Introduction
Colorectal cancer is the third most common
malignancy in Europe and the USA (1,2). The
treatment of patients with rectal cancer has improved significantly
the past decades, where meticulous surgery with clear resection
margins is important for cure. Besides improved surgery,
preoperative radiotherapy and chemoradiotherapy add to improved
treatment results. However, local recurrence after resection of
rectal cancer is still a substantial clinical problem. Dedicated
centers report local recurrence rates in selected series of less
than 4% but population-based results are rather close to 10%
(3–6). Such treatment failures lead to
severe, often intractable symptoms and premature death in the
majority of patients (7,8). Thus, it is important to gain further
knowledge of factors that determine increased risks for local
recurrence following primary operation of rectal carcinoma aimed at
cure. The present study evaluates whole genomic array-CGH
(comparative genomic hybridization) analysis comparing more than
55,000 DNA sites in primary rectal tumors from patients with tissue
in a large bio-bank with linked clinical information. DNA from
tumors that recurred locally and non-recurrent tumors were analyzed
with the aim to link structural DNA-alterations to isolated local
recurrence following R0-resections.
Materials and methods
Selection of patients
The Sahlgrenska University Hospital is a non-profit
institution serving a population of approximately 500,000. All
patients with adenocarcinoma of the colon or rectum treated in this
institution are prospectively registered in a database with
clinopathological variables. Tissue samples are also collected at
the time of surgery from all patients operated in office hours.
Samples from the tumor and mucosa >10 cm from the tumor are
collected at the time of specimen extraction, instantly frozen
liquid in nitrogen and stored in −80°C. The linked database is
continuously updated with clinical data such as variables from the
pathology report, postoperative oncological treatment and possible
recurrent disease. Data are also cross-checked with official
demographic registries to assure adequate registration of
survival.
A total of 2,576 patients were registered in the
database during the decade preceding data extraction (January
1999–September 2009). In 969 patients the tumor was located in the
rectum. From this cohort two groups of patients were selected for
comparative DNA-analysis. Patients in one group were diagnosed with
early, isolated local recurrence, while patients in the other group
had no sign of local or systemic recurrence at long-term follow-up.
Inclusion criteria were: adenocarcinoma in the rectum (<15 cm
from the anal verge), resectional surgery (anterior resection,
abdominoperineal resection, Hartmann’s procedure), clear resection
margins (R0), and tumor stage I–III [TNM 7 (9)]. Exclusion criteria were: preoperative
radiotherapy, distant metastases at the time of surgery and
T4-tumors [TNM 7 (9)].
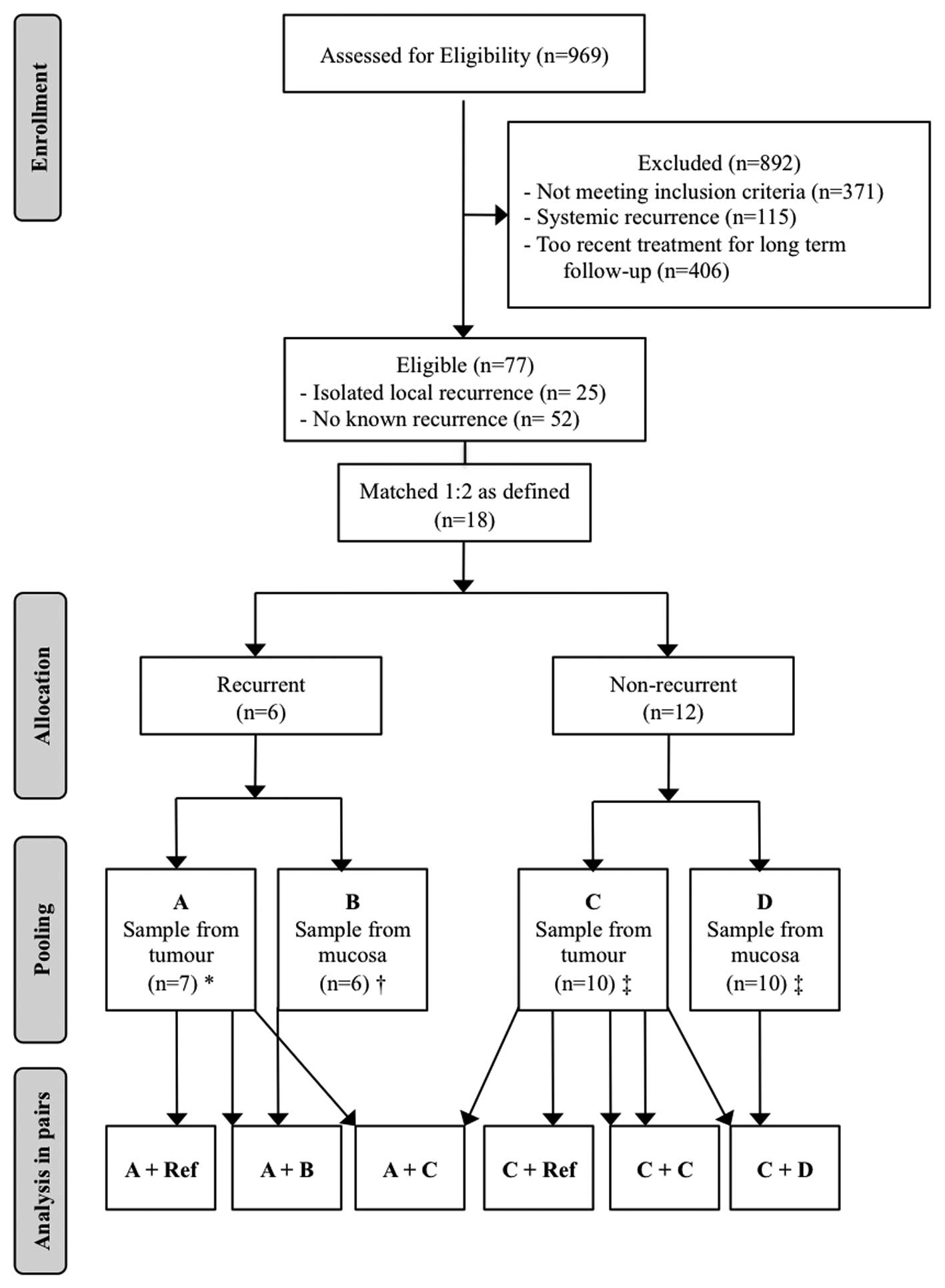
Of 77 eligible patients, 25 had isolated local
recurrence and 52 had no sign of recurrent disease on long-term
follow-up. The matched control group (non-recurrent) was stepwise
selected on a group characteristics level by selection against
study patients (recurrent) for the following variables in order of
priority: T-stage, N-stage, differentiation grade, type of surgery,
age and gender. Patients with early isolated local recurrence were
operated during the years 2002–2006 and the median and mean time to
