Introduction
The prevalence and mortality of cardiovascular
disease (CVD) have exceeded those of malignant tumors, making it
the most important threat to human health and quality of life
worldwide (1). Patients with CVD
are at increased risk of developing end-stage heart failure (HF).
With the rapid progress in medical research, the understanding of
the etiology and pathogenesis of HF has gradually improved.
However, there is currently no effective treatment for end-stage
HF. Therefore, the prevention and treatment of HF are the main
areas of research in the field of CVD (2). Cardiac remodeling is characterized
by an increased burden in CVD and is regulated by various cytokines
and neurohumoral factors. The heart has an adaptive compensatory
mechanism for maintaining cardiac structure and function. Such
compensation usually appears as an increase in the thickness,
weight and capacity of the ventricle, as well as microscopic
changes in terms of hypertrophy of the cardiac myocytes, increased
myocardial fibrosis and interstitial cell proliferation, among
others. Long-term persistence of these burdens results in
decompensation and, eventually, HF (3,4).
Genes are the functional units of DNA, which store
all the information necessary for the reproduction and functioning
of all forms of life (5). Gene
expression refers to the process of protein synthesis under the
guidance of genes. Several gene expression changes are involved in
the occurrence and development of various diseases (6). The gene chip, also referred to as
the DNA chip or DNA microarray, uses the principle of hybridization
to detect the existence and quantity of the corresponding fragments
for the functional and genomic research of genes; it has been
widely used in scientific research and for the clinical diagnosis
of various diseases, including CVD (6,7).
Successful establishment of animal models is the
most critical step in the study of the pathophysiology and
molecular mechanisms underlying myocardial remodeling. Common
models of cardiac remodeling are established by injection of
catecholamines, such as isoproterenol, phenylephrine and
angiotensin II, as well as by transverse aortic constriction (TAC)
in spontaneously hypertensive rats. TAC increases the afterload by
constricting the aorta and increasing the aortic pressure (8). In the present study, we downloaded
and analyzed four original microarray datasets in the Gene
Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo), namely GSE5500,
GSE18224, GSE36074 and GSE56348, in order to explore the potential
mechanisms underlying cardiac pathological remodeling induced by
TAC-mediated pressure overload. The correlation between cardiac
pathological remodeling and differentially expressed genes (DEGs)
was filtered using the limma package of R software (R version
3.3.3). We subsequently identified numerous potential DEGs related
to cardiac pathological remodeling and explored their function via
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes
(KEGG) pathway enrichment analyses using the Database for
Annotation, Visualization and Integrated Discovery (DAVID) online
tool, version 6.8 (https://david.ncifcrf.gov/home.jsp). Finally, a mouse
model of cardiac remodeling induced by TAC was established, and
reverse transcription-quantitative polymerase chain reaction
(RT-PCR) analysis was used to verify the expression of DEGs,
thereby providing new insights into potential molecular targets for
the treatment of pathological cardiac remodeling.
Materials and methods
Microarray data information and
identification of DEGs
GEO is a free public repository of gene microarray
data (http://www.ncbi.nlm.nih.gov/geo/) (9), from which the gene expression
profiles of GSE5500, GSE18224, GSE36074 and GSE56348 datasets were
determined in the left ventricular (LV) myocardial tissue of TAC or
sham-operated mice. GSE5500 included 6 TAC and 4 sham-operated LV
myocardial tissue samples (submission date: August 10, 2006)
(10); GSE18224 included 8 TAC
and 8 sham-operated LV myocardial tissue samples (submission date:
September 23, 2009) (11);
GSE36074 included 14 TAC and 5 sham-operated LV myocardial tissue
samples (submission date: February 24, 2012) (12); and GSE56348 included 5 pairs of
TAC and sham-operated LV myocardial tissue samples (submission
date: March 29, 2014) (13). The
GSE5500, GSE18224 and GSE36074 data were based on the GPL1261
platforms (Affymetrix Mouse Genome 430 2.0 Array, Affymetrix;
Thermo Fisher Scientific, Inc.); the GSE56348 data were based on
the GPL6480 platforms (Affymetrix Mouse Gene 1.0 ST Array)
(Table I).
 | Table IGEO Datasets used for the
bioinformatics analysis. |
Table I
GEO Datasets used for the
bioinformatics analysis.
| Study | Genus | Tissue | GEO | Platform | Sham | TAC | (Refs.) |
|---|
| Bisping et
al | Mus
musculus | Left
ventricular | GSE5500 | GPL1261 | 4 | 6 | (10) |
| Fliegner et
al | Mus
musculus | Left
ventricular | GSE18224 | GPL1261 | 8 | 8 | (11) |
| Skrbic et
al | Mus
musculus | Left
ventricular | GSE36074 | GPL1261 | 5 | 14 | (12) |
| Lai et
al | Mus
musculus | Left
ventricular | GSE56348 | GPL6246 | 10 | 10 | (13) |
Raw data (TXT format) of genomic expression were
integrated for analysis, and data preprocessing, including
background correction, normalization, logarithmic conversion and
screening of DEGs were subsequently performed using R software with
the limma package. Statistically significant DEGs were defined as
log2FC>1 or log2FC<−1 (FC, fold change)
and an adjusted P-value of <0.05. Volcano plots and heatmaps of
these screened DEGs were constructed using R software.
GO terms and pathway enrichment
analysis
DAVID version 6.8 is an open-source website for
scientists to explore the biological functions of DEGs (14). GO analysis was conducted using the
sub-databases GOTERM_BP_DIRECT, GOTERM_CC_DIRECT and
GOTERM_MF_DIRECT, and KEGG pathway analysis was conducted using
DAVID version 6.8 to analyze the DEGs at the functional level.
P<0.01 was set as the cut-off indicating statistically
significant differences.
Integration of protein-protein
interaction (PPI) network and identification of hub genes
PPIs include direct physical interactions and
indirect functions (15). DEGs
were selected from the four data series (i.e., GSE5500, GSE18224,
GSE36074 and GSE56348), and a PPI network was constructed. The
online database STRING (http://string-db.org) was used to construct the PPI
network and analyze the functional interactions based on the above
screened DEG-encoded proteins. The hub genes were screened by
calculating the number of interconnections. Proteins located at the
center nodes may be considered as core proteins with important
physiological functions, and the corresponding gene is a key
candidate gene.
Animals and animal models
All animal care and experimental procedures were
approved by the Animal Care and Use Committee of Renmin Hospital of
Wuhan University (Wuhan, China), and conformed to the Guidelines
for the Care and Use of Laboratory Animals published by the United
States National Institutes of Health (16). Adult male specific pathogen-free
(SPF) grade C57BL/6 mice (n=40, 8-10 weeks old, weighing 23.5-25.5
g) and adult male and female SPF grade Sprague-Dawley (SD) rats
were purchased from the Institute of Laboratory Animal Science,
Chinese Academy of Medical Sciences, Beijing, China. The TAC model
was selected mainly because of the TAC model of original chip data
analysis. In the study of TAC-mediated cardiac remodeling, the most
commonly used time points are 4 and 8 weeks. Previous studies have
reported that significant changes in cardiac remodeling and
decreased cardiac function are observed 4 weeks after TAC, whereas
at 8 weeks after TAC, end-stage HF is more likely (17,18). Furthermore, most of the original
chip data we analyzed were samples analyzed for ~4 weeks.
Therefore, testing at 4 weeks after TAC was selected. The mice were
randomly divided into two groups (TAC, n=25; sham, n=15) and
allowed to acclimatize for 7 days prior to the experiments. The
animals were anesthetized with 80 mg/kg sodium pentobarbital
(Sigma-Aldrich; Merck KGaA) by intra-peritoneal injection, and
subsequently subjected to TAC or sham operation, as described
previously (17). A total of 6
mice in the TAC group and 3 mice in the sham group died during or
after the operation. The animals were euthanized via 200 mg/kg
sodium pentobarbital by intraperitoneal injection.
Echocardiographic measurement
At 4 weeks after the operation, echocardiography was
used to measure the LV structure and function in mice that were
anesthetized by inhalation of 1.5% isoflurane. Echocardiography was
performed using a MyLab 30CV system (Esaote SpA) equipped with a
10-MHz linear array ultrasound transducer, as previously described
(18). Two-dimensional images
were captured from the LV parasternal long axis and parasternal
short axis at the level close to the papillary muscles at a frame
rate of 120 Hz. LV end-systolic diameter (LVESD), LV end-diastolic
dimension (LVEDD), LV ejection fraction (EF) and LV fractional
shortening (FS) were obtained by LV M-mode tracing with a sweep
speed of 50 mm/sec. LVESD, LVEDD, EF and FS were analyzed for
>10 beats per heart.
Histological analysis
Following completion of echocardiographic and
hemodynamic measurements, the mice were immediately euthanized.
Body weight, heart and lung weight, and tibial length were measured
for each mouse. Heart weight/body weight (HW/BW, mg/g), lung
weight/body weight (LW/BW, mg/g) and heart weight/tibial length
(HW/TL, mg/mm) ratios in each group were calculated using these
data. The heart was harvested and immediately placed in 10% KCl
solution to induce diastolic arrest. Subsequently, the hearts were
fixed in 4% paraformaldehyde for 3 days. Fixed myocardial tissues
were dehydrated, embedded in paraffin, and cut into 5-µm
sections as described previously (19). Sections from the middle segment of
each heart were subjected to hematoxylin and eosin (H&E)
staining for the observation of overall morphology.
For immunohistochemistry, fixation, embedding and
sectioning of cardiac specimens were performed in a similar manner
as for H&E staining. The heart sections were heated for antigen
retrieval using a pressure cooker. The sections were incubated with
anti-integrin subunit beta-like 1 (Itgbl1; Santa Cruz
Biotechnology, Inc.; sc-365162, 1:50) and anti-Asporin (Abcam;
ab58741, 1:100), and developed using peroxidase-coupled secondary
antibodies and 3,3′-diaminobenzidine as a substrate. Images were
captured at a magnification of ×400 from 10 fields/heart.
