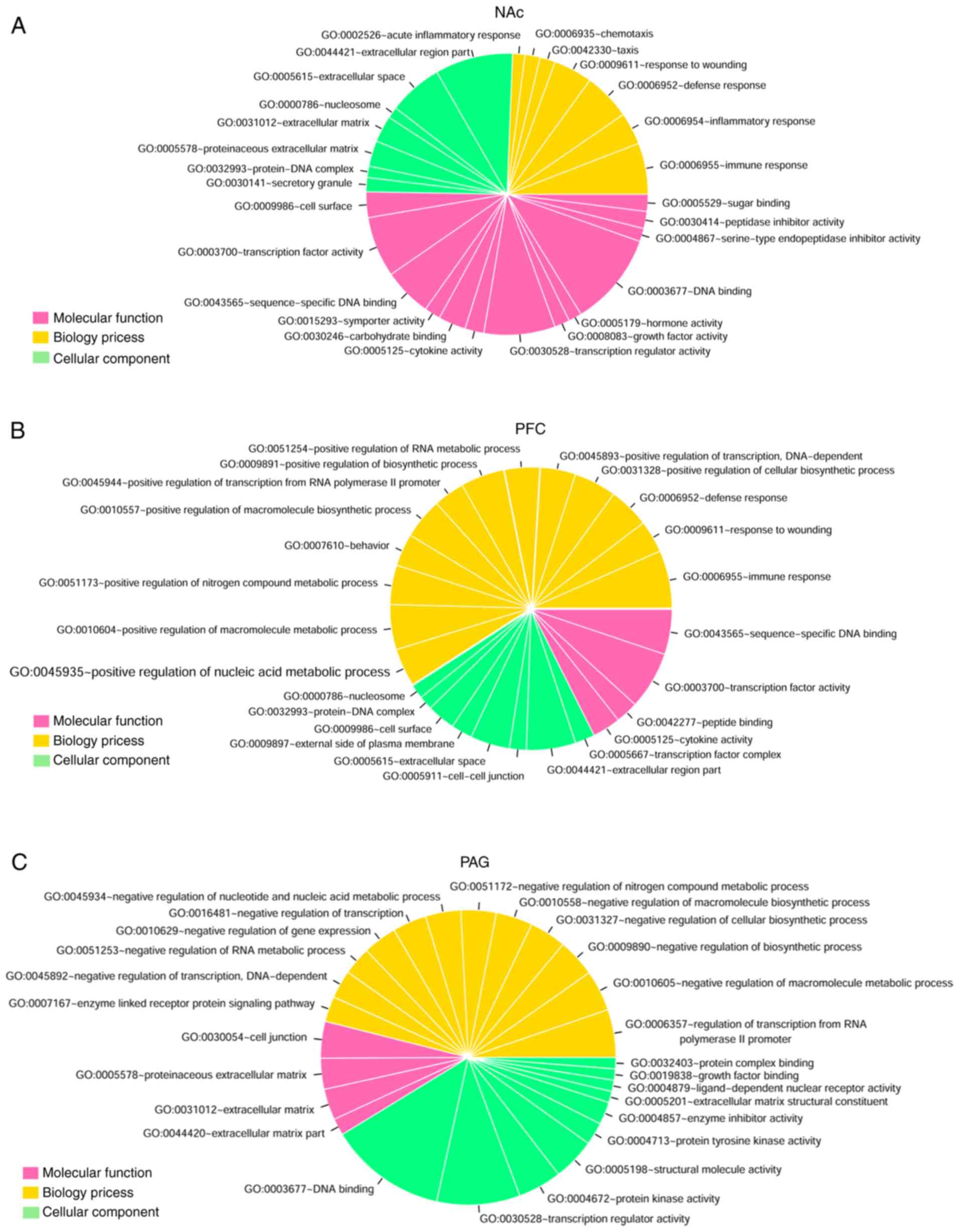
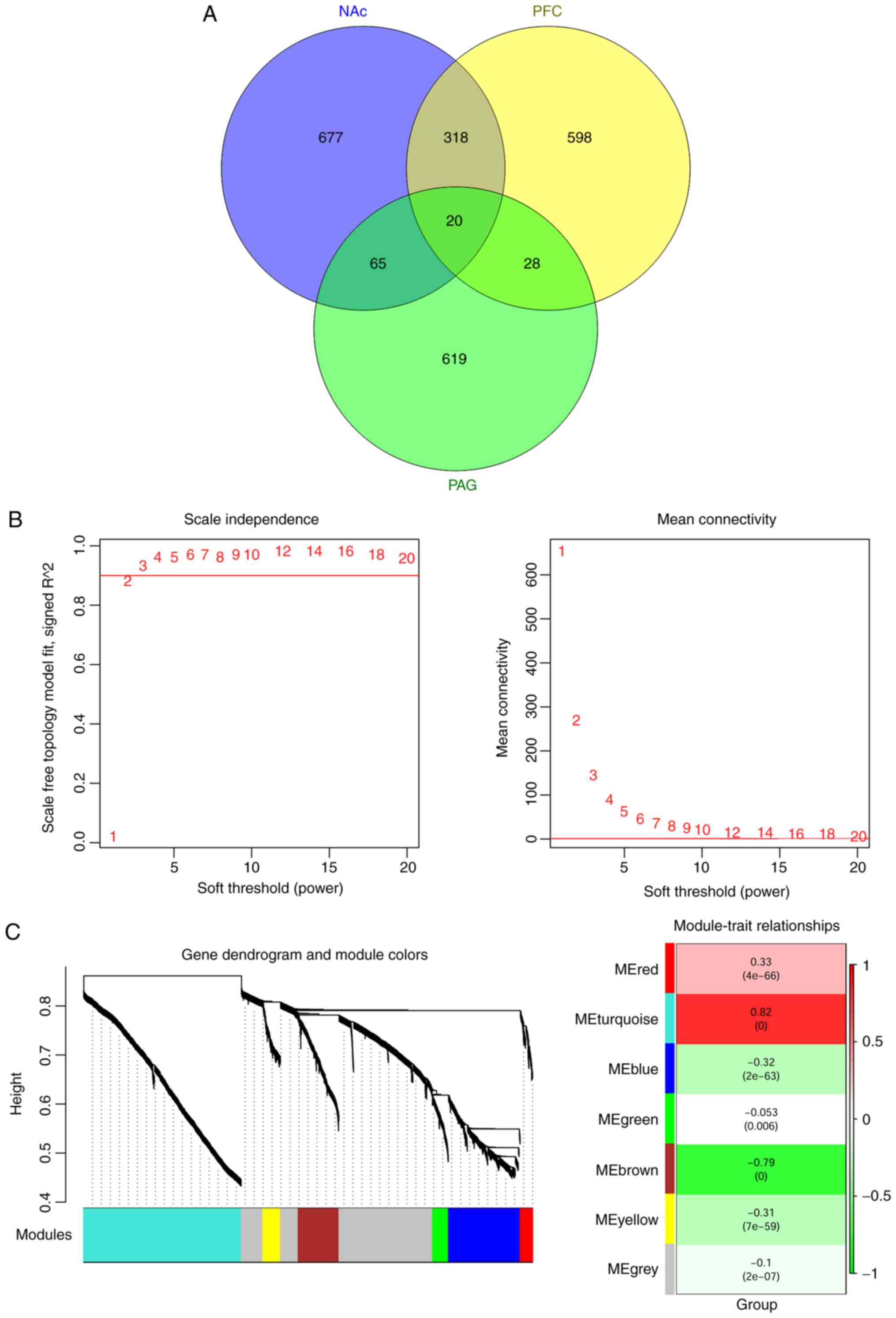
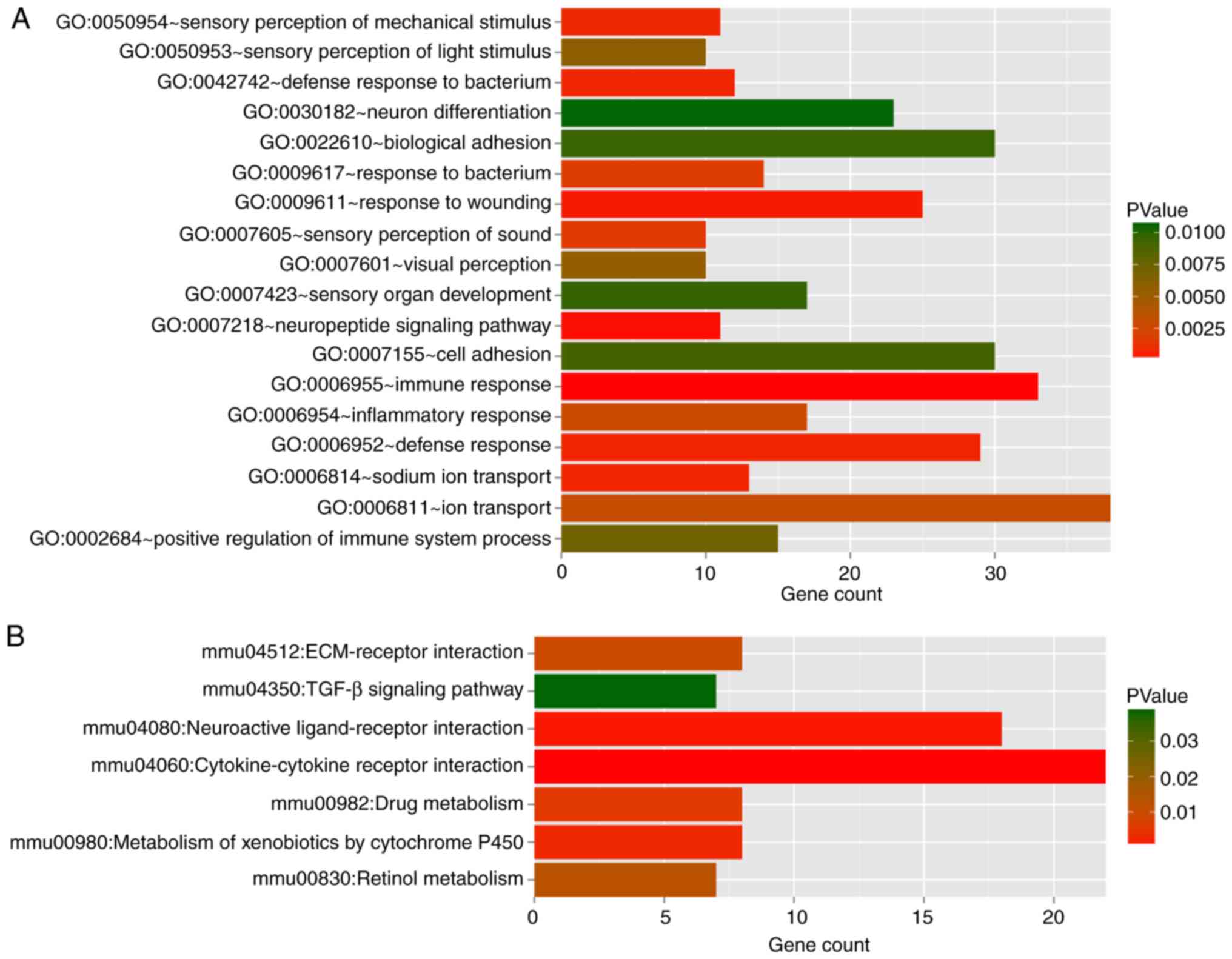
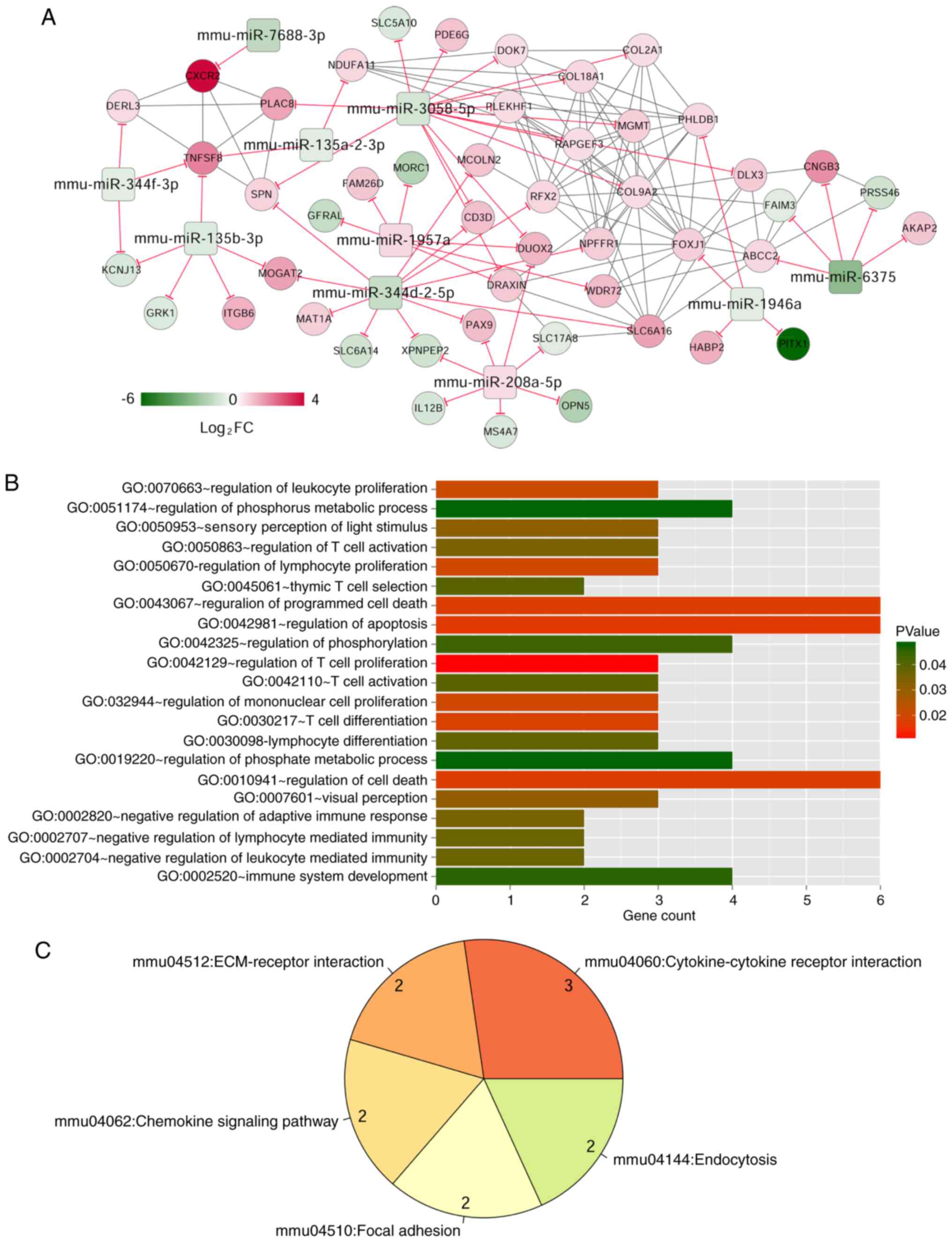