recurrence were 15 and 16 months, respectively. Patients with
long-term follow-up were operated from 2002 to 2003 with a minimum
time of follow-up of 93 months. See also Consort flow diagram for
the study (Fig. 1).
DNA extraction
Samples of tumor and mucosa were retrieved from the
bio-bank storage at −80°C. Genomic DNA (gDNA) was extracted with
Qiagen DNeasy Blood and Tissue Kit, cat no. 69504, following
mechanical bead-disaggregation of tissue in a TissueLyser (Qiagen)
with an RNase treatment included in the method. Concentration and
purity of gDNA was measured in a NanoDrop ND-1000A
spectrophotometer (NanoDrop Technologies, Inc.). Fractions were
separated electrophoretically in 1% agarose gel for check of
quality.
Sample analyses
Genomic DNA was pooled in four groups on basis of
origin: tumor from the recurrent group (A), mucosa from the
recurrent group (B), tumor from the non-recurrent group (C) and
mucosa from the non-recurrent group (D). Pooled gDNA was hybridized
in pairs as outlined in Fig. 1.
Data analyses from comparisons of DNA from recurrent tumors (A) to
DNA from non-recurrent tumors (C) were not regarded as conclusive
primary information since copy number gain and loss may cancel each
other out. Therefore a commercially available reference DNA was
always used as standard DNA in all comparisons, NA
10851*8 (Coriell). Tissue DNA from two patients in the
non-recurrent group (C and D) was not possible to analyze due to
low DNA quality in tissue specimens resulting in a study population
as depicted in Table I.
 | Table ITumor stage and clinical
characteristics of included patients. |
Table I
Tumor stage and clinical
characteristics of included patients.
| Study patients
(recurrent) (n=6) | Controls
(non-recurrent) (n=10) | p-value |
|---|
| Sex | | | |
| Female | 3 (50) | 5 (50) | |
| Male | 3 (50) | 5 (50) | |
| T-stage | | | 0.87 |
| T1 | 0 (0) | 0 (0) | |
| T2 | 1 (17) | 2 (20) | |
| T3 | 5 (83) | 8 (80) | |
| N-stage | | | 0.92 |
| N0 | 3 (50) | 5 (50) | |
| N1 | 2 (33) | 4 (40) | |
| N2 | 1 (17) | 1 (10) | |
| Differentiation,
grade | | | |
| Moderate, G2 | 6 (100) | 10 (100) | |
| Poor, G3 | 0 (0) | 0 (0) | |
| Preoperative
radiotherapy | | | |
| Yes | 0 (0) | 0 (0) | |
| No | 6 (100) | 10 (100) | |
| Operation | | | 0.15 |
| AR | 2 (33) | 8 (80) | |
| APR | 3 (50) | 1 (10) | |
| HA | 1 (17) | 1 (10) | |
| Age (years) | | | 0.97 |
| Mean ± SD | 74.7±11.0 | 74.5±9.1 | |
| Median | 74.5 | 78.5 | |
| Range | 58–92 | 60–84 | |
Labeling and hybridization
Five hundred nanograms of pooled gDNA were labelled
with either Cyanine 5-dUTP (test) or Cyanine 3-dUTP (reference)
according to the manufacturer’s instructions (Agilent Genomic DNA
Enzymatic Labeling Kit). Competitive hybridization was performed on
Agilent SurePrint G3 Human CGH Microarray Kit 8x60K Oligo, Design
ID 021924, according to ‘Agilent Oligonucleotide Array-Based CGH
for Genomic DNA Analysis. Enzymatic Labelling for Blood, Cells or
Tissues’ Protocol, version 6.2. Procedure A was chosen for post
washes. Images were scanned and quantified on Agilent G2565 AA
microarray scanner and fluorescence intensity was extracted using
the Feature Extraction (FE, v10.7.1.1) software (Agilent
Technologies, USA). At least two of four technical replicates for
each pair combination described in Fig. 1 passed the Feature Extraction
quality control and were used for analysis. Median spacing of the
probes was 33.3 kb in coding sequences and 78.9 kb in non-coding
sequences of the genome.
