Introduction
Ovarian cancer is the most lethal gynecological
malignancy worldwide; in Japan alone it is estimated that there
were 7913 new cases of ovarian cancer and 4415 cases of mortality
due to this disease in 2006 (Center for Cancer Control and
Information Services, National Cancer Center, Japan). Overall,
ovarian cancer patients respond to cytoreductive surgery and
platinum and taxane based combination chemotherapy, however,
advanced cases have a high recurrence rate and the 5-year survival
rate has remained largely unchanged since the 1980s, being close to
30% (1). This, however, is skewed,
given the mass of recent evidence suggesting that histological
subtypes of ovarian cancer represent unique diseases, that the
majority of cases (50–70%) are of serous histology, and that all
ovarian carcinoma subtypes are still treated with a ‘one size fits
all’ approach. Serous cancers seem to initially respond well to
platinum and taxane chemotherapy, whereas other subtypes respond
poorly or not at all (2–5). These data are finally initiating
separate clinical trials for unique subtypes such as clear cell
carcinoma (CCC) (6) and mucinous
adenocarcinoma (MA) (7). Even
within histologically defined groups, such as high-grade serous
tumors, molecular subtypes are emerging. Defining molecular
mechanisms involved in the development and progression of ovarian
cancer and their interaction with host defenses is integral to the
development of novel therapeutic approaches and successful
treatment stratification.
Factors intrinsic to the tumor and the host
influence progression, host defense against tumors is controlled by
several immunological mediators that play an important role in the
host-tumor immune system conflict (8). The host-tumor immune response is in
part regulated by CD4 helper T-lymphocytes, Th1 and Th2 cells. Th1
cells produce proinflammatory cytokines and drive the cell-mediated
immune response, while Th2 cells regulate humoral immunity by
expressing anti-inflammatory cytokines. Alterations of cytokine
expression and an imbalance in Th1/Th2 cytokine response have
previously been observed in ovarian carcinomas (9–11).
Serous adenocarcinoma (SA) is frequently subject to infiltration by
activated T cells (TILs; tumor infiltrating lymphocytes), and
patients with dense infiltrates of CD3+ CD8+
T cells had a more favorable prognosis while infiltration of other
cell types, including CD4+ CD25+
FoxP3+ regulatory T cells, opposed antitumor immunity
(9).
Seike et al reported that a unique cytokine
gene expression signature of noncancerous lung tissue and
corresponding tumor tissue in lung adenocarcinoma predicted
metastasis and disease progression (12). The prognostic signature consisted
mainly of cytokine genes that were expressed in Th1 and Th2 cells.
Taken together, these findings suggest that alterations in cytokine
gene expression of tumors could be involved in the pathogenesis of
other types of cancer, including ovarian cancer.
In this study, we sought to clarify whether the
cytokine gene expression profile could have clinical associations
with ovarian cancer by focusing on the expression profile of 12
cytokine genes in 50 ovarian cancers. We found that a cytokine gene
expression signature of ovarian cancer could distinguish the
histological subtype, and a unique expression pattern found in CCC
may be involved in the pathogenesis of this ovarian cancer subtype.
We further evaluated the biological significance of IL-6
overexpression in CCC using siRNA and found that the drug
resistance phenotype in CCC might be regulated by the activated
IL-6 signaling pathway including Stat3 activation.
Materials and methods
Clinical samples and cell lines
Tumor specimens were surgically obtained from
patients with primary ovarian carcinoma who were treated at the
Department of Obstetrics and Gynecology, The Jikei University
School of Medicine. The Jikei University School of Medicine Ethics
Review Committee approved the study protocol and informed consent
was obtained from all patients. Tumors were staged in accordance
with the International Federation of Gynecology and Obstetrics
(FIGO) system (1988). Fifty tumor RNA specimens that passed quality
control standards were used to identify a gene signature. Clinical
and pathological characteristics of the 50 patients are shown in
Table I. Forty-seven patients
received first-line platinum-based chemotherapy.
 | Table IClinical and pathological
characteristics of the 50 ovarian cancer patients. |
Table I
Clinical and pathological
characteristics of the 50 ovarian cancer patients.
| Parameters | No. of
patients | % of patients |
|---|
| Patient age | | |
| ≤60 years | 41 | 82 |
| >60 years | 9 | 18 |
| FIGO stage | | |
| I | 19 | 38 |
| II | 3 | 6 |
| III | 22 | 44 |
| IV | 6 | 12 |
| Histological
type | | |
| SA | 24 | 48 |
| EA | 5 | 10 |
| CCC | 21 | 42 |
| Residual tumor | | |
| ≤1 cm | 32 | 64 |
| >1 cm | 18 | 36 |
Seven human CCC cell lines (JHOC-5, JHOC-7, JHOC-8,
JHOC-9, HAC-2, RMG-I, and RMG-II) and five human ovarian non-CCC
cell lines were used in this study. JHOC-5, JHOC-7, JHOC-8, and
JHOC-9 were obtained from Riken BioResource center (Tsukuba,
Japan). HAC-2 was kindly provided by Dr M. Nishida (Tsukuba
University, Tsukuba, Japan). RMG-I and RMG-II were kindly provided
by Dr D. Aoki (Keio University, Tokyo, Japan). A2780
(undifferentiated carcinoma) was provided by Dr E. Reed (NCI,
Bethesda, MD) and 2008 (SA) was provided by Dr S.B. Howell (UCSD,
San Diego, CA). SKOV3 (SA), MCAS (MA), and Tyk-nu (undifferentiated
carcinoma) were obtained from ATCC (Rockville, MD). RMG-I and
RMG-II were maintained in Ham’s F12 medium (Sigma-Aldrich, Tokyo,
Japan) with 10% fetal bovine serum (FBS), and the other cells were
cultured in RPMI-1640 (Sigma-Aldrich) containing 10% FBS.