RT-qPCR
The mRNA expression level of the DEGs with research
value was measured using qPCR, as described previously (19). Total RNA was isolated from LV
myocardium using TRIzol reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) and mRNA was reverse-transcribed to cDNA using
oligo(dT) primers. Gene expression levels were analyzed using the
Light Cycler 480 Q-PCR System and Light Cycler 480 SYBR-Green 1
Master Mix (Roche Diagnostics). Target gene mRNA expression was
normalized to the internal control glyceraldehyde-3-phosphate
dehydrogenase (GAPDH). The forward and reverse primer pairs used
for RT-PCR are shown in Table
II.
 | Table IIForward and reverse primer pairs. |
Table II
Forward and reverse primer pairs.
| Gene names | Forward | Reverse |
|---|
| Mfap4 |
GCAACCCCTGGACTGTGATG |
TTGTCATGTCGCAGAAGACGG |
| Ltbp2 |
AACAGCACCAACCACTGTATC CC |
TGGCATTCTGAGGGTCAAA |
| Retnla |
CCAATCCAGCTAACTATCCCTCC |
ACCCAGTAGCAGTCATCCCA |
| Cthrc1 |
TGGACCAAGGAAGCCCTGAGT |
TGAACAGGTGCCGACCCAGA |
| Pamr1 |
ATGGAGCTAGACAGATGGGC |
GACCGTGTACTCTCTTGGCAA |
| Itgbl1 |
GTGGAAACTGTTACTGCGAGG |
TGGCAAGAACATTCACCACATAC |
| MFAP5 |
GTCTTGGCAATCAGCATCCC |
CCAGATTAGGGTCGTCTGTGAAT |
| Meox1 |
GAAACCCCCACTCAGAAGATAGC |
TCGTTGAAGATTCGCTCAGTC |
| Aqp8 |
TGTGTAGTATGGACCTACCTGAG |
ACCGATAGACATCCGATGAAGAT |
| Aspn |
AAGGAGTATGTGATGCTACTGCT |
ACATTGGCACCCAAATGGACA |
| Srpx2 |
ATGGTACGCAGGCTCAGGTTA |
TGAGTAGCATGTGGCTTCTCC |
| Col8a |
AGGAGAAGTACCGTTAGCCAG |
TACCGGGCTTTCCAATTCCTG |
| Cilp |
ATGGCAGCAATCAAGACTTGG |
AGGCTGGACTCTTCTCACTGA |
| Frzb |
CACAGCACCCAGGCTAACG |
TGCGTACATTGCACAGAGGAA |
| Fstl1 |
CACGGCGAGGAGGAACCTA |
TCTTGCCATTACTGCCACACA |
Western blotting
The protein expression level was measured by western
blotting as described previously. The cardiac LV tissue was lysed
in RIPA lysis buffer, and the protein concentration was measured
using the BCA Protein Assay Kit (Thermo Fisher Scientific, Inc.;
23227). Protein lysates (50 µg) were electrophoresed on 10%
sodium dodecyl sulfate-polyacrylamide gels, and subsequently
transferred onto polyvinylidene fluoride (PVDF) membranes (EMD
Millipore; IPFL00010) by a gel transfer device. The PVDF membranes
were blocked with 5% non-fat milk for 1 h and incubated with
primary antibodies against Itgbl1 (Santa Cruz Biotechnology, Inc.;
sc-365162, 1:200) and Asporin (Abcam, ab58741, 1:400), overnight at
4°C. After washing three times with Tris-buffered saline containing
Tween 20, the membrane was stained with anti-mouse or anti-rabbit
IgG (1:10,000) for 1 h. The blots were scanned and analyzed using
an Odyssey infrared imaging system (LI-COR Biosciences). Specific
protein expression levels were normalized to GAPDH (Cell Signaling
Technology, Inc.; 5174, 1:2,000).
Cultured neonatal rat ventricular
myocytes (CMs) and cardiac fibroblasts (CFs)
Neonatal rat CMs and CFs were prepared based on our
published literature (20,21).
Briefly, adequate neonatal hearts from 1- to 2-day-old
Sprague-Dawley (SD) rats were cut into ~1-mm3 pieces and
digested in enzyme solutions. Isolation of CMs and CFs was
performed by differential time adhesion. BrdU (0.1 mM) was used to
inhibit the proliferation of the CFs present in the CM fraction.
The CMs and CFs were cultured in DMEM/F12 containing fetal bovine
serum (15% for CMs or 10% for CFs), streptomycin (100 mg/ml), and
penicillin (100 IU/ml) at 37°C in a humidified incubator with 5%
CO2. CMs and CFs were stimulated by angiotensin (Ang) II
(1 µM) and transforming growth factor (TGF)β (10 ng/ml),
respectively. The effect of stimulation was evaluated by
immunofluorescence staining. The CM fraction was detected with
anti-α-actinin staining. CFs were stained with anti-α-smooth muscle
actin (SMA) to observe fluorescence intensity.
Immunofluorescence staining
Immunofluorescence staining was performed as
previously described (17).
Briefly, cell coverslips were washed three times with PBS, fixed
with 4% formaldehyde, permeabilized in 0.2% Triton X-100, and
stained with anti-α-actinin (1:100 in 1% goat serum) or anti-α-SMA
(1:100 in 1% goat serum) overnight after blocking with 10% goat
