Introduction
Breast cancer is one of the most common solid
malignant tumors among women worldwide. Breast cancer is a
heterogeneous disease that is currently classified based on the
expression of estrogen receptor (ER), progesterone receptor (PgR),
and the human epidermal growth factor receptor 2 (HER2) (1,2). For
patients with ER- or PgR-positive breast cancer, approximately five
years of adjuvant endocrine therapy reduces the annual breast
cancer death rate by approximately 30% (3). The addition of HER2-antagonist
trastuzumab to adjuvant chemotherapy has improved the prognosis of
HER2-positive breast cancer patients (4–6). In
contrast, triple negative breast cancer (TNBC), defined as tumors
that are negative for ER, PgR and HER2 overexpression, accounts for
at least 15–20% of all breast cancers, and the prognosis for TNBC
patients is poor because of its propensity for recurrence and
metastasis and a lack of clinically-established targeted therapies
(7,8). Therefore, only neoadjuvant
chemotherapy with conventional cytotoxic agents yield an excellent
outcome for TNBC patients who have a complete pathological
response, but the outcome for the vast majority with residual
disease after chemotherapy is relatively poor compared to non-TNBC
patients (6,7). Thus, because the heterogeneity of
breast cancer makes it difficult to treat many subtypes, including
TNBC, the molecular mechanisms of the carcinogenesis of TNBC must
be elucidated to develop novel molecular-targeted therapies that
improve the clinical outcome of TNBC patients.
Current ‘omics’ technology including DNA microarray
analysis can provide very helpful information that can be used to
categorize the characteristics of various malignant tumors and
identify genes that may be applicable for the development of novel
molecular targets for therapeutic modalities (9). To this end, we analyzed the gene
expression profile of 30 TNBC cells and normal breast ductal cells
that were purified by laser-microbeam microdissection and
identified a number of cancer-specific genes that might contribute
to the carcino genesis of TNBC. TNBC gene expression profiling
analysis can provide comprehensive information on the molecular
mechanism underlying the carcinogenesis of TNBC and possibly lead
to the development of novel effective therapies.
Materials and methods
Clinical samples and cell lines
A total of 48 TNBC (18 cases did not entry DNA
microarray analysis) and 13 normal mammary tissues were obtained
with informed consent from patients who were treated at Tokushima
Breast Care Clinic, Tokushima, Japan. This study, as well as the
use of all clinical materials described above, was approved by the
Ethics Committee of The University of Tokushima. Clinical
information was obtained from medical records and tumors were
diagnosed as triple-negative by pathologists when
immunohistochemical staining was ER-negative, PR-negative, and HER2
(0 or 1+). The clinicopathological features of each patient are
summarized in Table I. Samples
were immediately embedded in TissueTek OCT compound (Sakura, Tokyo,
Japan), frozen, and stored at −80°C. Human TNBC cell lines
MDA-MB-231, BT-20, BT-549, HCC1143, and HCC1937 were purchased from
the American Type Culture Collection (ATCC, Rockville, MD, USA).
The human normal breast epithelial cell line, MCF10A, was purchased
from Cambrex Bioscience, Inc. All cells were cultured under the
conditions recommended by their respective depositors.
 | Table I.Clinicopathological features of 48
TNBC patients. |
Table I.
Clinicopathological features of 48
TNBC patients.
| ID | Age | Histology | TNM | Stage | ER/PgR/HER2 | Microarray | RT-PCR |
|---|
| 1 | 44 |
Papillo-tubular | T0N3M1 | IV | −/−/0 | Done | Done |
| 8 | 79 | DCIS | T1N0M0 | I | −/−/0 | Not done | Done |
| 10 | 57 |
Papillo-tubular | T1N0M0 | I | −/−/1+ | Not done | Done |
| 19 | 63 | Solid-tubular | T1N0M0 | I | −/−/0 | Not done | Done |
| 27 | 60 | Solid-tubular | T2N1M0 | II | −/−/0 | Done | Done |
| 42 | 59 | Solid-tubular | T2N0M0 | II | −/−/0 | Not done | Done |
| 44 | 79 |
Papillo-tubular | Recurrence | - | −/−/1+ | Not done | Done |
| 53 | 55 |
Papillo-tubular | T1N0M0 | I | −/−/0 | Not done | Done |
| 54 | 77 | Solid-tubular | T1N1M0 | II | −/−/0 | Not done | Done |
| 56 | 28 | Scirrhous | T2N1M0 | II | −/−/0 | Done | Done |
| 57 | 58 | Solid-tubular | T1N1M0 | II | −/−/0 | Not done | Done |
| 60 | 54 | Solid-tubular | T2N1M0 | II | −/−/0 | Done | Done |
| 64 | 60 |
Papillo-tubular | T2N0M0 | II | −/−/0 | Not done | Done |
| 66 | 59 | Special type | T2N1M0 | II | −/−/0 | Not done | Done |
| 78 | 45 | Solid-tubular | T2N1M0 | II | −/−/0 | Done | Done |
| 89 | 44 |
Papillo-tubular | Recurrence | - | −/−/0 | Not done | Done |
| 95 | 60 | Solid-tubular | T1N0M0 | I | −/−/0 | Not done | Done |
| 101 | 60 | Scirrhous | T2N1M0 | II | −/−/0 | Not done | Done |
| 110 | 77 | Scirrhous | T2N1M0 | II | −/−/1+ | Not done | Done |
| 116 | 70 | Solid-tubular | T2N1M0 | II | −/−/0 | Done | Done |
| 155 | 36 | Solid-tubular | T1N1M0 | II | −/−/0 | Done | Done |
| 225 | 49 |
Papillo-tubular | T2N1M0 | II | −/−/1+ | Not done | Done |
| 252 | 49 | Solid-tubular | T2N1M0 | II | −/−/1+ | Done | Done |
| 253 | 49 | Scirrhous | T2N1M0 | II | −/−/0 | Done | Done |
| 265 | 80 | Scirrhous | T1N1M0 | II | −/−/0-1+ | Done | Done |
| 313 | 53 | Scirrhous | T3N2M0 | III | −/−/0 | Done | Done |
| 337 | 42 | Solid-tubular | T2N1M0 | II | −/−/1+ | Done | Done |
| 359 | 55 |
Papillo-tubular | T2N0M0 | II | −/−/0 | Done | Done |
| 362 | 37 |
Papillo-tubular | T2N1M0 | II | −/−/0 | Done | Done |
| 363 | 69 |
Papillo-tubular | T2N0M0 | II | −/−/0 | Done | Done |
| 366 | 61 | Special type | T2N1M0 | II | −/−/0-1+ | Done | Done |
| 384 | 32 |
Papillo-tubular | T3N0M0 | II | −/−/0 | Done | Done |
| 392 | 46 |
Papillo-tubular | T1N1M0 | II | −/−/0 | Done | Done |
| 414 | 60 |
Papillo-tubular | T2N1M0 | II | −/−/1+ | Not done | Done |
| 415 | 54 | Solid-tubular | T2N0M0 | II | −/−/1+ | Done | Done |
| 420 | 41 | Solid-tubular | T3N0M0 | II | −/−/0 | Done | Done |
| 423 | 70 | Solid-tubular | T2N0M0 | II | −/−/0 | Done | Done |
| 438 | 63 | Solid-tubular | T3N0M0 | II | −/−/0 | Done | Done |
| 445 | 39 | Solid-tubular | T2N1M0 | II | −/−/0 | Done | Done |
| 453 | 50 | Solid-tubular | T2N1M0 | II | −/−/0 | Done | Done |
| 481 | 59 | Solid-tubular | T3N1M0 | III | −/−/0 | Done | Done |
| 528 | 55 | Solid-tubular | T2N1M0 | II | −/−/0 | Done | Done |
| 535 | 58 | Solid-tubular | T2N1M0 | II | −/−/0 | Not done | Done |
| 553 | 71 | Solid-tubular | T0N1M0 | II | −/−/1+ | Not done | Done |
| 558 | 56 | Solid-tubular | T2N1M0 | II | −/−/0 | Done | Done |
| 562 | 64 | Scirrhous | T2N0M0 | II | −/−/0 | Done | Done |
| 566 | 52 | Solid-tubular | T3N1M0 | III | −/−/0 | Done | Done |
| 651 | 45 | Scirrhous | T2N1M0 | II | −/−/0 | Done | Done |
Laser-microbeam microdissection (LMM),
RNA extraction, RNA amplification, and hybridization
Frozen specimens were serially sectioned in
8-μm slices with a cryostat (Leica, Herborn, Germany) and
stained with hematoxylin and eosin to define the analyzed regions.
We purified 48 TNBC and 13 normal ductal cells using the LMM system
(Carl Zeiss, Jena, Germany) according to the manufacturer’s
instructions. Dissected cancer and normal ductal cells were
dissolved in RLT lysis buffer (Qiagen, Valencia, CA, USA)
containing 1% β-mercaptoethanol. The extracted total RNA was
purified with an RNeasy Mini kit (Qiagen) according to the
manufacturer’s instructions. For RNA amplification and labeling, we
used an Agilent Low-Input QuickAmp labeling kit according to the
manufacturer’s instructions. Briefly, 100 ng of total RNA from each
sample was amplified using T7 RNA polymerase with simultaneous
Cy3-labeled CTP incorporation. Then, 2 μg of Cy3-labeled
cRNA was fragmented, hybridized onto the Agilent Whole Human Genome
Microarray 4×44K slide (Agilent Technologies, Palo Alto, CA, USA)
and then incubated with rotation at 65°C for 18 h. Then slides were
washed and scanned by the Agilent Microarray scanner system in an
ozone protection fume hood.
Microarray analysis
The features of scanned image files containing the
Cy3-fluorescence signals of the hybridized Agilent Microarrays were
extracted using the Agilent Feature Extraction (version 9.5)
(Agilent Technologies). The data were analyzed using GeneSpring
(version 11.5). We normalized the microarray data across all chips
and genes by quantile normalization, and baseline transformed the
signal values to the median in all samples. Finally, we performed
quality control and filtering steps based on flags and expression
levels. To identify genes that were significantly alternated
between TNBC and normal ductal cells the mean signal intensity
values in each analysis were compared. In this experiment, we
applied Mann-Whitney (unpaired) t-test and random permutation test
10,000 times for each comparison and adjusted for multiple
comparisons using the Benjamini Hochberg false discovery rate
(FDR). Gene expression levels were considered significantly
different when the FDR (corrected P-value) <5×10−4
(when comparing normal ductal cells and TNBC) and the fold change
was ≥5.0. Data from this microarray analysis has been submitted to
the NCBI Gene Expression Omnibus (GEO) archive as series
GSE38959.
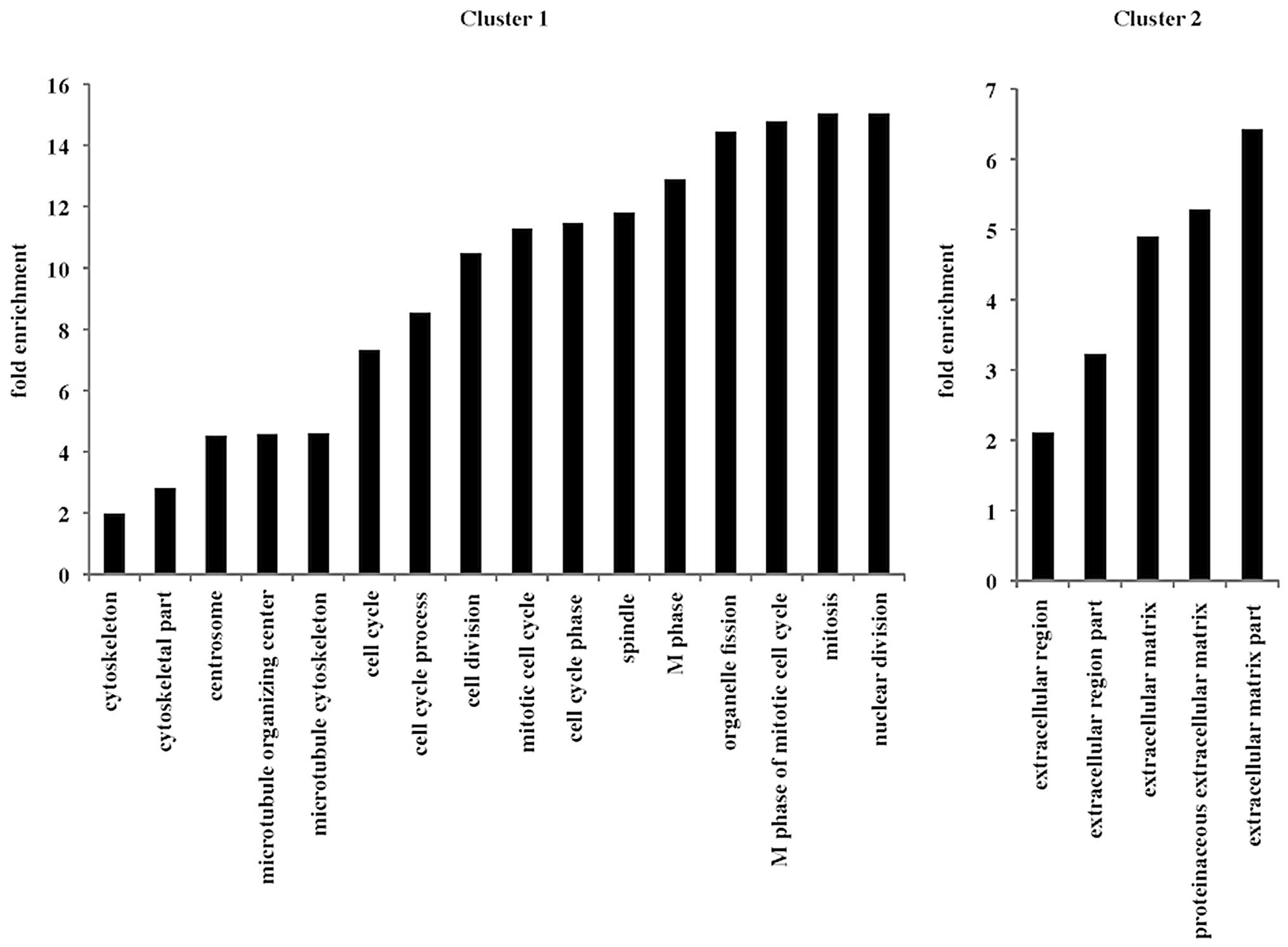
Functional gene annotation
clustering
The Database for Annotation, Visualization and
Integrated Discovery (DAVID 6.7) was approved to detect functional
gene annotation clusters based on gene expression profiling by gene
annotation enrichment analysis (http://david.abcc.ncifcrf.gov/) (10,11).
The clusters from the gene annotation enrichment analysis were
selected in this study based on a previous report (12).
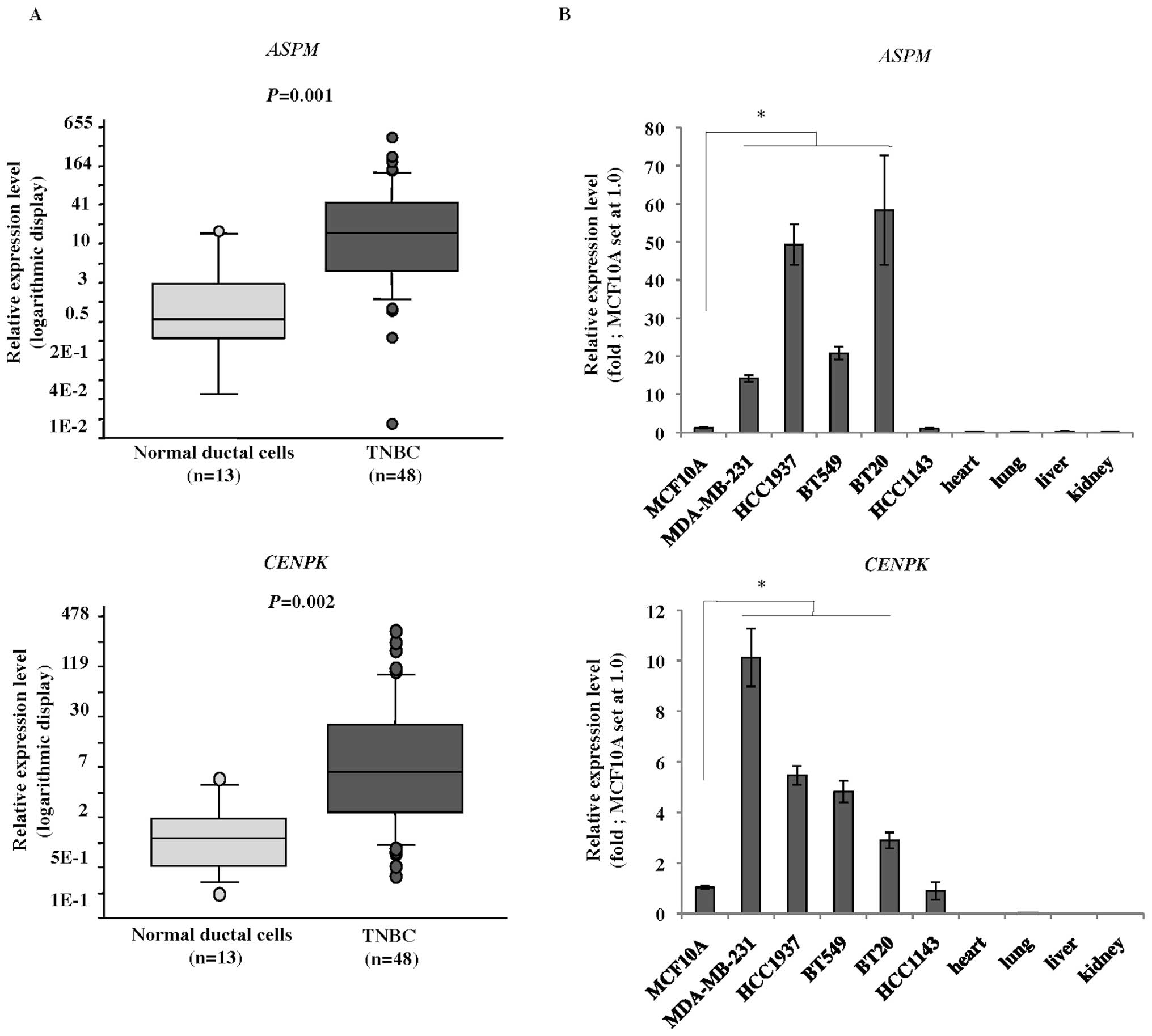
Quantitative reverse transcription-PCR
(qRT-PCR) analysis
Total RNA was extracted from each TNBC cell line and
clinical sample using an RNeasy mini kit (Qiagen) according to the
manufacturer’s instructions. Purified RNA from each clinical sample
and cell line, as well as poly-A RNA from normal human heart, lung,
liver, and kidney (Takara, Otsu, Japan) was reverse transcribed for
single-stranded cDNA using oligo(dT)12–18 primers with Superscript
II reverse transcriptase (Invitrogen, Life Technologies, Carlsbad,
CA, USA). qRT-PCR analysis was performed using an ABI PRISM 7500
Real-Time PCR system (Applied Biosystems, Life Technologies,
Carlsbad, CA, USA) and SYBR Premix Ex Taq (Takara) according to the
manufacturer’s instructions. The PCR primer sequences were as
follows: 5′-GCAGGTCTCC TTTCCTTTGCT-3′ and 5′-CTCGGCCTTCTTTGAGT
GGT-3′ for ASPM; 5′-CACTCACCGATTCAAATG CTC-3′ and
5′-ACCACCGTTGTTCCCTTTCT-3′ for CENPK; 5′-AAC
TTAGAGGTGGGGAGCAG-3′ and 5′-CACAACCATGCC TTACTTTATC-3′ for β2
microglobulin (β2-MG) as a quantitative control.
