Introduction
MicroRNAs (miRNAs, miRs) are a family of small (19
to 25 nucleotides in length) non-coding RNAs that regulate gene
expression by sequence-selective targeting of mRNAs (1–4),
leading to a translational repression or mRNA degradation,
depending on the degree of complementarity between miRNAs and the
target sequences (2). Since a
single miRNA can target several mRNAs and a single mRNA may contain
several signals for miRNA recognition, it is calculated that at
least 10–40% of human mRNAs are targets of microRNAs (2). In general, a low expression of a
given miRNA is expected to be potentially linked with an
accumulation of targets mRNAs; conversely, a high expression of
miRNAs is expected to be the cause of a low expression of the
target mRNAs.
miRNAs play a double role in cancer, behaving both
as tumor promoters or tumor suppressors. In general,
cancer-promoting miRNAs target mRNA coding for tumor-suppression
proteins, while miRNAs exhibiting tumor-suppression properties
usually target mRNAs coding oncoproteins. MicroRNAs which have been
demonstrated to play a crucial role in the initiation and
progression of human cancer are defined as oncogenic miRNAs
(oncomiRs) (5). Moreover, miRNAs
have been firmly demonstrated to be involved in cancer metastasis
(metastamiRs) (6). Thus,
therapeutic strategies involving miRNA silencing have been proposed
based on the roles of these small non-coding RNAs as oncogenes.
One of the most interesting microRNAs possibly
involved in cancer is miR-221. This miRNA has been found to be
upregulated in breast cancer (7),
glioma (8), hepatocellular
carcinoma (9), pancreatic
adenocarcinoma (10), melanoma
(11), chronic lymphocytic
leukemia (12), thyroid papillary
carcinoma (13). Possible target
molecules of miR-221 are DVL2 (14), PUMA (15), PTEN (16), p27Kip1 (17–19).
In this specific context, of great interest is the study published
by Galardi et al identifying p27Kip1 mRNA as a
possible target of miR-221 (20).
This finding is very intriguing, since p27Kip1 has been
proposed as a tumor suppressor gene, which is downregulated in
several types of tumors.
In consideration of the involvement of miRNAs in
cancer (7–13), the possibility to regulate gene
expression by interfering with the fundamental mechanisms mediated
by miRNAs is one of the most intriguing challenges in the
development of new types of drugs (miRNA therapeutics) in cancer.
In this respect peptide nucleic acid (PNA)-based molecules are
appealing (21–23).
PNAs are DNA mimics extensively used for the
pharmacological regulation of gene expression in a variety of
cellular and molecular systems (21–23).
In PNAs the pseudo-peptide backbone is composed of
N-(2-aminoethyl)glycine units (24). PNAs are resistant to both nucleases
and proteases (25–27) and, more importantly, hybridize with
high affinity to complementary sequences of single-stranded RNA and
DNA, forming Watson-Crick double helices (28–31).
For these reasons, PNAs were found to be excellent candidates for
antisense and antigene therapies (32–34).
In addition, PNA-based molecules (such as PNA-DNA chimeras) can act
as transcription factor decoys as demonstrated in the case of NF-κB
and Sp1 (34).
The major limit in the use of PNAs for alteration of
gene expression is the low uptake by eukaryotic cells (35). In order to solve this drawback,
several approaches have been considered, including the delivery of
PNA analogues with liposomes and microspheres (25,36,37).
One possible strategy is to link the PNAs to polylysine (K) or a
polyarginine (R) tails, based on the observation that these
cell-membrane penetrating oligopeptides are able to facilitate
uptake of conjugated molecules (38). Peptide-PNA conjugates have been
shown to be efficiently incorporated in cells by gymnosis, i.e.
without the need of transfecting agents, showing high uptake
efficiency (39).
At present, data on the use of PNAs as molecules
targeting miRNAs are accumulating. Fabani et al reported two
studies, one on PNAs against miR-122, the other on PNAs against
miR-155, demonstrating the potential role of PNA for future
therapeutic applications as well as for studying microRNA functions
(40,41).
A further example has been recently reported by Yan
et al (42), who addressed
the potential effects of PNA-antimiR-21 in vivo on the
growth of breast cancer cells. In their experiments, MCF-7 cells
treated with PNA-antimiR-21 or PNA-control were subcutaneously
injected into female nude mice and detectable tumor masses were
seen in only 5/8 of mice in the MCF/PNA-antimiR-21 group, while
much larger tumors were detected in all mice in the MCF/PNA-control
group. Both tumor weight and number showed that MCF/PNA-control
cells formed larger tumors more rapidly than MCF/PNA-antimiR-21
cells in nude mice (42).
Regarding possible effects of PNAs against microRNAs on biological
functions, our group recently reported a study on the effect of PNA
molecules targeting miR-210 in K562 cells. We have previously found
that this microRNA is upregulated during induced erythroid
differentiation (43). Treatment
of K562 cells with this PNA molecule leads, as expected, to a sharp
decrease of differentiation levels (44). An octa-arginine-PNA conjugate was
found to be efficiently delivered within the cells and showed high
inhibitory effects on miRNA-210 bioavailability.
The aim of the present study was to determine the
activity of PNA designed according to the same model, and targeted
against miR-221 on the biological activity of this miRNA, in
particular on its effects on p27Kip1. As a model system,
we employed the human breast cancer cell line MDA-MB-231, in which
miR-221 is upregulated and p27Kip1 downregulated
(45). In addition, this cell line
is suitable to address specific effects on miR-221, since it
accumulates far more miR-221 in respect to miR-222.
We first describe the designed and synthesized PNA
against miR-221. Second, we described and validated the delivery
strategy to maximize PNA uptake by target MDA-MB-231 cells. Third,
we analyzed the effects of anti-miR PNA on miR-221 accumulation and
biological effects on MDA-MB-231 cells.
Materials and methods
Synthesis and characterization of
PNAs
The synthesis of peptide Fl-Rpep was performed as
previously reported (46). The
PNAs were synthesized with standard manual Boc-based chemistry
using commercially available monomers (ASM Research Chemicals,
Hannover, Germany) with HBTU/DIPEA coupling as described elsewhere
(46). All the PNAs were
synthesized in a 5 μmol scale using MBHA resin loaded with
Boc-PNA-T-OH as first monomer. The R8 tail of
Rpep-PNA-a221 was introduced using the same coupling procedures.
5(6)-carboxyfluorescein (Sigma-Aldrich) was introduced using
DIC/DhBtOH coupling after the coupling of the PNA or PNA-peptide
conjugate with 2-(2-(Fmoc-amino) ethoxy)ethoxyacetic acid (AEEA)
spacer (Applied Biosystems, Foster City, CA, USA).
PNA purification was performed by RP-HPLC with UV
detection at 260 nm using a semi-prep column C18 (for unlabelled
PNA: 5 microns, 250×10 mm, Jupiter Phenomenex, 300 Å; for
fluorescein labeled PNAs: 10 microns, 300×7.7 mm, Xterra Waters,
300 Å), eluting with water containing 0.1% TFA (eluent A) and
acetonitrile containing 0.1% TFA (eluent B); elution gradient: from
100% A to 50% B in 30 min, flow: 4 ml/min. Purity and identity of
the purified PNAs were checked by HPLC-MS (Micromass Quattro micro
API with QqQ Detector) using a Phenomenex Jupiter C18; 250×4.6 mm;
5 μm column.