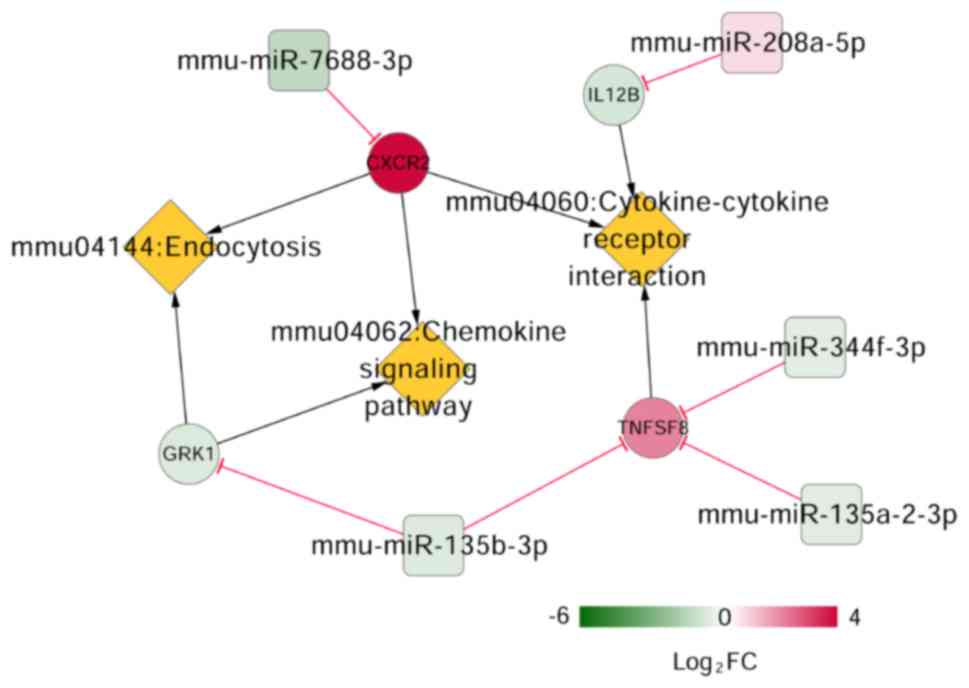
|
1
|
Bennett GJ and Xie YK: A peripheral
mononeuropathy in rat that produces disorders of pain sensation
like those seen in man. Pain. 33:87–107. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Institute of Medicine (US) Committee on
Advancing Pain Research Care, and Education: Relieving pain in
America: A blueprint for transforming prevention, care, education,
and research. Institute of Medicine (US); 2011
|
|
3
|
Baron R: Mechanisms of disease:
Neuropathic pain-a clinical perspective. Nat Clin Pract Neurol.