Analysis of array data
Dye-normalized, outlier- and background subtracted
values were imported into Agilent Genomic Workbench 6.5.0.18
(Agilent Technologies). The result files were filtered at the
feature level with Default Feature Filter. Technical replicates
were combined and normalized with centralization (treshold 6.0, bin
size 10). Aberration analysis was performed with ADM-1 algorithm
(treshold 6.0, nesting filter 0, fuzzy zero on) and filtered with
default aberration filter v2 (minimum number of probes 3, minimum
absolute average of log ratio for region 0.20). The statistical
confidence interval was ±0.2 log(2) ratio as determined in our earlier
study on DNA aberrations in colorectal carcinoma (10). Affected genes in an area of
particular interest on chromosome 4 (4q31.1-31.22) were identified
with the software algorithm in Genomic Workbench 6.5.0.18 (Agilent
Technologies). Identified genes were manually searched for in the
NCBI Gene-database and known functions evaluated in relevance to
the present context.
Confirmation with qPCR
Quantitative real-time polymerase chain reaction
(qPCR) was performed on gDNA from individual tumor tissue samples
of all studied patients. Primers were chosen to include five areas
on chromosome 4 where copy number gain was noted only in the
recurrent group (genes SETD7, MGST2, HHIP, SMAD1 and ANAPC10). For
comparison, primers were also chosen for analysis of areas on
chromosomes 13 and 20 where both patient groups displayed
significant copy number gains, with a statistically significant
difference between the groups in assay analyses. PCR data were
analysed with an efficiency adjusted comparative Cq method, where a
normal DNA area on chromosome 10 served as internal reference. The
commercially available reference DNA NA 10851*8
(Coriell) was used as standard DNA.
All primer design was performed with Primer BLAST
(http://www.ncbi.nlm.nih.gov/tools/primer-blast/index.cgi?LINK_LOC=BlastHome).
Details on primers are presented in Table II. An assay targeting chromosome 10
was already available (TATAA Biocenter, Gothenburg, Sweden).
Criteria for good performing assays were: linearity, high
efficiency (>80%) and negative NTC:s (no template control). All
qPCR assays were tested on reference gDNA. For all assays a
five-point standard curve was generated with four replicates in
each point, run in 5-fold dilution series with primer concentration
200 nM and gDNA template concentrations between 3020-5 pg/μl. PCR
products from designed assays were analyzed on a 2.2% FlashGel
(Lonza) according to the manufacturer’s instructions for control of
correct product size and specificity. No unspecific product could
be observed in gel analysis (data not shown).
 | Table IIDetails on qPCR-primers and
targets. |
Table II
Details on qPCR-primers and
targets.
| Target | Ch13 | Ch20 | Ch4
Gene SETD7 | Ch4
Gene MGST2 | Ch4
Gene HHIP | Ch4
Gene SMAD1 | Ch4
Gene ANAPC10 |
|---|
| Accession no. | NT_024524.14 | NT_011387.8 | NC_000004 | NC_000004 | NC_000004 | NC_000004 | NC_000004 |
| Region in
chromosome | - | - |
140427192..140477577 |
140586922..140661899 |
145617147-145667146 |
146402951..146480328 |
145916310-145966309 |
| Amplicon length | 184 | 123 | 105 | 120 | 109 | 130 | 149 |
| Name forward
primer | Ch13_5p_Fw01 | Ch20_5p_Fw01 | Ch4_SETD_Fw01 | Ch4_MGST_Fw01 | Ch4_HHIP_Fw02 | Ch4_SMAD1_Fw01 |
Ch4_ANAPC10_Fw01 |
| Name reverse
primer | Ch13_5p_Rv01 | Ch20_5p_Rv01 | Ch4_SETD_Rv01 | Ch4_MGST_Rv01 | Ch4_HHIP_Rv02 | Ch4_SMAD1_Rv01 |
Ch4_ANAPC10_Rv01 |
| Sequencing forward
primer |
ACTTCTGTTGTTGCTGTCCTTCTTGG |
TCGTATTCCTTTCCTTCCCTCCCACA |
ACATCCTGCCCTACTGACTTCTCGT |
CGGTGTTTTGTTTCCTACTTGCCCT |
GAGTTATTGGAGGGGAATGG |
TGAAGTTACCAAACAAGGGT |
CAGTCCTTTGCCTCACTAAC |
| Sequencing reverse
primer |
TTCCTCTGCTCCACCCCCGT |
TTGCCCCCACCTGTCACTACC |
TGCCCCAGCCCTTCACACCT |
TGTTCTCAGTCTCTGCTTGCCCCT |
CCGAAGGCTAAGAAGTCAAG |
CAGTGTTTCCCTCTGACTTG |
CTGGTTGAAAGTCTACAGGG |
All qPCR analyses were performed with 10 μl reaction
volume in triplicates on the LightCycler-480 instrument (Roche) in
384 or 96-well plate format using IQ™ SYBR Green Supermix cat no.