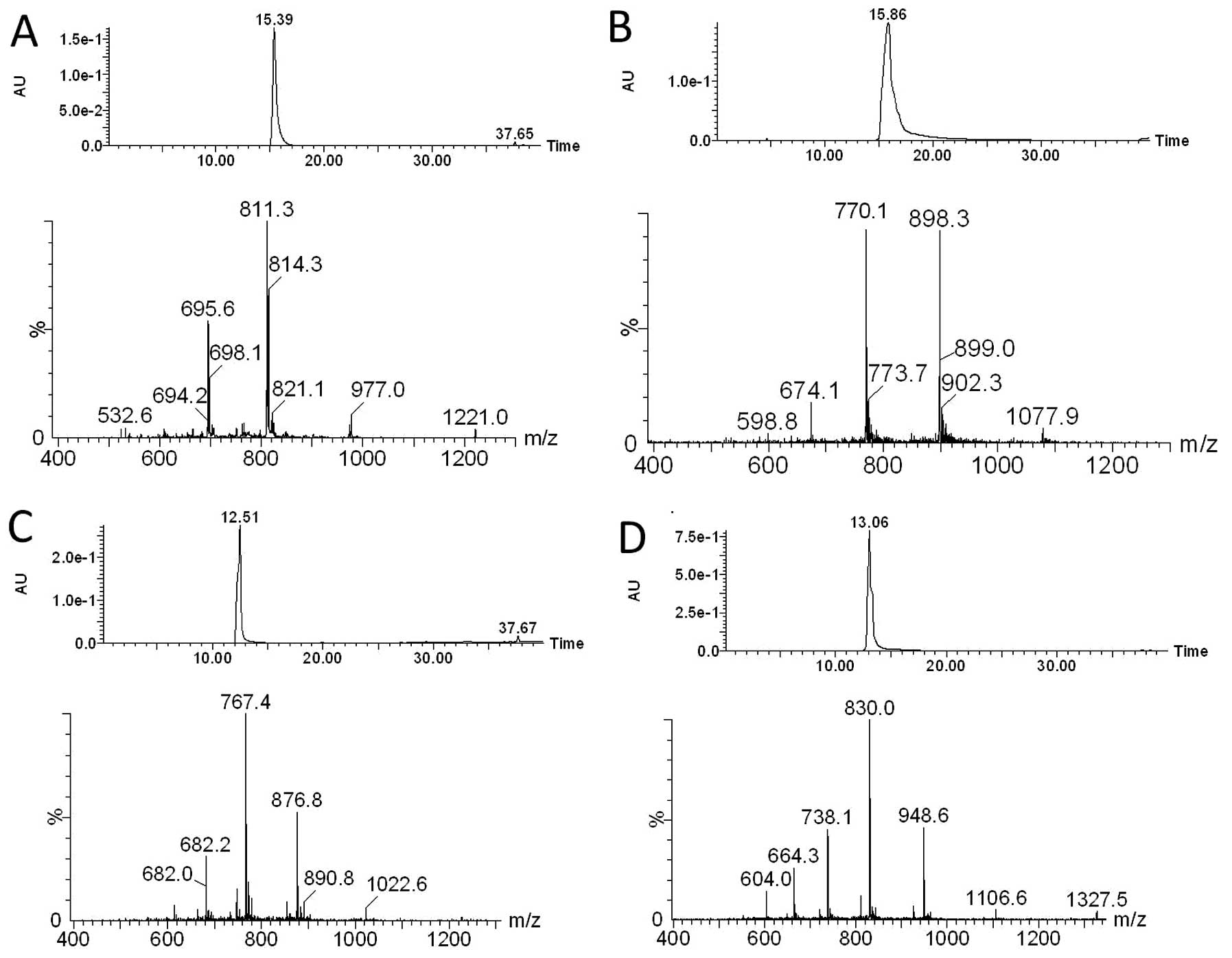
PNA-a221
Yield: 16%; calculated MW: 4881.0; ESI-MS: m/z found
(calculated): 122.0 (1221.9) [MH44+], 976.9
(977.7) [MH55+], 814.3 (815.0)
[MH66+], 698.2 (698.7)
[MH77+].
Rpep-PNA-a221
Yield: 47%; calculated MW: 5386.8; ESI-MS: m/z found
(calculated): 1077.9 (1078.9) [MH55+], 898.3
(899.3) [MH66+], 770.1 (771.0)
[MH77+], 674.1 (674.7)
[MH88+].
Fl-PNA-a221
Yield: 27%; calculated MW: 6129,8; ESI-MS: m/z found
(calculated): 1022.6 (1023.2) [MH6+], 876.8
(877.2) [MH7+], 767.4 (767.7)
[MH8+], 682.2 (682.5)
[MH9+], 614.0 (614.3)
[MH1010+].
Fl-Rpep-PNA-a221
Yield: 29%; calculated MW: 6635.6; ESI-MS: m/z found
(calculated): 1327.5 (1328.8) [MH55+], 1106.6
(1107.5) [MH66+], 948.6 (949.5)
[MH77+], 830.0 (830.9)
[MH88+], 738.1 (738.7)
[MH99+], 664.3 (664.9)
[MH1010+], 604.0 (604.6)
[MH1111+].
UV measurements
Stock solutions of Rpep-PNA-a221, and of
complementary DNA or RNA synthetic oligonucleotides (Thermo Fisher
Scientific, Ulm, Germany, HPLC grade), with the following
sequences: full match: 5′-ACATTGTCTGCTGGGTTT-3′; mismatch:
5′-ACATTGTCAGCTGGGTTT-3′; scrambled 5′-GTTCGTATGCTATTTGGC-3′, were
prepared in double distilled water and the PNA concentration
calculated by UV absorbance using the following ε260
(M−1 cm−1) for the nucleobases: T 8600, C
6600, A 13700, G 11700. For DNA and RNA the data provided by the
producer were used. From these, solutions containing single
stranded PNA, DNA, RNA or PNA:DNA and PNA:RNA duplexes were
prepared. Measurement condition: [PNA] = [DNA] or [RNA] = 5
μM in pH 7.0 PBS buffer (100 mM NaCl, 10 mM
NaH2PO4•H2O, 0.1 mM EDTA) or in
the same buffer containing 5 M urea. All the samples were first
incubated at 90°C for 5 min, then slowly cooled to room
temperature. Thermal denaturation profiles (Abs vs. T) of the
hybrids were measured at 260 nm with a UV/Vis Lambda Bio 20
Spectrophotometer equipped with a Peltier Temperature Programmer
PTP6 interfaced to a personal computer, in the range, 18–90°C
(0.1°C step resolution). A melting curve was recorded for each
duplex. The melting temperature (Tm) was determined from the
maximum of the first derivative of the melting curves.
Human cell lines and culture
conditions
Human breast cancer MCF-7 and MDA-MB-231 cells
(47) were cultured in humidified
atmosphere of 5% CO2/air in DMEM medium (Life
Technologies, Monza, Italy) supplemented with 10% fetal bovine
serum (FBS) (Celbio, Milan, Italy), and 2 mM L-glutamine
(Sigma-Aldrich, St. Louis, MO, USA). To determine the effects on
proliferation, cell growth was monitored by determining the cell
number/ml using a Z1 Coulter Counter (Coulter Electronics, Hialeah,
FL, USA).
RNA extraction
Cells were trypsinized and collected by
centrifugation at 1,500 rpm for 10 min at 4°C, washed with PBS, and
lysed with Tri-Reagent™ (Sigma-Aldrich), according to
manufacturer’s instructions. The isolated RNA was washed once with
cold 75% ethanol, dried and dissolved in nuclease-free pure water
before use.
Real-time quantitative PCR
For microRNA quantification using real-time RT-qPCR
reagents, the primers and probes were obtained from Applied
Biosystems. Reverse transcriptase (RT) reactions were performed
using the TaqMan® MicroRNA Reverse Transcription Kit
(Applied Biosystems); real-time PCR was performed according to the
manufacturer’s protocols. For each sample 20 ng were used for the
assays. All RT reactions, including no-template controls and
RT-minus controls, were performed in duplicate using the 7700
Sequence Detection System version 1.7 (Applied Biosystems). The
relative expression was calculated using the comparative cycle
threshold method and as reference U6 snRNA was used to normalize
all RNA samples, since it remains constant in the assayed samples
by miR-profiling and quantitative RT-PCR analysis, as previously
reported (43,44).