2:95–106. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dworkin RH, O'Connor AB, Backonja M,
Farrar JT, Finnerup NB, Jensen TS, Kalso EA, Loeser JD, Miaskowski
C, Nurmikko TJ, et al: Pharmacologic management of neuropathic
pain: Evidence-based recommendations. Pain. 132:237–251. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chang PC, Pollema-Mays SL, Centeno MV,
Procissi D, Contini M, Baria AT, Martina M and Apkarian AV: Role of
nucleus accumbens in neuropathic pain: Linked multi-scale evidence
in the rat transitioning to neuropathic pain. Pain. 155:1128–1139.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Weaver KE and Richardson AG: Medial
prefrontal cortex, secondary hyperalgesia, and the default mode
network. J Neurosci. 29:11424–11425. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Seifert F, Bschorer K, De Col R, Filitz J,
Peltz E, Koppert W and Maihöfner C: Medial prefrontal cortex
activity is predictive for hyperalgesia and pharmacological
antihyperalgesia. J Neurosci. 29:6167–6175. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Oh MY, Abosch A, Kim SH, Lang AE and
Lozano AM: Long-term hardware-related complications of deep brain
stimulation. Neurosurgery. 50:1268–1274. 2002.PubMed/NCBI
|
|
9
|
Du L, Wang SJ, Cui J, He WJ and Ruan HZ:
The role of HCN channels within the periaqueductal gray in
neuropathic pain. Brain Res. 1500:36–44. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Griffin RS, Costigan M, Brenner GJ, Ma CH,
Scholz J, Moss A, Allchorne AJ, Stahl GL and Woolf CJ: Complement
induction in spinal cord microglia results in anaphylatoxin
C5a-mediated pain hypersensitivity. J Neurosci. 27:8699–8708. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Descalzi G, Mitsi V, Purushothaman I,
Gaspari S, Avrampou K, Loh YE, Shen L and Zachariou V: Neuropathic
pain promotes adaptive changes in gene expression in brain networks
involved in stress and depression. Sci Signal. 10:2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu H, Xia T, Xu F, Ma Z and Gu X:
Identification of the key genes associated with neuropathic pain.
Mol Med Rep. 17:6371–6378. 2018.PubMed/NCBI
|
|
13
|
Chen CJ, Liu DZ, Yao WF, Gu Y, Huang F,
Hei ZQ and Li X: Identification of key genes and pathways
associated with neuropathic pain in uninjured dorsal root ganglion
by using bioinformatic analysis. J Pain Res. 10:2665–2674. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cline MS, Smoot M, Cerami E, Kuchinsky A,
Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross
B, et al: Integration of biological networks and gene expression
data using Cytoscape. Nat Protoc. 2:2366–2382. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Habibi I, Emamian ES and Abdi A:
Quantitative analysis of intracellular communication and signaling
errors in signaling networks. BMC Syst Biol. 8:892014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Barrett T, Wilhite SE, Ledoux P,
Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH,
Sherman PM, Holko M, et al: NCBI GEO: Archive for functional
genomics data sets-update. Nucleic Acids Res. 39:D991–D995.