170–8882 (Bio-Rad Laboratories Inc.). Detection was performed in
the FAM channel. Template gDNA samples were normalized to 1,000
pg/μl prior to qPCR analysis. All qPCR experiments were performed
according to the manufacturer’s instructions and all pipetting was
performed by robot (EpMotion 5070, Eppendorf, Germany).
Statistical analysis
The statistical confidence interval was set to ±0.2
log(2) ratio in analysis of
array-CGH data (10). Independent
samples t-test was used to compare log(2) ratios among groups. Data from qPCR
were analysed with the comparative Cq-method. Average Cq-values in
groups were analyzed with ANOVA. Individual Cq-values from all
assays with technical triplicates were used in analysis of
aberrations on chromosome 4. Results are presented as mean and SEM.
p-value <0.05 was considered statistically significant.
Comparison of group characteristics was performed with Pearson’s
χ2 except for age where independent samples t-test was
used.
The bio-bank was instituted and maintained in
accordance to national regulations. The present study was approved
by the regional ethics review board (Dnr 261-08).
Results
The selection of 6 patients with early, isolated
local recurrence after primary cancer surgery and 12 matched
non-recurrent control patients out of 77 eligible patients is
outlined in Fig. 1. Study and
control patients showed comparable clinical characteristics,
although they were selected and matched according to clinical
characteristics on group basis (Table
I).
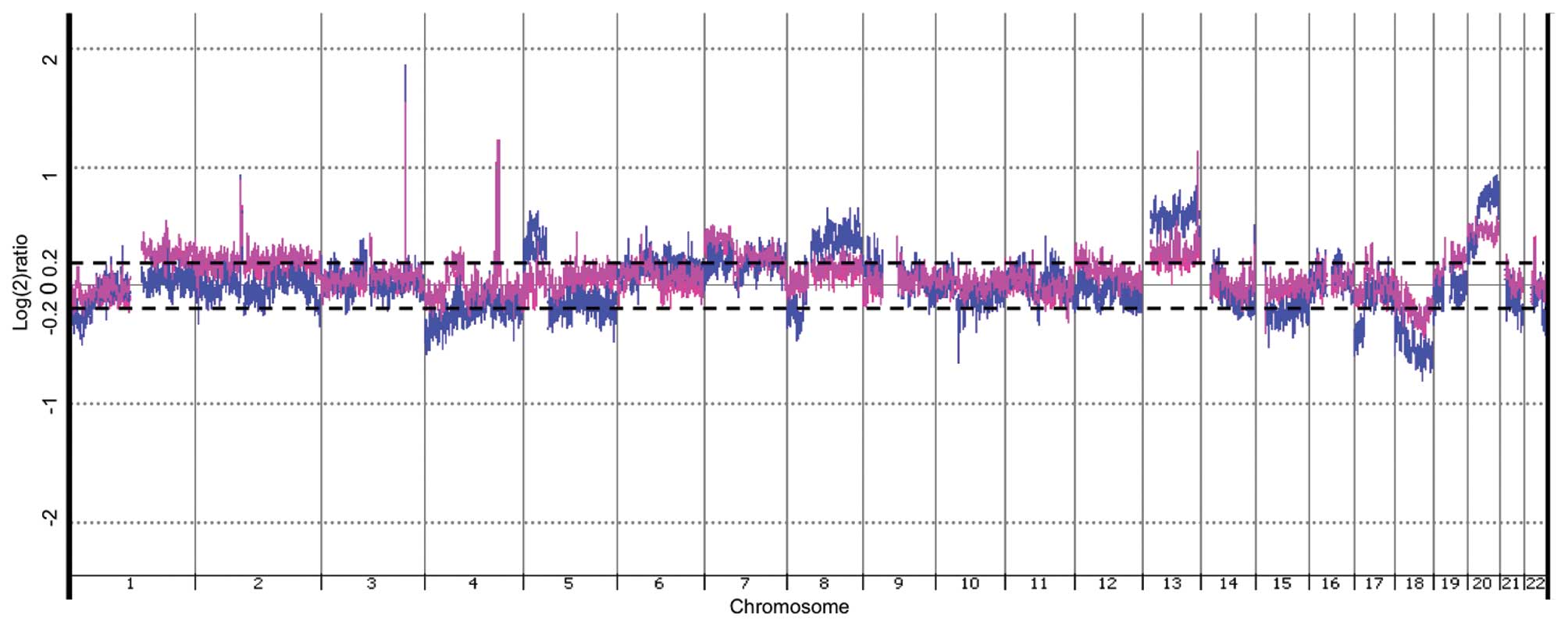
Genomic DNA from rectal carcinomas had DNA sequences
with statistically significant aberrations compared with reference
DNA. As depicted in Fig. 2,
several DNA areas were significantly affected in both recurrent and
non-recurrent tumors, assessed by CGH-array analysis (p<0.05).