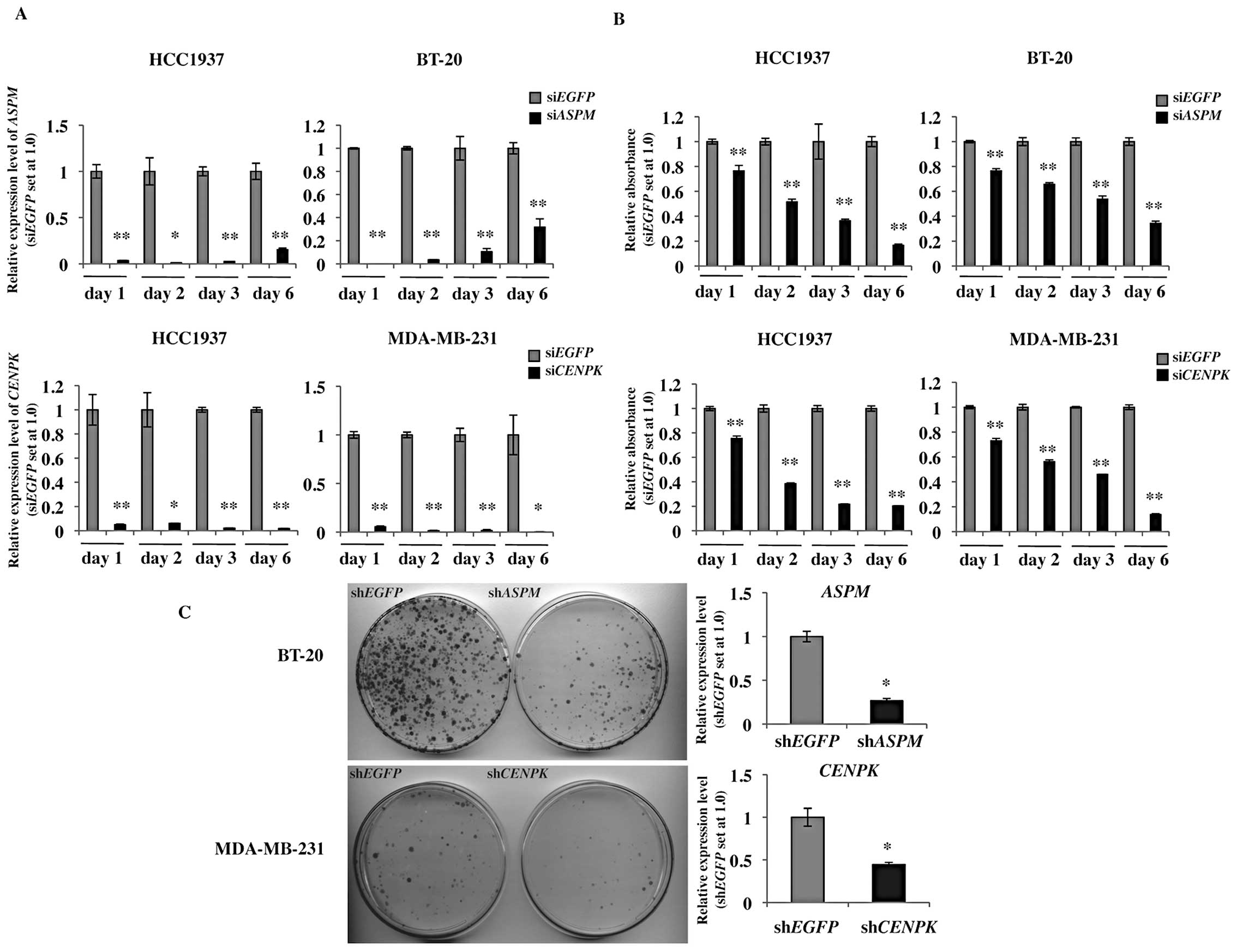
Gene-silencing effect by RNA
interference
Targeted sequences for ASPM and CENPK
were determined using an siRNA Targeted Finder (Applied Biosystems,
Life Technologies; http://www.ambion.com/techlib/misc/siRNA_finder.html).
The siRNA targeting sequences were 5′-CATACAGAAGT GCGAGAAA-3′ for
ASPM, 5′-CTCAGTCAATGGC AGAAAA-3′ for CENPK and
5′-GCAGCACGACTTCT TCAAG-3′ for EGFP as a control siRNA.
Human TNBC cell lines, HCC1937, MDA-MB-231 and BT-20, were plated
at a density of 1×104 cells per well in 12-wells for the
MTT assay and 3×104 cells per well in 6-well plates for
flow cytometry and RT-PCR analyses. Cells were transfected with
16.6 nM of each siRNA using Lipofectamine RNAiMAX Reagent
(Invitrogen). To evaluate the gene-silencing effects of the siRNAs
by qRT-PCR, total RNA was extracted from the siRNA-transfected
cells as described above after the indicated times. The following
specific qRT-PCR primer sets were used: 5′-CGGAAAAGAAAGAGCGATGG-3′
and 5′-ACCACCAAGTGAAGCCCTGT-3′ for ASPM and 5′-GG
GTGCCATCATTTTCTGGT-3′ and 5′-CCACCGTTGTT CCCTTTCTAAG-3′ for
CENPK. To evaluate cell viability, the MTT assay was
performed using the cell counting kit-8 reagent (Dojindo, Kumamoto,
Japan) according to the manufacturer’s instructions. Absorbance at
450 nm was measured with a micro-plate reader infinite 200 (Tecan,
Männedorf, Switzerland). These experiments were performed in
triplicate.
Colony formation assay
Vector-based shRNAs and the psiU6BX3 expression
system were constructed as previously described (13). The shRNA target sequences were the
same as those of the siRNA oligonucleotides. The DNA sequences of
all constructs were confirmed by DNA sequencing. BT-20 and
MDA-MB-231 cells were plated in 10-cm dishes (1×106
cells/dish) and transfected with 6 μg of
psiU6BX3.0-ASPM or psiU6BX3.0-CENPK and
psiU6BX3.0-EGFP as a control using Fugene-6 (Roche, Basel,
Switzerland) according to the manufacturer’s instructions.
Forty-eight hours after transfection, cells were re-seeded for a
colony formation assay (5.0×105 cells/10-cm dish) and
RT-PCR (5.0×105 cells/10-cm dish). We selected
psiU6BX3.0-transfected cells using selection medium containing 0.6
mg/ml of neomycin for BT-20 cells and 1.4 mg/ml for MDA-MB-231
cells. Total RNA was extracted from the cells after a 7-day
incubation with neomycin, and then the knockdown effects of the
siRNAs were examined by qRT-PCR. The specific primer sets for
quantitative RT-PCR were the same as those for the siRNA
oligonucleotides. Nineteen days after transfection, the cells were
fixed with 4% paraformaldehyde for 10 min and stained with Giemsa
solution (Merck, Darmstadt, Germany).
Cell cycle analysis
For flow cytometric analysis, adherent and detached
cells were harvested and fixed with 70% ethanol at room temperature
for 30 min. After washing with PBS (−), the cells were incubated at
37°C for 30 min with 1 mg/ml RNase I in PBS (−) and stained with 20
μg propidium iodide at room temperature for 30 min in the
dark. A total of 10,000 cells were analyzed for DNA content using
flow cytometry and CellQuest software (FACSCalibur; BD Biosciences,
Franklin Lakes, NJ, USA). Assays were performed in duplicate.
Immunocytochemical staining analysis
HCC1937 and MDA-MB-231 cells were plated onto a
2-well glass slide (Thermo Fisher Scientific, Rochester, NY, USA)
at a density of 1.0×104/well and incubated for 24 h
before siRNA trans fection. Forty-eight hours post-transfection,
the cells were fixed with 4% paraformaldehyde for 30 min at 4°C and
then permeablized with 0.1% Triton X-100 for 2 min at room
temperature. Subsequently, the cells were covered with 3% bovine
serum albumin for 60 min at room temperature and then incubated
with an anti-α/β tubulin antibody (Cell Signaling, Beverly, MA,
USA) diluted 1:50 for 1 h. After washing with PBS (−), the cells
were stained with an Alexa 488-conjugated anti-rabbit secondary
antibody (Molecular Probes, Eugene, OR, USA) diluted 1:1,000 for 1
h. The nuclei were counterstained with
4′,6′-diamidine-2′-phenylindole dihydrochloride (DAPI). Fluorescent
images were obtained using an IX71 microscope (Olympus, Tokyo,
Japan).
Statistical analysis
Statistical significance was calculated by
Mann-Whitney t-test using Stat View 5.0 J software (SAS Institute,
Inc., Cary, NC, USA) to compare the gene expression levels between
TNBC cells and normal ductal cells, and by Student’s two-sided
t-test using Microsoft® Excel 2008 to assess cell
proliferation, gene expression, and alteration of cell cycle. A
difference of P<0.05 was considered statistically
significant.
Results
Identification of genes upregulated or
downregulated in TNBCs
To obtain precise expression profiles of TNBC cells,
we used LMM to avoid contamination of non-cancer cells, such as
adipocytes, fibroblasts, and inflammatory cells from the tissue
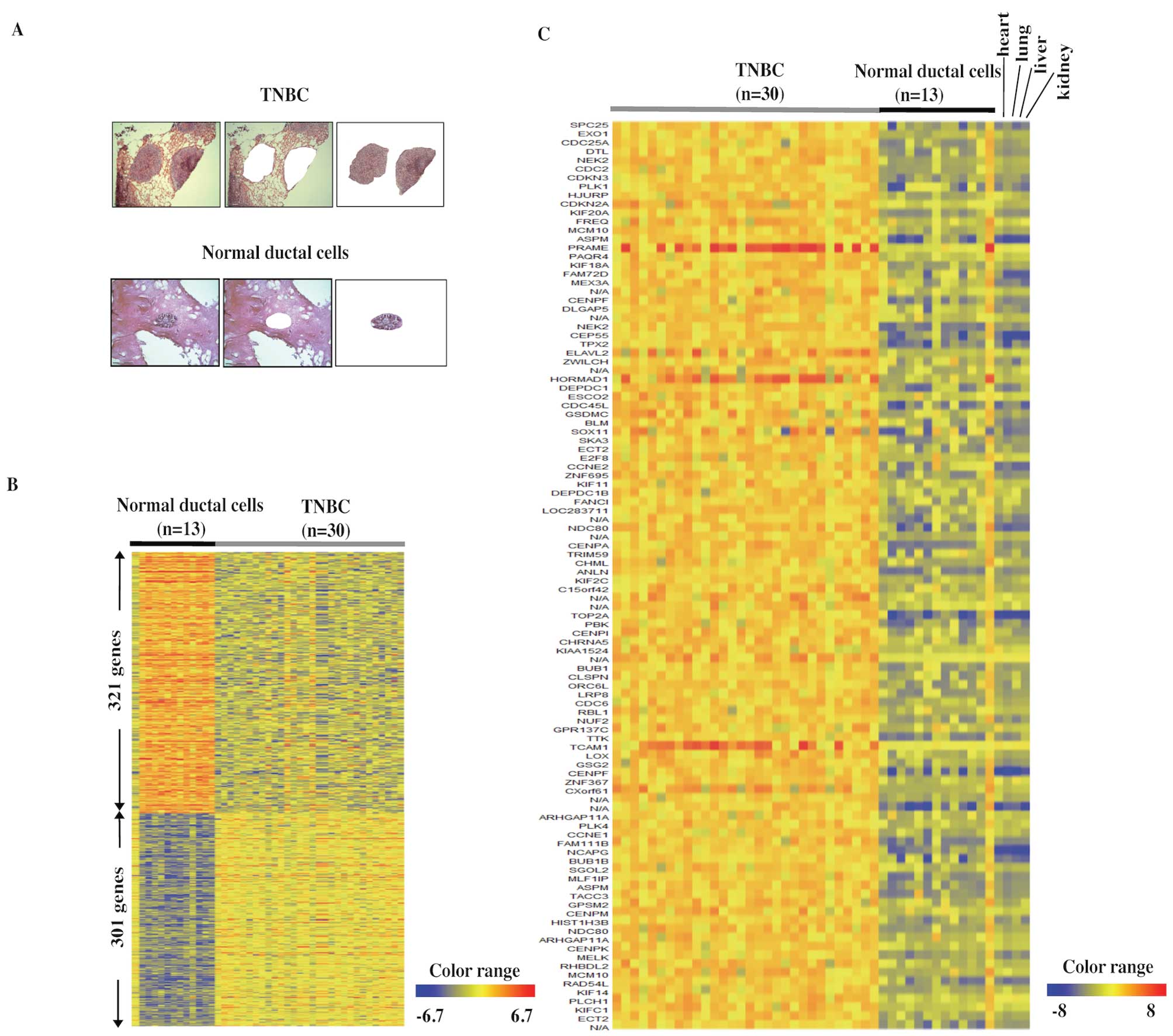
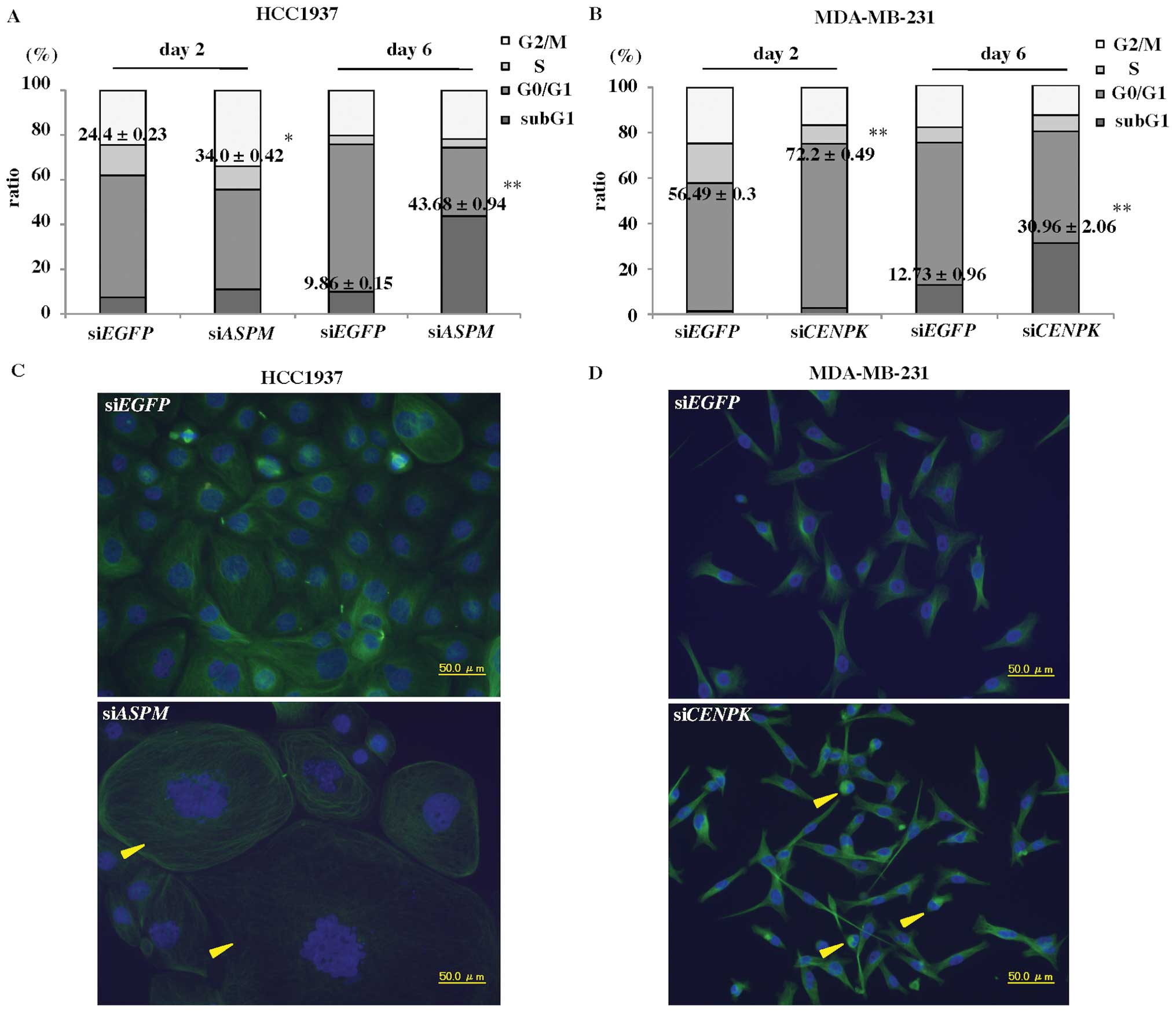
sections (Fig. 1A, upper panels).