2013.
|
|
19
|
Yan M, Song M, Bai R, Cheng S and Yan W:
Identification of potential therapeutic targets for colorectal
cancer by bioinformatics analysis. Oncol Lett. 12:5092–5098. 2016.
View Article : Google Scholar
|
|
20
|
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW,
Shi W and Smyth GK: Limma powers differential expression analyses
for RNA-sequencing and microarray studies. Nucleic Acids Res.
43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Diao C, Xi Y and Xiao T: Identification
and analysis of key genes in osteosarcoma using bioinformatics.
Oncol Lett. 15:2789–2794. 2018.PubMed/NCBI
|
|
22
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar
|
|
24
|
Chen H and Boutros PC: VennDiagram: A
package for the generation of highly-customizable Venn and Euler
diagrams in R. BMC Bioinformatics. 12:352011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Oldham MC, Konopka G, Iwamoto K,
Langfelder P, Kato T, Horvath S and Geschwind DH: Functional
organization of the transcriptome in human brain. Nat Neurosci.
11:1271–1282. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liao Q, Liu C, Yuan X, Kang S, Miao R,
Xiao H, Zhao G, Luo H, Bu D, Zhao H, et al: Large-scale prediction
of long non-coding RNA functions in a coding-non-coding gene
co-expression network. Nucleic Acids Res. 39:3864–3878. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Langfelder P and Horvath S: WGCNA: An R
package for weighted correlation network analysis. BMC
Bioinformatics. 9:5592008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Fromm B, Billipp T, Peck LE, Johansen M,
Tarver JE, King BL, Newcomb JM, Sempere LF, Flatmark K, Hovig E and
Peterson KJ: A uniform system for the annotation of vertebrate
microRNA genes and the evolution of the human microRNAome. Annu Rev
Genet. 49:213–242. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Davis AP, King BL, Mockus S, Murphy CG,
Saraceni-Richards C, Rosenstein M, Wiegers T and Mattingly CJ: The
comparative toxicogenomics database: Update 2011. Nucleic Acids
Res. 39:D1067–D1072. 2011. View Article : Google Scholar :
|
|
31
|
Mattingly CJ, Rosenstein MC, Davis AP,
Colby GT, Forrest JN Jr and Boyer JL: The comparative
toxicogenomics database: A cross-species resource for building
chemical-gene interaction networks. Toxicol Sci. 92:587–595. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mattingly CJ, Colby GT, Rosenstein MC,
Forrest JN Jr and Boyer JL: Promoting comparative molecular studies
in environmental health research: An overview of the comparative
toxicogenomics database (CTD). Pharmacogenomics J. 4:5–8. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lackie J: A Dictionary of Biomedicine. 1st
ed. Oxford University Press Inc; New York, NY: 2010
|
|
34
|
Rittner HL, Brack A and Stein C: Pain and
the immune system. Br J Anaesth. 101:40–44. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Okamoto K, Martin DP, Schmelzer JD, Mitsui
Y and Low PA: Pro- and anti-inflammatory cytokine gene expression
in rat sciatic nerve chronic constriction injury model of
neuropathic pain. Exp Neurol. 169:386–391. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Huang D, Cancilla MR and Morahan G:
Complete primary structure, chromosomal localisation, and
definition of polymorphisms of the gene encoding the human
interleukin-12 p40 subunit. Genes Immun. 1:515–520. 2000.
View Article : Google Scholar
|
|
37
|
Randolph AG, Lange C, Silverman EK,
Lazarus R, Silverman ES, Raby B, Brown A, Ozonoff A, Richter B and
Weiss ST: The IL12B gene is associated with asthma. Am J Hum Genet.
75:709–715. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lees JG, Duffy SS, Perera CJ and
Moalem-Taylor G: Depletion of Foxp3+ regulatory T cells
increases severity of mechanical allodynia and significantly alters
systemic cytokine levels following peripheral nerve injury.