TaqMan RT-qPCR
For gene expression analysis 1 μg of the
total-RNA were reverse transcribed by using random hexamers.
Quantitative real-time PCR assays were carried out using
gene-specific double fluorescently labeled probes. Primers and
probes used to assay p27Kip1 (assay ID: Hs00153277.m1)
were purchased from Applied Biosystems. The relative expression was
calculated using the comparative cycle threshold method and, as
reference genes, the endogenous control human 18S rRNA (43).
Western blotting
Cytoplasmic extracts (20 μg) were denatured
for 5 min at 98°C in 1X SDS sample buffer [62.5 mM Tris-HCl pH 6.8,
2% SDS, 50 mM dithiotreithol (DTT), 0.01% bromophenol blue, 10%
glicerol] and loaded on SDS-PAGE gel (10×8 cm) in Tris-glycine
buffer (25 mM Tris, 192 mM glycine, 0.1% SDS). A biotinylated
protein ladder (size range of 9–200 kDa) (Cell Signaling
Technology, Euroclone S.p.A., Pero, Italy) was used as standard to
determine molecular weight. The electrotransfer to 20 microns
nitrocellulose membrane (Pierce, Euroclone S.p.A.) was performed
overnight at 360 mA and 4°C in electrotransfer buffer (25 mM Tris,
192 mM glycine, 5% methanol). The membrane were prestained in
Ponceau S Solution (Sigma) to verify the transfer, washed with 25
ml TBS (10 mM Tris-HCl pH 7.4, 150 mM NaCl) for 10 min at room
temperature and incubated in 25 ml of blocking buffer for 2 h at
room temperature. The membranes were washed three times for 5 min
each with 25 ml of TBS/T (TBS, 0.1% Tween-20) and incubated with
primary rabbit monoclonal antibody (1:1000) (Cell Signaling
Technology) in 15 ml primary antibody dilution buffer with gentle
agitation overnight at 4°C. The day after, the membrane were washed
three times for 5 min each with 20 ml of TBS/T and incubated in 15
ml of blocking buffer, in gentle agitation for 2 h at room
temperature, with an appropriate HRP-conjugated secondary antibody
(1:2,000) and an HRP-conjugated anti-biotin antibody (1:1,000) used
to detect biotinylated protein marker. Finally, after three washes
each with 20 ml of TBS/T for 5 min, the membranes were incubated
with 10 ml LumiGLO® (0.5 ml 20X LumiGLO, 0.5 ml 20X
peroxide and 9.0 ml Milli-Q water) (Cell Signaling Technology) in
gentle agitation for 5 min at room temperature and exposed to X-ray
film (Pierce). As necessary, after stripping procedure using the
Restore™ Western Blot Stripping Buffer (Pierce) membranes were
reprobed with primary and secondary antibodies.
X-ray film for chemiluminescent blots was analyzed
by Gel Doc 2000 (Bio-Rad Laboratoires, Milan, Italy) using Quantity
One program to elaborate the intensity data of our specific protein
targets. Ponceau S staining was used as normalization control, but
also others marker proteins were taken as reference and
specifically reported. The rabbit mAb against p27Kip1
and β-actin were purchase from Cell Signaling Technology.
Statistics
Results are expressed as mean ± standard error of
the mean (SEM). Comparisons between groups were made by using
paired Student’s t-test and a one-way analysis of variance (ANOVA).
Statistical significance was defined with p<0.05.
Results
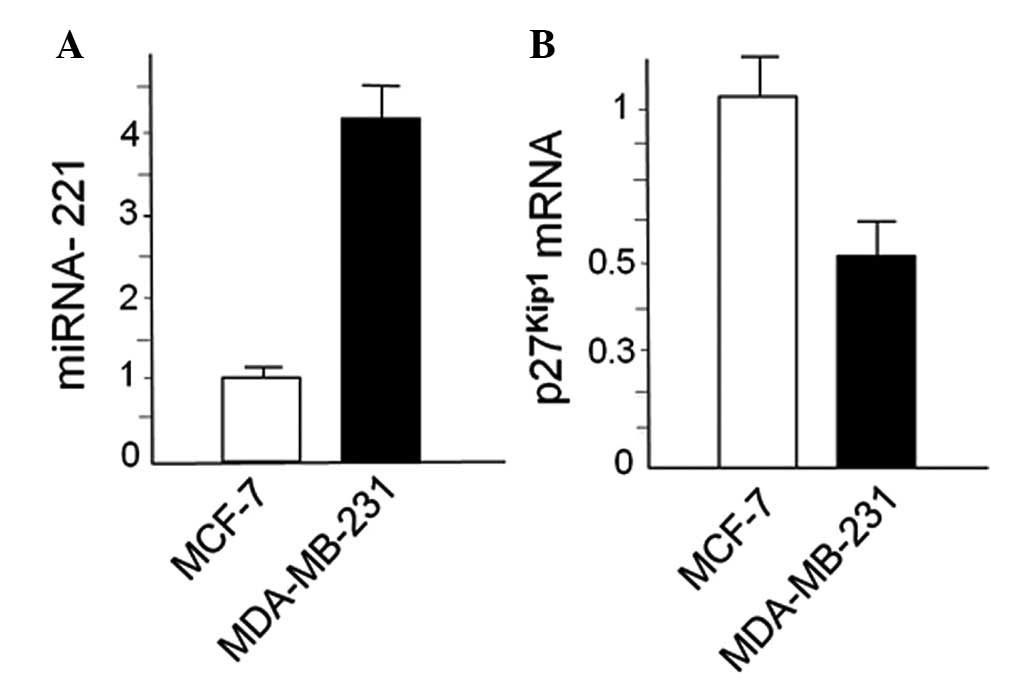
Expression of miR-221 and
p27Kip1 in breast cancer cell lines
MDA-MB-231 and MCF-7 cells were first characterized
with respect to expression of miR-221 and p27Kip1. This
was done by RT-qPCR, obtaining the results shown in Fig. 1. We confirmed that miR-221 is
upregulated in MDA-MB-231 cells in respect to MCF-7 cells and,
conversely, p27Kip1 is downregulated, as elsewhere
reported by several authors (19,45,48).