Many of these affected regions are known to be involved in
colorectal carcinogenesis, e.g., on chromosomes 5, 8, 13, 17, 18
and 20 (11–14). However, DNA aberrations seemed
overall more pronounced in DNA from non-recurrent tumors. This
appeared in both copy number gains (chromosomes 5, 8, 13, 20) and
copy number losses (chromosomes 1, 4, 5, 17, 18, 22) (Fig. 2), although, qPCR quantification on
sequences in chromosomes 13 and 20 did not confirm statistically
different alterations between the groups (Table IV).
 | Table IVQuantitative PCR analyses on gDNA
from recurrent and non-recurrent individual tumors. |
Table IV
Quantitative PCR analyses on gDNA
from recurrent and non-recurrent individual tumors.
| Ch13
5p01 | Ch20
5p01 | Ch4
SETD7 | Ch4
MGST2 | Ch4
HHIP | Ch4
SMAD1 | Ch4
ANAPC10 | Ch4
Sum of all probes |
|---|
| Recurrent
(n=7) | 1.16±0.13 | 1.33±0.15 | 2.05±1.13 | 1.13±0.61 | 0.92±0.07 | 2.12±1.19 | 1.71±0.83 | 1.62±0.37 |
| Non-recurrent
(n=10) | 1.24±0.21 | 1.23±0.13 | 1.04±0.19 | 0.64±0.04 | 0.78±0.04 | 0.91±0.06 | 0.82±0.05 | 0.84±0.04 |
| p-valuea | ns | ns | ns | ns | ns | ns | ns | 0.0147 |
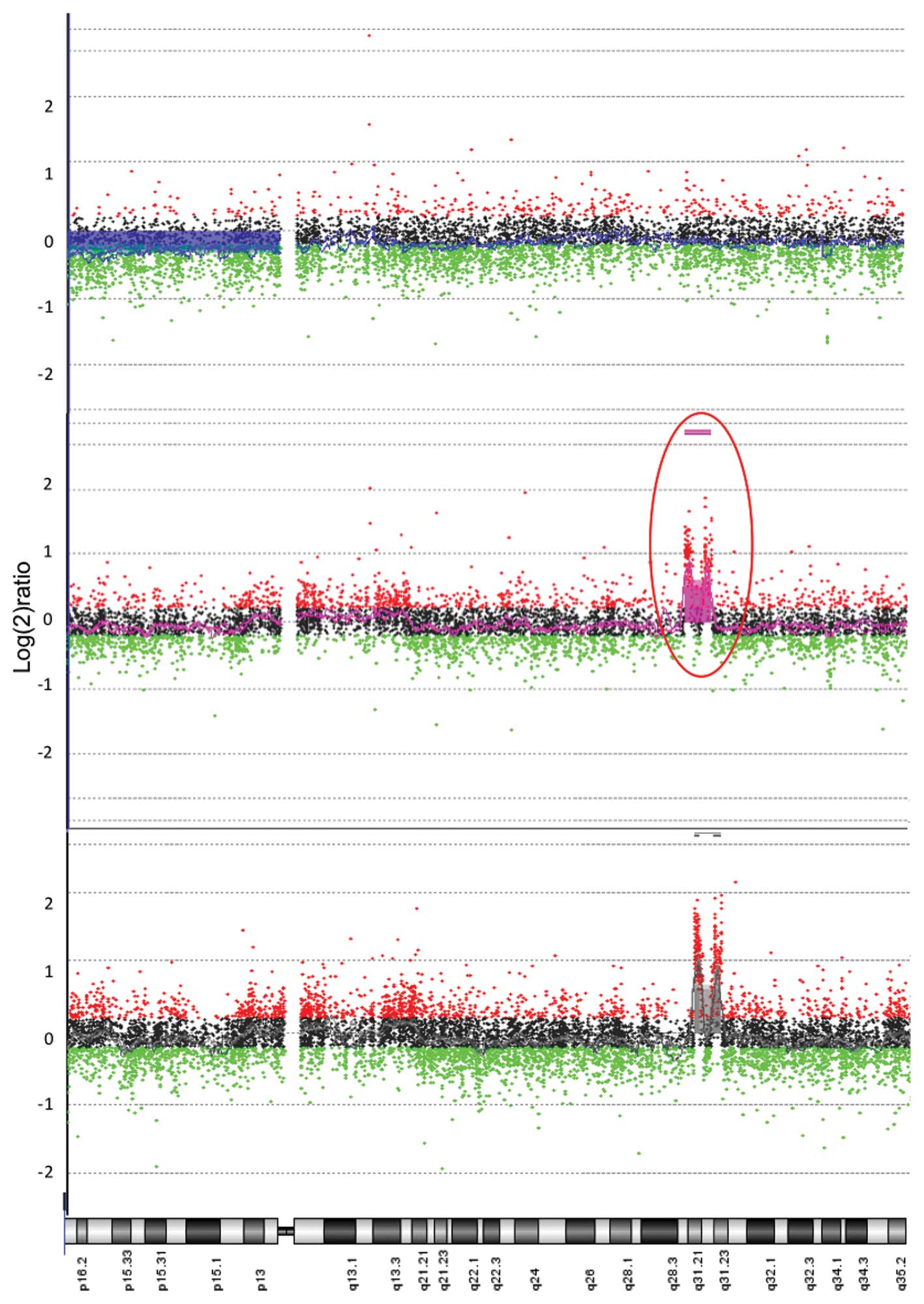
In contrast, an aberration on chromosome 4 with
great magnitude was noted in DNA from tumors in the locally
recurrent group. This gain on chromosome 4 (4q31.1-31.22) was thus
detected in DNA from locally recurrent tumors when compared to
reference DNA, p<0.0001, with no such alterations in the
non-recurrent group compared to reference DNA (Figs. 2 and 3). The difference in copy number DNA
between groups in this region on chromosome 4 was also
statistically significant when comparing tumor tissue versus mucosa
from patients with locally recurrent disease. This indicates a
genotype specific alteration in tumor DNA and not in host DNA,
p<0.0001 (Fig. 3).
Detailed analysis of relevant regions on chromosome
4 showed that it contained over 100 genes, of which 22 were
significantly affected. These 22 genes encode proteins with several
known functions including p53 regulation, progression through cell
cycle as well as regulation of transcription (Table III).
 | Table IIIGenes located in the 4q31.1-31.22
region with examples of known function. |
Table III
Genes located in the 4q31.1-31.22
region with examples of known function.
| Gene | Known function |
|---|
| SETD7 | Involved in p53
regulation |
| MGST2 | Involved in
production of leukotrienes and PGE2 |
| MAML3 | Mastermind-like 3,
involved in regulation of Notch signalling |
| SCOC | Interacts with
ADP-ribosylation factor-like proteins |
| CLGN | May play a role in
spermatogenesis and infertility |
| ELMOD2 | Antiviral
response |
| UCP1 | Only expressed in
brown adipose tissue |
| TBC1D9 (MDR1) | Transmembrane
efflux pump, decreased intracellular accumulation and multidrug