serum for 60 min at 37°C. The coverslips were then incubated with
Alexa Fluor 488-goat anti-rabbit secondary antibody for 60 min at
37°C, with DAPI (Invitrogen; Thermo Fisher Scientific, Inc.;
S36939) for nuclear staining. Images were captured by a special
Olympus DX51 fluorescence microscope (Olympus Corporation).
Statistical analysis
Data are presented as the mean ± standard error of
the mean from at least six independent experiments. To determine
statistical significance, the results were compared using Student's
two-tailed t-tests. All statistical analyses were performed using
GraphPad Prism 5.0 software (GraphPad Software, Inc.). A P-value of
<0.05 at a 95% confidence level was considered to indicate
statistically significant differences.
Results
Identification of DEGs
As shown in Table
I, 4 datasets (GSE5500, GSE18224, GSE36074 and GSE56348) were
included in the present study. Following data normalization and DEG
analysis using |log2FC| >1 and P<0.05 as cut-offs,
a total of 188, 293, 253 and 67 DEGs were identified from the
GSE5500, GSE18224, GSE36074 and GSE56348 datasets, respectively.
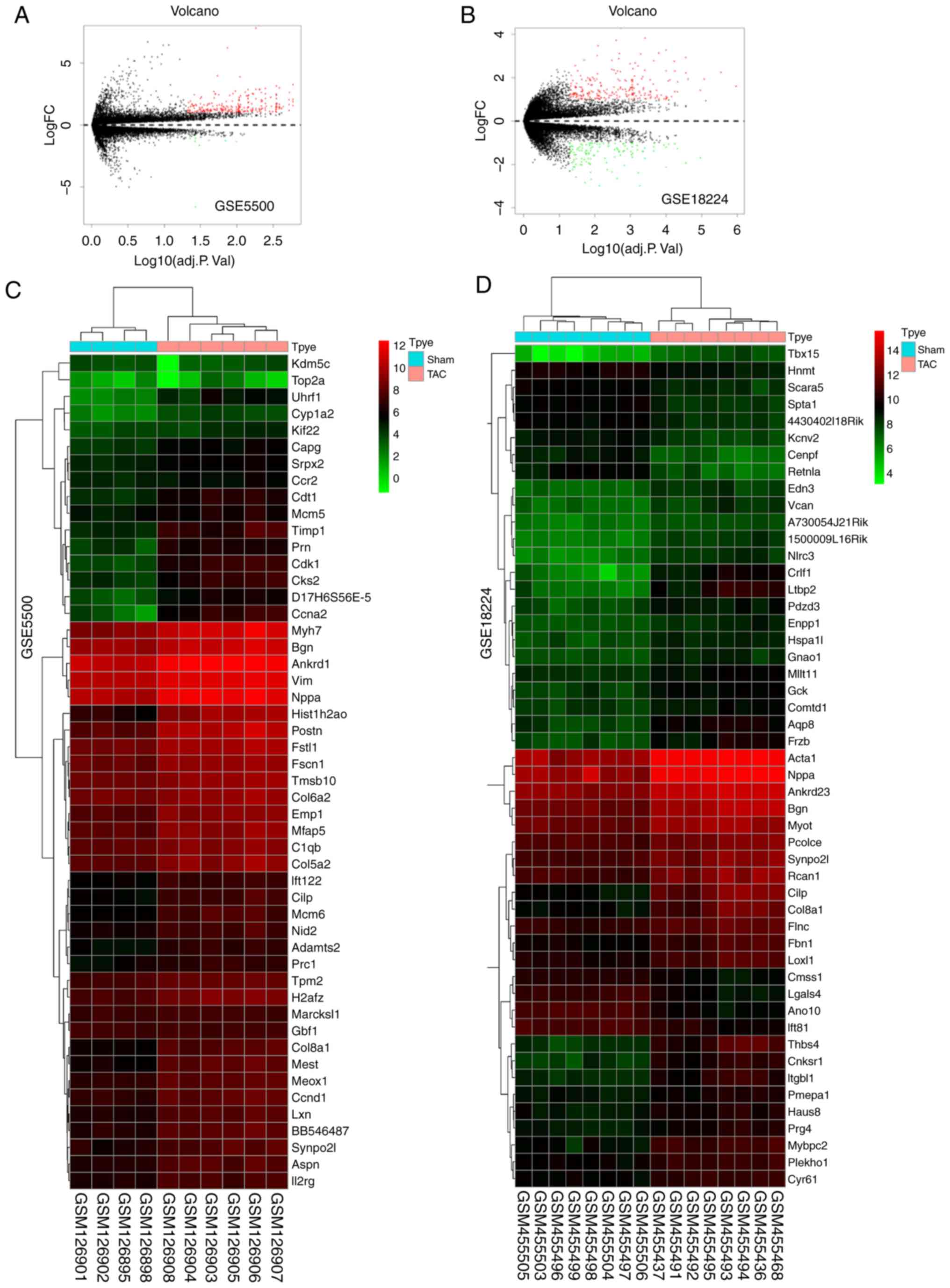
The volcano plots and heatmaps of the first 50 DEGs are displayed
in Fig. 1. In bioinformatics, the
method of collecting and intersecting multiple profile datasets is
commonly used. The selection criterion of 'all four positive'
methods is commonly used and the results obtained are considered as
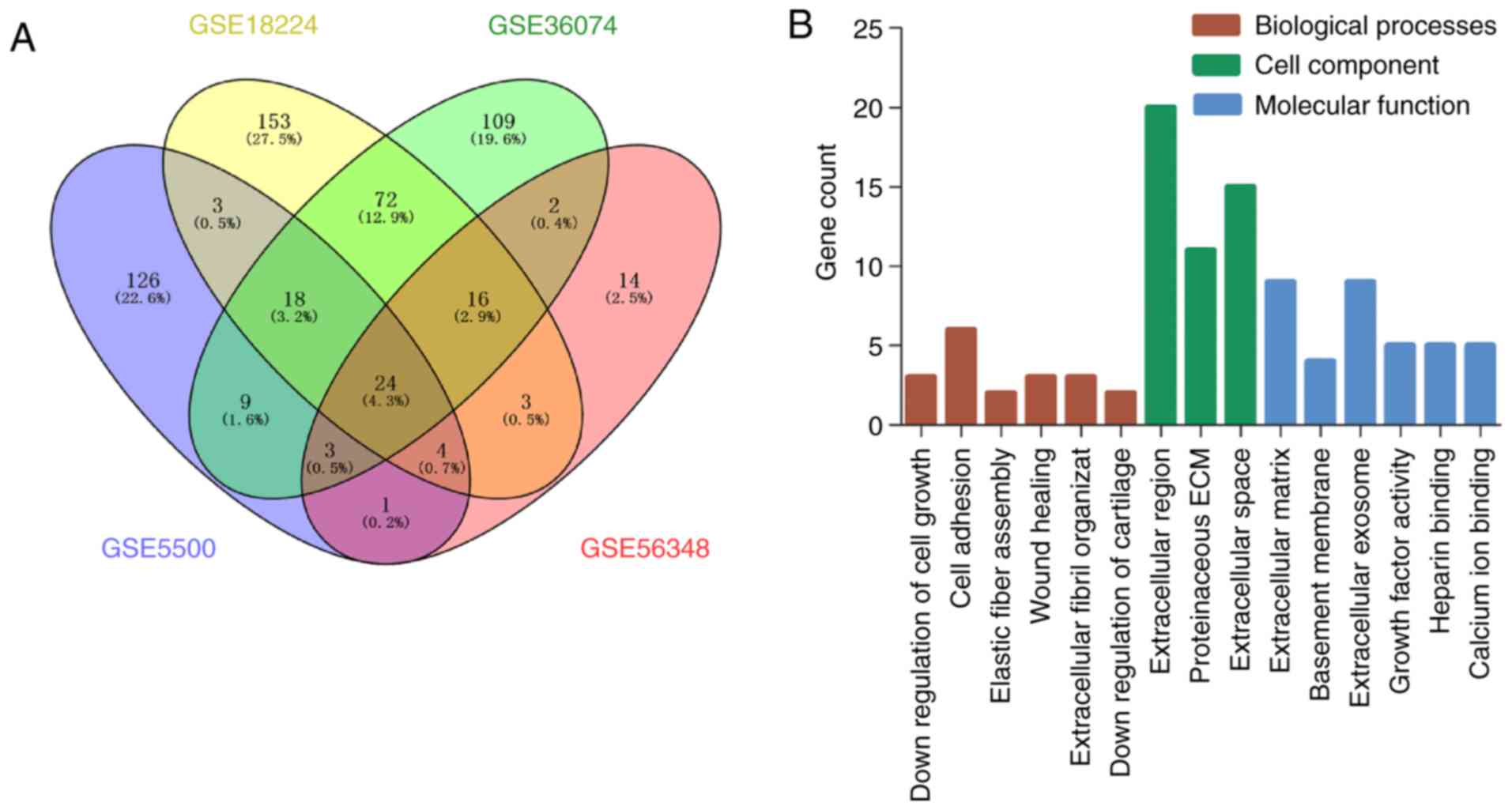
reliable. After using the Venny website (http://bioinfogp.cnb.csic.es/tools/venny/index.html)
for integrated bioinformatics, a total of 24 consistent DEGs were
identified from the four profile datasets (Fig. 2), including 23 upregulated and 1
downregulated genes in the mouse model of cardiac remodeling
induced by TAC vs. sham operation (Fig. 2A, Table III).
 | Table IIIDEGs identified from the four
profiles (n=24). |
Table III
DEGs identified from the four
profiles (n=24).
| DEGs | Genes |
|---|
| Upregulated | Ltbp2, Postn, Cilp,
Mfap4, Thbs4, Col8a1, Timp1, Nppa, Gdf15, Pamr1, Lox, Acta1,
Itgbl1, Ctgf, Cnksr1, Frzb, Mfap5, Meox1, Tgfb2, Aqp8, Fstl1, Aspn,
Srpx2 |
| Downregulated | Retnla |
DEG GO analysis in TAC-induced cardiac
remodeling
To explore the biological function of DEGs in a
mouse model of TAC-mediated pressure overload-induced LV
remodeling, the 24 screened DEGs were uploaded to DAVID version
6.8. The DEGs were found to be enriched in biological processes,
including extracellular fibril organization, cell adhesion, wound
healing, elastic fiber assembly, and negative regulation of cell
growth and cartilage development. For cell component, the DEGs were
mainly enriched in extracellular region, protein-aceous
extracellular matrix, extracellular space, extracellular matrix,
basement membrane, and extracellular exosome. Molecular function
analysis revealed that the upregulated DEGs were mainly enriched in
growth factor activity, heparin binding, and calcium ion binding
(Fig. 2B and Table IV). No DEGs were found to be
significantly enriched in KEGG pathway enrichment analysis.
 | Table IVSignificant enriched analysis of
differentially expressed genes. |
Table IV
Significant enriched analysis of
differentially expressed genes.
| Category | Term | Description | Count | P-value |
|---|
|
GOTERM_BP_DIRECT | GO:0043206 | Extracellular
fibril organization | 3 | 0.000070 |
|
GOTERM_BP_DIRECT | GO:0007155 | Cell adhesion | 6 | 0.000194 |
|
GOTERM_BP_DIRECT | GO:0042060 | Wound healing | 3 | 0.005265 |
|
GOTERM_BP_DIRECT | GO:0048251 | Elastic fiber
assembly | 2 | 0.008103 |
|
GOTERM_BP_DIRECT | GO:0030308 | Negative regulation
of cell growth | 3 | 0.009136 |
|
GOTERM_BP_DIRECT | GO:0061037 | Negative regulation
of cartilage development | 2 | 0.009255 |
|
GOTERM_CC_DIRECT | GO:0005576 | Extracellular
region | 20 | <0.000001 |
|
GOTERM_CC_DIRECT | GO:0005578 | Proteinaceous
extracellular matrix | 11 | <0.000000 |
|
GOTERM_CC_DIRECT | GO:0005615 | Extracellular
space | 15 | <0.000000 |
|
GOTERM_CC_DIRECT | GO:0031012 | Extracellular