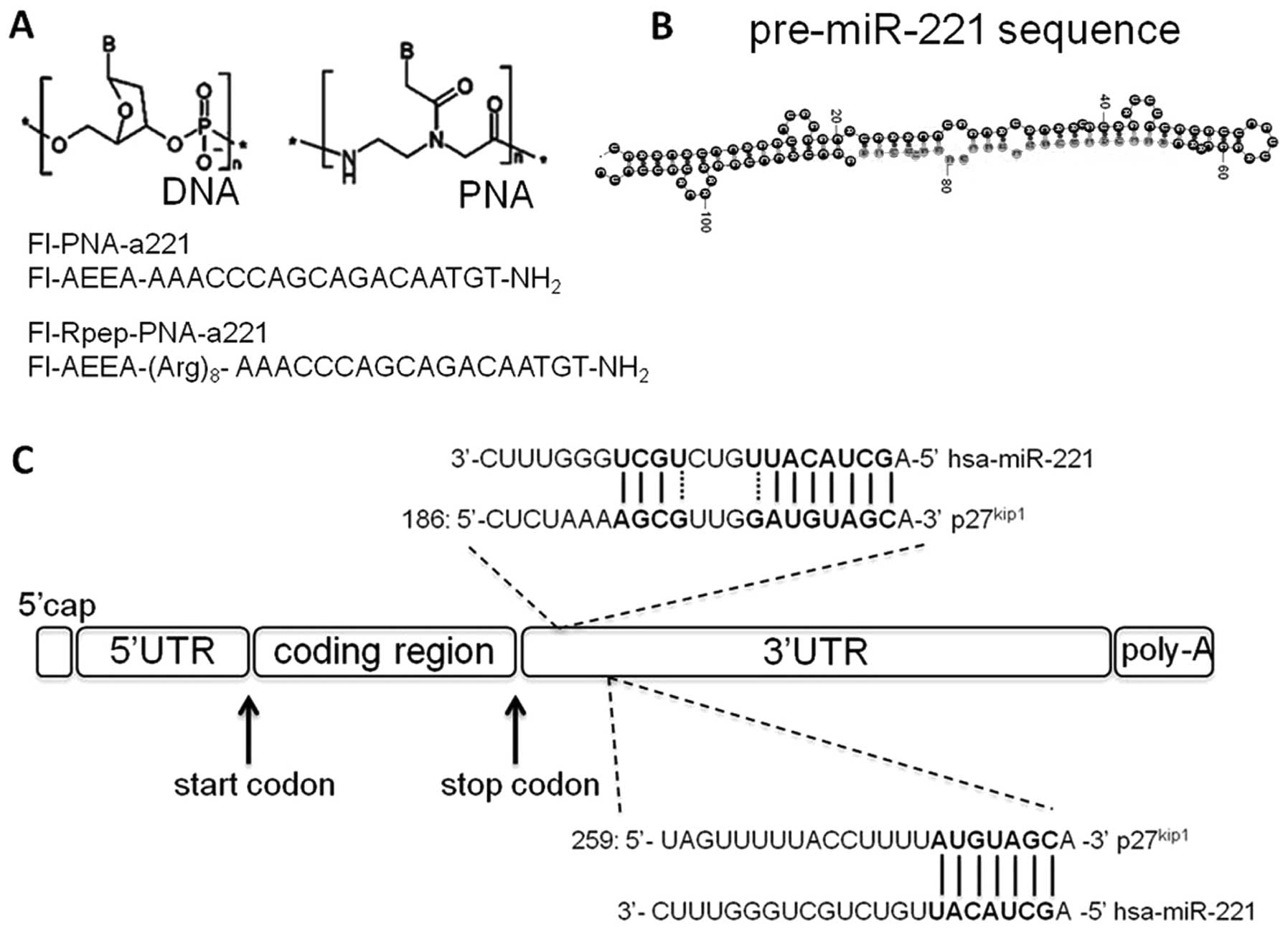
Synthesis and characterization of
PNA-a221 and Rpep-PNA-a221
In Fig. 2A the PNA
structure and the PNA sequences used for the present study are
shown. In Fig. 2B the structure of
pre-miR-221 is depicted, together with the possible miR-221
interaction with the 3′UTR sequences of human p27Kip1
mRNA (Fig. 2C). In Fig. 2C the PNA structure and the PNA
sequences used for the present study are shown. These PNA molecules
are expected to bind to pre-miR-221 as well as to mature miR-221.
The sequence of the PNA was chosen in order to present the lowest
number of off-target binding with mRNA, as evaluated by BLAST
search. In order to increase the uptake by the target cells, an
octa-arginine peptide (Rpep) was conjugated to the PNA-a221. The
synthesis was carried out manually using standard protocols of
solid-phase synthesis of PNAs and peptides. The fluorescein tag was
linked to the PNA or peptide through a spacer, the products were
all purified with RP-HPLC and their purity and identity was
confirmed by HPLC/ESI-MS analysis (Fig. 3).
The formation of duplexes of the two PNAs was
studied by UV melting analysis of the PNA:DNA duplexes, since for
simple conjugated PNAs, the stability of PNA:RNA duplexes, although
higher, followed the same trend as PNA:DNA duplexes. Both PNAs
showed very high melting temperatures with complementary DNA
(Table I), which was significantly
higher for Rpep-PNA-a221 (Tm≥90°C). This effect can be explained by
the contribution of electrostatic interactions between the
positively charged (Arg)8 tail and the phosphates of
DNA, as previously observed in the case of PNA conjugated to a
cationic NLS peptide (49).
 | Table IMelting temperatures (°C) of the PNA
with full-match (FM), mismatched (MM) and scrambled (SCR) DNA in
PBS buffer and in PBS buffer containing 5 M urea. |
Table I
Melting temperatures (°C) of the PNA
with full-match (FM), mismatched (MM) and scrambled (SCR) DNA in
PBS buffer and in PBS buffer containing 5 M urea.
| PNA | DNA | Tm (PBS) | Tm (PBS with 5 M
urea) |
|---|
| PNA-a221 | FM | 79 | 67 |
| PNA-a221 | MM | 73 | 64 |
| PNA-a221 | SCR | 43a | 21a |
| Rpep-PNA-a221 | FM | >90 | 79 |
| Rpep-PNA-a221 | MM | 77 | 65 |
| Rpep-PNA-a221 | SCR | 67 | 38 |
The formation of a PNA:DNA duplex in the case of
Rpep-PNA-a221 (which do not show a clear melting transition in the
range experimentally accessible) was confirmed by the measurements
performed under strongly denaturing conditions (5 M urea). The very
high melting temperatures observed at 5 μM concentration
(and even under strongly denaturing conditions for Rpep-PNA-a221)
ensure that the target miR-221 RNA can be efficiently bound by the
PNA used also in cellular systems, if the PNA is delivered to the
same cellular compartment of the miRNA.
Sequence specificity was tested using a scrambled
sequence (SCR, with the same base composition of full-match DNA,
but a maximum of 4 consecutive complementary bases, with a total of
8 possible pairs) and a DNA containing a single mismatch (MM). The
melting temperature decreased for both Rpep-PNA-a221 and PNA-a221
in the order FM>MM>SCR. Comparing the binding abilities of
Rpep-PNA-a221 and PNA-a221, shows that the polycationic
octa-arginine tail has the effect of increasing the melting
temperatures by aspecific electrostatic interactions with the
polyanionic nucleic acid target. It is worth noting that even under
extremely denaturing conditions, such as 5 M urea, the PNAs:DNA
duplexes show remarkably high melting temperatures (77°C for
Rpep-PNA-a221 and 67°C for PNA-a221).