Quantitative reverse
transcription-polymerase chain reaction analysis
All tissue was freshly collected during surgery and
stored at −80°C. Cryostat sections containing >80% cancer cells
were prepared as tumor specimens. Total RNA was isolated with
TRIzol reagent (Invitrogen, Carlsbad, CA). Total RNA (3 μg)
was converted to complementary DNA (cDNA) with random hexamers and
SuperScript III First-Strand Synthesis kit (Invitrogen). The cDNAs
were then used for qRT-PCR analysis of 16-gene expression profile.
Normal ovarian tissue from a patient who had undergone surgical
resection was used as a reference for each tissue sample. Several
studies have suggested that normal ovarian tissue and even surface
ovarian epithelium may not be the tissue of origin for many ovarian
epithelial malignancies (13),
therefore the use of ovarian tissue as a reference for qRT-PCR is
arbitrary and not meant for a direct comparison of subtypes to
normal ovarian tissue but rather for an intercomparison of tumor
samples. The expression profiles of 12 cytokine genes [i.e.,
interleukin 1α (IL-1α), interleukin 1β (IL-1β),
interleukin 2 (IL-2), interleukin 4 (IL-4),
interleukin 5 (IL-5), interleukin 8 (IL-8),
interleukin 10 (IL-10), interleukin 12 p35
(IL-12p35), interleukin 12 p40 (IL-12p40),
interleukin 15 (IL-15), interferon γ (IFN-γ)
and tumor necrosis factor-α (TNF-α)] were quantified with
the use of TaqMan Cytokine Gene Expression Plates (Applied
Biosystems, Foster City, CA). Additional individual Taqman assays
(Applied Biosystems) were also used to measure expression of
interleukin 6 (IL-6), major histocompatibility complex (MHC)
class II antigen DR α (HLA-DRA), MHC class II antigen DP
α 1 (HLA-DPA1), and colony stimulating factor 1
(CSF1). All PCR reactions were performed with a StepOnePlus
Real-Time PCR System (Applied Biosystems), Human 18S ribosomal RNA
(rRNA) labeled with VIC reporter dye (Applied Biosystems) was used
as an endogenous control. Gene expression was quantified using the
comparative method (2−ΔΔCT), where CT =
threshold cycle, ΔΔCT = (CT cytokine −
CT 18S rRNA) − (CT reference − CT
18S rRNA), as previously described (14). To ensure RNA of sufficient quality
and eliminate spurious amplification artifacts, all samples were
required to have average CT values for cytokine genes >35
cycles, as previously described (12).
Small interfering RNA and cytotoxicity
assays
Small interfering RNA (siRNA) transfection was
performed using Lipofectamine™ RNAiMAX (Invitrogen). Briefly, HAC-2
and JHOC-5 cells were seeded at 1.5×105 in a 6 cm dish,
validated Stealth RNAi™ siRNA for IL-6 (Invitrogen), or stealth
RNAi siRNA negative control (Invitrogen), were transfected at a
final concentration of 2.5 nM. After 24 h of transfection, cells
were re-seeded in 96-well plates with various concentrations of
cisplatin or paclitaxel. In vitro cytotoxicity was measured
after 96 h by means of the MTS assay using CellTiter 96 AQueous One
Solution (Promega, Madison, WI). MTS solution was added to each
well and incubated for 4 h. Absorbance was measured at 490 nm using
a microplate reader. Data were collected as the average absorbance
of three wells in any one experiment and is presented from three
independent experiments; mean ± 1 standard deviation.
Western blot analysis
Total cell lysates were prepared in 1X RIPA lysis
buffer and protein concentration was determined by the DC Protein
Assay (Bio-Rad, Hercules, CA). Total protein (40 μg) was
resolved on gradient NuPage 4–12% Bis-Tris gels (Invitrogen) and
immunoblotted with specific antibodies: anti-Stat3 (clone 79D7;
1:2000), pStat3 (clone D3A7; 1:500), and β-actin (clone 13E5;
1:1000) from Cell Signaling Technology (Beverly, MA); anti-IL-6Rα
(clone C-20; 1:1000) from Santa Cruz Biotechnology (Santa Cruz,
CA). All blots were incubated with primary antibodies diluted in
TBS with 0.1% Tween-20 and 5% bovine serum albumin overnight at 4
°C with gentle agitation. Horseradish peroxidase (HRP)-conjugated
secondary antibody (Cell Signaling Technology; 1:10000) was diluted
in TBS with 0.1% Tween-20 and 5% nonfat milk for 1 h at room
temperature with gentle agitation. Positive immunoreactions were
detected using ImmunoStar LD chemiluminescence system (Wako, Tokyo,
Japan).