resistance. MDR1-negative samples were most common among tumor
types known to be relatively responsive to chemotherapy |
| RNF150 | Ring finger
protein |
| ZNF330 | Zinc finger
protein |
| GAB1 | Important mediator
of branching tubulogenesis and plays a central role in cellular
growth response, transformation and apoptosis |
| SMARCA5 | Regulate
transcription |
| LOC441046 | Pseudogene |
| GYPE | Glycophorin E, M
blood group antigen |
| GYPB | Glycophorin B, M
blood group antigen |
| GYPA | Glycophorin A, M
blood group antigen |
| HHIP | Inhibitor of
Hedgehog signalling, involved in developmental process, implicated
in COPD and lung cancer |
| ANAPC10 | Progression through
cell cycle |
| ABCE1 | Transport across
extra- and intra-cellular membranes |
| OTUD4 | Expressed at HIV-1
infection |
| SMAD1 | Signal transducer
and transcriptional modulator in for example cell growth,
apoptosis, immune responses, morphogenesis and development |
| MMAA | Translocation of
cobalamin in mitochondria |
Confirmation with quantitative real-time PCR of gDNA
from individual tumor tissue samples including all studied DNA
probes showed a statistically significant difference between
recurrent and non-recurrent tumors targeting the region of interest
on chromosome 4, which analytically confirms the significant
array-CGH findings on a patient group level (Table IV).
Discussion
Local recurrence after rectal cancer surgery is a
main quality indicator in the management of patients with rectal
cancer. It is well established that surgical technique is of
crucial importance and the concept of TME (total mesorectal
excision) is today universally accepted. Radiotherapy, especially
preoperatively, and in selected cases preoperative
chemoradiotherapy, can further improve the clinical outcome.
However, as Marijnen et al has pointed out (15), radiotherapy cannot compensate for
positive tumor resection margins. Inability to achieve clear
resection margins (R0) entails such a high risk of local recurrence
that many surgeons regard this as persistent rather than recurrent
disease. Established and suggested risk factors for local
recurrence related to the surgical quality include inadequate
distal margin, circumferential resection margin <1 mm,
perioperative perforation of the rectum, inadequate clearance of
intraluminal viable tumor cells before transection of the bowel,
breaching of the mesorectal fascia and producing a non-cylindrical
specimen in abdominoperineal resection. Other risk factors include
tumor size, stage, location, grade of differentiation, tumor
budding, and lymphatic, perineural or vascular invasion. Thus, with
this knowledge in mind, stepwise selection of study and control
patients (1:2) retrieved two groups with comparable clinical
characteristics for DNA analysis.