Because breast cancer originates from normal breast ductal cells,
we used similarly purified populations of normal duct cells as
controls (Fig. 1A, lower panels).
The precise gene-expression profiles of TNBC by DNA microarray
identified 301 genes that were upregulated >5-fold in TNBC
compared to 13 normal ductal cells, and 321 genes that were
downregulated to <1/5 of the normal ductal cells (Fig. 1B). Table II lists the 301 upregulated genes
in TNBC, including ubiquitin-conjugating enzyme E2C (UBE2C)
(14), S100 calcium binding
protein P (S100P) (15),
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase)
(UCHL1) (16), pituitary
tumor-transforming 1 (PTTG1) (17), ubiquitinconjugating enzyme E2T
(UBE2T) (13),
ubiquitin-like with PHD and ring finger domains 1 (UHRF1)
(18), SIX homeobox 1
(SIX1) (19), and protein
regulator of cytokinesis 1 (PRC1) (20), which were previously reported to be
overexpressed in breast cancer and involved in mammary
carcinogenesis. In particular, topoisomerase (DNA) IIα
(TOP2A) (21,22), HORMA domain containing 1
(HORMAD1) (23), ATPase
family, Fatty acid binding protein 5 (psoriasis-associated)
(FABP5) (24), and AAA
domain containing 2 (ATAD2) (25) were previously reported to be
potentially involved in the carcinogenesis of TNBC, and to serve as
prognostic markers or therapeutic targets for TNBC.
 | Table II.Genes significantly upregulated in
TNBC compared with normal ductal cells. |
Table II.
Genes significantly upregulated in
TNBC compared with normal ductal cells.
| Probe ID | Accession no. | Symbol | Gene name | Fold change
(log) | P-value |
|---|
| A_24_P334130 | NM_054034 | FN1 | Fibronectin 1 | 5.33 | 1.26E-04 |
| A_24_P940678 | N/A | N/A | | 5.07 | 1.26E-04 |
| A_23_P367618 | NM_003412 | ZIC1 | Zic family member 1
(odd-paired homolog, Drosophila) | 5.01 | 1.26E-04 |
| A_23_P118834 | NM_001067 | TOP2A | Topoisomerase (DNA)
IIα 170 kDa | 4.76 | 1.26E-04 |
| A_32_P119154 | BE138567 | N/A | | 4.75 | 1.26E-04 |
| A_23_P35219 | NM_002497 | NEK2 | NIMA (never in
mitosis gene a)-related kinase 2 | 4.67 | 1.26E-04 |
| A_23_P166360 | NM_206956 | PRAME | Preferentially
expressed antigen in melanoma | 4.64 | 1.26E-04 |
| A_24_P332314 | NM_198947 | FAM111B | Family with
sequence similarity 111, member B | 4.63 | 1.26E-04 |
| A_24_P413884 | NM_001809 | CENPA | Centromere protein
A | 4.59 | 1.26E-04 |
| A_23_P68610 | NM_012112 | TPX2 | TPX2,
microtubule-associated, homolog (Xenopus laevis) | 4.58 | 1.26E-04 |
| A_23_P58266 | NM_005980 | S100P | S100 calcium
binding protein P | 4.57 | 1.26E-04 |
| A_24_P297539 | NM_181803 | UBE2C |
Ubiquitin-conjugating enzyme E2C | 4.49 | 1.26E-04 |
| A_23_P401 | NM_016343 | CENPF | Centromere protein
F, 350/400 ka (mitosin) | 4.44 | 1.26E-04 |
| A_23_P57379 | NM_003504 | CDC45L | CDC45 cell division
cycle 45-like (S. cerevisiae) | 4.44 | 1.26E-04 |
| A_23_P118815 | NM_001012271 | BIRC5 | Baculoviral IAP
repeat-containing 5 | 4.43 | 1.26E-04 |
| A_23_P210853 | NM_021067 | GINS1 | GINS complex
subunit 1 (Psf1 homolog) | 4.41 | 1.26E-04 |
| A_23_P258493 | NM_005573 | LMNB1 | Lamin B1 | 4.31 | 1.26E-04 |
| A_24_P119745 | NM_212482 | FN1 | Fibronectin 1 | 4.31 | 1.26E-04 |
| A_24_P680947 | BC044933 | KIF18B | Kinesin family
member 18B | 4.3 | 1.26E-04 |
| A_32_P92642 | N/A | N/A | | 4.3 | 1.26E-04 |
| A_23_P356684 | NM_018685 | ANLN | Anillin, actin
binding protein | 4.29 | 1.26E-04 |
| A_24_P314571 | BU616832 | N/A | | 4.24 | 1.26E-04 |
| A_23_P98580 | NM_004265 | FADS2 | Fatty acid
desaturase 2 | 4.2 | 1.26E-04 |
| A_23_P52017 | NM_018136 | ASPM | asp (abnormal
spindle) homolog, microcephaly associated (Drosophila) | 4.17 | 1.26E-04 |
| A_24_P20607 | NM_005409 | CXCL11 | Chemokine (C-X-C
motif) ligand 11 | 4.16 | 2.33E-04 |
| A_32_P199884 | NM_032132 | HORMAD1 | HORMA domain
containing 1 | 4.13 | 2.33E-04 |
| A_23_P70007 | NM_012484 | HMMR | Hyaluronan-mediated
motility receptor (RHAMM) | 4.11 | 1.26E-04 |
| A_23_P22378 | NM_003108 | SOX11 | SRY (sex
determining region Y)-box 11 | 4.1 | 1.26E-04 |
| A_23_P259586 | NM_003318 | TTK | TTK protein
kinase | 4.09 | 1.26E-04 |
| A_23_P200310 | NM_017779 | DEPDC1 | DEP domain
containing 1 | 4.08 | 1.26E-04 |
| A_24_P378331 | NM_170589 | CASC5 | Cancer
susceptibility candidate 5 | 4.06 | 1.26E-04 |
| A_23_P111888 | NM_138455 | CTHRC1 | Collagen triple
helix repeat containing 1 | 4.05 | 1.26E-04 |
| A_23_P48835 | NM_138555 | KIF23 | Kinesin family
member 23 | 4.05 | 1.26E-04 |
| A_23_P115872 | NM_018131 | CEP55 | Centrosomal protein
55 kDa | 4.03 | 1.26E-04 |
| A_23_P132956 | NM_004181 | UCHL1 | Ubiquitin
carboxyl-terminal esterase L1 (ubiquitin thiolesterase) | 4.03 | 1.26E-04 |
| A_24_P911179 | NM_018136 | ASPM | asp (abnormal
spindle) homolog, microcephaly associated (Drosophila) | 4.02 | 1.26E-04 |
| A_23_P408955 | NM_004091 | E2F2 | E2F transcription
factor 2 | 4.02 | 1.26E-04 |
| A_23_P7636 | NM_004219 | PTTG1 | Pituitary
tumor-transforming 1 | 4 | 1.26E-04 |
| A_23_P204941 | NM_004004 | GJB2 | Gap junction
protein, β2, 26 kDa | 4 | 1.26E-04 |
| A_23_P18452 | NM_002416 | CXCL9 | Chemokine (C-X-C
motif) ligand 9 | 3.94 | 2.33E-04 |
| A_24_P96780 | NM_016343 | CENPF | Centromere protein
F, 350/400 ka (mitosin) | 3.92 | 1.26E-04 |
| A_23_P69537 | NM_006681 | NMU | Neuromedin U | 3.9 | 1.26E-04 |
| A_24_P14156 | NM_006101 | NDC80 | NDC80
homolog, kinetochore complex component (S. cerevisiae) | 3.86 | 1.26E-04 |
| A_23_P254733 | NM_024629 | MLF1IP | MLF1 interacting
protein | 3.85 | 1.26E-04 |
| A_23_P74115 | NM_003579 | RAD54L | RAD54-like (S.
cerevisiae) | 3.84 | 1.26E-04 |
| A_23_P50108 | NM_006101 | NDC80 | NDC80 homolog,
kinetochore complex component (S. cerevisiae) | 3.84 | 1.26E-04 |
| A_24_P150160 | NM_004265 | FADS2 | Fatty acid
desaturase 2 | 3.83 | 1.26E-04 |
| A_23_P155815 | NM_022346 | NCAPG | Non-SMC condensin I
complex, subunit G | 3.82 | 1.26E-04 |
| A_23_P125278 | NM_005409 | CXCL11 | Chemokine (C-X-C
motif) ligand 11 | 3.81 | 1.26E-04 |
| A_23_P51085 | NM_020675 | SPC25 | SPC25, NDC80
kinetochore complex component, homolog (S. cerevisiae) | 3.81 | 1.26E-04 |
| A_23_P133123 | NM_032117 | MND1 | Meiotic nuclear
divisions 1 homolog (S. cerevisiae) | 3.8 | 1.26E-04 |
| A_32_P62997 | NM_018492 | PBK | PDZ binding
kinase | 3.8 | 1.26E-04 |
| A_23_P256956 | NM_005733 | KIF20A | Kinesin family
member 20A | 3.79 | 1.26E-04 |
| A_24_P933613 | N/A | N/A | | 3.78 | 1.26E-04 |
| A_23_P212844 | NM_006342 | TACC3 | Transforming,
acidic coiled-coil containing protein 3 | 3.78 | 1.26E-04 |
| A_24_P254705 | NM_020394 | ZNF695 | Zinc finger protein
695 | 3.76 | 1.26E-04 |
| A_23_P115482 | NM_014176 | UBE2T |
Ubiquitin-conjugating enzyme E2T
(putative) | 3.75 | 1.26E-04 |
| A_32_P201723 | N/A | N/A | | 3.73 | 1.26E-04 |
| A_23_P256425 | NM_014479 |
ADAMDEC1 | ADAM-like, decysin
1 | 3.73 | 1.26E-04 |
| A_23_P432352 | NM_001017978 | CXorf61 | Chromosome X open
reading frame 61 | 3.73 | 1.26E-04 |
| A_23_P208880 | NM_013282 | UHRF1 | Ubiquitin-like with
PHD and ring finger domains 1 | 3.72 | 1.26E-04 |
| A_23_P323751 | NM_030919 | FAM83D | Family with
sequence similarity 83, member D | 3.71 | 1.26E-04 |
| A_23_P48669 | NM_005192 | CDKN3 | Cyclin-dependent
kinase inhibitor 3 | 3.71 | 1.26E-04 |
| A_24_P234196 | NM_001034 | RRM2 | Ribonucleotide
reductase M2 | 3.69 | 1.26E-04 |
| A_23_P253791 | NM_004345 | CAMP | Cathelicidin
antimicrobial peptide | 3.69 | 1.26E-04 |
| A_23_P76914 | NM_005982 | SIX1 | SIX homeobox 1 | 3.67 | 4.43E-04 |
| A_23_P94571 | NM_004432 | ELAVL2 | ELAV (embryonic
lethal, abnormal vision, Drosophila)-like 2 (Hu antigen
B) | 3.67 | 1.26E-04 |
| A_23_P200222 | NM_033300 | LRP8 | Low density
lipoprotein receptor-related protein 8, apolipoprotein E
receptor | 3.67 | 1.26E-04 |
| A_24_P416079 | NM_016359 | NUSAP1 | Nucleolar and
spindle associated protein 1 | 3.66 | 1.26E-04 |
| A_23_P104651 | NM_080668 | CDCA5 | Cell division cycle
associated 5 | 3.65 | 1.26E-04 |
| A_23_P150667 | NM_031217 | KIF18A | Kinesin family
member 18A | 3.64 | 1.26E-04 |
| A_24_P859859 | N/A | N/A | | 3.63 | 4.43E-04 |
| A_23_P312150 | NM_001956 | EDN2 | Endothelin 2 | 3.61 | 1.26E-04 |
| A_23_P375 | NM_018101 | CDCA8 | Cell division cycle
associated 8 | 3.59 | 1.26E-04 |
| A_32_P68525 | BC035392 | N/A | | 3.58 | 1.26E-04 |
| A_23_P43490 | NM_058197 | CDKN2A | Cyclin-dependent
kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | 3.56 | 1.26E-04 |
| A_23_P1691 | NM_002421 | MMP1 | Matrix
metallopeptidase 1 (interstitial collagenase) | 3.55 | 1.26E-04 |
| A_23_P117852 | NM_014736 |
KIAA0101 | KIAA0101 | 3.54 | 1.26E-04 |
| A_24_P319613 | NM_002497 | NEK2 | NIMA (never in
mitosis gene a)-related kinase 2 | 3.53 | 1.26E-04 |
| A_23_P10385 | NM_016448 | DTL | Denticleless
homolog (Drosophila) | 3.53 | 1.26E-04 |
| A_32_P1173 | NM_138441 |
C6orf150 | Chromosome 6 open
reading frame 150 | 3.51 | 1.26E-04 |
| A_23_P94422 | NM_014791 | MELK | Maternal embryonic
leucine zipper kinase | 3.5 | 1.26E-04 |
| A_23_P340909 | BC013418 | SKA3 | Spindle and
kinetochore associated complex subunit 3 | 3.48 | 1.26E-04 |
| A_23_P385861 | NM_152562 | CDCA2 | Cell division cycle
associated 2 | 3.47 | 1.26E-04 |
| A_23_P124417 | NM_004336 | BUB1 | Budding uninhibited
by benzimidazoles 1 homolog (yeast) | 3.47 | 1.26E-04 |
| A_24_P257099 | NM_018410 | HJURP | Holliday junction
recognition protein | 3.43 | 1.26E-04 |
| A_24_P270460 | NM_005532 | IFI27 | Interferon,
α-inducible protein 27 | 3.41 | 2.33E-04 |
| A_23_P206059 | NM_003981 | PRC1 | Protein regulator
of cytokinesis 1 | 3.39 | 1.26E-04 |
| A_23_P74349 | NM_145697 | NUF2 | NUF2, NDC80
kinetochore complex component, homolog (S. cerevisiae) | 3.36 | 1.26E-04 |
| A_24_P302584 | NM_003108 | SOX11 | SRY (sex
determining region Y)-box 11 | 3.36 | 4.43E-04 |
| A_24_P68088 | NR_002947 | TCAM1 | Testicular cell
adhesion molecule 1 homolog (mouse) | 3.35 | 2.33E-04 |
| A_24_P605612 | NM_003247 | THBS2 | Thrombospondin
2 | 3.34 | 1.26E-04 |
| A_24_P366033 | NM_018098 | ECT2 | Epithelial cell
transforming sequence 2 oncogene | 3.34 | 1.26E-04 |
| A_23_P93258 | NM_003537 |
HIST1H3B | Histone cluster 1,
H3b | 3.33 | 1.26E-04 |
| A_23_P211762 | N/A | COL8A1 | Collagen, type
VIII, α1 | 3.29 | 4.43E-04 |
| A_23_P77493 | NM_006086 | TUBB3 | Tubulin, β3 | 3.29 | 1.26E-04 |
| A_23_P204947 | NM_004004 | GJB2 | Gap junction
protein, β2, 26 kDa | 3.29 | 1.26E-04 |
| A_23_P149668 | NM_014875 | KIF14 | Kinesin family
member 14 | 3.29 | 1.26E-04 |
| A_23_P34325 | NM_033300 | LRP8 | Low density
lipoprotein receptor-related protein 8, apolipoprotein E
receptor | 3.28 | 1.26E-04 |
| A_32_P56154 | N/A | N/A | | 3.28 | 1.26E-04 |
| A_32_P10403 | BU618641 |
SERPINE1 | Serpin peptidase
inhibitor, clade E (nexin, plasminogen activator inhibitor type 1),
member 1 | 3.27 | 1.26E-04 |
| A_23_P138507 | NM_001786 | CDC2 | Cell division cycle
2, G1→S and G2→M | 3.24 | 1.26E-04 |
| A_23_P48513 | NM_005532 | IFI27 | Interferon,
α-inducible protein 27 | 3.23 | 1.26E-04 |
| A_23_P49972 | NM_001254 | CDC6 | Cell division cycle
6 homolog (S. cerevisiae) | 3.22 | 1.26E-04 |
| A_24_P306896 | XR_040656 |
LOC283711 | Hypothetical
protein LOC283711 | 3.22 | 1.26E-04 |
| A_23_P44684 | NM_018098 | ECT2 | Epithelial cell
transforming sequence 2 oncogene | 3.21 | 1.26E-04 |
| A_24_P161773 | N/A | N/A | | 3.2 | 1.26E-04 |
| A_23_P100344 | NM_014321 | ORC6L | Origin recognition
complex, subunit 6 like (yeast) | 3.2 | 1.26E-04 |
| A_32_P162183 | NM_000063 | C2 | Complement
component 2 | 3.18 | 1.26E-04 |
| A_23_P163481 | NM_001211 | BUB1B | Budding uninhibited
by benzimidazoles 1 homolog β (yeast) | 3.17 | 1.26E-04 |
| A_32_P113784 | N/A | N/A | | 3.16 | 1.26E-04 |