Statistical analysis
Unsupervised hierarchical clustering analysis was
performed with Gene Cluster 3.0 (http://genexpress.stanford.edu/tutorials/cluster_view.html;
Stanford University, Palo Alto, CA) and visualized with Tree View
(http://genexpress.stanford.edu/tutorials/tree_view.html;
Stanford University). Tissue samples and genes were analyzed for
the calculated centered correlation distance and average linkage
according to the ratios of their abundance to the median abundance
of all genes among all samples as previously described (12). The correlation between sample
clusters and clinical parameters was analyzed using the Fisher’s
exact test, with p<0.05 considered to indicate statistically
significant differences.
In class comparison analysis using BRB-ArrayTools,
we identified genes that were differently expressed among groups
using t-test with univariate permutation tests to evaluate the
significance of individual genes and correct for multiple
hypothesis testing. The proportion of the permutations of the class
label giving a t-test p-value as small as obtained with the true
class labels is the univariate permutation p-value for that gene.
The false discovery rate (FDR) associated with a row of the table
is an estimate of the proportion of the genes with univariate
p-values less than or equal to the one in that row that represent
false positives.
A 2-sided Student’s t-test was used to evaluate the
sensitivity of cytotoxic agents in ovarian cancer cell lines with
p<0.05 considered to indicate statistically significant
differences.
Results
Hierarchical clustering analysis of
ovarian cancer
To investigate the role of cytokine gene expression
in ovarian cancer, we first analyzed the expression of 16 cytokine
genes in tumor specimens from 50 primary ovarian cancers. These 16
genes were previously shown to be part of a unique
inflammation/immune response-related signature in lung
adenocarcinoma patients (12).
Expression of IL-2, IL-4, IL-5, and
IL-12p40 genes was below the detectable threshold across our
sample set and were therefore not carried forward in subsequent
analysis. Unsupervised hierarchical clustering analysis of the 50
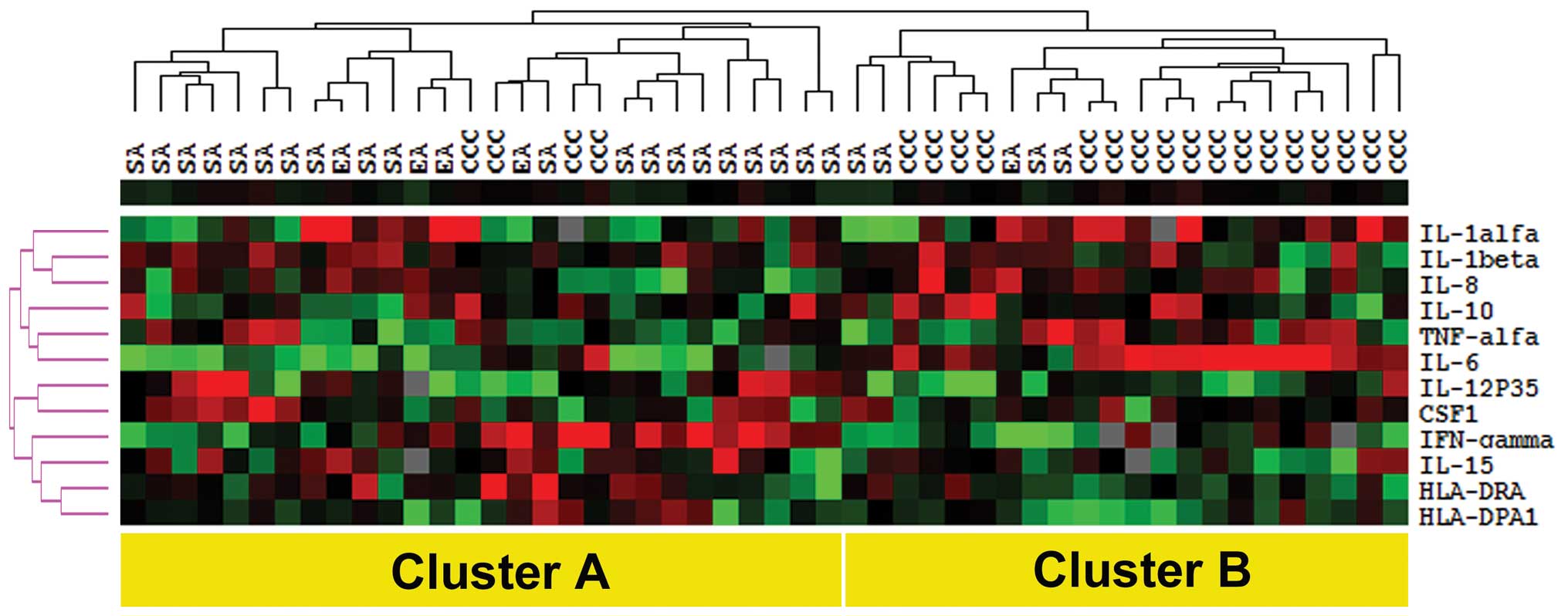
ovarian cancer samples, which was based on the similarities in
expression patterns of the remaining 12-gene panel, revealed two
distinct main clusters, namely cluster A (n=28) and cluster B
(n=22) (Fig. 1). Examination of
the relationship between the two clusters and clinicopathological
features revealed statistically significant differences between
cluster A and cluster B with respect to histological type (Table II). Cluster A consisted of 20 SA
cases (83%), 4 EA cases (80%) and 4 CCC cases (19%), and the
cluster B included 4 cases of SA (17%), one EA case (20%) and 17
cases of CCC (81%). In addition, there was a marginally significant
difference between the two clusters and FIGO stage. However, there
was no statistically significant difference in age, residual tumor
size, and patient prognosis between the clusters (data not shown).