Despite recent advances in the management of rectal
cancer patients, local recurrences do occur. Dedicated centers
report local recurrence rates below 4% in selected series (3–5) and
population-based recurrence rates of 7–9% are not uncommon
(6). The local recurrence rate in
the present population-based cohort is comparable to the national
average for the relevant period, i.e., less than 10%. This included
all patients with a curative or palliative resections.
Approximately 45% of patients in our database were operated with
anterior resection, 30% with abdominoperineal resection, 10%
Hartmann’s procedure and 10% with local excision or TEM (transanal
endoscopic microsurgery). Thus, the cohort in our database appears
relevant in general perspectives of rectal cancer surgery.
Although, our present material represents low statistical power, it
is important to know that recruiting a double number of patients,
would require 15–20 years continuous collection of tissue samples
from all patients operated in the largest colorectal department in
Scandinavia. The main reasons for this is declining local
recurrence rates and increasing proportions of patients treated
with preoperative radiotherapy.
The importance of ‘lateral’ lymph nodes in the
context of local recurrence is unclear. Our present material
appears well matched relative to lymph node metastases encountered
during pathological examination of the operative specimens. The
presence of remaining tumor tissue after rectal surgery may not be
enough for a local recurrence to be established. This conclusion is
supported by the fact that not all patients with R1-resections
(microscopically involved resection margin) develop local
recurrences (16,17). Also, not all patients in whom the
operative field has been contaminated by tumor cells, due to
perioperative perforation of the tumor, will develop local
recurrences (18). Furthermore,
not all patients develop anastomotic recurrence despite the fact
that there are viable cells in the lumen of the bowel following
resection (19). Thus it appears,
there are both known and unknown factors that determine the
patients who will experience local recurrences. Such factors may be
host- and immune-related, or primarily related to tumor cell
biology. Therefore, efforts have also been made to identify risk
factors at tumor cell level but such potential findings remain to
be confirmed and clinically applied in rectal cancer treatment
(16,17). Possibly, there are rectal tumors
that are more prone to recur locally than others and such
characteristics may thus be related to aberrations in the tumor
genome. It has been proposed that colorectal adenocarcinomas can be
divided in at least five subgroups based on genetic characteristics
(12,20,21).
Our previous studies with array-CGH have also shown that there are
aberrations in the tumor genome of colorectal malignancies that are
stage-dependent (14). It is also
well recognized that tumors are heterogeneous and contain several
clones and perhaps different cancer stem cells (13,21).
By evolutionary clonal selection some clones may prevail (13). Thus, it is likely that different
tumors should vary in characteristics by several clinical aspects
such as ability to metastasize and generate local recurrence.
Accordingly, genomic DNA from both recurrent and
non-recurrent rectal tumors in the present study showed aberrations
in DNA areas that have previously been described to be frequently
involved in colorectal carcinogenesis [e.g., on chromosomes 5, 8,
13, 17, 18 and 20 (11–14)]. Interestingly, the copy number
gains and losses were generally less pronounced in the present
tumors prone to local recurrence; an observation which may not be
expected in the light of numerous reports on the relationship
between copy number gains and poor prognosis in colon carcinomas.
Again, such observations may point to the possibility that systemic
spread and local recurrent growth may indicate different genetic
background. Thus, several well-recognized DNA alterations are
unlikely of major importance for the development of local
recurrences in rectal carcinoma following R0 resections.
The area on chromosome 4 (4q31.1-31.22), where
locally recurrent tumors displayed significant copy number gains
compared to non-recurrent tumors, has not been reported earlier in
colorectal cancer. A similar gain pattern in comparison of tumor
DNA to mucosa DNA in recurrent patients supports that observed DNA
alterations were genotype specific for tumor cells and not a
patient DNA phenomenon, even though low statistical power in our
present material should exclude firm conclusions. However, affected
genes located in the DNA area on chromosome 4 code for at least 22
products, some of which have known functions that are of high
relevance in tumor biology, such as regulation of p53, cellular
growth, cellular transformation, progression through cell cycle,
regulation of apoptosis as well as arachnoid acid metabolism with
production of leukotriens and prostaglandins. Whether few or
several genes in this area act in concert to promote recurrent
growth must await future research.
In conclusion, the availability of a large bio-bank
on rectal carcinoma with linked clinopathological data was a
prerequisite for careful matching of groups of non-irradiated
patients and subsequent evaluation of possible genetic differences
between locally recurrent and non-recurrent rectal carcinomas. The
surgical management and clinical outcome in our study population
was in agreement with international standards. Most well recognized
aberrations in colorectal carcionogenesis were identified in both
recurrent and non-recurrent tumors, but seemed to be of less
importance for local recurrences. However, the 4q31.1-31.22 region
represents a potential area for further investigations in larger
patient cohorts.