matrix | 9 | <0.000000 |
|
GOTERM_CC_DIRECT | GO:0005604 | Basement
membrane | 4 | 0.000192 |
|
GOTERM_CC_DIRECT | GO:0070062 | Extracellular
exosome | 9 | 0.008497 |
|
GOTERM_MF_DIRECT | GO:0008083 | Growth factor
activity | 5 | 0.000016 |
|
GOTERM_MF_DIRECT | GO:0008201 | Heparin
binding | 5 | 0.000019 |
|
GOTERM_MF_DIRECT | GO:0005509 | Calcium ion
binding | 5 | 0.006123 |
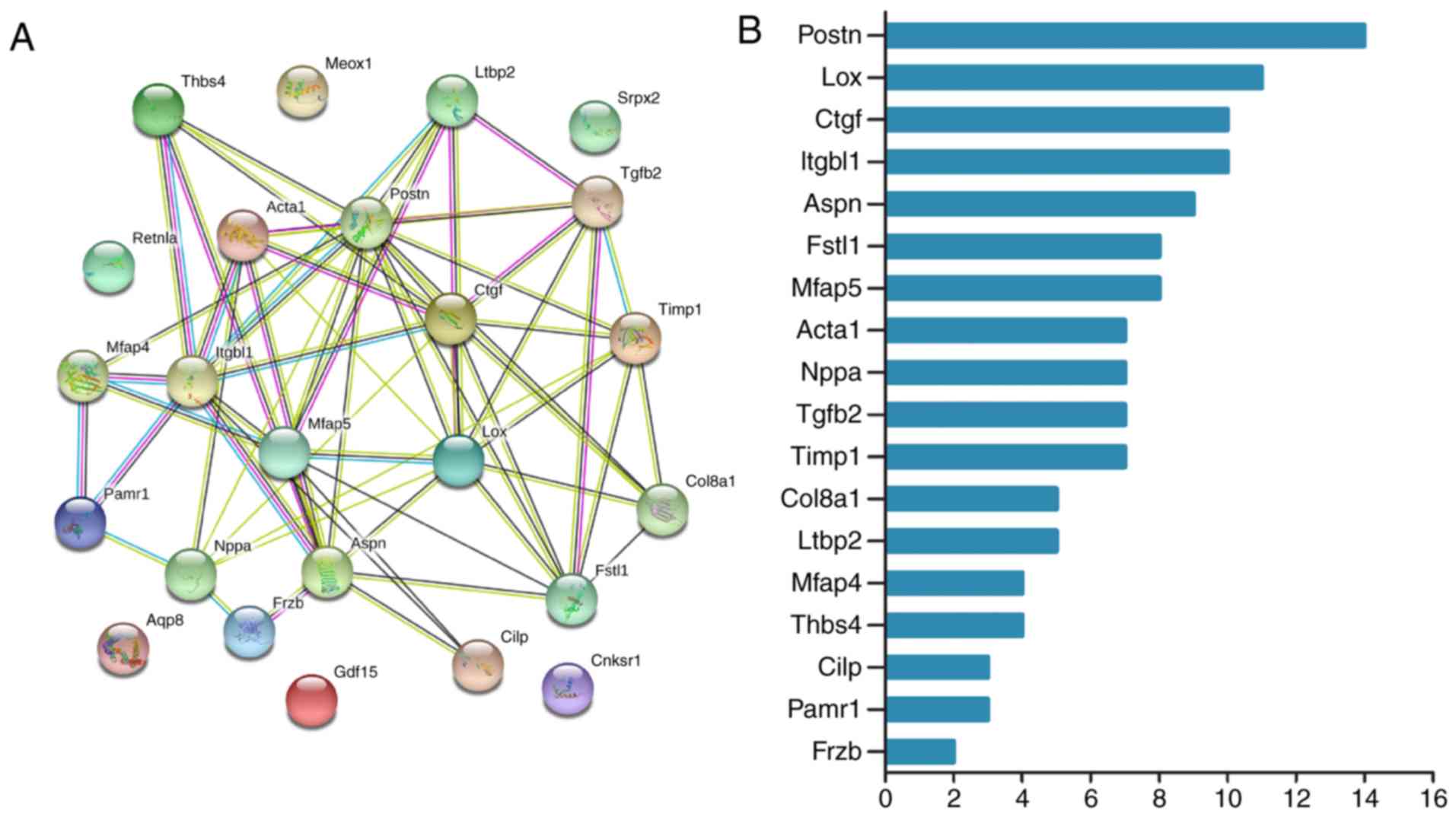
Integration of PPI network and
identification of hub genes
The 24 screened DEGs from these four datasets were
selected to construct a PPI network using the STRING online
database. A total of 18 DEGs (all upregulated) of the 24 commonly
altered DEGs were filtered into the DEG PPI network complex. These
18 DEGs included (in descending order) Postn, Lox, Ctgf, Itgbl1,
Aspn, Fstl1, Mfap5, Acta1, Nppa, Tgfb2, Timp1, Col8a1, Ltbp2,
Mfap4, Thbs4, Cilp, Pamr1 and Frzb sequentially (Fig. 3).
Establishment of a cardiac pathological
remodeling model in mice
For 4 weeks after TAC, the remodeling responses of
these mice in response to pressure overload were detected. TAC mice
exhibited a significant cardiac pathological remodeling phenotype,
including increased HW/BW, LW/BW and HW/TL ratios (Fig. 4A-C). To investigate cardiac
function, echocardiographic examination was performed. TAC mice
exhibited deteriorated cardiac remodeling and dysfunction compared
with sham-operated mice, as indicated by LVEDD, LVESD, FS and EF
(Fig. 4D-H). H&E staining and
cardiomyocyte cross-sectional area also confirmed the adverse
effect of TAC surgery on cardiac remodeling (Fig. 4I and J). Several sections of the
heart were stained with picrosirius red for assessment of
interstitial fibrosis (Fig. 4K and
L). Based on these results, the successful establishment of a
TAC-induced cardiac pathological remodeling model in mice was
confirmed.
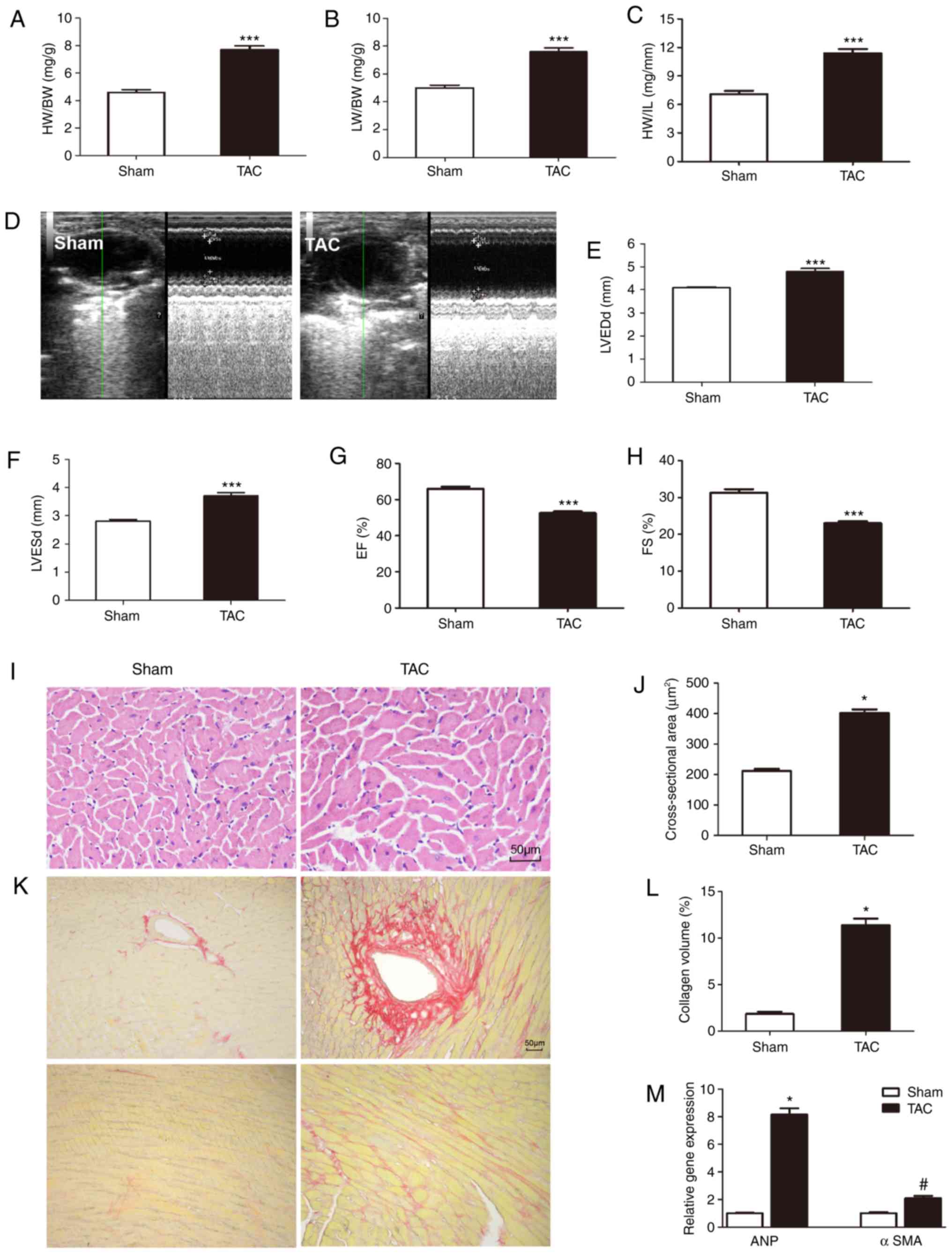 | Figure 4Established TAC-induced mouse model
of cardiac remodeling. (A-C) Statistical results of the HW/BW,
LW/BW and HW/TL ratios (n=11). (D-H) Echocardiography pictures and
parameters (LVESD, LVEDD, EF and FS) (n=10). (I and J)
Representative hematoxylin and eosin staining and statistical
results for the cross-sectional area (n=6 sample, 100-150 cells per
sample). (K and L) Representative picrosirius red staining on
histological sections and statistical results (n=6). (M) Reverse
transcription-quantitative polymerase chain reaction analysis
validation of mRNA expression levels of cardiac hypertrophy marker
ANP and cardiac fibrosis marker αSMA (n=6 sample).
*P<0.05, #P<0.05,
***P<0.001. TAC, transverse aortic constriction; HW,
heart weight; BW, body weight; LW, lung weight; TL, tibial length;
LVESD, left ventricular (LV) end-systolic diameter; LVEDD, LV
end-diastolic dimension; EF, LV ejection fraction; FS, LV
fractional shortening; ANP, atrial natriuretic peptide; SMA, smooth
muscle actin. |
Validation of gene expression levels of
DEGs in a mouse model of cardiac remodeling
The mRNA expression levels of DEGs were detected
using RT-PCR in a mouse model of TAC-induced cardiac remodeling,
excluding the 9 published genes Postn, Lox, Ctgf, Nppa, ANP, Acta1,
Timp1, Tgfb2 and Clip, which have been previously confirmed to be
directly involved in the occurrence and development of myocardial
remodeling (these 9 genes have been found to be upregulated, which
is consistent with the results of a large number of published
studies) (22-30). Hence, the mRNA expression level of
the remaining 16 DEGs was determined by RT-PCR. As shown in
Fig. 5, compared with the sham
group, the expression of Itgbl1, Aspn, Fstl1, Mfap5, Col8a1, Ltbp2,
Mfap4, Pamr1, Cnksr1, Aqp8, Meox1, Gdf15 and Srpx was upregulated,
and that of Retnla was downregulated in the mouse model of cardiac
pathological remodeling, which is consistent with the results of
the bioinformatics analysis. Frzb was not found to be significantly
differentially expressed, as determined by PCR. The reason for the
inconsistency is that the sensitivity of different detection
methods varies.