| A_32_P87849 | N/A | N/A | | 3.16 | 1.26E-04 |
| A_24_P397107 | NM_001789 | CDC25A | Cell division cycle
25 homolog A (S. pombe) | 3.15 | 1.26E-04 |
| A_23_P209200 | NM_001238 | CCNE1 | Cyclin E1 | 3.15 | 1.26E-04 |
| A_32_P16625 | N/A | N/A | | 3.15 | 1.26E-04 |
| A_23_P58321 | NM_001237 | CCNA2 | Cyclin A2 | 3.15 | 1.26E-04 |
| A_24_P37903 | N/A | LOX | Lysyl oxidase | 3.12 | 1.26E-04 |
| A_32_P64919 | NM_001042517 | DIAPH3 | Diaphanous homolog
3 (Drosophila) | 3.12 | 1.26E-04 |
| A_23_P379614 | NM_007280 | OIP5 | Opa interacting
protein 5 | 3.12 | 1.26E-04 |
| A_23_P206441 | NM_000135 | FANCA | Fanconi anemia,
complementation group A | 3.09 | 1.26E-04 |
| A_23_P16915 | NM_012413 | QPCT | Glutaminyl-peptide
cyclotransferase | 3.09 | 1.26E-04 |
| A_23_P137173 | NM_021992 | TMSB15A | Thymosin β 15a | 3.07 | 1.26E-04 |
| A_24_P313504 | NM_005030 | PLK1 | Polo-like kinase 1
(Drosophila) | 3.07 | 1.26E-04 |
| A_23_P251421 | NM_031942 | CDCA7 | Cell division cycle
associated 7 | 3.06 | 1.26E-04 |
| A_23_P252292 | NM_006733 | CENPI | Centromere protein
I | 3.04 | 1.26E-04 |
| A_23_P158725 | NM_001042422 | SLC16A3 | Solute carrier
family 16, member 3 (monocarboxylic acid transporter 4) | 3.04 | 1.26E-04 |
| A_23_P57417 | NM_005940 | MMP11 | Matrix
metallopeptidase11 (stromelysin 3) | 3.03 | 1.26E-04 |
| A_24_P291044 | N/A | N/A | | 3.02 | 1.26E-04 |
| A_23_P343927 | NM_175065 |
HIST2H2AB | Histone cluster 2,
H2ab | 3.01 | 1.26E-04 |
| A_23_P63789 | NM_032997 | ZWINT | ZW10
interactor | 3.01 | 1.26E-04 |
| A_23_P123596 | NM_000170 | GLDC | Glycine
dehydrogenase (decarboxylating) | 3 | 1.26E-04 |
| A_23_P88731 | NM_002875 | RAD51 | RAD51 homolog (RecA
homolog, E. coli) (S. cerevisiae) | 3 | 1.26E-04 |
| A_23_P161474 | NM_182751 | MCM10 | Minichromosome
maintenance complex component 10 | 2.99 | 1.26E-04 |
| A_24_P303354 | NM_021064 |
HIST1H2AG | Histone cluster 1,
H2ag | 2.98 | 1.26E-04 |
| A_23_P10518 | NM_016521 | TFDP3 | Transcription
factor Dp family, member 3 | 2.98 | 1.26E-04 |
| A_24_P247660 | NM_001002033 | HN1 | Hematological and
neurological expressed 1 | 2.97 | 1.26E-04 |
| A_23_P134910 | NM_003878 | GGH | γ-glutamyl
hydrolase (conjugase, folylpolygammaglutamyl hydrolase) | 2.97 | 1.26E-04 |
| A_32_P7193 | N/A | N/A | | 2.97 | 1.26E-04 |
| A_23_P49878 | NM_019013 | FAM64A | Family with
sequence similarity 64, member A | 2.96 | 1.26E-04 |
| A_24_P359231 | BC014312 |
HIST1H2BJ | Histone cluster 1,
H2bj | 2.95 | 1.26E-04 |
| A_32_P140262 | N/A | N/A | | 2.95 | 1.26E-04 |
| A_23_P55270 | NM_002988 | CCL18 | Chemokine (C-C
motif) ligand 18 (pulmonary and activation-regulated) | 2.95 | 1.26E-04 |
| A_24_P462899 | NM_001012507 |
C6orf173 | Chromosome 6 open
reading frame 173 | 2.94 | 1.26E-04 |
| A_23_P502520 | NM_172374 | IL4I1 | Interleukin 4
induced 1 | 2.94 | 1.26E-04 |
| A_23_P253762 | N/A | N/A | | 2.94 | 1.26E-04 |
| A_23_P214908 | AY374131 | N/A | | 2.94 | 1.26E-04 |
| A_24_P225534 | NM_017821 | RHBDL2 | Rhomboid,
veinlet-like 2 (Drosophila) | 2.94 | 1.26E-04 |
| A_23_P203419 | NM_013402 | FADS1 | Fatty acid
desaturase 1 | 2.94 | 1.26E-04 |
| A_23_P150935 | NM_005480 | TROAP | Trophinin
associated protein (tastin) | 2.94 | 1.26E-04 |
| A_24_P412088 | NM_182751 | MCM10 | Minichromosome
maintenance complex component 10 | 2.94 | 1.26E-04 |
| A_23_P71727 | NM_001827 | CKS2 | CDC28 protein
kinase regulatory subunit 2 | 2.93 | 1.26E-04 |
| A_23_P217236 | NM_005342 | HMGB3 | High-mobility group
box 3 | 2.92 | 1.26E-04 |
| A_32_P109296 | NM_152259 |
C15orf42 | Chromosome 15 open
reading frame 42 | 2.91 | 1.26E-04 |
| A_23_P89509 | NM_006461 | SPAG5 | Sperm associated
antigen 5 | 2.91 | 1.26E-04 |
| A_24_P563068 | N/A | N/A | | 2.91 | 1.26E-04 |
| A_23_P416468 | NM_025049 | PIF1 | PIF1 5′-to-3′ DNA
helicase homolog (S. cerevisiae) | 2.91 | 1.26E-04 |
| A_24_P38895 | NM_002105 | H2AFX | H2A histone family,
member X | 2.9 | 1.26E-04 |
| A_23_P52278 | NM_004523 | KIF11 | Kinesin family
member 11 | 2.89 | 1.26E-04 |
| A_24_P144543 | N/A | N/A | | 2.89 | 1.26E-04 |
| A_24_P71468 | NM_012413 | QPCT | Glutaminyl-peptide
cyclotransferase | 2.88 | 2.33E-04 |
| A_23_P116123 | NM_001274 | CHEK1 | CHK1 checkpoint
homolog (S. pombe) | 2.88 | 1.26E-04 |
| A_32_P106235 | N/A | N/A | | 2.87 | 1.26E-04 |
| A_24_P139152 | AL359062 | COL8A1 | Collagen, type
VIII, α1 | 2.87 | 4.43E-04 |
| A_23_P36831 | NM_003979 | GPRC5A | G protein-coupled
receptor, family C, group 5, member A | 2.87 | 1.26E-04 |
| A_23_P387471 | NM_005931 | MICB | MHC class I
polypeptide-related sequence B | 2.85 | 1.26E-04 |
| A_23_P9574 | NM_018098 | ECT2 | Epithelial cell
transforming sequence 2 oncogene | 2.84 | 1.26E-04 |
| A_24_P535256 | AK001903 | INHBA | Inhibin, βA | 2.84 | 1.26E-04 |
| A_24_P76521 | AK056691 | GSG2 | germ cell
associated 2 (haspin) | 2.83 | 1.26E-04 |
| A_23_P103795 | NM_138959 | VANGL1 | vang-like 1 (van
gogh, Drosophila) | 2.83 | 1.26E-04 |
| A_32_P74409 | NM_001145033 |
LOC387763 | Hypothetical
protein LOC387763 | 2.83 | 1.26E-04 |
| A_23_P100632 | NM_001002033 | HN1 | Hematological and
neurological expressed 1 | 2.83 | 1.26E-04 |
| A_23_P126212 | NM_022111 | CLSPN | Claspin homolog
(Xenopus laevis) | 2.83 | 1.26E-04 |
| A_24_P659113 | NM_152523 | CCNYL1 | Cyclin Y-like
1 | 2.83 | 1.26E-04 |
| A_24_P367227 | NM_001144755 | MYBL1 | v-myb
myeloblastosis viral oncogene homolog (avian)-like 1 | 2.82 | 1.26E-04 |
| A_23_P162719 | NM_030932 | DIAPH3 | Diaphanous homolog
3 (Drosophila) | 2.81 | 1.26E-04 |
| A_32_P221799 | NM_003514 |
HIST1H2AM | Histone cluster 1,
H2am | 2.81 | 1.26E-04 |
| A_23_P60120 | NM_031415 | GSDMC | Gasdermin C | 2.81 | 2.33E-04 |
| A_24_P902509 | NM_018193 | FANCI | Fanconi anemia,
complementation group I | 2.8 | 1.26E-04 |
| A_23_P50096 | NM_001071 | TYMS | Thymidylate
synthetase | 2.79 | 1.26E-04 |
| A_32_P143245 | NM_001012507 |
C6orf173 | Chromosome 6 open
reading frame 173 | 2.79 | 1.26E-04 |
| A_23_P155969 | NM_014264 | PLK4 | Polo-like kinase 4
(Drosophila) | 2.79 | 1.26E-04 |
| A_23_P62021 | N/A | N/A | | 2.78 | 1.26E-04 |
| A_32_P183218 | NM_153695 | ZNF367 | Zinc finger protein
367 | 2.77 | 1.26E-04 |
| A_23_P46118 | NM_001821 | CHML | Choroideremia-like
(Rab escort protein 2) | 2.76 | 2.33E-04 |
| A_23_P327643 | N/A | N/A | | 2.75 | 1.26E-04 |
| A_23_P375104 | NM_018193 | FANCI | Fanconi anemia,
complementation group I | 2.75 | 1.26E-04 |
| A_23_P1823 | NM_000280 | PAX6 | Paired box 6 | 2.75 | 1.26E-04 |
| A_23_P168014 | NM_021066 |
HIST1H2AJ | Histone cluster 1,
H2aj | 2.74 | 1.26E-04 |
| A_24_P413126 | NM_020182 | PMEPA1 | Prostate
transmembrane protein, androgen induced 1 | 2.74 | 1.26E-04 |
| A_23_P80032 | NM_005225 | E2F1 | E2F transcription
factor 1 | 2.74 | 1.26E-04 |
| A_23_P215976 | NM_057749 | CCNE2 | Cyclin E2 | 2.72 | 2.33E-04 |
| A_32_P231415 | AF132203 | SCD | Stearoyl-CoA
desaturase (δ-9-desaturase) | 2.72 | 1.26E-04 |
| A_23_P370989 | NM_005914 | MCM4 | Minichromosome
maintenance complex component 4 | 2.72 | 1.26E-04 |
| A_23_P216429 | NM_017680 | ASPN | Asporin | 2.71 | 1.26E-04 |
| A_24_P195621 | NR_027288 |
LOC341056 | SUMO-1 activating
enzyme subunit 1 pseudogene | 2.71 | 1.26E-04 |
| A_32_P151800 | NM_207418 | FAM72D | Family with
sequence similarity 72, member D | 2.7 | 1.26E-04 |
| A_23_P122197 | NM_031966 | CCNB1 | Cyclin B1 | 2.7 | 1.26E-04 |
| A_23_P34788 | NM_006845 | KIF2C | Kinesin family
member 2C | 2.7 | 1.26E-04 |
| A_32_P206698 | NM_001826 | CKS1B | CDC28 protein
kinase regulatory subunit 1B | 2.7 | 1.26E-04 |
| A_23_P99292 | NM_006479 |
RAD51AP1 | RAD51 associated
protein 1 | 2.7 | 1.26E-04 |
| A_23_P133956 | NM_002263 | KIFC1 | Kinesin family
member C1 | 2.69 | 1.26E-04 |
| A_32_P143496 | N/A | N/A | | 2.69 | 1.26E-04 |
| A_32_P163858 | NM_005063 | SCD | Stearoyl-CoA
desaturase (δ-9-desaturase) | 2.69 | 1.26E-04 |
| A_32_P175557 | R01145 | N/A | | 2.69 | 1.26E-04 |
| A_23_P63618 | NM_005063 | SCD | Stearoyl-CoA
desaturase (δ-9-desaturase) | 2.69 | 1.26E-04 |
| A_23_P88630 | NM_000057 | BLM | Bloom syndrome,
RecQ helicase-like | 2.68 | 1.26E-04 |
| A_24_P276102 | NM_183404 | RBL1 | Retinoblastoma-like
1 (p107) | 2.68 | 1.26E-04 |
| A_23_P135385 | N/A | N/A | | 2.68 | 1.26E-04 |
| A_23_P57658 | NM_020386 | HRASLS | HRAS-like
suppressor | 2.67 | 1.26E-04 |
| A_23_P23303 | NM_003686 | EXO1 | Exonuclease 1 | 2.67 | 1.26E-04 |
| A_23_P88691 | NM_000745 | CHRNA5 | Cholinergic
receptor, nicotinic, α5 | 2.67 | 1.26E-04 |
| A_24_P923381 | NR_002219 | EPR1 | Effector cell
peptidase receptor 1 (non-protein coding) | 2.66 | 1.26E-04 |
| A_23_P24444 | NM_001360 | DHCR7 |
7-dehydrocholesterol reductase | 2.65 | 1.26E-04 |
| A_23_P43157 | NM_001080416 | MYBL1 | v-myb
myeloblastosis viral oncogene homolog (avian)-like 1 | 2.65 | 2.33E-04 |
| A_23_P88740 | NM_018455 | CENPN | Centromere protein
N | 2.64 | 1.26E-04 |
| A_23_P131866 | NM_198433 | AURKA | Aurora kinase
A | 2.64 | 1.26E-04 |
| A_23_P259641 | NM_004456 | EZH2 | Enhancer of zeste
homolog 2 (Drosophila) | 2.64 | 1.26E-04 |
| A_32_P72341 | NM_173084 | TRIM59 | Tripartite
motif-containing 59 | 2.62 | 1.26E-04 |
| A_24_P227091 | NM_004523 | KIF11 | Kinesin family
member 11 | 2.61 | 1.26E-04 |
| A_23_P145238 | NM_080593 |
HIST1H2BK | Histone cluster 1,
H2bk | 2.61 | 1.26E-04 |
| A_23_P136805 | NM_014783 |
ARHGAP11A | Rho GTPase
activating protein 11A | 2.6 | 1.26E-04 |
| A_23_P167997 | NM_003518 |
HIST1H2BG | Histone cluster 1,
H2bg | 2.6 | 1.26E-04 |
| A_23_P63402 | NM_013296 | GPSM2 | G-protein signaling
modulator 2 (AGS3-like, C. elegans) | 2.6 | 1.26E-04 |
| A_24_P192994 | NM_013402 | FADS1 | Fatty acid
desaturase 1 | 2.59 | 1.26E-04 |
| A_23_P25559 | NM_005845 | ABCC4 | ATP-binding
cassette, sub-family C (CFTR/MRP), member 4 | 2.59 | 3.41E-04 |
| A_23_P309381 | NM_001040874 |
HIST2H2AA4 | Histone cluster 2,
H2aa4 | 2.59 | 1.26E-04 |
| A_23_P35871 | NM_024680 | E2F8 | E2F transcription
factor 8 | 2.58 | 1.26E-04 |
| A_23_P207307 | N/A | N/A | | 2.58 | 1.26E-04 |
| A_24_P399888 | NM_001002876 | CENPM | Centromere protein
M | 2.58 | 1.26E-04 |
| A_23_P360754 | NM_005099 | ADAMTS4 | ADAM
metallopeptidase with thrombospondin type 1 motif, 4 | 2.57 | 3.41E-04 |
| A_23_P21706 | NM_001905 | CTPS | CTP synthase | 2.57 | 1.26E-04 |
| A_24_P174924 | NM_003537 |
HIST1H3B | Histone cluster 1,
H3b | 2.57 | 1.26E-04 |
| A_23_P155989 | NM_022145 | CENPK | Centromere protein
K | 2.57 | 1.26E-04 |
| A_23_P103981 | NM_001040874 |
HIST2H2AA4 | Histone cluster 2,
H2aa4 | 2.56 | 1.26E-04 |
| A_23_P571 | NM_006516 | SLC2A1 | Solute carrier
family 2 (facilitated glucose transporter), member 1 | 2.56 | 1.26E-04 |
| A_23_P420551 | NM_007174 | CIT | Citron
(rho-interacting, serine/threonine kinase 21) | 2.56 | 1.26E-04 |
| A_23_P411335 | NM_152524 | SGOL2 | Shugoshin-like 2
(S. pombe) | 2.54 | 1.26E-04 |
| A_32_P147090 | NM_199357 |
ARHGAP11A | Rho GTPase
activating protein 11A | 2.54 | 1.26E-04 |
| A_23_P70448 | NM_005325 |
HIST1H1A | Hstone cluster 1,
H1a | 2.53 | 1.26E-04 |
| A_23_P43484 | NM_058197 | CDKN2A | Cyclin-dependent
kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | 2.52 | 1.26E-04 |
| A_24_P85539 | NM_212482 | FN1 | Fibronectin 1 | 2.52 | 1.26E-04 |
| A_32_P28704 | N/A | N/A | | 2.52 | 1.26E-04 |
| A_23_P107421 | NM_003258 | TK1 | Thymidine kinase 1,
soluble | 2.51 | 1.26E-04 |
| A_23_P502425 | NM_020409 | MRPL47 | Mitochondrial
ribosomal protein L47 | 2.5 | 1.26E-04 |
| A_24_P351466 | NM_020890 |
KIAA1524 | KIAA1524 | 2.5 | 1.26E-04 |
| A_23_P211910 | NM_182943 | PLOD2 | Procollagen-lysine,
2-oxoglutarate 5-dioxygenase 2 | 2.5 | 1.26E-04 |
| A_24_P9321 | NM_003533 |
HIST1H3I | Histone cluster 1,
H3i | 2.49 | 1.26E-04 |