Acknowledgements
This work was supported by grants from
the Swedish Cancer Society, the Swedish State under the LUA/ALF
agreement, the Göteborg Medical Society and the Anna-Lisa and Bror
Björnsson Foundation. We acknowledge the expert technical skill of
Ingrid Palmgren, Marianne Åkerström, Marianne Andersson and
Jacqueline Flach. We thank Hillevi Björkqvist and Ann-Louise
Helminen for handling surgical samples and Lena Munro and Birgitta
Sjöberg for excellent administration of the clinical database. The
authors are not aware of any affiliations, memberships, funding or
financial holdings that might be perceived as affecting the
objectivity of this paper.
References
|
1
|
European Cancer Observatory (ECO): Cancer
fact sheet 2011. http://eu-cancer.iarc.fr/country-930-european-union-27.html,en.
[Accessed: 2011, Nov 8].
|
|
2
|
American Cancer Society: Cancer Facts and
Figures 2011. http://www.cancer.org/acs/groups/content/@epidemiologysurveilance/documents/document/acspc-029771.pdf.
[Accessed: 2011, Nov 8].
|
|
3
|
Edwards DP, Sexton R, Heald RJ and Moran
BJ: Long-term results show triple stapling facilitates safe low
colorectal and coloanal anastomosis and is associated with low
rates of local recurrence after anterior resection for rectal
cancer. Tech Coloproctol. 11:17–21. 2007. View Article : Google Scholar
|
|
4
|
MacFarlane JK, Ryall RD and Heald RJ:
Mesorectal excision for rectal cancer. Lancet. 341:457–460. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Moore E, Heald RJ, Cecil TD, Sharpe GD,
Sexton R and Moran BJ: Almost all five year disease free survivors
are cured following rectal cancer surgery, but longer term
follow-up detects some late local and systemic recurrences.
Colorectal Dis. 7:403–405. 2005.
|
|
6
|
Pahlman L, Bohe M, Cedermark B, et al: The
Swedish rectal cancer registry. Br J Surg. 94:1285–1292. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Camilleri-Brennan J and Steele RJ: The
impact of recurrent rectal cancer on quality of life. Eur J Surg
Oncol. 27:349–353. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Palmer G, Martling A, Cedermark B and Holm
T: A population-based study on the management and outcome in
patients with locally recurrent rectal cancer. Ann Surg Oncol.
14:447–454. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Sobin L, Gospodarowicz M and Wittekind C:
TNM Classification of Malignant Tumours. 7th edition.
Wiley-Blackwell; 2010
|
|
10
|
Lagerstedt KK, Staaf J, Jonsson G, et al:
Tumor genome wide DNA alterations assessed by array CGH in patients
with poor and excellent survival following operation for colorectal
cancer. Cancer Inform. 3:341–355. 2007.PubMed/NCBI
|
|
11
|
Brenner BM and Rosenberg D:
High-throughput SNP/CGH approaches for the analysis of genomic
instability in colorectal cancer. Mutat Res. 693:46–52. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cunningham D, Atkin W, Lenz HJ, et al:
Colorectal cancer. Lancet. 375:1030–1047. 2010. View Article : Google Scholar
|
|
13
|
Fearon ER: Molecular genetics of
colorectal cancer. Annu Rev Pathol. 6:479–507. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lagerstedt KK, Kristiansson E, Lonnroth C,
et al: Genes with relevance for early to late progression of colon
carcinoma based on combined genomic and transcriptomic information
from the same patients. Cancer Inform. 9:79–91. 2010.
|
|
15
|
Marijnen CA, Nagtegaal ID, Kapiteijn E, et
al: Radiotherapy does not compensate for positive resection margins
in rectal cancer patients: report of a multicenter randomized
trial. Int J Radiat Oncol Biol Phys. 55:1311–1320. 2003. View Article : Google Scholar
|
|
16
|
Enriquez-Navascues JM, Borda N, Lizerazu
A, et al: Patterns of local recurrence in rectal cancer after a
multidisciplinary approach. World J Gastroenterol. 17:1674–1684.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Den Dulk M, Marijnen CA, Putter H, et al:
Risk factors for adverse outcome in patients with rectal cancer
treated with an abdominoperineal resection in the total mesorectal
excision trial. Ann Surg. 246:83–90. 2007.
|
|
18
|
Jorgren F, Johansson R, Damber L and
Lindmark G: Oncological outcome after incidental perforation in
radical rectal cancer surgery. Int J Colorectal Dis. 25:731–740.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kodeda K, Holmberg E, Jorgren F, Nordgren
S and Lindmark G: Rectal washout and local recurrence of cancer
after anterior resection. Br J Surg. 97:1589–1597. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jass JR: Classification of colorectal
cancer based on correlation of clinical, morphological and
molecular features. Histopathology. 50:113–130. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Markowitz SD and Bertagnolli MM: Molecular
origins of cancer: molecular basis of colorectal cancer. N Engl J
Med. 361:2449–2460. 2009. View Article : Google Scholar : PubMed/NCBI
|