Of particular note, cases in cluster B showed higher expression of
Th2-related cytokine genes including IL-6, IL-8, and
IL-10.
 | Table IIClinicopathological characteristics
of the 50 ovarian cancer samples by unsupervised hierarchical
cluster group. |
Table II
Clinicopathological characteristics
of the 50 ovarian cancer samples by unsupervised hierarchical
cluster group.
| Parameters | Cluster A
(n=28) | Cluster B
(n=22) | p-value |
|---|
| Patient age | | | |
| ≤60 years | 23 | 18 | 0.733 |
| >60 years | 5 | 4 | |
| Histological
type | | | |
| SA+EA | 24 | 5 | <0.001 |
| CCC | 4 | 17 | |
| FIGO stage | | | |
| I+II | 9 | 13 | 0.105 |
| III+IV | 19 | 9 | |
| Residual tumor | | | |
| ≤1 cm | 17 | 16 | 0.373 |
| >1 cm | 11 | 6 | |
Given a trend for CCC and non-CCC across the two
clusters we further examined the normalized expression data
comparing histological classes of ovarian carcinoma, CCC vs.
non-CCC. Four of the twelve genes had a statistically significant
difference in expression between groups at p<0.05 with
univariate permutation test (Table
III). Among them, the expression level of IL-6 was the
most significant gene in CCC compared to non-CCC.
 | Table IIICytokine genes differentially
expressed among CCC vs. non-CCC. |
Table III
Cytokine genes differentially
expressed among CCC vs. non-CCC.
| Gene | p-value | FDR | Permutation
p-value |
|---|
| IL-6 | <1e-07 | <1e-07 | <1e-07 |
|
IL-12p35 | 0.0255652 | 0.105 | 0.0237 |
| IL-1β | 0.0262116 | 0.105 | 0.0249 |
| IL-10 | 0.0384981 | 0.115 | 0.0375 |
IL-6 expression and IL-6 siRNA
transfection in ovarian cancer cell lines
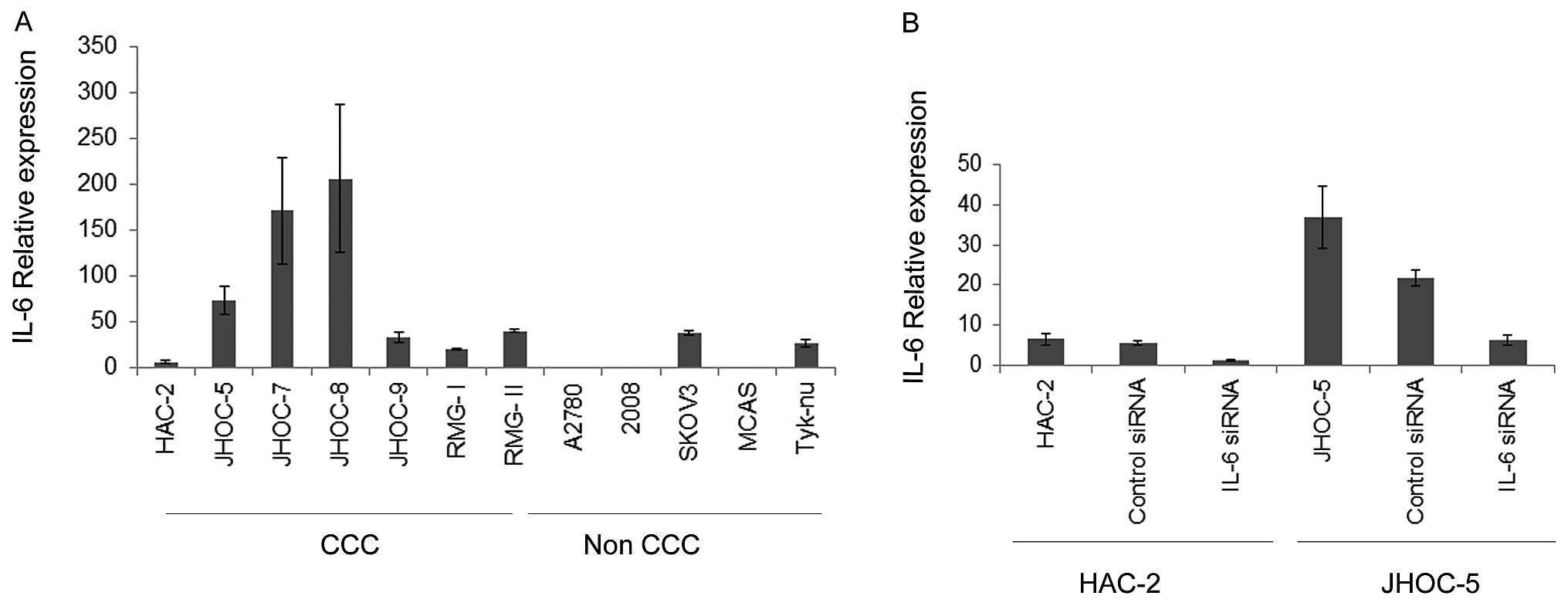
Next, we analyzed the IL-6 gene expression in
twelve ovarian cancer cell lines including seven CCC cell lines.