Cytokine. 71:207–214. 2015. View Article : Google Scholar
|
|
39
|
Chen IF, Khan J, Noma N, Hadlaq E, Teich
S, Benoliel R and Eliav E: Anti-nociceptive effect of IL-12 p40 in
a rat model of neuropathic pain. Cytokine. 62:401–406. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Callis TE, Pandya K, Seok HY, Tang RH,
Tatsuguchi M, Huang ZP, Chen JF, Deng Z, Gunn B, Shumate J, et al:
MicroRNA-208a is a regulator of cardiac hypertrophy and conduction
in mice. J Clin Invest. 119:2772–2786. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Boon H, Sjögren RJ, Massart J, Egan B,
Kostovski E, Iversen PO, Hjeltnes N, Chibalin AV, Widegren U and
Zierath JR: MicroRNA-208b progressively declines after spinal cord
injury in humans and is inversely related to myostatin expression.
Physiol Rep. 3:2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zinovieva E, Kadi A, Letourneur F, Cagnard
N, Izac B, Vigier A, Said-Nahal R, Elewaut D, de Vlam K,
Pimentel-Santos F, et al: Systematic candidate gene investigations
in the SPA2 locus (9q32) show an association between TNFSF8 and
susceptibility to spondylarthritis. Arthritis Rheum. 63:1853–1859.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fava VM, Cobat A, Van Thuc N, Latini AC,
Stefani MM, Belone AF, Ba NN, Orlova M, Manry J, Mira MT, et al:
Association of TNFSF8 regulatory variants with excessive
inflammatory responses but not leprosy per se. J Infect Dis.
211:968–977. 2015. View Article : Google Scholar
|
|
44
|
Shi J, Jiang K and Li Z: MiR-145
ameliorates neuropathic pain via inhibiting inflammatory responses
and mTOR signaling pathway by targeting Akt3 in a rat model.
Neurosci Res. 134:10–17. 2018. View Article : Google Scholar
|
|
45
|
Lu X, Yin D, Zhou B and Li T: MiR-135a
promotes inflammatory responses of vascular smooth muscle cells
from db/db mice via downregulation of FOXO1. Int Heart J.
59:170–179. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Du XJ and Lu JM: MiR-135a represses
oxidative stress and vascular inflammatory events via targeting
toll-like receptor 4 in atherogenesis. J Cell Biochem.
119:6154–6161. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Xu H, Wu Y, Li L, Yuan W, Zhang D, Yan Q,
Guo Z and Huang W: MiR-344b-1-3p targets TLR2 and negatively
regulates TLR2 signaling pathway. Int J Chron Obstruct Pulmon Dis.
12:627–638. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Kiguchi N, Kobayashi Y, Maeda T, Fukazawa
Y, Tohya K, Kimura M and Kishioka S: Epigenetic augmentation of the
macrophage inflammatory protein 2/C-X-C chemokine receptor type 2
axis through histone H3 acetylation in injured peripheral nerves
elicits neuropathic pain. J Pharmacol Exp Ther. 340:577–587. 2012.
View Article : Google Scholar
|
|
49
|
Silva RL, Lopes AH, Guimarães RM and Cunha
TM: CXCL1/CXCR2 signaling in pathological pain: Role in peripheral
and central sensitization. Neurobiol Dis. 105:109–116. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Manjavachi MN, Costa R, Quintão NL and
Calixto JB: The role of keratinocyte-derived chemokine (KC) on
hyperalgesia caused by peripheral nerve injury in mice.
Neuropharmacology. 79:17–27. 2014. View Article : Google Scholar
|
|
51
|
Chen G, Park CK, Xie RG, Berta T,
Nedergaard M and Ji RR: Connexin-43 induces chemokine release from
spinal cord astrocytes to maintain late-phase neuropathic pain in
mice. Brain. 137:2193–2209. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Bayburt TH, Vishnivetskiy SA, McLean MA,
Morizumi T, Huang CC, Tesmer JJ, Ernst OP, Sligar SG and Gurevich
VV: Monomeric rhodopsin is sufficient for normal rhodopsin kinase
(GRK1) phosphorylation and arrestin-1 binding. J Biol Chem.
286:1420–1428. 2011. View Article : Google Scholar :
|
|
53
|
Elenkov IJ, Iezzoni DG, Daly A, Harris AG
and Chrousos GP: Cytokine dysregulation, inflammation and
well-being. Neuroimmunomodulation. 12:255–269. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Bellinger DL and Lorton D: Sympathetic
nerve hyperactivity in the spleen: Causal for nonpathogenic-driven
chronic immune-mediated inflammatory diseases (IMIDs)? Int J Mol
Sci. 19:2018. View Article : Google Scholar : PubMed/NCBI
|