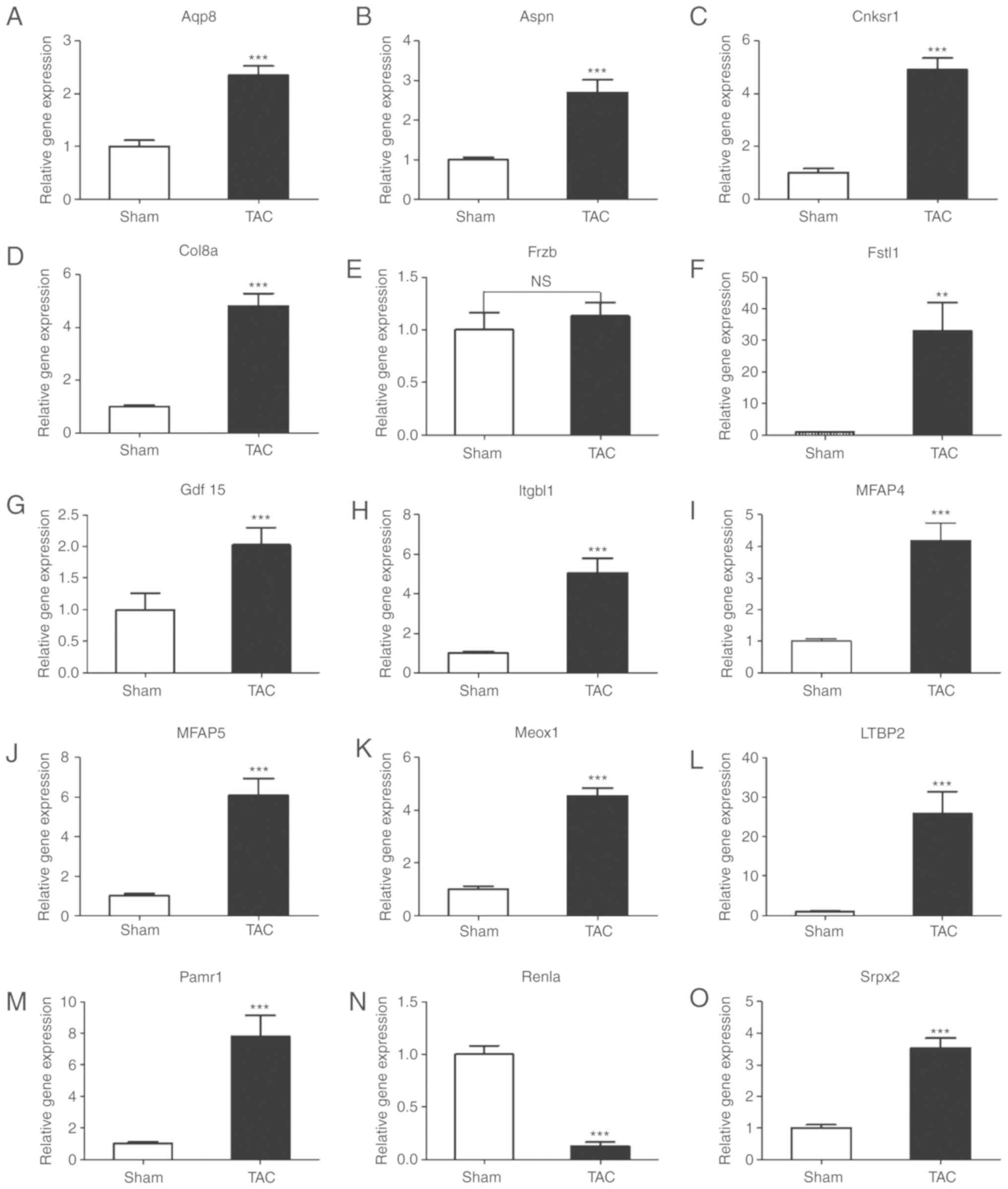 | Figure 5Reverse transcription-quantitative
polymerase chain reaction analysis validation of mRNA expression
levels of the selected common differentially expressed genes. (A-O)
mRNA levels of Aqp8, Aspn, Cnksr1, Col8a1, Frzb, Fstl1, Gdf15,
Itgbl1, Mfap4, Mfap5, Meox1, Ltbp2, Retnla and Pamr1 in heart
tissues of mice in the TAC and sham-operated groups. Values shown
are normalized to GAPDH (n=6); **P<0.01,
***P<0.001; NS, not significant; TAC, transverse
aortic constriction. |
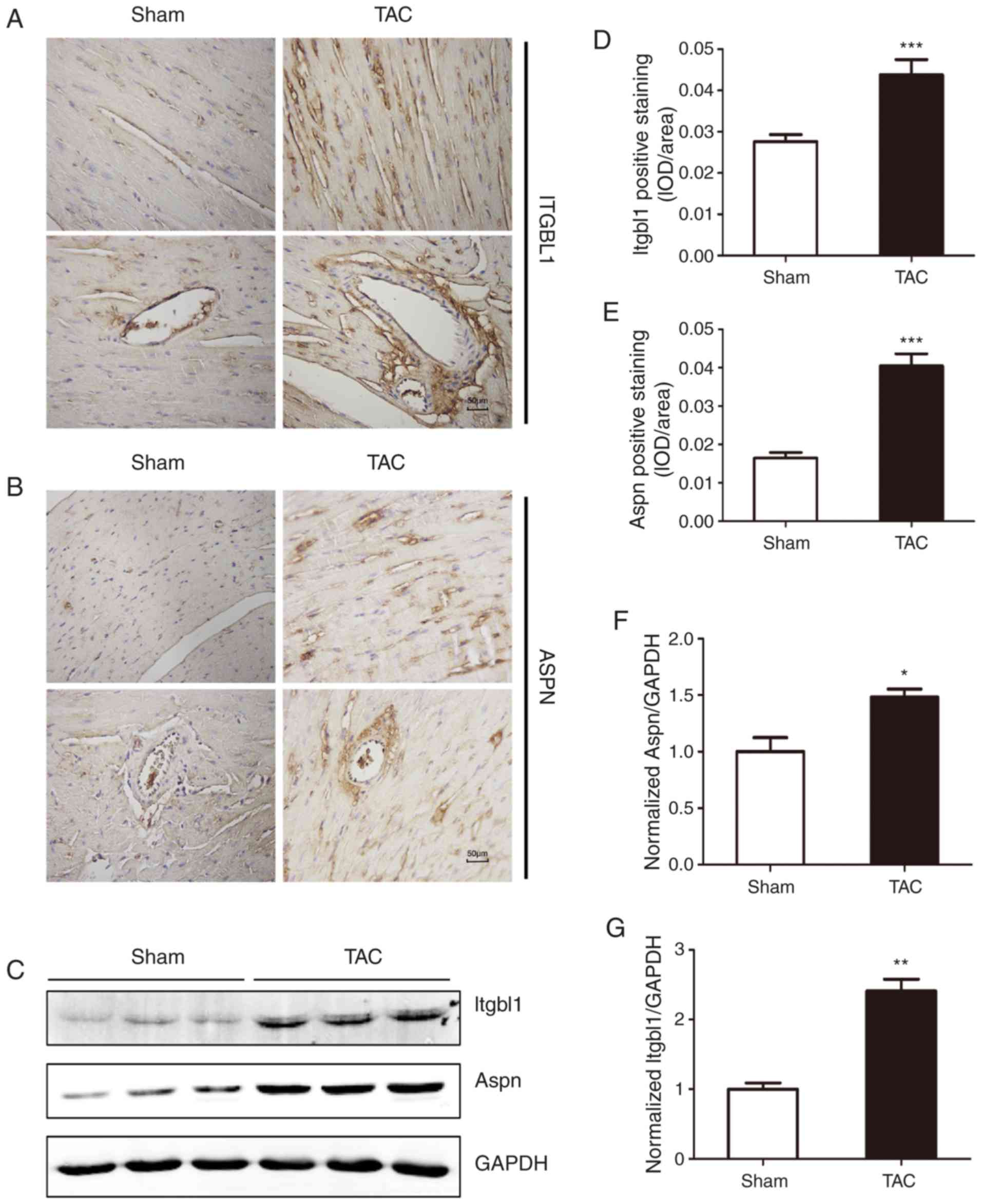
Detection of the protein level of key
DEGs by western blotting and immunohistochemistry
According to the results of the PCR and combined
with the hub genes by PPI network analysis, Itgbl1 and Aspn were
selected, and their protein expression level was determined by
western blotting and immunohistochemistry. As shown in Fig. 6, western blotting and
immunohistochemistry confirmed that TAC surgery significantly
increased the protein expression level of Itgbl1 and Asporin
(protein encoded by Aspn gene), consistently with the results of
PCR and bioinformatics analysis.