| A_24_P334248 | NM_014996 | PLCH1 | Phospholipase C,
eta 1 | 2.48 | 1.26E-04 |
| A_24_P819890 | NM_001005210 | LRRC55 | Leucine rich repeat
containing 55 | 2.48 | 4.43E-04 |
| A_23_P146456 | NM_001333 | CTSL2 | Cathepsin L2 | 2.48 | 2.33E-04 |
| A_24_P242440 | NM_003780 | B4GALT2 | UDP-Gal:βGlcNAc β
1,4-galactosyltransferase, polypeptide 2 | 2.47 | 1.26E-04 |
| A_23_P88331 | NM_014750 | DLGAP5 | Discs, large
(Drosophila) homolog-associated protein 5 | 2.47 | 1.26E-04 |
| A_23_P216068 | NM_014109 | ATAD2 | ATPase family, AAA
domain containing 2 | 2.46 | 1.26E-04 |
| A_32_P31021 | N/A | N/A | | 2.46 | 1.26E-04 |
| A_23_P373119 | NR_002165 | HMGB3L1 | High-mobility group
box 3-like 1 | 2.46 | 1.26E-04 |
| A_23_P361419 | NM_018369 | DEPDC1B | DEP domain
containing 1B | 2.45 | 1.26E-04 |
| A_23_P10870 | NM_014908 | DOLK | Dolichol
kinase | 2.44 | 1.26E-04 |
| A_23_P420692 | NM_015053 | PPFIA4 | Protein tyrosine
phosphatase, receptor type, f polypeptide (PTPRF), interacting
protein (liprin), α4 | 2.43 | 1.26E-04 |
| A_23_P146284 | NM_003129 | SQLE | Squalene
epoxidase | 2.43 | 1.26E-04 |
| A_32_P159254 | AK123584 | N/A | | 2.43 | 2.33E-04 |
| A_23_P25626 | NM_024808 |
C13orf34 | Chromosome 13 open
reading frame 34 | 2.43 | 1.26E-04 |
| A_23_P59005 | NM_000593 | TAP1 | Transporter 1,
ATP-binding cassette, sub-family B (MDR/TAP) | 2.43 | 2.33E-04 |
| A_24_P49747 | XM_929965 |
LOC646993 | Similar to high
mobility group box 3 | 2.43 | 1.26E-04 |
| A_23_P252740 | NM_024094 | DSCC1 | Defective in sister
chromatid cohesion 1 homolog (S. cerevisiae) | 2.42 | 1.26E-04 |
| A_23_P397341 | NM_152341 | PAQR4 | Progestin and
adipoQ receptor family member IV | 2.42 | 1.26E-04 |
| A_23_P59045 | NM_021052 |
HIST1H2AE | Histone cluster 1,
H2ae | 2.42 | 1.26E-04 |
| A_23_P140316 | NM_001099652 | GPR137C | G protein-coupled
receptor 137C | 2.42 | 1.26E-04 |
| A_23_P207520 | Z74615 | COL1A1 | Collagen, type I,
α1 | 2.41 | 1.26E-04 |
| A_24_P920968 | NM_182625 | GEN1 | Gen homolog 1,
endonuclease (Drosophila) | 2.41 | 1.26E-04 |
| A_23_P366216 | NM_003524 |
HIST1H2BH | Histone cluster 1,
H2bh | 2.41 | 1.26E-04 |
| A_23_P217049 | NM_014286 | FREQ | Frequenin homolog
(Drosophila) | 2.41 | 2.33E-04 |
| A_32_P194264 | NM_001008708 | CHAC2 | ChaC, cation
transport regulator homolog 2 (E. coli) | 2.4 | 2.33E-04 |
| A_32_P35839 | N/A | N/A | | 2.4 | 1.26E-04 |
| A_23_P154894 | NM_000100 | CSTB | Cystatin B (stefin
B) | 2.4 | 1.26E-04 |
| A_24_P340066 | NM_001421 | ELF4 | E74-like factor 4
(ets domain transcription factor) | 2.4 | 1.26E-04 |
| A_24_P857404 | NM_001093725 | MEX3A | mex-3 homolog A
(C. elegans) | 2.4 | 1.26E-04 |
| A_24_P133488 | NM_017955 | CDCA4 | Cell division cycle
associated 4 | 2.4 | 1.26E-04 |
| A_23_P339240 | NM_014996 | PLCH1 | Phospholipase C,
eta 1 | 2.39 | 2.33E-04 |
| A_23_P52410 | NM_145307 | RTKN2 | Rhotekin 2 | 2.39 | 1.26E-04 |
| A_23_P59877 | NM_001444 | FABP5 | Fatty acid binding
protein 5 (psoriasis-associated) | 2.39 | 1.26E-04 |
| A_23_P29594 | NM_052969 | RPL39L | Ribosomal protein
L39-like | 2.38 | 1.26E-04 |
| A_23_P11984 | NM_201649 | SLC6A9 | Solute carrier
family 6 (neurotransmitter transporter, glycine), member 9 | 2.38 | 2.33E-04 |
| A_23_P200866 | NM_203401 | STMN1 | Stathmin 1 | 2.37 | 1.26E-04 |
| A_32_P182135 | N/A | N/A | | 2.36 | 1.26E-04 |
| A_24_P323598 | NM_001017420 | ESCO2 | Establishment of
cohesion 1 homolog 2 (S. cerevisiae) | 2.36 | 1.26E-04 |
| A_23_P39574 | NM_001080539 | CCDC150 | Coiled-coil domain
containing 150 | 2.36 | 1.26E-04 |
| A_24_P275386 | AK025766 | BRI3BP | BRI3 binding
protein | 2.36 | 1.26E-04 |
| A_23_P85460 | NM_078626 | CDKN2C | Cyclin-dependent
kinase inhibitor 2C (p18, inhibits CDK4) | 2.35 | 1.26E-04 |
| A_23_P57306 | NM_005441 | CHAF1B | Chromatin assembly
factor 1, subunit B (p60) | 2.35 | 1.26E-04 |
| A_23_P335329 | NM_004485 | GNG4 | Guanine nucleotide
binding protein (G protein), γ4 | 2.35 | 2.33E-04 |
| A_23_P92441 | NM_002358 | MAD2L1 | MAD2 mitotic arrest
deficient-like 1 (yeast) | 2.35 | 1.26E-04 |
| A_24_P13390 | NM_032814 | RNFT2 | Ring finger
protein, transmembrane 2 | 2.35 | 1.26E-04 |
| A_23_P362046 | NM_138779 |
C13orf27 | Chromosome 13 open
reading frame 27 | 2.34 | 1.26E-04 |
| A_23_P24716 | NM_017870 |
TMEM132A | Transmembrane
protein 132A | 2.34 | 1.26E-04 |
| A_23_P91900 | NM_005496 | SMC4 | structural
maintenance of chromosomes 4 | 2.33 | 1.26E-04 |
| A_24_P105102 | NM_182687 | PKMYT1 | Protein kinase,
membrane associated tyrosine/threonine 1 | 2.33 | 1.26E-04 |
| A_24_P244420 | NM_018367 | ACER3 | alkaline ceramidase
3 | 2.33 | 2.33E-04 |
| A_23_P112673 | NM_017975 | ZWILCH | Zwilch, kinetochore
associated, homolog (Drosophila) | 2.33 | 1.26E-04 |
| A_23_P87769 | NM_017915 |
C12orf48 | Chromosome 12 open
reading frame 48 | 2.33 | 1.26E-04 |
| A_24_P296254 | NM_014783 |
ARHGAP11A | Rho GTPase
activating protein 11A | 2.32 | 1.26E-04 |
| A_23_P166306 | NM_000071 | CBS |
Cystathionine-β-synthase | 2.32 | 1.26E-04 |
On the other hand, Table III lists the 321 genes that were
downregulated to <1/5 of normal ductal cells. Among these
significantly downregulated genes, prolactin-induced protein
(PIP) and dynein, axonemal, light intermediate chain 1
(DNALI1) were previously shown to be downregulated in TNBC
(26). In particular, suppression
of WNT inhibitory factor 1 (WIF1) (27) and signal peptide, CUB domain,
EGF-like (SCUBE2) (28),
both of which function as tumor suppressors, were among the genes
that were downregulated as malignancy progressed. These data
suggest that silencing or depletion of these genes might lead to
the carcinogenesis of TNBC.
 | Table III.Significantly downregulated genes in
TNBC compared with normal ductal cells. |
Table III.
Significantly downregulated genes in
TNBC compared with normal ductal cells.
| Probe ID | Accession no. | Symbol | Gene name | Fold change
(log) | P-value |
|---|
| A_23_P127781 | NM_006552 | SCGB1D1 | Secretoglobin,
family 1D, member 1 | −6.77 | 1.26E-04 |
| A_32_P234405 | CK570316 | N/A | | −6.62 | 1.26E-04 |
| A_23_P150555 | NM_006551 | SCGB1D2 | Secretoglobin,
family 1D, member 2 | −6.51 | 1.26E-04 |
| A_23_P12533 | NM_052997 |
ANKRD30A | Ankyrin repeat
domain 30A | −6.44 | 1.26E-04 |
| A_23_P8702 | NM_002652 | PIP | Prolactin-induced
protein | −6.34 | 1.26E-04 |
| A_23_P501010 | NM_000494 | COL17A1 | Collagen, type
XVII, α1 | −5.69 | 1.26E-04 |
| A_24_P844984 | NM_002644 | PIGR | Polymeric
immunoglobulin receptor | −5.55 | 1.26E-04 |
| A_32_P216520 | NM_007191 | WIF1 | WNT inhibitory
factor 1 | −5.53 | 1.26E-04 |
| A_23_P71364 | NM_015886 | PI15 | Peptidase inhibitor
15 | −5.33 | 1.26E-04 |
| A_24_P273756 | NM_003722 | TP63 | Tumor protein
p63 | −5.11 | 1.26E-04 |
| A_23_P132619 | NM_000916 | OXTR | Oxytocin
receptor | −4.89 | 1.26E-04 |
| A_32_P111873 | BQ432543 | N/A | | −4.88 | 1.26E-04 |
| A_32_P23272 | N/A | N/A | | −4.85 | 1.26E-04 |
| A_24_P643776 | N/A | N/A | | −4.74 | 1.26E-04 |
| A_23_P136777 | NM_001647 | APOD | Apolipoprotein
D | −4.71 | 1.26E-04 |
| A_23_P9711 | NM_006040 | HS3ST4 | Heparan sulfate
(glucosamine) 3-O-sulfotransferase 4 | −4.58 | 1.26E-04 |
| A_23_P305292 | NR_027180 |
LOC728264 | Hypothetical
LOC728264 | −4.57 | 1.26E-04 |
| A_23_P159974 | NM_033495 | KLHL13 | Kelch-like 13
(Drosophila) | −4.55 | 1.26E-04 |
| A_23_P105144 | NM_020974 | SCUBE2 | Signal peptide, CUB
domain, EGF-like 2 | −4.51 | 1.26E-04 |
| A_32_P14253 | N/A | N/A | | −4.47 | 1.26E-04 |
| A_23_P327380 | NM_003722 | TP63 | Tumor protein
p63 | −4.45 | 1.26E-04 |
| A_23_P337270 | AK057247 | N/A | | −4.43 | 1.26E-04 |
| A_23_P420442 | NM_153618 | SEMA6D | Sema domain,
transmembrane domain (TM), and cytoplasmic domain, (semaphorin)
6D | −4.34 | 1.26E-04 |
| A_23_P8812 | N/A | N/A | | −4.3 | 1.26E-04 |
| A_23_P160377 | NM_003462 | DNALI1 | Dynein, axonemal,
light intermediate chain 1 | −4.26 | 1.26E-04 |
| A_24_P92680 | AK093340 |
LOC100132116 | Hypothetical
LOC100132116 | −4.23 | 1.26E-04 |
| A_23_P216779 | NM_001007097 | NTRK2 | Neurotrophic
tyrosine kinase, receptor, type 2 | −4.23 | 1.26E-04 |
| A_23_P148249 | NM_024817 | THSD4 | Thrombospondin,
type I, domain containing 4 | −4.18 | 1.26E-04 |
| A_23_P206920 | NM_001040114 | MYH11 | Myosin, heavy chain
11, smooth muscle | −4.13 | 1.26E-04 |
| A_32_P154473 | NM_004522 | KIF5C | Kinesin family
member 5C | −4.13 | 1.26E-04 |
| A_23_P128362 | NM_206819 | MYBPC1 | Myosin binding
protein C, slow type | −4.11 | 3.41E-04 |
| A_23_P83381 | NM_001143962 | CAPN8 | Calpain 8 | −4.08 | 1.26E-04 |
| A_23_P397208 | NM_000848 | GSTM2 | Glutathione
S-transferase mu 2 (muscle) | −4.07 | 1.26E-04 |
| A_23_P503072 | NM_148672 | CCL28 | Chemokine (C-C
motif) ligand 28 | −4.03 | 1.26E-04 |
| A_23_P143068 | NM_024726 | IQCA1 | IQ motif containing
with AAA domain 1 | −4.01 | 1.26E-04 |
| A_24_P829209 | AK096334 | LOC285944 | Hypothetical
protein LOC285944 | −3.99 | 2.33E-04 |
| A_23_P394246 | | GPR81 | G protein-coupled
receptor 81 | −3.96 | 1.26E-04 |
| A_24_P34186 | NM_004010 | DMD | Dystrophin | −3.96 | 1.26E-04 |
| A_23_P303087 | NM_002825 | PTN | Pleiotrophin | −3.95 | 1.26E-04 |
| A_24_P243749 | NM_002612 | PDK4 | Pyruvate
dehydrogenase kinase, isozyme 4 | −3.94 | 1.26E-04 |
| A_32_P39944 | AK095791 | N/A | | −3.82 | 1.26E-04 |
| A_23_P217379 | NM_033641 | COL4A6 | Collagen, type IV,
α6 | −3.8 | 1.26E-04 |
| A_23_P407565 | NM_001337 | CX3CR1 | Chemokine (C-X3-C
motif) receptor 1 | −3.76 | 1.26E-04 |
| A_23_P373464 | NM_002285 | AFF3 | AF4/FMR2 family,
member 3 | −3.75 | 1.26E-04 |
| A_32_P183765 | NM_005235 | ERBB4 | v-erb-a
erythroblastic leukemia viral oncogene homolog 4 (avian) | −3.75 | 1.26E-04 |
| A_23_P145514 | NM_014432 | IL20RA | Interleukin 20
receptor, α | −3.75 | 1.26E-04 |
| A_24_P870620 | NM_002825 | PTN | Pleiotrophin | −3.74 | 2.33E-04 |
| A_32_P154361 | N/A | N/A | | −3.73 | 1.26E-04 |
| A_24_P330633 | NM_000353 | TAT | Tyrosine
aminotransferase | −3.72 | 1.26E-04 |
| A_23_P360777 | NM_013960 | NRG1 | Neuregulin 1 | −3.72 | 1.26E-04 |
| A_23_P253982 | NM_002141 | HOXA4 | Homeobox A4 | −3.69 | 1.26E-04 |
| A_32_P114475 | N/A | N/A | | −3.68 | 1.26E-04 |
| A_32_P221774 | BX099483 | N/A | | −3.66 | 1.26E-04 |
| A_23_P212608 | NM_022131 | CLSTN2 | Calsyntenin 2 | −3.66 | 2.33E-04 |
| A_23_P254165 | NM_021785 | RAI2 | Retinoic acid
induced 2 | −3.65 | 1.26E-04 |
| A_24_P794447 | NR_024430 |
LOC399959 | Hypothetical
LOC399959 | −3.64 | 1.26E-04 |
| A_23_P149517 | NM_002644 | PIGR | Polymeric
immunoglobulin receptor | −3.64 | 1.26E-04 |
| A_24_P904484 | NR_024344 |
LOC283174 | Hypothetical
LOC283174 | −3.62 | 1.26E-04 |
| A_32_P194423 | N/A | N/A | | −3.62 | 1.26E-04 |
| A_23_P371495 | NM_175861 | TMTC1 | Transmembrane and
tetratricopeptide repeat containing 1 | −3.6 | 2.33E-04 |
| A_23_P134162 | NM_016356 | DCDC2 | Doublecortin domain
containing 2 | −3.58 | 1.26E-04 |
| A_32_P232455 | NM_178840 | C1orf64 | Chromosome 1 open
reading frame 64 | −3.58 | 1.26E-04 |
| A_24_P318160 | NM_014903 | NAV3 | Neuron navigator
3 | −3.57 | 1.26E-04 |
| A_23_P59388 | NM_001723 | DST | Dystonin | −3.56 | 1.26E-04 |
| A_23_P399217 | NM_153445 | OR5P3 | Olfactory receptor,
family 5, subfamily P, member 3 | −3.56 | 1.26E-04 |
| A_23_P309739 | NM_000125 | ESR1 | Estrogen receptor
1 | −3.53 | 1.26E-04 |
| A_24_P608007 | AK022390 | N/A | | −3.53 | 1.26E-04 |
| A_23_P501538 | NM_153631 | HOXA3 | Homeobox A3 | −3.52 | 1.26E-04 |
| A_24_P602871 | NM_001030060 | SAMD5 | Sterile α motif
domain containing 5 | −3.52 | 1.26E-04 |
| A_23_P136433 | N/A | N/A | | −3.51 | 1.26E-04 |
| A_23_P30294 | NM_001801 | CDO1 | Cysteine
dioxygenase, type I | −3.48 | 1.26E-04 |
| A_23_P218928 | NM_016613 | FAM198B | Family with