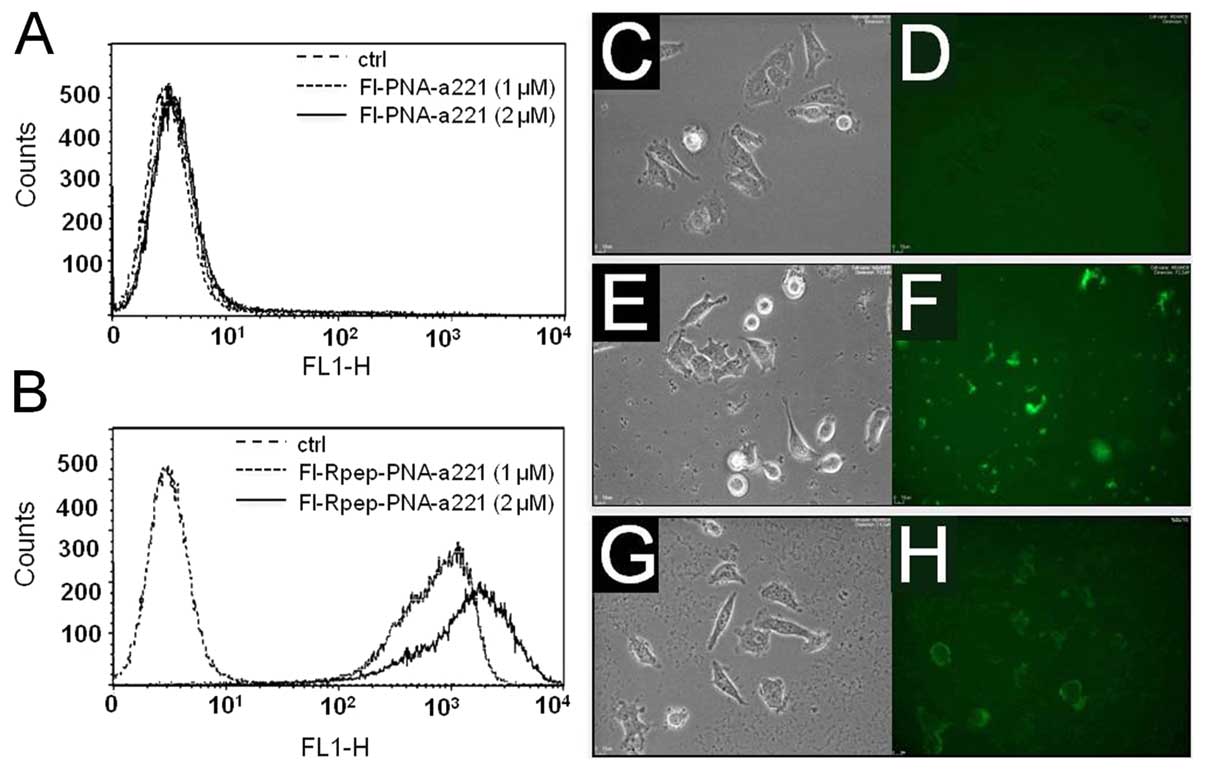
Uptake of PNA-a221 and Rpep-PNA-a221 by
MDA-MB-231 cells
In order to investigate the uptake of PNA-a221 and
Rpep-PNA-a221 by breast cancer MDA-MB-231 cells, 1.5×105
cells were incubated in the presence of increasing concentrations
of fluorescein labeled PNAs (Fl-PNA-a221 and Fl-Rpep-PNA-a221) for
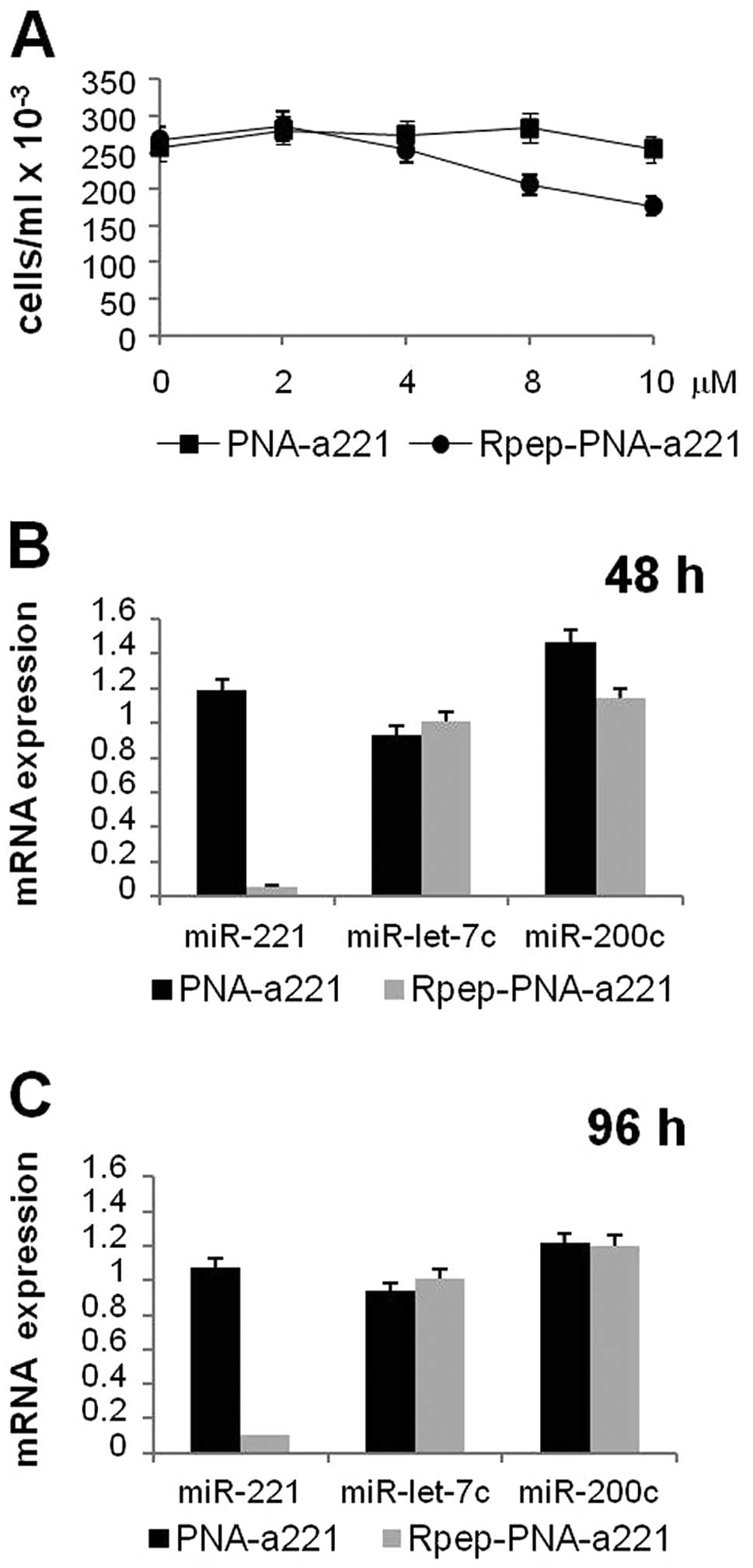
24 h, obtaining the results shown in Fig. 4A and B. As expected, efficient
binding of the Fl-Rpep (data not shown) and low binding of the
Fl-PNA-a221 (Fig. 4A) to target
MDA-MB-231 cells were observed. On the contrary, Fl-Rpep-PNA-a221
displayed efficient binding to MDA-MB-231 cells (Fig. 4B). FACS analyses, performed several
times and obtaining consistent results, are compatible with uptake
of Fl-Rpep-PNA-a221 by target cells, but cannot exclude the
possibility that the fluorescence signal is due at least partially
to cell-surface interactions, caused by the positive charged
polyarginine peptide, which might interact strongly with negative
charged protein components. Therefore, the intracellular
distribution of Fl-Rpep, Fl-PNA-a221 and Fl-Rpep-PNA-a221 was
analyzed using BioStation instrument, a compact cell incubation and
monitoring system (BioStation IM, Nikon Instruments Europe B.V.,
Florence, Italy). Representative results shown in Fig. 4C–H, indicate the differential
uptake (fully consistent with the FACS analysis) obtained using
Fl-Rpep, Fl-PNA-a221 and Fl-Rpep-PNA-a221 fluorescent compounds.
The highest uptake was obtained when fluorescein-labeled
Fl-Rpep-PNA-a221 was used (Fig.
4H). A low level of fluorescence was detectable with Rpep and
PNA-a221 (Fig. 4D and F,
respectively). Taken together, the FACS analyses and the studies
employing fluorescence microscopy support the conclusion that
Fl-Rpep-PNA-a221 is internalized within target cells.
In order to determine the concentrations of Fl-Rpep,
Fl-PNA-a221 and Fl-Rpep-PNA-a221 to be employed for in vitro
studies on MDA-MB-231 cells, the IC50 after three days
treatment was determined. While Fl-PNA-a221 displayed an
IC50 value higher that 15 μM, the IC50
value of Fl-Rpep-PNA-a221 was found to be 7.5±1.75 μM
(Fig. 5A). Accordingly, in order
to avoid the use of antiproliferative (and possibly cytotoxic)
concentrations, Fl-Rpep, Fl-PNA-a221 and Fl-Rpep-PNA-a221 were used
at 2 μM.