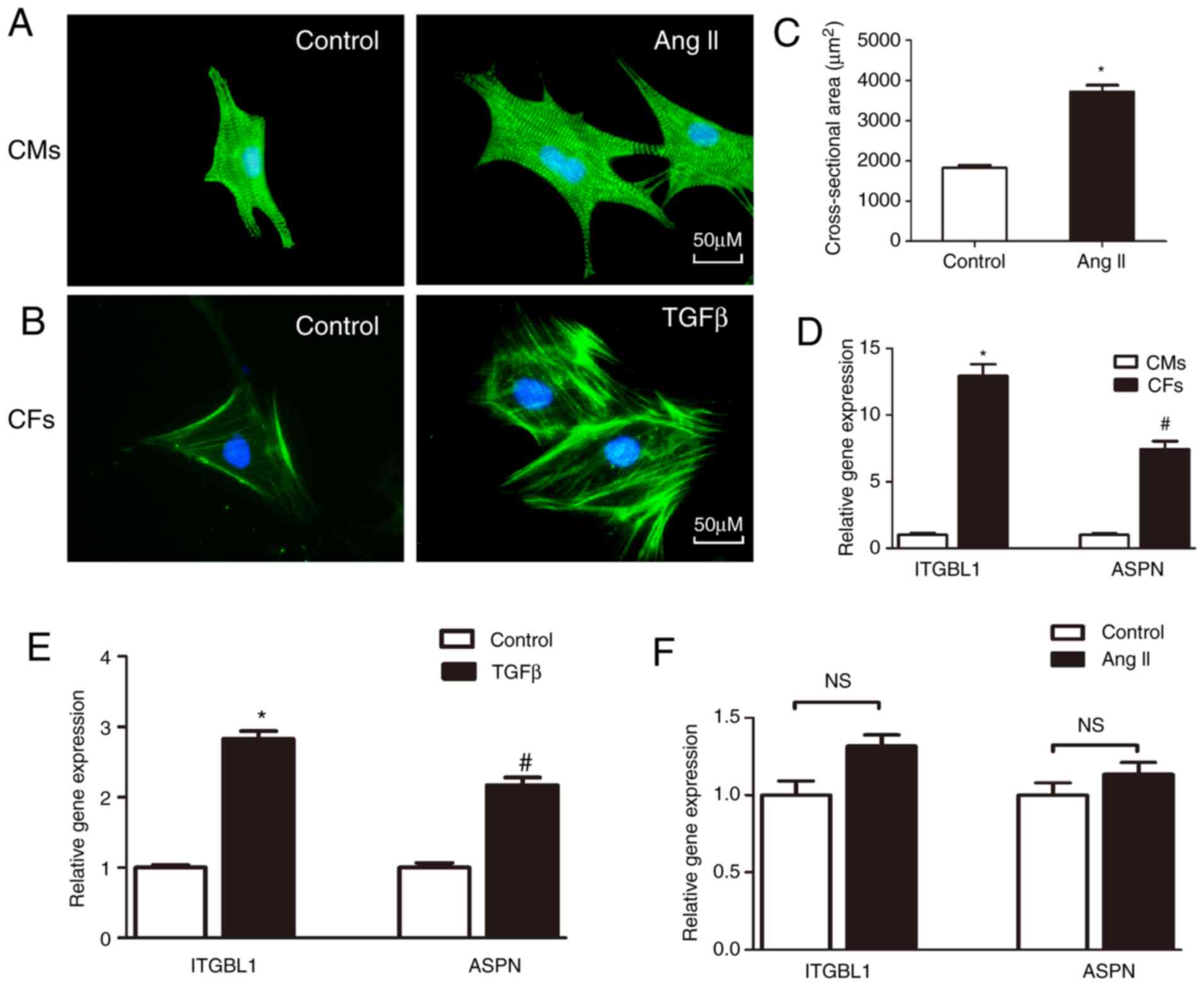
Expression of Itgbl1 and ASPN in CMs and
CFs
CMs and CFs were extracted from neonatal rats, and
stimulated by Ang II (1 µM) and TGF-β (10 ng/ml),
respectively. As shown in Fig. 7A and
B, CMs were stained with anti-α-actinin and CFs were stained
with anti-α-SMA. Ang II (1 µM) increased the cross-sectional
area of CMs (Fig. 7A and C), and
TGF-β (10 ng/ml) increased the fluorescence intensity of α-SMA in
CFs (Fig. 7B). The mRNA
expression of Itgbl1 and ASPN in CMs and CFs was also detected. It
was observed that Itgbl1 and ASPN were mainly expressed in CFs
(Fig. 7D), and their expression
was markedly increased following TGF-β stimulation (Fig. 7E), while the expression of Itgbl1
and ASPN in CMs was very low, and there was no significant
difference following stimulation by Ang II (Fig. 7F).
Discussion
In recent decades, ongoing research has unveiled
that cardiac remodeling plays an important role in the
pathophysiology of HF. In recent years, the emergence and rapid
development of gene chip (microarray) and high-throughput
sequencing technology has provided valuable targets for early
detection, diagnosis, treatment and prognosis of various diseases,
including HF. Comprehensive analysis of various microarray and
high-throughput sequencing technologies using bioinformatics
methods increases the sample size and reduces the impact of
research error on the results, thus effectively improving their
reliability.
In the present study, four microarray datasets
(GSE5500, GSE18224, GSE36074 and GSE56348) were collected and
downloaded for TAC-induced cardiac remodeling. These datasets were
analyzed using limma package of R software, common DEGs (defined as
log2FC>1 or log2FC<−1 and an adjusted
P-value <0.05) were obtained, and 24 commonly altered DEGs (23
upregulated and 1 downregulated) were identified in the first step.
Subsequently, these 24 DEGs were separated by GO terms into three
groups, namely cellular component, molecular functions and
biological process, using DAVID version 6.8. The result of GO term
analysis indicated that these 24 DEGs datasets were mainly enriched
in extracellular fibril organization, cell adhesion, wound healing,
elastic fiber assembly, negative regulation of cell growth and
cartilage development, extracellular region, proteinaceous
extracellular matrix, extracellular space, extracellular matrix,
basement membrane, extracellular exosome, growth factor activity,
heparin binding and calcium ion binding. These functions are
closely associated with the development of cardiac remodeling.
In the third step, a DEG PPI network complex was
developed and a total of 18 DEGs (all upregulated) of the 24
commonly altered DEGs were filtered into this DEG complex,
including (in descending order of the number of nodes) Postn, Lox,
Ctgf, Itgbl1, Aspn, Fstl1, Mfap5, Acta1, Nppa, Tgfb2, Timp1,
Col8a1, Ltbp2, Mfap4, Thbs4, Cilp, Pamr1 and Frzb.
Of these 24 DEGs, 9 (Postn, Lox, Ctgf, ANP, Acta1,
Timp1, Tgfb2, Clip and Thbs4) have been previously reported to be
directly involved in the development of cardiac remodeling
(22-30); therefore, the level of expression
of these genes was not examined in subsequent analyses. To verify
the expression of the remaining 15 DEGs, a TAC-induced pressure
overload model was constructed to elucidate the mechanism
underlying cardiac remodeling. To verify whether the cardiac
pathological remodeling model in mice was successfully established,
echocardiography was used to detect cardiac function in the
experimental and control groups, and gross morphological evaluation
of the hearts was performed by H&E staining. RT-PCR was used to
verify that the expression of Itgbl1, Aspn, Fstl1, Mfap5, Col8a1,
Ltbp2, Mfap4, Pamr1, Cnksr1, Aqp8, Meox1, Gdf15 and Srpx was
upregulated and that of Retnla was downregulated in the mouse model
of cardiac pathological remodeling, which was consistent with the
results of the bioinformatics analysis. Frzb was not significantly
differentially expressed, as determined by RT-PCR. The reason for
this inconsistency is that the sensitivity of different detection
methods varies.
The Itgbl1 gene encodes a β-integrin-related
protein. The Itgbl1 protein is an extracellular matrix protein that
promotes ovarian cancer cell migration and adhesion through Wnt/PCP
signaling and the FAK/SRC pathway (30). The Itgbl1 protein may promote bone
metastasis of breast cancer by activating the TGF-β signaling
pathway (31). The Wnt/PCP,
FAK/SRC and TGF-β signaling pathways play an important role in the
process of cardiac remodeling. Similarly, Itgbl1 is a key regulator
of fibrosis in patients with hepatitis B virus-related liver
fibrosis (32). This evidence
suggests that Itgbl1 may be involved in the pathogenesis of cardiac
remodeling. Aspn encodes Asporin, a cartilage extracellular protein
that belongs to the small leucine-rich proteoglycan family. Asporin
regulates chondrogenesis through regulating TGF-β signaling; it can
also bind to collagen and calcium and may induce collagen
mineralization (33). There are
currently no studies on the effects of Aspn on remodeling of
various tissues.
The Fstl1 gene encodes a protein similar to
follistatin. A previous study reported that the Fstl1 levels in a
patient with asthma were associated with increased smooth muscle
mass, and suggested that Fstl1 may contribute to airway remodeling
in such patients (34). In
addition, another study also confirmed that Fstl1 is involved in
TGF-β1-mediated pulmonary fibrosis (35). Fstl1 may also be used as a
potential mediator of exercise-induced cardioprotection
post-myocardial infarction (36).
However, there has yet been no study that elucidates whether Fstl1
participates in pressure overload-mediated cardiac remodeling.
The microfibril-associated protein (Mfap)4 and Mfap5
genes encode extracellular matrix proteins that are involved in
cell adhesion and intercellular interaction. A number of studies
have confirmed the involvement of Mfap4 in vascular remodeling
after vascular injury and airway remodeling induced by bronchial
asthma (37,38). Furthermore, the expression of
Mfap4 in the serum and liver of patients with liver cirrhosis was
found to be significantly increased (39). Mfap5 is associated with
obesity-associated adipose tissue and extracellular matrix
remodeling and regulates adipose tissue inflammation (40).
Collagen type VIII alpha 1 chain (Col8a1) encodes
one of the two α chains of type VIII collagen, which is a key
component of the basement membrane of the corneal endothelium. A
previous study demonstrated that Col8a1 mediates vessel wall
remodeling following arterial injury and fibrous cap formation in
atherosclerosis. The latent transforming growth factor beta binding
protein 2 (Ltbp2) gene encodes a protein that belongs to the latent
TGF-β-binding protein (LTBP) family, and this protein shares
similarities with the fibrillins. Ltbp2 is a potential prognostic
blood biomarker that may reflect the level of differentiation of
lung fibroblasts into myofibroblasts in idiopathic pulmonary
fibrosis (41). Peptidase domain
containing associated with muscle regeneration 1 (Pamr1) is a
putative tumor suppressor that is frequently inactivated by
promoter hyper-methylation in breast cancer tissues (42). However, there is little research
on Pamr1, so its role remains elusive.