sequence similarity 198, member B | −3.47 | 1.26E-04 |
| A_23_P154627 | XM_002345419 | TSHZ2 | Teashirt zinc
finger homeobox 2 | −3.47 | 1.26E-04 |
| A_23_P303833 | NM_174934 | SCN4B | Sodium channel,
voltage-gated, type IV, β | −3.45 | 1.26E-04 |
| A_24_P930088 | XM_002342181 |
LOC100286909 | Hypothetical
protein LOC100286909 | −3.45 | 1.26E-04 |
| A_32_P81623 | AA514833 | N/A | | −3.42 | 1.26E-04 |
| A_24_P923028 | BC020707 | TAT | Tyrosine
aminotransferase | −3.41 | 1.26E-04 |
| A_23_P58869 | NR_002932 |
LOC442245 | Glutathione
S-transferase mu 2 pseudogene | −3.4 | 1.26E-04 |
| A_23_P2271 | NM_198965 | PTHLH | Parathyroid
hormone-like hormone | −3.4 | 1.26E-04 |
| A_32_P43664 | | | | −3.39 | 1.26E-04 |
| A_32_P16007 | NM_207355 | POTEB | POTE ankyrin domain
family, member B | −3.39 | 1.26E-04 |
| A_23_P94840 | NM_130897 | DYNLRB2 | Dynein, light
chain, roadblock-type 2 | −3.38 | 1.26E-04 |
| A_24_P5153 | NM_024817 | THSD4 | Thrombospondin,
type I, domain containing 4 | −3.38 | 1.26E-04 |
| A_32_P223675 | N/A | N/A | | −3.37 | 1.26E-04 |
| A_24_P904845 | AK095791 | N/A | | −3.37 | 1.26E-04 |
| A_23_P403209 | N/A | N/A | | −3.36 | 1.26E-04 |
| A_23_P215382 | N/A | N/A | | −3.35 | 3.41E-04 |
| A_24_P209710 | NM_004816 |
FAM189A2 | Family with
sequence similarity 189, member A2 | −3.35 | 1.26E-04 |
| A_23_P167168 | NM_144646 | IGJ | Immunoglobulin J
polypeptide, linker protein for immunoglobulin α and mu
polypeptides | −3.34 | 1.26E-04 |
| A_24_P70183 | NM_001040113 | MYH11 | Myosin, heavy chain
11, smooth muscle collagen, type XIV, α1 | −3.32 | 1.26E-04 |
| A_23_P216361 | NM_021110 | COL14A1 | | −3.32 | 1.26E-04 |
| A_23_P113351 | NM_004684 | SPARCL1 | SPARC-like 1
(hevin) | −3.31 | 1.26E-04 |
| A_32_P17145 | N/A | N/A | | −3.31 | 1.26E-04 |
| A_23_P35414 | NM_005398 | PPP1R3C | Protein phosphatase
1, regulatory (inhibitor) subunit 3C | −3.29 | 1.26E-04 |
| A_23_P31945 | NM_033439 | IL33 | Interleukin 33 | −3.27 | 1.26E-04 |
| A_23_P204630 | NM_021229 | NTN4 | Netrin 4 | −3.26 | 1.26E-04 |
| A_23_P501831 | NM_032385 | C5orf4 | Chromosome 5 open
reading frame 4 | −3.26 | 1.26E-04 |
| A_23_P200015 | NM_174858 | AK5 | Adenylate kinase
5 | −3.26 | 1.26E-04 |
| A_24_P802145 | NM_005544 | IRS1 | Insulin receptor
substrate 1 | −3.26 | 1.26E-04 |
| A_24_P251969 | NM_000800 | FGF1 | Fibroblast growth
factor 1 (acidic) | −3.24 | 1.26E-04 |
| A_32_P228618 | NM_001003793 | RBMS3 | RNA binding motif,
single stranded interacting protein | −3.23 | 1.26E-04 |
| A_23_P125233 | NM_001299 | CNN1 | Calponin 1, basic,
smooth muscle | −3.22 | 2.33E-04 |
| A_23_P500998 | NM_152739 | HOXA9 | Homeobox A9 | −3.19 | 2.33E-04 |
| A_23_P83838 | NM_004056 | CA8 | Carbonic anhydrase
VIII | −3.19 | 1.26E-04 |
| A_24_P911950 | N/A | N/A | | −3.17 | 1.26E-04 |
| A_23_P159952 | NM_018476 | BEX1 | Brain expressed,
X-linked 1 | −3.17 | 1.26E-04 |
| A_23_P45185 | NM_004469 | FIGF | c-fos induced
growth factor (vascular endothelial growth factor D) | −3.16 | 2.33E-04 |
| A_23_P14083 | NM_181847 | AMIGO2 | Adhesion molecule
with Ig-like domain 2 | −3.16 | 1.26E-04 |
| A_24_P920366 | N/A | N/A | | −3.14 | 1.26E-04 |
| A_24_P167668 | NM_000428 | LTBP2 | Latent transforming
growth factor β binding protein 2 | −3.12 | 1.26E-04 |
| A_32_P161033 | BC043411 | N/A | | −3.11 | 1.26E-04 |
| A_23_P348159 | NM_020388 | DST | Dystonin | −3.11 | 1.26E-04 |
| A_32_P89415 | N/A | N/A | | −3.1 | 1.26E-04 |
| A_23_P165778 | NM_024101 | MLPH | Melanophilin | −3.08 | 1.26E-04 |
| A_32_P168701 | N/A | N/A | | −3.07 | 3.41E-04 |
| A_32_P78491 | NM_004956 | ETV1 | ets variant 1 | −3.06 | 1.26E-04 |
| A_24_P87036 | NM_018043 | ANO1 | Anoctamin 1,
calcium activated chloride channel | −3.06 | 1.26E-04 |
| A_24_P912799 | NM_003966 | SEMA5A | Sema domain, seven
thrombospondin repeats (type 1 and type 1-like), transmembrane
domain (TM) and short cytoplasmic domain, (semaphorin) 5A | −3.06 | 1.26E-04 |
| A_23_P315364 | NM_002089 | CXCL2 | Chemokine (C-X-C
motif) ligand 2 | −3.05 | 1.26E-04 |
| A_24_P71341 | NM_001461 | FMO5 | Flavin containing
monooxygenase 5 | −3.05 | 2.33E-04 |
| A_32_P199796 | NM_004023 | DMD | Dystrophin | −3.05 | 2.33E-04 |
| A_32_P179998 | NM_033053 | DMRTC1 | DMRT−like family
C1 | −3.04 | 1.26E-04 |
| A_32_P17984 | N/A | N/A | | −3.04 | 1.26E-04 |
| A_23_P138938 | NM_000926 | PGR | Progesterone
receptor | −3.04 | 1.26E-04 |
| A_23_P18559 | NM_003866 | INPP4B | Inositol
polyphosphate-4-phosphatase, type II, 105 kDa | −3.03 | 1.26E-04 |
| A_23_P124946 | NM_153610 | CMYA5 | Cardiomyopathy
associated 5 | −3.03 | 1.26E-04 |
| A_23_P212241 | NM_006614 | CHL1 | Cell adhesion
molecule with homology to L1CAM (close homolog of L1) | −3.03 | 1.26E-04 |
| A_23_P156402 | NM_003551 | NME5 | Non-metastatic
cells 5, protein expressed in (nucleoside-diphosphate kinase) | −3.02 | 1.26E-04 |
| A_23_P150053 | NM_001613 | ACTA2 | Actin, α2, smooth
muscle, aorta | −3.02 | 1.26E-04 |
| A_32_P58912 | N/A | N/A | | −3.02 | 1.26E-04 |
| A_32_P216841 | NM_145263 | SPATA18 | Spermatogenesis
associated 18 homolog (rat) | −3.01 | 2.33E-04 |
| A_23_P257087 | NM_002612 | PDK4 | Pyruvate
dehydrogenase kinase, isozyme 4 | −3.01 | 1.26E-04 |
| A_23_P110686 | NM_003714 | STC2 | Stanniocalcin
2 | −3 | 1.26E-04 |
| A_23_P369994 | NM_004734 | DCLK1 | Doublecortin-like
kinase 1 | −2.99 | 2.33E-04 |
| A_23_P422831 | NM_004816 |
FAM189A2 | Family with
sequence similarity 189, member A2 | −2.98 | 1.26E-04 |
| A_24_P325992 | NM_002310 | LIFR | Leukemia inhibitory
factor receptor α | −2.98 | 1.26E-04 |
| A_23_P387000 | NM_173683 | XKR6 | XK, Kell blood
group complex subunit-related family, member 6 | −2.98 | 3.41E-04 |
| A_32_P83811 | NM_001136570 | FAM47E | Family with
sequence similarity 47, member E | −2.98 | 1.26E-04 |
| A_32_P44210 | BX538299 | N/A | | −2.97 | 1.26E-04 |
| A_24_P918317 | NM_015881 | DKK3 | Dickkopf homolog 3
(Xenopus laevis) | −2.97 | 4.43E-04 |
| A_23_P203957 | NM_175861 | TMTC1 | Transmembrane and
tetratricopeptide repeat containing 1 | −2.96 | 3.41E-04 |
| A_23_P30217 | NM_052863 | SCGB3A1 | Secretoglobin,
family 3A, member 1 | −2.96 | 1.26E-04 |
| A_23_P77066 | NM_022807 | SNRPN | Small nuclear
ribonucleoprotein polypeptide N | −2.94 | 1.26E-04 |
| A_32_P109242 | AK055302 | CSRNP3 |
Cysteine-serine-rich nuclear protein
3 | −2.91 | 1.26E-04 |
| A_24_P937265 | N/A | N/A | | −2.91 | 1.26E-04 |
| A_32_P97968 | N/A | N/A | | −2.9 | 1.26E-04 |
| A_32_P85684 | AA069768 | N/A | | −2.89 | 1.26E-04 |
| A_23_P385067 | NM_053277 | CLIC6 | Chloride
intracellular channel 6 | −2.89 | 4.43E-04 |
| A_23_P82868 | NM_000930 | PLAT | Plasminogen
activator, tissue | −2.88 | 1.26E-04 |
| A_32_P108396 | N/A | N/A | | −2.88 | 1.26E-04 |
| A_23_P148345 | NM_194463 | RNF128 | Ring finger protein
128 | −2.87 | 1.26E-04 |
| A_24_P314477 | NM_178012 | TUBB2B | Tubulin, β 2B | −2.87 | 1.26E-04 |
| A_24_P895836 | N/A | N/A | | −2.87 | 1.26E-04 |
| A_23_P171074 | NM_004867 | ITM2A | Integral membrane
protein 2A | −2.85 | 1.26E-04 |
| A_23_P9135 | NM_033655 | CNTNAP3 | Contactin
associated protein-like 3 | −2.85 | 4.43E-04 |
| A_23_P372234 | NM_001218 | CA12 | Carbonic anhydrase
XII | −2.83 | 1.26E-04 |
| A_23_P393099 | NM_003226 | TFF3 | Trefoil factor 3
(intestinal) | −2.82 | 2.33E-04 |
| A_23_P113701 | NM_002607 | PDGFA | Platelet-derived
growth factor α polypeptide | −2.82 | 1.26E-04 |
| A_23_P10995 | NM_014483 | RBMS3 | RNA binding motif,
single stranded interacting protein | −2.82 | 1.26E-04 |
| A_24_P269006 | NM_001182 | ALDH7A1 | Aldehyde
dehydrogenase 7 family, member A1 | −2.81 | 1.26E-04 |
| A_23_P415533 | AK054879 | N/A | | −2.81 | 1.26E-04 |
| A_23_P216225 | NM_004430 | EGR3 | Early growth
response 3 | −2.8 | 1.26E-04 |
| A_24_P101282 | N/A | N/A | | −2.8 | 1.26E-04 |
| A_32_P72541 | N/A | N/A | | −2.8 | 2.33E-04 |
| A_24_P299474 | NM_001122679 | ODZ2 | odz, odd Oz/ten-m
homolog 2 (Drosophila) | −2.8 | 1.26E-04 |
| A_23_P416395 | NM_003714 | STC2 | Stanniocalcin
2 | −2.8 | 1.26E-04 |
| A_23_P40415 | NM_007038 | ADAMTS5 | ADAM
metallopeptidase with thrombospondin type 1 motif, 5 | −2.8 | 1.26E-04 |
| A_32_P3545 | XM_002345868 |
LOC100131504 | Hypothetical
LOC100131504 | −2.79 | 4.43E-04 |
| A_23_P106405 | NM_002487 | NDN | Necdin homolog
(mouse) | −2.79 | 1.26E-04 |
| A_23_P405129 | NM_000428 | LTBP2 | Latent transforming
growth factor β binding protein 2 | −2.79 | 1.26E-04 |
| A_24_P237804 | NM_174981 | POTED | POTE ankyrin domain
family, member D | −2.78 | 1.26E-04 |
| A_23_P89780 | NM_198129 | LAMA3 | Laminin, α3 | −2.78 | 1.26E-04 |
| A_23_P213415 | NM_003966 | SEMA5A | Sema domain, seven
thrombospondin repeats (type 1 and type 1-like), transmembrane
domain (TM) and short cytoplasmic domain, (semaphorin) 5A | −2.77 | 3.41E-04 |
| A_24_P397386 | NM_002310 | LIFR | Leukemia inhibitory
factor receptor α | −2.77 | 1.26E-04 |
| A_23_P73297 | NM_004742 | MAGI1 | Membrane associated
guanylate kinase, WW and PDZ domain containing 1 | −2.77 | 1.26E-04 |
| A_23_P165783 | NM_024101 | MLPH | Melanophilin | −2.76 | 1.26E-04 |
| A_23_P212061 | NM_007289 | MME | Membrane
metallo-endopeptidase | −2.76 | 1.26E-04 |
| A_23_P75056 | NM_001002295 | GATA3 | GATA binding
protein 3 | −2.76 | 1.26E-04 |
| A_24_P748377 | CR749529 | | | −2.75 | 2.33E-04 |
| A_24_P810476 | | NTRK3 | Neurotrophic
tyrosine kinase, receptor, type 3 | −2.74 | 3.41E-04 |
| A_32_P60606 | AL713753 |
DKFZp667F0711 | Hypothetical
protein DKFZp667F0711 | −2.74 | 1.26E-04 |
| A_32_P200697 | NM_181709 | FAM101A | Family with
sequence similarity 101, member A | −2.73 | 4.43E-04 |
| A_24_P84220 | NR_027995 |
LOC284232 | Ankyrin repeat
domain 20 family, member A2 pseudogene | −2.73 | 1.26E-04 |
| A_23_P157914 | NM_153267 | MAMDC2 | MAM domain
containing 2 | −2.71 | 1.26E-04 |
| A_24_P393596 | N/A | N/A | | −2.71 | 1.26E-04 |
| A_32_P25419 | N/A | N/A | | −2.7 | 1.26E-04 |
| A_24_P169873 | N/A | N/A | | −2.7 | 1.26E-04 |
| A_24_P358534 | N/A | N/A | | −2.69 | 3.41E-04 |
| A_32_P34750 | AV702101 | N/A | | −2.69 | 1.26E-04 |
| A_32_P9941 | NM_007191 | WIF1 | WNT inhibitory
factor 1 | −2.68 | 2.33E-04 |
| A_23_P335143 | U81001 | SNRPN | Small nuclear
ribonucleoprotein polypeptide N | −2.67 | 1.26E-04 |
| A_23_P56855 | NM_001137671 | POTEC | POTE ankyrin domain
family, member C | −2.67 | 1.26E-04 |
| A_32_P59837 | AK091914 | N/A | | −2.65 | 1.26E-04 |
| A_24_P737553 | AK023774 | N/A | | −2.65 | 2.33E-04 |
| A_23_P204286 | NM_000900 | MGP | Matrix Gla
protein | −2.65 | 1.26E-04 |
| A_24_P725895 | BE218249 | N/A | | −2.63 | 1.26E-04 |
| A_32_P4337 | N/A | N/A | | −2.63 | 1.26E-04 |
| A_23_P154400 | NM_001042467 | MLPH | Melanophilin | −2.62 | 1.26E-04 |
| A_23_P29800 | NM_005602 | CLDN11 | Claudin 11 | −2.61 | 1.26E-04 |
| A_23_P156025 | NM_033267 | IRX2 | Iroquois homeobox
2 | −2.61 | 1.26E-04 |
| A_32_P193091 | N/A | N/A | | −2.61 | 1.26E-04 |
| A_23_P83857 | NM_000240 | MAOA | Monoamine oxidase
A | −2.6 | 1.26E-04 |
| A_32_P355396 | NM_014844 | TECPR2 | Tectonin
β-propeller repeat containing 2 | −2.6 | 1.26E-04 |
| A_32_P214565 | BU928689 | N/A | | −2.6 | 1.26E-04 |
| A_24_P468950 | AK021439 | N/A | | −2.6 | 1.26E-04 |
| A_24_P683583 | N/A | N/A | | −2.6 | 1.26E-04 |
| A_23_P203558 | NM_000518 | HBB | Hemoglobin, β | −2.6 | 2.33E-04 |
| A_32_P140153 | N/A | N/A | | −2.6 | 1.26E-04 |
| A_32_P124461 | AK129743 | N/A | | −2.59 | 1.26E-04 |
| A_23_P136026 | AK128476 | N/A | | −2.59 | 1.26E-04 |
| A_23_P28295 | NM_004525 | LRP2 | Low density
lipoprotein-related protein 2 | −2.59 | 4.43E-04 |
| A_24_P586712 | NM_198485 | TPRG1 | Tumor protein p63
regulated 1 | −2.58 | 1.26E-04 |
| A_23_P139500 | NM_030762 | BHLHE41 | Basic
helix-loop-helix family, member e41 | −2.58 | 1.26E-04 |