Consistent with the data obtained from surgical samples, the
IL-6 gene expression of CCC cell lines was typically higher
than that of non-CCC cell lines (Fig.
2A). To begin to address whether the endogenous production of
IL-6 by CCC could have biological significance of CCC, we inhibited
expression of IL-6 in CCC cells using the siRNA approach and
examined the effects of IL-6 expression on the cell growth
and drug sensitivity. Expression of the IL-6 gene was
significantly decreased 24 h after IL-6 siRNA transfection
in HAC-2 and JHOC-5 (Fig. 2B). The
biological effect on cell proliferation was analyzed by cell count
after transfection of IL-6 siRNA in CCC cell lines. No
significant effect on cell proliferation was observed when compared
to transfection of negative control siRNA (data not shown).
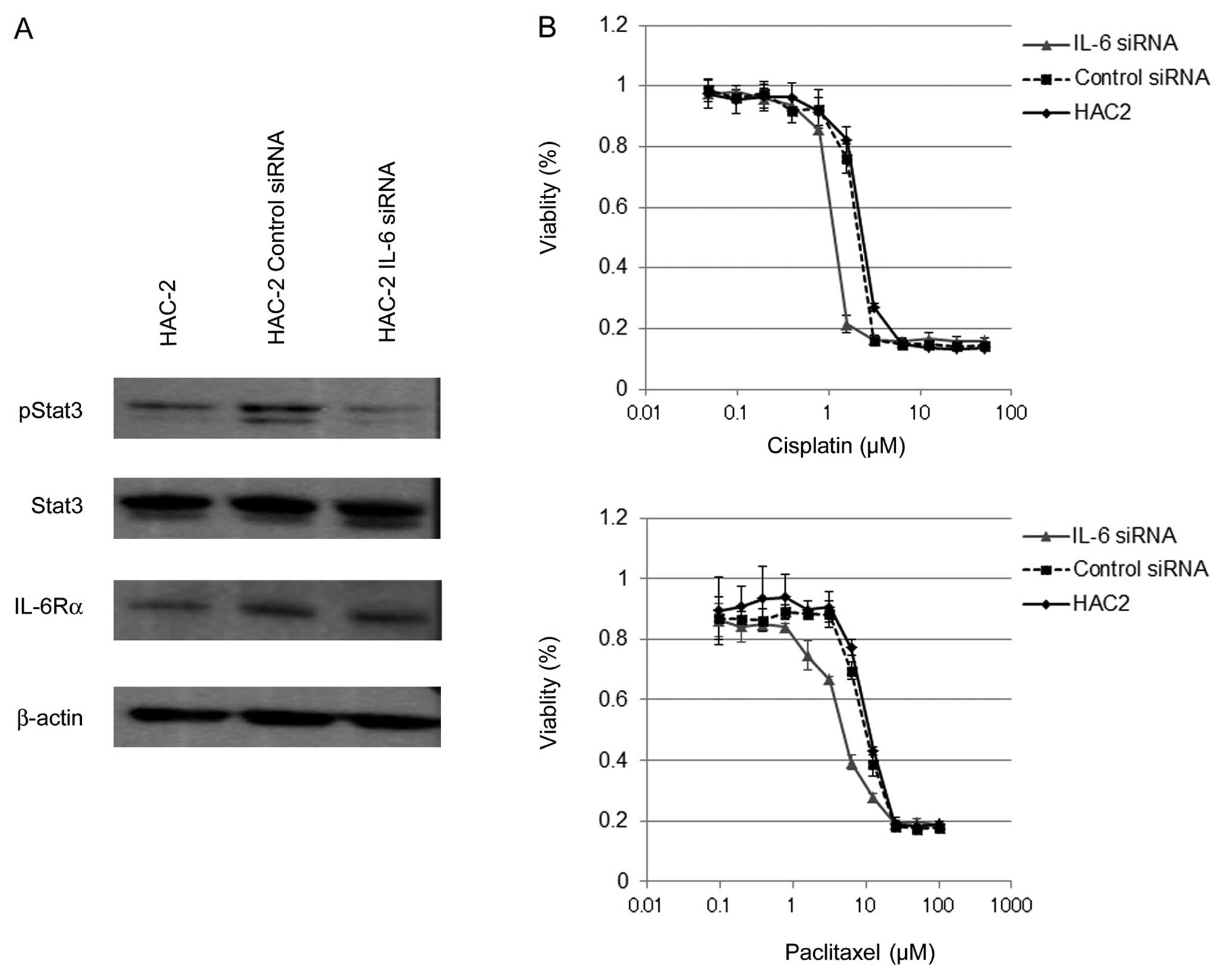
Although there was no change in proliferation, IL-6
inhibition did modulate downstream signaling. Stat3 is a major
signaling effector of IL-6 and has been shown to be required for
survival in multiple cancer models (15). Suppression of IL-6 reduced
activation of Stat3 (pStat3) without affecting overall levels of
Stat3 or other components of the signaling pathway (IL-6Rα;
Fig. 3A). Given these effects on a
pro-survival pathway we next evaluated the susceptibility of CCC
cell lines to cytotoxic agents after suppression of IL-6.