The connector enhancer of kinase suppressor of Ras 1
(Cnksr1) gene encodes a protein containing several motifs involved
in PPI. The Cnksr1 protein can mediate the association among
different signaling pathways (43). Similar to the Pamr1 gene, there is
little research on the exact role of the Cnksr1 gene. The aquaporin
8 (Aqp8) gene encodes a protein that belongs to a family of small
integral membrane proteins, and functions as a water channel
(44). The sushi
repeat-containing protein X-linked (Srpx) gene encodes the Srpx
protein, which is involved in phagocytosis during disk shedding,
cell adhesion to cells other than the pigment epithelium, or signal
transduction (45). At present,
it remains unknown whether Aqp8 and Srpx are involved in the
remodeling of cardiac or other tissues. The mesenchyme homeobox 1
(Meox1) gene encodes a member of a subfamily of antennapedia-like
homeobox-containing proteins. Meox1 is part of a regulatory circuit
that serves an essential, non-redundant function in the maintenance
of rostrocaudal sclerotome polarity and leads to remodeling of the
cranio-cervical joints of the axial skeleton (46). Meox1 promotes vascular smooth
muscle cell phenotypic modulation and balloon injury-induced
vascular remodeling via regulating the FAK-ERK1/2 signaling cascade
(47).
The growth differentiation factor 15 (Gdf15) gene
encodes a secreted ligand of the TGF-β superfamily of proteins,
which bind various TGF-β receptors (TGF-βR) and leads to activation
of Smad family transcription factors. The TGF-β/Smad signaling
pathway is closely associated with cardiac remodeling.
Resistin-like alpha (Retnla) is also referred to as hypoxia-induced
mitogenic factor (Himf). In a model of lung inflammation, Retnla
knockout led to lung inflammation and higher Th2 cell cytokine
production compared with that observed in wild-type mice. Retnla
strongly activates Akt phosphorylation, and treatment with Retnla
has been shown to result in a significant reduction of apoptosis in
cultured embryonic lung cells (48,49). Retnla is involved in immune
response-induced pulmonary vascular remodeling and enhanced
inflammatory response typically observed after intermittent
ovalbumin challenge (50).
There were certain limitations to the present study.
First, the amount of data obtained from the GEO database is not
sufficiently comprehensive, as some of the data samples are too
small to screen for common DEGs, and were thus excluded. Second,
DEGs represent only one type of molecular changes that occur in the
mouse model of TAC-induced cardiac remodeling. Other factors,
including non-coding RNAs (such as microRNAs, long non-coding RNAs
and circular RNAs), gene mutations and metabolomic changes, should
also be considered to provide a comprehensive understanding of the
development of cardiac remodeling. Third, of the 15 DEGs, only the
protein levels of Itgbl1 and Asporin were verified by western
blotting and immunohistochemistry. Fourth, the findings of the
present study only indicated that the expression of some genes was
altered during cardiac remodeling, but did not elucidate the
underlying mechanism. This study is only part of our research group
on HF. Through bioinformatics analysis, the DEGs were screened and
preliminarily verified. By screening the candidate genes, related
gene knockout and overexpression mouse models are constructed in
order to provide a solid early basis for exploring the pathogenesis
and treatment of HF in the future.
In conclusion, the present study identified DEGs in
mouse hearts with TAC-induced cardiac remodeling and sham samples
by bioinformatics analysis, thereby indicating the crucial role of
DEGs in the process of cardiac remodeling. The study findings
provide a set of candidate genes for future investigation of the
mechanisms and selection of biomarkers for cardiac remodeling. In
future research, knockout or transgenic mice will be used to
further elucidate the pathophysiological mechanisms underlying the
involvement of these genes in cardiac remodeling in vivo and
in vitro.
Acknowledgments
Not applicable.
Funding
The present was supported by grants from the
National Natural Science Foundation of China (no. 81470516) and the
Key Project of the National Natural Science Foundation (no.
81530012).
Availability of data and materials
The datasets generated and/or analyzed during the
present study are available from the corresponding author on
reasonable request. The URLs to all datasets used in this study as
follows: GSE5500 datasets: https://www.ncbi.nlm.nih.gov/sites/GDSbrowser?acc=GDS2316;
GSE18224 datasets: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE18224;
GSE36074 datasets: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE36074;
GSE56348 datasets: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE56348.
Authors' contributions
HBW and QZT conceived and designed the study; HBW,
KY and RH performed data analysis; MX, DF, MXL and LBL performed
the experiments; SHH and HMW wrote the paper. All the authors have
read and approved the final version of this manuscript for
publication.
Ethics approval and consent to
participate
All experiments have been approved by the Ethics
Committee of The Renmin Hospital of Wuhan University (Wuhan,
China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Yusuf S, Wood D, Ralston J and Reddy KS:
The World Heart Federation's vision for worldwide cardiovascular
disease prevention. Lancet. 386:399–402. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sandri M, Viehmann M, Adams V, Rabald K,
Mangner N, Höllriegel R, Lurz P, Erbs S, Linke A, Kirsch K, et al:
Chronic heart failure and aging-effects of exercise training on
endothelial function and mechanisms of endothelial regeneration:
Results from the leipzig exercise intervention in chronic heart
failure and aging (LEICA) study. Eur J Prev Cardiol. 23:349–358.
2016. View Article : Google Scholar
|
|
3
|
Schirone L, Forte M, Palmerio S, Yee D,
Nocella C, Angelini F, Pagano F, Schiavon S, Bordin A, Carrizzo A,
et al: A review of the molecular mechanisms underlying the
development and progression of cardiac remodeling. Oxid Med Cell
Longev. 2017:39201952017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wu QQ, Xiao Y, Yuan Y, Ma ZG, Liao HH, Liu
C, Zhu JX, Yang Z, Deng W and Tang QZ: Mechanisms contributing to
cardiac remodelling. Clin Sci (Lond). 131:2319–2345. 2017.
View Article : Google Scholar
|
|
5
|
Keeler AM and Flotte TR: Cell and gene
therapy for genetic diseases: Inherited disorders affecting the
lung and those mimicking sudden infant death syndrome. Hum Gene
Ther. 23:548–556. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Barbosa C, Peixeiro I and Romao L: Gene
expression regulation by upstream open reading frames and human
disease. PLoS Genet. 9:e10035292013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Patel A and Cheung SW: Application of DNA
Microarray to clinical diagnostics. Methods Mol Biol. 1368:111–132.
2016. View Article : Google Scholar
|
|
8
|
Hanif W, Alex L, Su Y, Shinde AV, Russo I,
Li N and Frangogiannis NG: Left atrial remodeling, hypertrophy, and
fibrosis in mouse models of heart failure. Cardiovasc Pathol.
30:27–37. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Guo Y, Bao Y, Ma M and Yang W:
Identification of key candidate genes and pathways in colorectal
cancer by integrated bioinformatical analysis. Int J Mol Sci.
18:E7222017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bisping E, Ikeda S, Kong SW, Tarnavski O,
Bodyak N, McMullen JR, Rajagopal S, Son JK, Ma Q, Springer Z, et
al: Gata4 is required for maintenance of postnatal cardiac function
and protection from pressure overload-induced heart failure. Proc
Natl Acad Sci USA. 103:14471–14476. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Fliegner D, Schubert C, Penkalla A, Witt
H, Kararigas G, Dworatzek E, Staub E, Martus P, Ruiz Noppinger P,
Kintscher U, et al: Female sex and estrogen receptor-beta attenuate
cardiac remodeling and apoptosis in pressure overload. Am J Physiol
Regul Integr Comp Physiol. 298:R1597–R1606. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Skrbic B, Bjørnstad JL, Marstein HS,
Carlson CR, Sjaastad I, Nygård S, Bjørnstad S, Christensen G and
Tønnessen T: Differential regulation of extracellular matrix
constituents in myocardial remodeling with and without heart
failure following pressure overload. Matrix Biol. 32:133–142. 2013.
View Article : Google Scholar
|
|
13
|
Lai L, Leone TC, Keller MP, Martin OJ,
Broman AT, Nigro J, Kapoor K, Koves TR, Stevens R, Ilkayeva OR, et
al: Energy metabolic reprogramming in the hypertrophied and early
stage failing heart: A multisystems approach. Circ Heart Fail.
7:1022–1031. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Popik OV, Saik OV, Petrovskiy ED, Sommer
B, Hofestädt R, Lavrik IN and Ivanisenko VA: Analysis of signaling
networks distributed over intracellular compartments based on
protein-protein interactions. BMC Genomics. 15(Suppl 12): S72014.
View Article : Google Scholar
|
|
16
|
Yuan YP, Ma ZG, Zhang X, Xu SC, Zeng XF,
Yang Z, Deng W and Tang QZ: CTRP3 protected against
doxorubicin-induced cardiac dysfunction, inflammation and cell
death via activation of Sirt1. J Mol Cell Cardiol. 114:38–47. 2018.
View Article : Google Scholar
|
|
17
|
Ma ZG, Dai J, Zhang WB, Yuan Y, Liao HH,
Zhang N, Bian ZY and Tang QZ: Protection against cardiac
hypertrophy by genipo-side involves the GLP-1 receptor/AMPKα
signalling pathway. Br J Pharmacol. 173:1502–1516. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang X, Ma ZG, Yuan YP, Xu SC, Wei WY,
Song P, Kong CY, Deng W and Tang QZ: Rosmarinic acid attenuates
cardiac fibrosis following long-term pressure overload via
AMPKα/Smad3 signaling. Cell Death Dis. 9:1022018. View Article : Google Scholar
|
|
19
|
Wang HB and Yang J, Ding JW, Chen LH, Li
S, Liu XW, Yang CJ, Fan ZX and Yang J: RNAi-mediated downregulation
of CD47 protects against ischemia/reperfusion-induced myocardial
damage via activation of eNOS in a rat model. Cell Physiol Biochem.