Rpep-PNA-a221: inhibitory effects on
miR-221
When MDA-MB-231 cells are cultured in the presence
of Rpep, PNA-a221 and Rpep-PNA-a221 a very different effect was
observed. After RNA isolation, RT-qPCR was performed following
protocols reported and applied to PNAs against miRNAs (44), demonstrating that the miR-221
specific hybridization signal was strongly reduced only when RNA
was isolated from MDA-MB-231 cells cultured for 48 h (Fig. 5B) and 96 h (Fig. 5C) in the presence of Rpep-PNA-a221,
while no major effects were observed PNA-a221. Fig. 5 (panels B and C) shows that these
effects are restricted to miR-221, since, despite the fact that
some alteration of miRNA content occurs, no suppression of
accumulation of miR-200c and miR-let-7c was obtained. These data
demonstrate specificity of the PNA treatment.
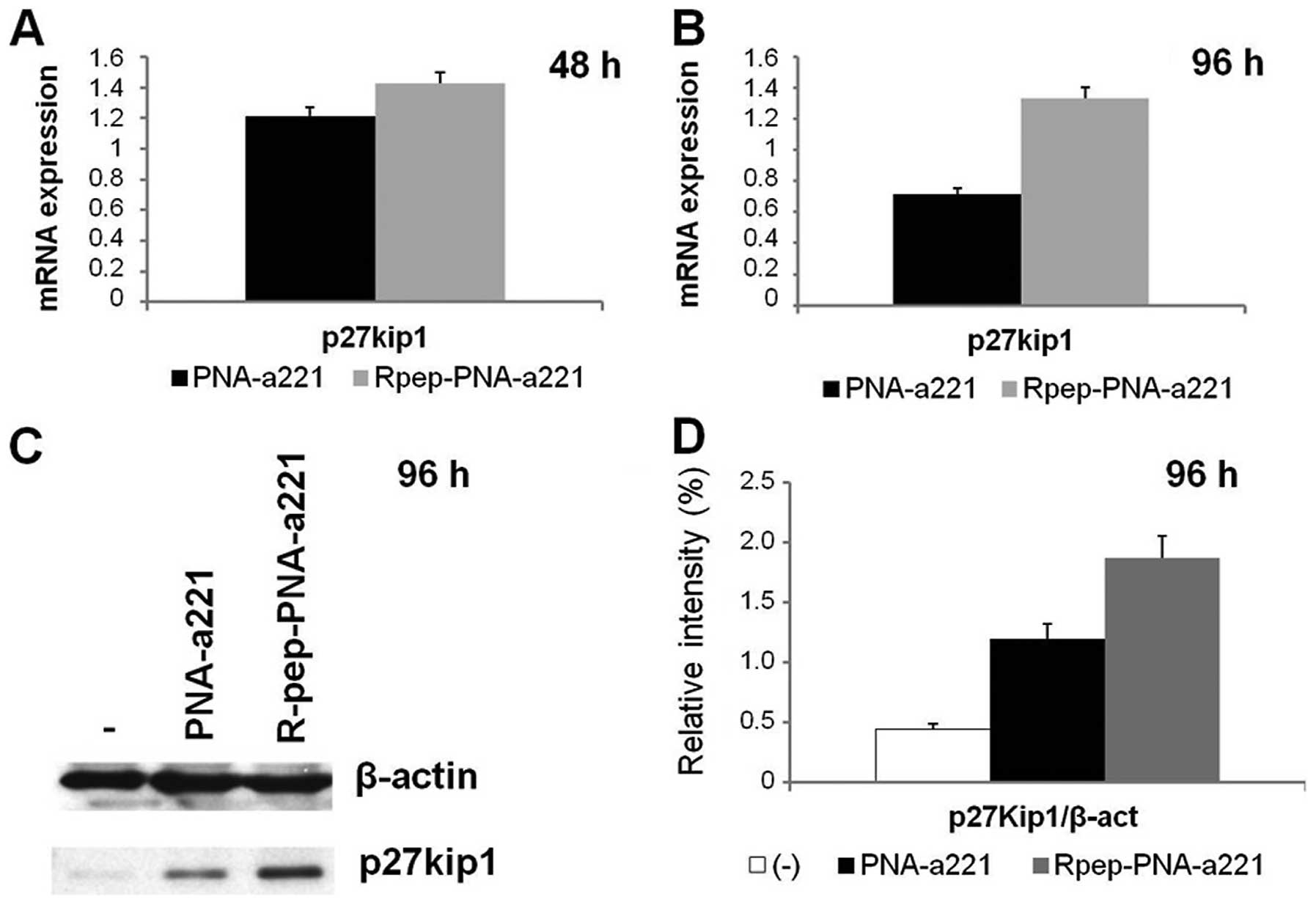
The expression of the p27Kip1
gene is upregulated in MDA-MB-231 cells treated with
Rpep-PNA-a221
The effects of Rpep-PNA-a221 were analyzed on the
expression of p27Kip1 in MDA-MB-231 cells, by RT-qPCR
and by western blotting. The effects on p27Kip1 mRNA,
shown in Fig. 6 (panels A and B),
indicate that no change of p27Kip1 mRNA content occurs
in MDA-MB-231 cells in the presence of PNA-a221, whereas
significant increase of p27Kip1 mRNA is observed with
the Rpep-PNA-a221 (p<0.05). These data were confirmed by western
blot assay (Fig. 6, panels C and
D); a clear increment of p27Kip1 protein expression
in the sample treated with Rpep-PNA-a221 is detectable. Fig. 6D shows the relative intensity of
the p27Kip1 spots, obtained from densitometric analysis
of the autoradiography film.
Discussion
MicroRNA-221 is deeply involved in cancer, and it
was found upregulated in glioma, hepatocellular carcinoma,
pancreatic adenocarcinoma, melanoma, chronic lymphocytic leukemia,
and thyroid papillary carcinoma. In breast cancer, miR-221 was
found to be upregulated in breast cancer cell lines and primary
tumor cell cultures exhibiting high metastatic potential. Taken
together, miR-221 should be considered as an oncomiR and, for this
reason, a strong candidate for miRNA-therapeutics based on
antagomiR molecules. Target molecules of miR-221 have been firmly
established, such as DVL2, PUMA, PTEN, p27Kip1. In the
context of breast tumors, of great interest is the study published
by Galardi et al identifying p27Kip1 mRNA as a
possible target of miR-221 (20).
This finding is very intriguing, since p27Kip1 has been
proposed as a tumor suppressor gene, which is downregulated in
several types of tumors. These data support the concept that
targeting miR-221 with antagomiR molecules might lead to an
increased expression of the tumor-suppressor p27Kip1,
bringing novel treatment options to anticancer therapy.
The major conclusions of this study are that a PNA
against miR-221 is efficiently internalized within target cells
only if linked to an arginine-rich peptide, strongly inhibits
miR-221 activity and deeply alters the expression of the
p27Kip1 gene. Unlike commercially available antagomiRs,
which need continuous administration, a single administration of
Rpep-PNA-a221 is sufficient to obtain the biological effects.
Interestingly, modifications allowing efficient uptake by target
cells are necessary to obtain the biological activity, since
PNA-a221, despite being able to hybridize to the target nucleotide
sequence (Fig. 3A) is not
internalized (Fig. 4A) and
displays a very low activity on cells (Figs. 5 and 6). Therefore, conjugation with the
octa-arginine peptide, according to a previously developed strategy
for K562 cells, turned out to be effective also in the MDA-MB-231
cellular system; we would like to underline that the delivery of
Rpep-PNA-a221 needs no transfection reagents (i.e. lipofectin,
lipofectamine or similar reagents) which, on the contrary, are
required when RNA or DNA based analogues are used. The
octa-arginine peptide has also beneficial effects in terms of
affinity for nucleic acid targets, by an additional contribution of
electrostatic interactions to the sequence specific base-pairing
interactions of the PNA.