IL-6 siRNA transfected HAC-2 exhibited a statistically
significant increase in cell death to both paclitaxal
(IC50 was 4.78±0.21 μM) and cisplatin
(IC50 was 1.14±0.02 μM) as compared with negative
control siRNA transfected cells (IC50 for paclitaxal was
9.82±0.41 μM and for cisplatin 2.12±0.01 μM,
respectively) (Fig. 3B). Similar
results were obtained from JHOC-5 cells. These results indicate
that inhibition of endogenous IL-6 in CCC cells decreases Stat3
activation and sensitizes these cells to cytotoxic stress.
Discussion
Cytokine expression within a tumor microenvironment
plays a fundamental role in cancer development and progression;
tumor cells that produce immunosuppressive cytokines can escape the
host immune response. In ovarian cancer, several cytokines
associated with cellular immunity were correlated to cancer
development and patient prognosis, including TNF-α, IFN-γ, IL-6,
and MHC molecules (9). We report
here that the expression profile of 12 pro- and anti-inflammatory
cytokine genes in tumor could reliably distinguish the CCC
histopathological classification of ovarian carcinoma from other
epithelial subtypes. Overall evidence suggested CCC to have a Th-2
cytokine dominant expression pattern, including high levels of
IL-6. In our series, 17 of 19 stage III/IV cases (89%) in cluster A
were SA and 12 of 13 stage I/II cases (92%) in cluster B were CCC.
It should also be noted that since >50% of CCC is diagnosed at
an early stage, as seen in this study (67%), the marginal
association between the expression signature and FIGO stage is
likely the result of confounded representation of CCC in cluster
B.
CCC differs considerably from the other histological
types of ovarian cancer with respect to its clinical and molecular
characteristics (5,16,19,20).
With respect to cytokine expression signature and pathway
activation, Yamaguchi et al identified a CCC cell line
specific gene signature from a large set of microarray data that
contained IL-6, consistent with data presented here, as well
as several previously known CCC markers such as HNF-1β
(20). Our data further supports
findings that implicate the activated Stat3 signaling pathway in
CCC cells (19,20), with specific overexpression IL-6 in
CCC compared to SA amongst the 12-cytokine panel examined and
suppression of Stat3 signaling when IL-6 was knocked down with
siRNA. These findings also suggest that the tumor cells, rather
than the host-microenvironment, is the source of IL-6 expression.
This is in contrast to previous work with high-grade SA where a
subset of this histological type is associated with tumor
infiltrating lymphocytes (TILs) (5,17,18).
In these SA-specific studies the dominant cytokine expression
pattern is presumed to be driven by the non-tumor microenvironment.
However, given the modest number of samples in this study and the
overall prevalence of TILs amongst high-grade SA, it seems unlikely
that TILs alone are influencing the expression pattern of cytokines
in the serous tumors examined here. Markedly, expression profiling
has suggested that high-grade SA with the highest levels of IL-6
are not those with TILs (18).
One of the most clinically important biological
features of CCC is its inherent chemoresistance, which is
associated with its poor prognosis particularly at advanced stages.
Several mechanisms involved in drug resistance in CCC have been
proposed, however, the precise mechanisms underlying
chemoresistance remain to be elucidated (16). In this study, siRNA-mediated IL-6
expression did not directly affect proliferation, however, we found
that IL-6 inhibition did reduce Stat3 activation and increase the
sensitivity of CCC cell lines to both cisplatin and paclitaxel.