| A_23_P121480 | NM_001004196 | CD200 | CD200 molecule | −2.58 | 1.26E-04 |
| A_23_P32577 | NM_080759 | DACH1 | Dachshund homolog 1
(Drosophila) | −2.58 | 1.26E-04 |
| A_23_P315815 | NM_004495 | NRG1 | Neuregulin 1 | −2.58 | 1.26E-04 |
| A_23_P93772 | NM_019102 | HOXA5 | Homeobox A5 | −2.58 | 1.26E-04 |
| A_32_P150748 | CR749529 | N/A | | −2.58 | 1.26E-04 |
| A_32_P204959 | N/A | N/A | | −2.58 | 1.26E-04 |
| A_23_P363149 | N/A | N/A | | −2.57 | 4.43E-04 |
| A_23_P41487 | NM_015130 | TBC1D9 | TBC1 domain family,
member 9 (with GRAM domain) | −2.57 | 1.26E-04 |
| A_23_P257296 | NM_003226 | TFF3 | Trefoil factor 3
(intestinal) | −2.56 | 3.41E-04 |
| A_23_P250735 | NM_175709 | CBX7 | Chromobox homolog
7 | −2.56 | 1.26E-04 |
| A_24_P189516 | NM_001609 | ACADSB | acyl-coenzyme A
dehydrogenase, short/branched chain | −2.56 | 1.26E-04 |
| A_23_P253012 | NM_017577 | GRAMD1C | GRAM domain
containing 1C | −2.56 | 1.26E-04 |
| A_24_P179244 | XM_001723863 |
LOC100128979 | Hypothetical
protein LOC100128979 | −2.55 | 1.26E-04 |
| A_32_P117846 | N/A | N/A | | −2.55 | 1.26E-04 |
| A_32_P42224 | BX097190 | N/A | | −2.55 | 2.33E-04 |
| A_24_P119665 | NM_001128933 |
SYNPO2 | Synaptopodin
2 | −2.54 | 1.26E-04 |
| A_32_P105825 | NM_001584 |
MPPED2 |
Metallophosphoesterase domain containing
2 | −2.54 | 3.41E-04 |
| A_24_P225679 | NM_005544 | IRS1 | Insulin receptor
substrate 1 | −2.54 | 1.26E-04 |
| A_32_P226907 | N/A | N/A | | −2.54 | 1.26E-04 |
| A_23_P356581 | NM_022370 | ROBO3 | Roundabout, axon
guidance receptor, homolog 3 (Drosophila) | −2.53 | 1.26E-04 |
| A_32_P221096 | AW015426 | N/A | | −2.53 | 1.26E-04 |
| A_23_P106016 | NM_002742 | PRKD1 | Protein kinase
D1 | −2.52 | 1.26E-04 |
| A_32_P210193 | N/A | N/A | | −2.52 | 1.26E-04 |
| A_32_P38436 | N/A | N/A | | −2.52 | 1.26E-04 |
| A_24_P512775 | N/A | N/A | | −2.52 | 1.26E-04 |
| A_23_P151529 | NR_023938 |
C14orf132 | Chromosome 14
open reading frame 132 | −2.52 | 1.26E-04 |
| A_32_P235568 | AK125221 | N/A | | −2.52 | 1.26E-04 |
| A_23_P71270 | NM_001185 | AZGP1 | α-2-glycoprotein
1, zinc-binding | −2.52 | 4.43E-04 |
| A_24_P650425 | N/A | N/A | Matrilin 2 | −2.51 | 1.26E-04 |
| A_23_P71328 | NM_030583 | MATN2 | ras homolog gene
family, member J | −2.51 | 2.33E-04 |
| A_24_P153803 | NM_020663 | RHOJ | | −2.51 | 1.26E-04 |
| A_24_P912730 | N/A | N/A | | −2.51 | 1.26E-04 |
| A_24_P347624 | NM_022804 | SNURF | SNRPN upstream
reading frame | −2.5 | 1.26E-04 |
| A_32_P52785 | NM_015345 | DAAM2 | Dishevelled
associated activator of morphogenesis 2 | −2.5 | 3.41E-04 |
| A_23_P61042 | N/A | N/A | | −2.5 | 1.26E-04 |
| A_23_P67661 | NM_001864 |
COX7A1 | Cytochrome c
oxidase subunit VIIa polypeptide 1 (muscle) | −2.49 | 1.26E-04 |
| A_23_P213486 | N/A | PARP8 | Poly(ADP-ribose)
polymerase family, member 8 | −2.49 | 1.26E-04 |
| A_23_P18713 | NM_004827 | ABCG2 | ATP-binding
cassette, sub-family G (WHITE), member 2 | −2.48 | 4.43E-04 |
| A_23_P76658 | NM_052818 |
N4BP2L1 | NEDD4 binding
protein 2-like 1 | −2.48 | 1.26E-04 |
| A_23_P96590 | NM_014710 |
GPRASP1 | G protein-coupled
receptor associated sorting protein 1 | −2.48 | 1.26E-04 |
| A_24_P460763 | AK022443 | N/A | | −2.48 | 1.26E-04 |
| A_23_P85672 | NM_006610 | MASP2 | Mannan-binding
lectin serine peptidase 2 | −2.48 | 1.26E-04 |
| A_24_P416489 | N/A | N/A | | −2.47 | 1.26E-04 |
| A_24_P321525 | NM_032918 | RERG | RAS-like,
estrogen-regulated, growth inhibitor | −2.47 | 1.26E-04 |
| A_24_P256526 | BC005914 | SP2 | Sp2 transcription
factor | −2.47 | 1.26E-04 |
| A_24_P261417 | NM_015881 | DKK3 | Dickkopf homolog
3 (Xenopus laevis) | −2.47 | 1.26E-04 |
| A_23_P98369 | NM_000829 | GRIA4 | Glutamate
receptor, ionotrophic, AMPA 4 | −2.47 | 1.26E-04 |
| A_23_P6818 | NM_020163 |
SEMA3G | Sema domain,
immunoglobulin domain (Ig), short basic domain, secreted,
(semaphorin) 3G | −2.46 | 3.41E-04 |
| A_32_P100379 | N/A | N/A | | −2.46 | 1.26E-04 |
| A_23_P30163 | NR_026804 |
FLJ13197 | Hypothetical
FLJ13197 | −2.46 | 1.26E-04 |
| A_24_P206328 | NM_005020 | PDE1C | Phosphodiesterase
1C, calmodulin-dependent 70 kDa | −2.46 | 1.26E-04 |
| A_24_P93948 | AB210045 | N/A | | −2.46 | 1.26E-04 |
| A_32_P52414 | N/A | N/A | | −2.45 | 1.26E-04 |
| A_23_P123228 | NM_000111 |
SLC26A3 | Solute carrier
family 26, member 3 | −2.45 | 1.26E-04 |
| A_24_P666553 | N/A | N/A | | −2.45 | 1.26E-04 |
| A_24_P916816 | N/A | N/A | | −2.44 | 1.26E-04 |
| A_23_P134734 | NM_017786 |
GOLSYN | Golgi-localized
protein | −2.44 | 1.26E-04 |
| A_24_P296772 | NM_033256 |
PPP1R14A | Protein
phosphatase 1, regulatory (inhibitor) subunit 14A | −2.43 | 1.26E-04 |
| A_24_P267523 | NM_144613 |
COX6B2 | Cytochrome c
oxidase subunit VIb polypeptide 2 (testis) | −2.43 | 1.26E-04 |
| A_23_P133517 | NM_002310 | LIFR | Leukemia
inhibitory factor receptor α | −2.43 | 1.26E-04 |
| A_24_P787680 | N/A | N/A | | −2.43 | 1.26E-04 |
| A_32_P52829 | N/A | N/A | | −2.43 | 3.41E-04 |
| A_23_P162047 | NM_015881 | DKK3 | Dickkopf homolog
3 (Xenopus laevis) | −2.43 | 1.26E-04 |
| A_32_P185140 | BX648171 | TPM1 | Tropomyosin 1
(α) | −2.43 | 1.26E-04 |
| A_24_P319892 | NM_198274 | SMYD1 | SET and MYND
domain containing 1 | −2.43 | 1.26E-04 |
| A_24_P226322 | NM_031469 |
SH3BGRL2 | SH3 domain
binding glutamic acid-rich protein like 2 | −2.42 | 1.26E-04 |
| A_23_P86012 | NM_001017402 | LAMB3 | Laminin, β3 | −2.42 | 1.26E-04 |
| A_23_P62255 | NM_005314 | GRPR | Gastrin-releasing
peptide receptor | −2.41 | 1.26E-04 |
| A_24_P141520 | N/A | N/A | | −2.41 | 2.33E-04 |
| A_23_P114883 | NM_002023 | FMOD | Fibromodulin | −2.41 | 1.26E-04 |
| A_23_P300033 | NM_006206 |
PDGFRA | Platelet-derived
growth factor receptor, α polypeptide | −2.41 | 2.33E-04 |
| A_24_P108311 | NM_015277 |
NEDD4L | Neural precursor
cell expressed, developmentally downregulated 4-like | −2.41 | 1.26E-04 |
| A_23_P345746 | NM_199261 | TPTE | Transmembrane
phosphatase with tensin homology | −2.41 | 1.26E-04 |
| A_23_P418083 | NM_181714 | LCA5 | Leber congenital
amaurosis 5 | −2.41 | 1.26E-04 |
| A_32_P208341 | N/A | N/A | | −2.41 | 1.26E-04 |
| A_24_P930337 | N/A | N/A | | −2.41 | 1.26E-04 |
| A_24_P915095 | NM_017577 |
GRAMD1C | GRAM domain
containing 1C | −2.4 | 1.26E-04 |
| A_32_P4792 | AK057820 | N/A | | −2.4 | 1.26E-04 |
| A_24_P82032 | NM_020663 | RHOJ | ras homolog gene
family, member J | −2.39 | 2.33E-04 |
| A_23_P204296 | NM_032918 | RERG | RAS-like,
estrogen-regulated, growth inhibitor | −2.38 | 1.26E-04 |
| A_24_P920712 | N/A | N/A | | −2.38 | 2.33E-04 |
| A_24_P401185 | NM_001042784 |
CCDC158 | Coiled-coil
domain containing 158 | −2.38 | 1.26E-04 |
| A_32_P109604 | XM_001715342 |
LOC100132733 | Similar to
FLJ00310 protein | −2.37 | 1.26E-04 |
| A_24_P131173 | NM_024709 |
C1orf115 | Chromosome 1 open
reading frame 115 | −2.37 | 2.33E-04 |
| A_24_P64241 | NM_001012421 |
ANKRD20A2 | Ankyrin repeat
domain 20 family, member A2 | −2.37 | 1.26E-04 |
| A_32_P58437 | N/A | N/A | | −2.37 | 1.26E-04 |
| A_24_P602348 | N/A | N/A | | −2.37 | 1.26E-04 |
| A_24_P135856 | NM_016124 | RHD | Rh blood group, D
antigen | −2.37 | 1.26E-04 |
| A_23_P333038 | NM_025145 |
C10orf79 | Chromosome 10
open reading frame 79 | −2.37 | 2.33E-04 |
| A_23_P352266 | NM_000633 | BCL2 | B-cell
CLL/lymphoma 2 | −2.36 | 1.26E-04 |
| A_23_P207699 | NM_016835 | MAPT |
Microtubule-associated protein tau | −2.36 | 1.26E-04 |
| A_23_P392529 | NR_027270 |
C21orf81 | Ankyrin repeat
domain 20 family, member A3 pseudogene | −2.36 | 1.26E-04 |
| A_23_P904 | NM_024603 | BEND5 | BEN domain
containing 5 | −2.36 | 1.26E-04 |
| A_23_P115785 | NM_145235 | FANK1 | Fibronectin type
III and ankyrin repeat domains 1 | −2.35 | 1.26E-04 |
| A_32_P146844 | N/A | N/A | | −2.35 | 1.26E-04 |
| A_23_P26865 | NM_002470 | MYH3 | Myosin, heavy
chain 3, skeletal muscle, embryonic | −2.35 | 1.26E-04 |
| A_32_P100641 | XM_001714998 |
LOC100128139 | Hypothetical
LOC100128139 | −2.35 | 2.33E-04 |
| A_24_P930727 | AK091677 | N/A | | −2.35 | 1.26E-04 |
| A_23_P406341 | NM_001001936 |
AFAP1L2 | Actin filament
associated protein 1-like 2 | −2.35 | 1.26E-04 |
| A_24_P54863 | NM_152400 |
C4orf32 | Chromosome 4 open
reading frame 32 | −2.34 | 1.26E-04 |
| A_23_P133120 | NM_018342 |
TMEM144 | Transmembrane
protein 144 | −2.34 | 1.26E-04 |
| A_32_P86705 | BC040577 | N/A | | −2.34 | 1.26E-04 |
| A_24_P833256 | N/A | N/A | | −2.33 | 1.26E-04 |
| A_23_P401106 | NM_002599 | PDE2A | Phosphodiesterase
2A, cGMP-stimulated | −2.33 | 1.26E-04 |
| A_24_P102119 | AF264623 | N/A | | −2.33 | 1.26E-04 |
| A_23_P358714 | NM_020775 |
KIAA1324 | KIAA1324 | −2.32 | 1.26E-04 |
| A_32_P162494 | N/A | N/A | | −2.32 | 3.41E-04 |
| A_23_P326931 | NM_145170 | TTC18 | Tetratricopeptide
repeat domain 18 | −2.32 | 1.26E-04 |
Identification of cancer-specific
genes
Next, to develop novel therapeutic targets for TNBC
with a minimum risk of adverse events, we performed a DNA
microarray analysis of normal human vital organs consisting of the
heart, lung, liver and kidney as well as TNBC cases and attempted
to identify genes whose expression was exclusively upregulated in
TNBC, but not expressed in normal vital organs. We identified 104
genes, which were specifically upregulated in TNBC, including
cancer-specific molecules such as NIMA-related kinase 2
(NEK2) (29,30), PDZ binding kinase (PBK)
(31), denticleless homolog
(Drosohila) (DTL) (32), maternal leucine zipper kinase
(MELK) (33), and kinesin
family member C (KIF2C) (34), which have previously been shown to
be involved in breast carcinogenesis (Fig. 1C and Table IV).
 | Table IV.Genes specifically expressed in
TNBC, but not expressed in normal human vital organs. |
Table IV.
Genes specifically expressed in
TNBC, but not expressed in normal human vital organs.
| Probe ID | Accession
no. | Symbol | Gene name | Fold change
(log) | P-value |
|---|
| A_23_P118834 | NM_001067 | TOP2A | Topoisomerase
(DNA) IIα 170 kDa | 4.76 | 1.26E-04 |
| A_32_P119154 | BE138567 | N/A | | 4.75 | 1.26E-04 |
| A_23_P35219 | NM_002497 | NEK2 | NIMA (never in
mitosis gene a)-related kinase 2 | 4.67 | 1.26E-04 |
| A_23_P166360 | NM_206956 | PRAME | Preferentially
expressed antigen in melanoma | 4.64 | 1.26E-04 |
| A_24_P332314 | NM_198947 |
FAM111B | Family with
sequence similarity 111, member B | 4.63 | 1.26E-04 |
| A_24_P413884 | NM_001809 | CENPA | Centromere
protein A | 4.59 | 1.26E-04 |
| A_23_P68610 | NM_012112 | TPX2 | TPX2,
microtubule-associated, homolog (Xenopus laevis) | 4.58 | 1.26E-04 |
| A_23_P401 | NM_016343 | CENPF | Centromere
protein F, 350/400 ka (mitosin) | 4.44 | 1.26E-04 |
| A_23_P57379 | NM_003504 |
CDC45L | CDC45 cell
division cycle 45-like (S. cerevisiae) | 4.44 | 1.26E-04 |
| A_23_P356684 | NM_018685 | ANLN | Anillin, actin
binding protein | 4.29 | 1.26E-04 |
| A_23_P52017 | NM_018136 | ASPM | asp (abnormal
spindle) homolog, microcephaly associated (Drosophila) | 4.17 | 1.26E-04 |
| A_32_P199884 | NM_032132 |
HORMAD1 | HORMA domain
containing 1 | 4.13 | 2.33E-04 |
| A_23_P259586 | NM_003318 | TTK | TTK protein
kinase | 4.09 | 1.26E-04 |
| A_23_P200310 | NM_017779 |
DEPDC1 | DEP domain
containing 1 | 4.08 | 1.26E-04 |
| A_23_P115872 | NM_018131 | CEP55 | Centrosomal
protein 55 kDa | 4.03 | 1.26E-04 |
| A_24_P911179 | NM_018136 | ASPM | asp (abnormal
spindle) homolog, microcephaly associated (Drosophila) | 4.02 | 1.26E-04 |
| A_24_P96780 | NM_016343 | CENPF | Centromere
protein F, 350/400 ka (mitosin) | 3.92 | 1.26E-04 |
| A_24_P14156 | NM_006101 | NDC80 | NDC80 homolog,
kinetochore complex component (S. cerevisiae) | 3.86 | 1.26E-04 |
| A_23_P254733 | NM_024629 |
MLF1IP | MLF1 interacting
protein | 3.85 | 1.26E-04 |
| A_23_P74115 | NM_003579 |
RAD54L | RAD54-like (S.