40:1163–1174. 2016. View Article : Google Scholar
|
|
20
|
Schorb W, Booz GW, Dostal DE, Conrad KM,
Chang KC and Baker KM: Angiotensin II is mitogenic in neonatal rat
cardiac fibroblasts. Circ Res. 72:1245–1254. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Toth A, Jeffers JR, Nickson P, Min JY,
Morgan JP, Zambetti GP and Erhardt P: Targeted deletion of Puma
attenuates cardiomyocyte death and improves cardiac function during
ischemia-reperfusion. Am J Physiol Heart Circ Physiol. 291:H52–H60.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Azhar M, Brown K, Gard C, Chen H, Rajan S,
Elliott DA, Stevens MV, Camenisch TD, Conway SJ and Doetschman T:
Transforming growth factor Beta2 is required for valve remod-eling
during heart development. Dev Dyn. 240:2127–2141. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Takawale A, Zhang P, Patel VB, Wang X,
Oudit G and Kassiri Z: Tissue inhibitor of matrix
metalloproteinase-1 promotes myocardial fibrosis by mediating
CD63-integrin β1 interaction. Hypertension. 69:1092–1103. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Schwartz K, de la Bastie D, Bouveret P,
Oliviéro P, Alonso S and Buckingham M: Alpha-skeletal muscle actin
mRNA's accumulate in hypertrophied adult rat hearts. Circ Res.
59:551–555. 1986. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Deschepper CF, Masciotra S, Zahabi A,
Boutin-Ganache I, Picard S and Reudelhuber TL: Functional
alterations of the Nppa promoter are linked to cardiac ventricular
hypertrophy in WKY/WKHA rat crosses. Circ Res. 88:223–228. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu X, Gai Y, Liu F, Gao W, Zhang Y, Xu M
and Li Z: Trimetazidine inhibits pressure overload-induced cardiac
fibrosis through NADPH oxidase-ROS-CTGF pathway. Cardiovasc Res.
88:150–158. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Hu C, Dandapat A, Chen J, Fujita Y, Inoue
N, Kawase Y, Jishage K, Suzuki H, Sawamura T and Mehta JL: LOX-1
deletion alters signals of myocardial remodeling immediately after
ischemia-reperfusion. Cardiovasc Res. 76:292–302. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Kaur H, Takefuji M, Ngai CY, Carvalho J,
Bayer J, Wietelmann A, Poetsch A, Hoelper S, Conway SJ, Möllmann H,
et al: Targeted ablation of periostin-expressing activated
fibroblasts prevents adverse cardiac remodeling in mice. Circ Res.
118:1906–1917. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang CL, Zhao Q, Liang H, Qiao X, Wang
JY, Wu D, Wu LL and Li L: Cartilage intermediate layer protein-1
alleviates pressure overload-induced cardiac fibrosis via
interfering TGF-β1 signaling. J Mol Cell Cardiol. 116:135–144.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Sun L, Wang D, Li X, Zhang L, Zhang H and
Zhang Y: Extracellular matrix protein ITGBL1 promotes ovarian
cancer cell migration and adhesion through Wnt/PCP signaling and
FAK/SRC pathway. Biomed Pharmacother. 81:145–151. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Li XQ, Du X, Li DM, Kong PZ, Sun Y, Liu
PF, Wang QS and Feng YM: ITGBL1 is a Runx2 transcriptional target
and promotes breast cancer bone metastasis by activating the TGFβ
signaling pathway. Cancer Res. 75:3302–3313. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang M, Gong Q, Zhang J, Chen L, Zhang Z,
Lu L, Yu D, Han Y, Zhang D, Chen P, et al: Characterization of gene
expression profiles in HBV-related liver fibrosis patients and
identification of ITGBL1 as a key regulator of fibrogenesis. Sci
Rep. 7:434462017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kaliakatsos M, Tzetis M, Kanavakis E,
Fytili P, Chouliaras G, Karachalios T, Malizos K and Tsezou A:
Asporin and knee osteoarthritis in patients of Greek origin.
Osteoarthritis Cartilage. 14:609–611. 2006. View Article : Google Scholar
|
|
34
|
Liu Y, Liu T, Wu J, Li T, Jiao X, Zhang H,
Zhao J, Wang J, Liu L, Cao L, et al: The Correlation between FSTL1
expression and airway remodeling in asthmatics. Mediators Inflamm.
2017:79184722017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zheng X, Qi C, Zhang S, Fang Y and Ning W:
TGF-β1 induces Fstl1 via the Smad3-c-Jun pathway in lung
fibroblasts. Am J Physiol Lung Cell Mol Physiol. 313:L240–L251.
2017. View Article : Google Scholar
|
|
36
|
Xi Y, Gong DW and Tian Z: FSTL1 as a
potential mediator of exercise-induced cardioprotection in
post-myocardial infarction rats. Sci Rep. 6:324242016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Schlosser A, Pilecki B, Hemstra LE,
Kejling K, Kristmannsdottir GB, Wulf-Johansson H, Moeller JB,
Füchtbauer EM, Nielsen O, Kirketerp-Møller K, et al: MFAP4 promotes
vascular smooth muscle migration, proliferation and accelerates
neointima formation. Arterioscler Thromb Vasc Biol. 36:122–133.
2016. View Article : Google Scholar
|
|
38
|
Pilecki B, Schlosser A, Wulf-Johansson H,
Trian T, Moeller JB, Marcussen N, Aguilar-Pimentel JA, de Angelis
MH, Vestbo J, Berger P, et al: Microfibrillar-associated protein 4
modulates airway smooth muscle cell phenotype in experimental
asthma. Thorax. 70:862–872. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Mölleken C, Sitek B, Henkel C, Poschmann
G, Sipos B, Wiese S, Warscheid B, Broelsch C, Reiser M, Friedman
SL, et al: Detection of novel biomarkers of liver cirrhosis by
proteomic analysis. Hepatology. 49:1257–1266. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Vaittinen M, Kolehmainen M, Ryden M,
Eskelinen M, Wabitsch M, Pihlajamäki J, Uusitupa M and Pulkkinen L:
MFAP5 is related to obesity-associated adipose tissue and
extracellular matrix remodeling and inflammation. Obesity (Silver
Spring). 23. pp. 1371–1378. 2015, View Article : Google Scholar
|
|
41
|
Enomoto Y, Matsushima S, Shibata K,
Aoshima Y, Yagi H, Meguro S, Kawasaki H, Kosugi I, Fujisawa T,
Enomoto N, et al: LTBP2 is secreted from lung myofibroblasts and is
a potential biomarker for idiopathic pulmonary fibrosis. Clin Sci
(Lond). 132:1565–1580. 2018. View Article : Google Scholar
|
|
42
|
Lo PH, Tanikawa C, Katagiri T, Nakamura Y
and Matsuda K: Identification of novel epigenetically inactivated
gene PAMR1 in breast carcinoma. Oncol Rep. 33:267–273. 2015.
View Article : Google Scholar
|
|
43
|
Fischer A, Muhlhauser WWD, Warscheid B and
Radziwill G: Membrane localization of acetylated CNK1 mediates a
positive feedback on RAF/ERK signaling. Sci Adv. 3:e17004752017.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Bestetti S, Medraño-Fernandez I, Galli M,
Ghitti M, Bienert GP, Musco G, Orsi A, Rubartelli A and Sitia R: A
persulfidation-based mechanism controls aquaporin-8 conductance.
Sci Adv. 4:eaar57702018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Meindl A, Carvalho MR, Herrmann K, Lorenz
B, Achatz H, Lorenz B, Apfelstedt-Sylla E, Wittwer B, Ross M and
Meitinger T: A gene (Srpx) encoding a sushi-repeat-containing
protein is deleted in patients with X-linked retinitis-pigmentosa.
Hum Mol Genet. 4:2339–2346. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Skuntz S, Mankoo B, Nguyen MT, Hustert E,
Nakayama A, Tournier-Lasserve E, Wright CV, Pachnis V, Bharti K and
Arnheiter H: Lack of the mesodermal homeodomain protein MEOX1
disrupts sclerotome polarity and leads to a remodeling of the
craniocervical joints of the axial skeleton. Dev Biol. 332:383–395.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhang Wu B, Zhu L, Zhang YH, Zheng YE,
Yang F, Guo JY, Li LY, Wang XY, Tang LJM, et al:
Mesoderm/mesenchyme homeobox gene l promotes vascular smooth muscle
cell phenotypic modulation and vascular remodeling. Int J Cardiol.
251:82–89. 2018. View Article : Google Scholar
|
|
48
|
Pesce JT, Ramalingam TR, Wilson MS,
Mentink-Kane MM, Thompson RW, Cheever AW, Urban JF Jr and Wynn TA:
Retnla (relmalpha/fizz1) suppresses helminth-induced Th2-type
immunity. PLoS Pathog. 5:e10003932009. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Nair MG, Du Y, Perrigoue JG, Zaph C,
Taylor JJ, Goldschmidt M, Swain GP, Yancopoulos GD, Valenzuela DM,
Murphy A, et al: Alternatively activated macrophage-derived
RELM-{alpha} is a negative regulator of type 2 inflammation in the
lung. J Exp Med. 206:937–952. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Angelini DJ, Su Q, Yamaji-Kegan K, Fan C,
Skinner JT, Poloczek A, El-Haddad H, Cheadle C and Johns RA:
Hypoxia-induced mitogenic factor (HIMF/FIZZ1/RELMα) in chronic
hypoxia- and antigen-mediated pulmonary vascular remodeling. Respir
Res. 14:12013. View Article : Google Scholar
|