From a theoretical point of view, these studies
fully support the concept that p27Kip1 mRNA might be
considered among possible targets of miR-221. In fact, in the
presence of Rpep-PNA-a221 we observed effects in MDA-MB-231 cells
compatible with a decrease of miR-221 (it should be underlined that
the effects of Rpep-PNA-a221 might be based on binding to mature
miR-221, but also to pre-miRNA sequences) and increase of
p27Kip1.
From a general point of view, our results allow to
propose PNA-based molecules as very promising reagents to modulate
the biological activity of microRNAs and to encourage further
research on PNA analogues to increase efficiency of delivery,
stability and change of intracellular distribution in view of the
selected miRNA targets, i.e. mature miRNA, pre-miRNA or pri-miRNA
sequences.
Despite the fact that in this study we focused our
attention on breast cancer cellular model systems, we like to
underline that p27Kip1/miR-221 are deeply involved in
other tumors for which PNA-based treatments are expected to be
appealing. One example is glioma, which expresses high levels of
miR-221 and downregulated p27Kip1, that should be
considered the major onco-suppressor protein in this tumor type.
Zhang et al first demonstrated that miR-221/222 promote
malignant progression of glioma through activation of the Akt
pathway and inhibition of p27Kip1; in a further study,
the same group reported that co-suppression of miR-221/222 cluster
suppresses human glioma cell growth by targeting p27Kip1
in vitro and in vivo. Since delivery systems of PNA
across the BBB have been described (50,51)
and uptake of PNA within neuronal cells has been demonstrated to be
more efficient than other cellular types (22,52),
our data can be a relevant starting point for the development of
therapeutic strategies using PNAs targeting miR-221 and restoring
p27Kip1 levels in gliomas.
Abbreviations:
|
AEEA
|
2-(2-aminoethoxy)ethoxyacetyl
spacer;
|
|
DhBtOH
|
3-hydroxy-1,2,3-benzotriazin-4-(3H)-one;
|
|
DIC
|
N,N′-diisopropylcarbodiimide;
|
|
DIPEA
|
N,N′-diisopropylethylamine;
|
|
FACS
|
fluorescence-activated cell
sorter;
|
|
FBS
|
fetal bovine serum;
|
|
Fl
|
fluorescein;
|
|
HBTU
|
O-benzotriazol-1-yl-N,N,N′,N′-tetramethyluronium
hexafluorophosphate;
|
|
MBHA
|
(4-methylbenhydryl)amine;
|
|
PBS
|
phosphate-buffered saline;
|
|
PNA
|
peptide nucleic acid;
|
|
3′UTR
|
3′-untranslated region;
|
|
RT-qPCR
|
retro transcription-quantitative
polymerase chain reaction;
|
|
TFA
|
trifluoroacetic acid;
|
|
EDTA
|
ethylenediaminetetraacetic acid;
|
|
SDS
|
sodium dodecyl sulfate;
|
|
HRP
|
horseradish peroxidase;
|
|
RISC
|
RNA-induced silencing complex
|
Acknowledgements
This study was partially supported by
a grant from MIUR [PRIN09 grant n. 20093N774P ‘Molecular
recognition of micro-RNA (miR) by modified PNA: from structure to
activity’]. R.G. is granted by Fondazione Cariparo (Cassa di
Risparmio di Padova e Rovigo), by UE ITHANET Project
(Infrastructure for the Thalassaemia Research Network), by Telethon
(contract GGP10214). This research is also supported by CIB
(Interuniversity Consortium for Biotechnologies) and by
Associazione Veneta per la Lotta alla Talassemia (AVLT),
Rovigo.
References
|
1.
|
Filipowicz W, Jaskiewicz L, Kolb FA and
Pillai RS: Post-transcriptional gene silencing by siRNAs and
miRNAs. Curr Opin Struct Biol. 15:331–341. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
He L and Hannon GJ: MicroRNAs: small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Kozomara A and Griffiths-Jones S: miRBase:
integrating microRNA annotation and deep-sequencing data. Nucleic
Acids Res. 39:D152–D157. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Krol J, Loedige I and Filipowicz W: The
widespread regulation of microRNA biogenesis, function and decay.
Nat Rev Genet. 11:597–610. 2010.PubMed/NCBI
|
|
5.
|
Cho WCS: OncomiRs: the discovery and
progress of microRNAs in cancers. Mol Cancer. 6:602007. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Edmonds MD, Hurst DR and Welch DR: Linking
metastasis suppression with metastamiR regulation. Cell Cycle.
17:2673–2675. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Shah MY and Calin GA: MicroRNAs miR-221
and miR-222: a new level of regulation in aggressive breast cancer.
Genome Med. 3:56–68. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Zhang CZ, Zhang JX, Zhang AL, et al:
MiR-221 and miR-222 target PUMA to induce cell survival in
glioblastoma. Mol Cancer. 9:2292010. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Fu X, Wang Q, Chen J, et al: Clinical
significance of miR-221 and its inverse correlation with
p27Kip1 in hepatocellular carcinoma. Mol Biol Rep.
38:3029–3035. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Park JK, Lee EJ, Esau C and Schmittgen TD:
Antisense inhibition of microRNA-21 or -221 arrests cell cycle,
induces apoptosis, and sensitizes the effects of gemcitabine in
pancreatic adenocarcinoma. Pancreas. 38:e190–e199. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Kanemaru H, Fukushima S, Yamashita J, et
al: The circulating microRNA-221 level in patients with malignant
melanoma as a new tumor marker. J Dermatol Sci. 3:187–193. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Frenquelli M, Muzio M, Scielzo C, et al:
MicroRNA and proliferation control in chronic lymphocytic leukemia:
functional relationship between miR-221/222 cluster and p27. Blood.
19:3949–3959. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Visone R, Russo L, Pallante P, et al:
MicroRNAs (miR)-221 and miR-222, both overexpressed in human
thyroid papillary carcinomas, regulate p27Kip1 protein levels and
cell cycle. Endocr Relat Cancer. 14:791–798. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Zheng C, Yinghao S and Li J: MiR-221
expression affects invasion potential of human prostate carcinoma
cell lines by targeting DVL2. Med Oncol. 29:815–822. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Zhang C, Zhang J, Zhang A, Wang Y, Han L,
You Y, Pu P and Kang C: PUMA is a novel target of miR-221/222 in
human epithelial cancers. Int J Oncol. 6:1621–1626. 2010.PubMed/NCBI
|
|
16.
|
Chun-Zhi Z, Lei H, An-Ling Z, et al:
MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell
proliferation and radioresistance by targeting PTEN. BMC Cancer.
10:3672010. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Lu X, Zhao P, Zhang C, et al: Analysis of
miR-221 and p27 expression in human gliomas. Mol Med Rep.
4:651–656. 2009.PubMed/NCBI
|
|
18.
|
Zhang C, Kang C, You Y, et al:
Co-suppression of miR-221/222 cluster suppresses human glioma cell
growth by targeting p27kip1 in vitro and in
vivo. Int J Oncol. 34:1653–1660. 2009.PubMed/NCBI
|
|
19.
|
Le Sage C, Nagel R, Egan DA, et al:
Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222
promotes cancer cell proliferation. EMBO J. 26:3699–3708.
2007.PubMed/NCBI
|
|
20.
|
Galardi S, Mercatelli N, Giorda E,
Massalini S, Frajese GV, Ciafrè SA and Farace MG: miR-221 and
miR-222 expression affects the proliferation potential of human
prostate carcinoma cell lines by targeting p27Kip1. J Biol Chem.