IL-6 is a pleiotropic cytokine that plays important roles in the
immune response and inflammation. Aberrant expression of
IL-6 has been implicated in many types of cancer, and the
role of IL-6 in chemoresistance has been addressed in several
malignancies, including multiple myeloma, renal cell carcinoma,
cholangiocarcinoma, prostate cancer, and breast cancer (21). A clear link between an IL-6 rich
tumor microenvironment and tumor progression or survival has been
observed in other tumor models (15). Recent evidence indicates that
autocrine and paracrine effects by constitutive production of IL-6
in ovarian cancer could be involved in the tumorigenic potential
through the regulation of angiogenesis (22) or MMP secretion (23). These results suggest that a drug
resistant phenotype in CCC may, in part, be explained by the
activated autocrine IL-6 signaling pathway including Stat3
activation. It is interesting to speculate on a biological link
between recently discovered endometriosis associated ovarian cancer
ARID1A mutations (24,25),
chemoresistance, and the cytokine profile specific to CCC. Though
not clearly established in this context, modulation of gene
expression controlled by the SWI/SNF chromatin binding complex,
specifically the BRG1 component, has been shown to be required for
induction of cytokine genes, including IL-6, during inflammatory
responses (26).
Finally, although SA and CCC behave like distinct
entities, a chemoresistant mechanism converging around the IL-6
pathway may be highly relevant to both CCC and at least some
high-grade SA. Wang et al showed that IL-6 secreted by
serous type ovarian cancer cells might contribute to the
chemoresistance through the downregulation of caspase-3 and
increased expression of both multidrug resistance-related genes and
apoptosis inhibitory proteins (27). High IL-6 levels in the serum and
ascites of ovarian cancer have been found to be associated with
poor prognosis and chemoresistance (21). Most studies on this topic, however,
have not differentiated between histological subtypes. More
recently, blockage of IL-6 signaling by a monoclonal anti-IL-6
antibody siltuximab (CNTO 328) with cytotoxic agent has been shown
to disrupt cancer progression in serous ovarian cancer cell lines
both in vitro and in mouse xenograft models (28). Siltuximab decreased Stat3
phosphorylation and protein levels of downstream effectors
including MCL-1, Bcl-XL, and Survivin in paclitaxel resistant
ovarian cancer cell lines. This is consistent with decreased levels
of phospho-Stat3 and chemosensitization observed in this report. In
the phase II clinical trial with platinum-resistant ovarian cancer,
siltuximab had some therapeutic activity (29). Exposure of ovarian cancer cells to
siltuximab had no effect on cell growth, also consistent with our
siRNA-mediate IL-6 knockdown experiments. This has led to
the hypothesis that the growth inhibitory effects of IL-6 knockdown
may be evident only with tumor-stromal influences. Functional in
vivo model systems paired with immunohistochemical and
molecular analysis are needed to further refine the roles of
various immune cells contributing to the molecular pathogenesis of
ovarian cancer. In particular, the IL-6 related mechanism of
chemoresistance in both CCC and high-grade SA, direct inhibition of
IL-6, IL-6R or other downstream signaling effectors, such as Stat3,
should not be overlooked. Nonetheless, the results obtained in this
study support the idea of targeting the IL-6 signaling pathway in a
combination therapy approach sensitizing CCC tumor cells in
particular to current gold-standard chemotherapies already in use
for ovarian carcinoma.
Acknowledgements
This work was supported in part by
Grants-in-Aid for Scientific Research from Japan Society for the
Promotion of Science to N.Y., A.O., K.Y., and S.T., and by the
Japanese Ministry of Health, Labour and Welfare to K.O. Analyses
were performed using BRB ArrayTools developed by Dr Richard Simon
and Amy Peng Lam.
References
|
1.
|
Vaughan S, Coward JI, Bast RC Jr, et al:
Rethinking ovarian cancer: recommendations for improving outcomes.
Nat Rev Cancer. 23:719–725. 2011. View
Article : Google Scholar
|
|
2.
|
Köbel M, Kalloger SE, Boyd N, et al:
Ovarian carcinoma subtypes are different diseases: implications for
biomarker studies. PLoS Med. 5:e2322008.PubMed/NCBI
|
|
3.
|
Swenerton KD, Santos JL, Gilks CB, et al:
Histotype predicts the curative potential of radiotherapy: the
example of ovarian cancers. Ann Oncol. 22:341–347. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Gilks CB, Ionescu DN, Kalloger SE, et al:
Tumor cell type can be reproducibly diagnosed and is of independent
prognostic significance in patients with maximally debulked ovarian
carcinoma. Hum Pathol. 39:1239–1251. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Anglesio MS, Carey MS, Köbel M, et al:
Clear cell carcinoma of the ovary: A report from the first Ovarian
Clear Cell Symposium, June 24th, 2010 (Review). Gynecol Oncol.
121:407–415. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Takakura S, Takano M, Takahashi F, et al
Japanese Gynecologic Oncology Group: Randomized phase II trial of
paclitaxel plus carboplatin therapy versus irinotecan plus
cisplatin therapy as first-line chemotherapy for clear cell
adenocarcinoma of the ovary: a JGOG study. Int J Gynecol Cancer.