282:23716–23724. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Marin VL, Roy S and Armitage BA: Recent
advances in the development of peptide nucleic acid as a
gene-targeted drug. Expert Opin Biol Ther. 4:337–348. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Pession A, Tonelli R, Fronza R, et al:
Targeted inhibition of NMYC by peptide nucleic acid (PNA) in N-myc
amplified human neuroblastoma cells: cell-cycle inhibition with
induction of neuronal cell differentiation and apoptosis. Int J
Oncol. 24:265–272. 2004.PubMed/NCBI
|
|
23.
|
Gambari R: Biological activity and
delivery of peptide nucleic acids (PNA)-DNA chimeras for
transcription factor decoy (TFD) pharmacotherapy. Curr Med Chem.
11:1253–1263. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Nielsen PE, Egholm M, Berg RH and Buchardt
O: Sequence-selective recognition of DNA by strand displacement
with a thymine-substituted polyamide. Science. 254:1497–1500. 1991.
View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Nastruzzi C, Cortesi R, Esposito E, et al:
Liposomes as carriers for DNA-PNA hybrids. J Control Release.
68:237–249. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Paulasova P and Pellestor F: The peptide
nucleic acids (PNAs): a new generation of probes for genetic and
cytogenetic analyses. Ann Genet. 47:349–358. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Karkare S and Bhatnagar D: Promising
nucleic acid analogs and mimics: characteristic features and
applications of PNA, LNA, and morpholino. Appl Microbiol
Biotechnol. 71:575–586. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Menchise V, De Simone G, Tedeschi T, et
al: Insights into peptide nucleic acid (PNA) structural features:
the crystal structure of a D-lysine-based chiral PNA-DNA duplex.
Proc Natl Acad Sci USA. 100:12021–12026. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Nielsen PE: Antisense peptide nucleic
acids. Curr Opin Mol Ther. 2:282–287. 2000.
|
|
30.
|
Soomets U, Hällbrink M and Langel U:
Antisense properties of peptide nucleic acids. Front Biosci.
4:D782–D786. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Ray A and Nordén B: Peptide nucleic acid
(PNA): its medical and biotechnical applications and promise for
the future. FASEB J. 14:1041–1060. 2000.PubMed/NCBI
|
|
32.
|
Tonelli R, Purgato S, Camerin C, Fronza S,
et al: Anti-gene peptide nucleic acid specifically inhibits MYCN
expression in human neuroblastoma cells leading to persistent cell
growth inhibition and apoptosis. Mol Cancer Ther. 4:779–786. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Nielsen PE: Targeting double stranded DNA
with peptide nucleic acid (PNA). Curr Med Chem. 8:545–550. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Borgatti M, Lampronti I, Romanelli A, et
al: Transcription factor decoy molecules based on a peptide nucleic
acid (PNA)-DNA chimera mimicking Sp1 binding sites. J Biol Chem.
278:7500–7509. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Rasmussen FW, Bendifallah N, Zachar V, et
al: Evaluation of transfection protocols for unmodified and
modified peptide nucleic acid (PNA) oligomers. Oligonucleotides.
16:43–57. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Cortesi R, Mischiati C, Borgatti M, et al:
Formulations for natural and peptide nucleic acids based on
cationic polymeric submicron particles. AAPS J. 6:10–21. 2004.
|
|
37.
|
Borgatti M, Breda L, Cortesi R, et al:
Cationic liposomes as delivery systems for double-stranded PNA-DNA
chimeras exhibiting decoy activity against NF-kappaB transcription
factors. Biochem Pharmacol. 64:609–616. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Abes R, Arzumanov A, Moulton H, et al:
Arginine-rich cell penetrating peptides: design, structureactivity,
and applications to alter pre-mRNA splicing by steric-block
oligonucleotides. J Pept Sci. 14:455–460. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Torres AG, Threlfall RN and Gait MJ:
Potent and sustained cellular inhibition of miR-122 by
lysine-derivatized peptide nucleic acids (PNA) and phosphorothioate
locked nucleic acid (LNA)/2′-O-methyl (OMe) mixmer anti-miRs in the
absence of transfection agents. Artificial DNA PNA XNA. 2:71–78.
2011.PubMed/NCBI
|
|
40.
|
Fabani MM and Gait MJ: miR-122 targeting
with LNA/2′-O-methyl oligonucleotide mixmers, peptide nucleic acids
(PNA), and PNA-peptide conjugates. RNA. 14:336–346. 2008.
|
|
41.
|
Fabani MM, Abreu-Goodger C, Williams D, et
al: Efficient inhibition of miR-155 function in vivo by peptide
nucleic acids. Nucleic Acids Res. 38:4466–4475. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42.
|
Yan LX, Wu QN, Zhang Y, et al: Knockdown
of miR-21 in human breast cancer cell lines inhibits proliferation,
in vitro migration and in vivo tumor growth. Breast Cancer Res.
13:R22011. View Article : Google Scholar : PubMed/NCBI
|
|
43.
|
Bianchi N, Zuccato C, Lampronti I,
Borgatti M and Gambari R: Expression of miR-210 during erythroid
differentiation and induction of gamma-globin gene expression. BMB
Rep. 42:493–499. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
44.
|
Fabbri E, Manicardi A, Tedeschi T, et al:
Modulation of the biological activity of microRNA-210 with peptide
nucleic acids (PNAs). Chem Med Chem. 6:2192–2202. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45.
|
Mizuma M, Katayose Y, Yamamoto K, et al:
Up-regulated p27Kip1 reduces matrix metalloproteinase-9 and
inhibits invasion of human breast cancer cells. Anticancer Res.
28(5A): 2669–2677. 2008.PubMed/NCBI
|
|
46.
|
Manicardi A, Calabretta A, Bencivenni M,
Tedeschi T, Sforza S, Corradini R and Marchelli R: Affinity and
selectivity of C2- and C5-substituted ‘chiral-box’ PNA in solution
and on microarrays. Chirality. 22(Suppl 1): E161–E172.
2010.PubMed/NCBI
|
|
47.
|
Cailleau R, Olivé M and Cruciger QV:
Long-term human breast carcinoma cell lines of metastatic origin:
preliminary characterization. In Vitro. 14:911–915. 1978.
View Article : Google Scholar : PubMed/NCBI
|
|
48.
|
Brown I, Shalli K, McDonald SL, Moir SE,
Hutcheon AW, Heys SD and Schofield AC: Reduced expression of p27 is
a novel mechanism of docetaxel resistance in breast cancer cells.
Breast Cancer Res. 6:R601–R607. 2004. View
Article : Google Scholar : PubMed/NCBI
|
|
49.
|
Faccini A, Tortori A, Tedeschi T, et al:
Circular dichroism study of DNA binding by a potential anticancer
peptide nucleic acid targeted against the MYCN oncogene. Chirality.
20:494–500. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
50.
|
Suzuki T, Wu D, Schlachetzki F, Li JY,
Boado RJ and Pardridge WM: Imaging endogenous gene expression in
brain cancer in vivo with 111In-peptide nucleic acid antisense
radiopharmaceuticals and brain drug-targeting technology. J Nucl
Med. 45:1766–1775. 2004.PubMed/NCBI
|
|
51.
|
Pardridge WM, Boado RJ and Kang YS:
Vector-mediated delivery of a polyamide (‘peptide’) nucleic acid
analogue through the blood-brain barrier in vivo. Proc Natl Acad
Sci USA. 92:5592–5596. 1995.PubMed/NCBI
|
|
52.
|
Adlerz L, Soomets U, Holmlund L, Viirlaid
S, Langel U and Iverfeldt K: Down-regulation of amyloid precursor
protein by peptide nucleic acid oligomer in cultured rat primary
neurons and astrocytes. Neurosci Lett. 336:55–59. 2003. View Article : Google Scholar : PubMed/NCBI
|