20:240–247. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Shimada M, Kigawa J, Ohishi Y, et al:
Clinicopathological characteristics of mucinous adenocarcinoma of
the ovary. Gynecol Oncol. 113:331–334. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Schreiber RD, Old LJ and Smyth MJ: Cancer
immunoediting: integrating immunity’s roles in cancer suppression
and promotion (Review). Science. 331:1565–1570. 2011.PubMed/NCBI
|
|
9.
|
Nelson BH: The impact of T-cell immunity
on ovarian cancer outcomes (Review). Immunol Rev. 222:101–116.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Kusuda T, Shigemasa K, Arihiro K, et al:
Relative expression levels of Th1 and Th2 cytokine mRNA are
independent prognostic factors in patients with ovarian cancer.
Oncol Rep. 13:1153–1158. 2005.PubMed/NCBI
|
|
11.
|
Marth C, Fiegl H, Zeimet AG, et al:
Interferon-gamma expression is an independent prognostic factor in
ovarian cancer. Am J Obstet Gynecol. 191:1598–1605. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Seike M, Yanaihara N, Bowman ED, et al:
Use of a cytokine gene expression signature in lung adenocarcinoma
and the surrounding tissue as a prognostic classifier. J Natl
Cancer Inst. 99:1257–1269. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Kurman RJ and Shih IeM: Molecular
pathogenesis and extraovarian origin of epithelial ovarian cancer -
shifting the paradigm. Hum Pathol. 42:918–931. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Bustin SA: Absolute quantification of mRNA
using real-time reverse transcription polymerase chain reaction
assays. J Mol Endocrinol. 25:169–193. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Li N, Grivennikov SI and Karin M: The
unholy trinity: inflammation, cytokines, and STAT3 shape the cancer
microenvironment. Cancer Cell. 12:429–431. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Itamochi H, Kigawa J and Terakawa N:
Mechanisms of chemoresistance and poor prognosis in ovarian clear
cell carcinoma (Review). Cancer Sci. 99:653–658. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Tsiatas ML, Gyftaki R, Liacos C, et al:
Study of T lymphocytes infiltrating peritoneal metastases in
advanced ovarian cancer: associations with vascular endothelial
growth factor levels and prognosis in patients receiving
platinum-based chemotherapy. Int J Gynecol Cancer. 19:1329–1334.
2009. View Article : Google Scholar
|
|
18.
|
Tothill RW, Tinker AV, George J, et al:
Novel molecular subtypes of serous and endometrioid ovarian cancer
linked to clinical outcome. Clin Cancer Res. 14:5198–5208. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Anglesio MS, George J, Kulbe H, et al:
IL6-STAT3-HIF signaling and therapeutic response to the
angiogenesis inhibitor sunitinib in ovarian clear cell cancer. Clin
Cancer Res. 17:2538–2548. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Yamaguchi K, Mandai M, Oura T, et al:
Identification of an ovarian clear cell carcinoma gene signature
that reflects inherent disease biology and the carcinogenic
processes. Oncogene. 29:1741–1752. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Hong DS, Angelo LS and Kurzrock R:
Interleukin-6 and its receptor in cancer: implications for
translational therapeutics (Review). Cancer. 110:1911–1928. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Nilsson MB, Langley RR and Fidler IJ:
Interleukin-6, secreted by human ovarian carcinoma cells, is a
potent proangiogenic cytokine. Cancer Res. 65:10794–10800. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Rabinovich A, Medina L, Piura B, Segal S
and Huleihel M: Regulation of ovarian carcinoma SKOV-3 cell
proliferation and secretion of MMPs by autocrine IL-6. Anticancer
Res. 27:267–272. 2007.PubMed/NCBI
|
|
24.
|
Jones S, Wang TL, Shih IeM, et al:
Frequent mutations of chromatin remodeling gene ARID1A in ovarian
clear cell carcinoma. Science. 330:228–231. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Wiegand KC, Shah SP, Al-Agha OM, et al:
ARID1A mutations in endometriosis-associated ovarian carcinomas. N
Engl J Med. 363:1532–1543. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Cullen SJ, Ponnappan S and Ponnappan U:
Catalytic activity of the proteasome fine-tunes Brg1-mediated
chromatin remodeling to regulate the expression of inflammatory
genes. Mol Immunol. 47:600–605. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Wang Y, Niu XL, Qu Y, et al: Autocrine
production of interleukin-6 confers cisplatin and paclitaxel
resistance in ovarian cancer cells. Cancer Lett. 295:110–123. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Guo Y, Nemeth J, O’Brien C, et al: Effects
of siltuximab on the IL-6-induced signaling pathway in ovarian
cancer. Clin Cancer Res. 16:5759–5769. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Coward J, Kulbe H, Chakravarty P, et al:
Interleukin-6 as a therapeutic target in human ovarian cancer. Clin
Cancer Res. 17:6083–6096. 2011. View Article : Google Scholar : PubMed/NCBI
|