cerevisiae) | 3.84 | 1.26E-04 |
| A_23_P50108 | NM_006101 | NDC80 | NDC80 homolog,
kinetochore complex component (S. cerevisiae) | 3.84 | 1.26E-04 |
| A_23_P155815 | NM_022346 | NCAPG | Non-SMC condensin
I complex, subunit G | 3.82 | 1.26E-04 |
| A_23_P51085 | NM_020675 | SPC25 | SPC25, NDC80
kinetochore complex component, homolog (S. cerevisiae) | 3.81 | 1.26E-04 |
| A_32_P62997 | NM_018492 | PBK | PDZ binding
kinase | 3.8 | 1.26E-04 |
| A_23_P256956 | NM_005733 |
KIF20A | Kinesin family
member 20A | 3.79 | 1.26E-04 |
| A_23_P212844 | NM_006342 | TACC3 | Transforming,
acidic coiled-coil containing protein 3 | 3.78 | 1.26E-04 |
| A_24_P254705 | NM_020394 |
ZNF695 | Zinc finger
protein 695 | 3.76 | 1.26E-04 |
| A_23_P432352 | NM_001017978 |
CXorf61 | Chromosome X open
reading frame 61 | 3.73 | 1.26E-04 |
| A_23_P48669 | NM_005192 | CDKN3 | Cyclin-dependent
kinase inhibitor 3 | 3.71 | 1.26E-04 |
| A_23_P94571 | NM_004432 |
ELAVL2 | ELAV (embryonic
lethal, abnormal vision, Drosophila)-like 2 (Hu antigen
B) | 3.67 | 1.26E-04 |
| A_23_P150667 | NM_031217 |
KIF18A | Kinesin family
member 18A | 3.64 | 1.26E-04 |
| A_32_P68525 | BC035392 | N/A | | 3.58 | 1.26E-04 |
| A_24_P319613 | NM_002497 | NEK2 | NIMA (never in
mitosis gene a)-related kinase 2 | 3.53 | 1.26E-04 |
| A_23_P10385 | NM_016448 | DTL | Denticleless
homolog (Drosophila) | 3.53 | 1.26E-04 |
| A_23_P94422 | NM_014791 | MELK | Maternal
embryonic leucine zipper kinase | 3.5 | 1.26E-04 |
| A_23_P340909 | BC013418 | SKA3 | Spindle and
kinetochore associated complex subunit 3 | 3.48 | 1.26E-04 |
| A_23_P124417 | NM_004336 | BUB1 | Budding
uninhibited by benzimidazoles 1 homolog (yeast) | 3.47 | 1.26E-04 |
| A_24_P257099 | NM_018410 | HJURP | Holliday junction
recognition protein | 3.43 | 1.26E-04 |
| A_23_P74349 | NM_145697 | NUF2 | NUF2, NDC80
kinetochore complex component, homolog (S. cerevisiae) | 3.36 | 1.26E-04 |
| A_24_P302584 | NM_003108 | SOX11 | SRY (sex
determining region Y)-box 11 | 3.36 | 4.43E-04 |
| A_24_P68088 | NR_002947 | TCAM1 | Testicular cell
adhesion molecule 1 homolog (mouse) | 3.35 | 2.33E-04 |
| A_24_P366033 | NM_018098 | ECT2 | Epithelial cell
transforming sequence 2 oncogene | 3.34 | 1.26E-04 |
| A_23_P93258 | NM_003537 |
HIST1H3B | Histone cluster
1, H3b | 3.33 | 1.26E-04 |
| A_23_P149668 | NM_014875 | KIF14 | Kinesin family
member 14 | 3.29 | 1.26E-04 |
| A_23_P34325 | NM_033300 | LRP8 | Low density
lipoprotein receptor-related protein 8, apolipoprotein E
receptor | 3.28 | 1.26E-04 |
| A_32_P56154 | N/A | N/A | | 3.28 | 1.26E-04 |
| A_23_P138507 | NM_001786 | CDC2 | Cell division
cycle 2, G1→S and G2→M | 3.24 | 1.26E-04 |
| A_23_P49972 | NM_001254 | CDC6 | Cell division
cycle 6 homolog (S. cerevisiae) | 3.22 | 1.26E-04 |
| A_24_P306896 | XR_040656 |
LOC283711 | Hypothetical
protein LOC283711 | 3.22 | 1.26E-04 |
| A_23_P44684 | NM_018098 | ECT2 | Epithelial cell
transforming sequence 2 oncogene | 3.21 | 1.26E-04 |
| A_23_P100344 | NM_014321 | ORC6L | Origin
recognition complex, subunit 6 like (yeast) | 3.2 | 1.26E-04 |
| A_23_P163481 | NM_001211 | BUB1B | Budding
uninhibited by benzimidazoles 1 homolog β (yeast) | 3.17 | 1.26E-04 |
| A_32_P87849 | N/A | N/A | | 3.16 | 1.26E-04 |
| A_24_P397107 | NM_001789 |
CDC25A | Cell division
cycle 25 homolog A (S. pombe) | 3.15 | 1.26E-04 |
| A_23_P209200 | NM_001238 | CCNE1 | Cyclin E1 | 3.15 | 1.26E-04 |
| A_32_P16625 | N/A | N/A | | 3.15 | 1.26E-04 |
| A_24_P37903 | N/A | LOX | Lysyl
oxidase | 3.12 | 1.26E-04 |
| A_24_P313504 | NM_005030 | PLK1 | Polo-like kinase
1 (Drosophila) | 3.07 | 1.26E-04 |
| A_23_P252292 | NM_006733 | CENPI | Centromere
protein I | 3.04 | 1.26E-04 |
| A_23_P161474 | NM_182751 | MCM10 | Minichromosome
maintenance complex component 10 | 2.99 | 1.26E-04 |
| A_23_P253762 | N/A | N/A | | 2.94 | 1.26E-04 |
| A_24_P225534 | NM_017821 |
RHBDL2 | Rhomboid,
veinlet-like 2 (Drosophila) | 2.94 | 1.26E-04 |
| A_24_P412088 | NM_182751 | MCM10 | Minichromosome
maintenance complex component 10 | 2.94 | 1.26E-04 |
| A_32_P109296 | NM_152259 |
C15orf42 | Chromosome 15
open reading frame 42 | 2.91 | 1.26E-04 |
| A_24_P76521 | AK056691 | GSG2 | Germ cell
associated 2 (haspin) | 2.83 | 1.26E-04 |
| A_23_P126212 | NM_022111 | CLSPN | Claspin homolog
(Xenopus laevis) | 2.83 | 1.26E-04 |
| A_23_P60120 | NM_031415 | GSDMC | Gasdermin C | 2.81 | 2.33E-04 |
| A_24_P902509 | NM_018193 | FANCI | Fanconi anemia,
complementation group I | 2.8 | 1.26E-04 |
| A_23_P155969 | NM_014264 | PLK4 | Polo-like kinase
4 (Drosophila) | 2.79 | 1.26E-04 |
| A_32_P183218 | NM_153695 |
ZNF367 | Zinc finger
protein 367 | 2.77 | 1.26E-04 |
| A_23_P46118 | NM_001821 | CHML |
Choroideremia-like (Rab escort protein
2) | 2.76 | 2.33E-04 |
| A_23_P327643 | N/A | N/A | | 2.75 | 1.26E-04 |
| A_23_P215976 | NM_057749 | CCNE2 | Cyclin E2 | 2.72 | 2.33E-04 |
| A_32_P151800 | NM_207418 |
FAM72D | Family with
sequence similarity 72, member D | 2.7 | 1.26E-04 |
| A_23_P34788 | NM_006845 | KIF2C | Kinesin family
member 2C | 2.7 | 1.26E-04 |
| A_23_P133956 | NM_002263 | KIFC1 | Kinesin family
member C1 | 2.69 | 1.26E-04 |
| A_23_P88630 | NM_000057 | BLM | Bloom syndrome,
RecQ helicase-like | 2.68 | 1.26E-04 |
| A_24_P276102 | NM_183404 | RBL1 |
Retinoblastoma-like 1 (p107) | 2.68 | 1.26E-04 |
| A_23_P23303 | NM_003686 | EXO1 | Exonuclease
1 | 2.67 | 1.26E-04 |
| A_23_P88691 | NM_000745 |
CHRNA5 | Cholinergic
receptor, nicotinic, α5 | 2.67 | 1.26E-04 |
| A_32_P72341 | NM_173084 |
TRIM59 | Tripartite
motif-containing 59 | 2.62 | 1.26E-04 |
| A_24_P227091 | NM_004523 | KIF11 | Kinesin family
member 11 | 2.61 | 1.26E-04 |
| A_23_P136805 | NM_014783 |
ARHGAP11A | Rho GTPase
activating protein 11A | 2.6 | 1.26E-04 |
| A_23_P63402 | NM_013296 | GPSM2 | G-protein
signaling modulator 2 (AGS3-like, C. elegans) | 2.6 | 1.26E-04 |
| A_23_P35871 | NM_024680 | E2F8 | E2F transcription
factor 8 | 2.58 | 1.26E-04 |
| A_23_P207307 | N/A | N/A | | 2.58 | 1.26E-04 |
| A_24_P399888 | NM_001002876 | CENPM | Centromere
protein M | 2.58 | 1.26E-04 |
| A_23_P155989 | NM_022145 | CENPK | Centromere
protein K | 2.57 | 1.26E-04 |
| A_23_P411335 | NM_152524 | SGOL2 | Shugoshin-like 2
(S. pombe) | 2.54 | 1.26E-04 |
| A_23_P43484 | NM_058197 |
CDKN2A | Cyclin-dependent
kinase inhibitor 2A (melanoma, p16, inhibits CDK4) | 2.52 | 1.26E-04 |
| A_32_P28704 | N/A | N/A | | 2.52 | 1.26E-04 |
| A_24_P351466 | NM_020890 |
KIAA1524 | KIAA1524 | 2.5 | 1.26E-04 |
| A_24_P334248 | NM_014996 | PLCH1 | Phospholipase C,
eta 1 | 2.48 | 1.26E-04 |
| A_23_P88331 | NM_014750 |
DLGAP5 | Discs, large
(Drosophila) homolog-associated protein 5 | 2.47 | 1.26E-04 |
| A_32_P31021 | N/A | N/A | | 2.46 | 1.26E-04 |
| A_23_P361419 | NM_018369 |
DEPDC1B | DEP domain
containing 1B | 2.45 | 1.26E-04 |
| A_23_P397341 | NM_152341 | PAQR4 | Progestin and
adipoQ receptor family member IV | 2.42 | 1.26E-04 |
| A_23_P140316 | NM_001099652 |
GPR137C | G protein-coupled
receptor 137C | 2.42 | 1.26E-04 |
| A_23_P217049 | NM_014286 | FREQ | Frequenin homolog
(Drosophila) | 2.41 | 2.33E-04 |
| A_32_P35839 | N/A | N/A | | 2.4 | 1.26E-04 |
| A_24_P857404 | NM_001093725 | MEX3A | mex-3 homolog A
(C. elegans) | 2.4 | 1.26E-04 |
| A_24_P323598 | NM_001017420 | ESCO2 | Establishment of
cohesion 1 homolog 2 (S. cerevisiae) | 2.36 | 1.26E-04 |
| A_23_P112673 | NM_017975 |
ZWILCH | Zwilch,
kinetochore associated, homolog (Drosophila) | 2.33 | 1.26E-04 |
| A_24_P296254 | NM_014783 |
ARHGAP11A | Rho GTPase
activating protein 11A | 2.32 | 1.26E-04 |
Functional gene annotation clustering
analysis
To elucidate the biological processes and pathways
characterized in TNBC, we performed a functional analysis of these
upregulated or downregulated genes in 30 TNBC cases using the gene
annotation clustering of the DAVID algorithm. We identified the
most prominent cluster (cluster 1; gene enrichment score, 29.90)
composed of various functional annotation terms consisting of 87
upregulated genes in TNBC (Table
V). Cluster 1 consisted almost entirely of cell
cycle-associated genes as represented by nuclear division (fold
enrichment, 15.04), mitosis (fold enrichment, 15.04), M phase of
the mitotic cell cycle (fold enrichment, 14.78), organelle fission
(fold enrichment, 14.45), and M phase (fold enrichment, 12.90)
(Fig. 2). These findings suggest
that most of the upregulated genes in TNBC might be functionally
responsible for cell cycle progression.
 | Table V.Genes listed in cluster 1 and
cluster 2. |
Table V.
Genes listed in cluster 1 and
cluster 2.
| No. of genes | Genes |
|---|
| Cluster 1
(enrichment score, 29.90) | |
| 87 | BLM, CKS1B,
CKS2, CHEK1, E2F1, E2F2, E2F8, FANCA, FANCI, H2AFX, HORMAD1, HJURP,
MAD2L1, NDC80, NEK2, NUF2, OIP5, PBK, RAD51, RAD54L, SPC25, TPX2,
TTK, ZWINT ZWILCH, ANLN, ASPM, AURKA, BIRC5, BUB1, BUB1B, CASC5,
CDC25A, CDC6, CDCA2, CDCA5, CDCA8, CENPA, CENPF, CEP55, CHAF1B,
SKA3, C13orf34, CIT, CLSPN, CCNA2, CCNB1, CCNE1, CCNE2, CDKN2A,
CDKN2C, CDKN3, DSCC1, DLGAP5, ESCO2, EXO1, FAM83D, GSG2, INHBA,
KIF11, KIF14, KIF18A, KIF18B, KIF20A, KIF23, KIF2C, KIFC1, LMNB1,
MND1, NCAPG, NUSAP1, PTTG1, PLK1, PLK4, PKMYT1, PRC1, RBL1, SGOL2
SPAG5, STMN1, SMC4, TMSB15A, TOP2A, TACC3, TUBB3, UBE2C,
UHRF1 |
| Cluster 2
(enrichment score, 6.43) | |
| 45 | ADAMTS5,
MAMDC2, SPARCL1, WIF1, AZGP1, APOD, FIGF, CHL1, CCL28, CXCL2,
COL4A6, COL14A1, COL17A1, CNTNAP3, DKK3, DST, FGF1, FMOD, HS3ST4,
IGJ, IL33, LAMA3, LAMAB, LTBP2, LIFR, LRP2, MASP2, MATN2, MGP,
NTN4, NRG1, PTHLH, PI15, PLAT, PDGFA, PTN, PIGR, PIP, SCGB1D1,
SCGB1D2, SCGB3A1, SEMA3G, STC2, THSD4, TFF3 |
On the other hand, we also identified the most
prominent cluster functionally deactivated in TNBC based on
down-regulated genes in TNBC (cluster 2; enrichment score, 6.43).
As shown in Table V and Fig. 2, cluster 2 consisted of functions
induced by extracellular matrix-cell adhesion-associated genes such
as latent transforming growth factor β binding protein 2
(LTBP2), laminin α3 (LAMA3) and cell adhesion
molecule with homology to L1CAM (close homolog of L1)
(CHL1), which have been reported to be downregulated in
various tumors (35–37). These results suggest that loss of
cell-cell or matrix-cell interactions might be a key mechanism in
TNBC progression.
Identification of ASPM and CENPK as novel
molecular targets for TNBC therapy
Because the upregulated genes were mainly included
in the cell cycle-associated gene cluster as described above, we
directed our focus to two cancer-specific genes that function as
cell cycle regulators, asp (abnormal spindle) homolog, microcephaly
associated (Drosophila) (ASPM), which is fundamental
for cytokinesis (38) and
centromere protein K (CENPK), which is essential for proper
kinetochore assembly during mitosis (39), as novel therapeutic targets for
TNBC. qRT-PCR experiments confirmed that ASPM and
CENPK genes were significantly upregulated in 48 clinical
TNBC cases (Fig. 3A) and five cell
lines derived from TNBC (Fig. 3B),
but undetectably expressed in a mixture of 13 microdissected normal
mammary ductal cells and the normal mammary epithelial cell line
MCF10A as well as normal human vital organs.
To ascertain the possible roles of ASPM and
CENPK in TNBC cell growth, we knocked down the expression of
endogenous ASPM and CENPK in three TNBC cell lines,
HCC1937, BT-20 and MDA-MB-231 cells, which highly express both of
these genes (Fig. 3), using RNAi.
qRT-PCR experiments showed that ASPM and CENPK were
significantly knocked down in cells transfected with siASPM
and siCENPK, but not with siEGFP as a control
(Fig. 4A). In concordance with
their knockdown, the MTT assay clearly revealed growth suppression
of breast cancer cells in a time-dependent manner by siASPM
and siCENPK, compared with a control siEGFP, which
showed no knockdown (Fig. 4B). In
addition, a colony formation assay also confirmed that introducing
both shRNA-ASPM and -CENPK constructs remarkably
suppressed the growth of BT-20 and MDA-MB-231 cells, respectively,
compared with shEGFP-transfected cells (Fig. 4C), suggesting that both genes are
likely indispensable for breast cancer cell growth. Furthermore, we
investigated the phenotypic alterations of TNBC cells transfected
with ASPM and CENPK siRNAs showing significant
knockdown effects. FACS analysis revealed that depleting
ASPM caused a cell cycle arrest at the G2/M phase in HCC1937
cells (siEGFP:siASPM, 24.4:34.0%) at 2 days after
transfection, and a subsequent increase in the sub-G1 population
(siEGFP:siASPM, 9.86:43.68%) at 6 days (Fig. 5A). On the other hand, reduced
CENPK expression resulted in an increase in the proportion
of G0/G1 phase cells (siEGFP:siCENPK, 56.49:72.2%) in
MDA-MB-231 after 2 days of transfection, and a subsequent increase
in the sub-G1 population (siEGFP:siCENPK,
12.73:30.96%) at 6 days (Fig. 5B).
Interestingly, we observed an enlarged size of HCC1937 cells, which
was likely due to abnormal tubulin formation due to decreased
ASPM expression (Fig. 5C,
arrowheads). In addition, we observed a disruption in the
structural integrity of tubulin in CENPK-depleted MDA-MB-231
cells (Fig. 5D, arrowheads),
compared with those in siEGFP-transfected cells.
These results suggest that the absence of
ASPM and CENPK caused an arrest in the G2/M and G0/G1
phases, respectively, and then induced cell death. Taken together,
these findings strongly suggest that ASPM and CENPK
have indispensable roles in cell proliferation and mitosis,
especially in the G2/M and G0/G1 phases, in TNBC cells.
Discussion
TNBC patients do not benefit from endocrine therapy
and trastuzumab. Conventional chemotherapy is currently the
mainstay of systemic medical treatment, although TNBC patients have
a worse outcome after chemotherapy than patients with other breast
subtypes. In particular, because cytotoxic drugs often cause severe
adverse effects, it is obvious that thoughtful selection of novel
target molecules based on the detailed molecular mechanisms of TNBC
carcinogenesis should be very helpful to develop effective
anticancer drugs with a minimum risk of side effects. To this end,
we performed DNA microarray using the microdissected TNBC and
normal ductal cells, and normal human vital organs including the
heart, lung, liver and kidney and identified 104 genes that were
significantly upregulated in TNBC compared to normal duct cells,
but not expressed in normal human vital organs. They included
cancer specific kinases, such as NEK2, PBK, and
MELK, which might serve as druggable targets for new
therapeutic agents against TNBC.
NEK2, a member of the NIMA-related
serine/threonine kinase family, is involved in cell division and
the mitotic regulation by centrosome splitting, and is upregulated
in a wide variety of human cancers including breast cancer
(40). siRNA-mediated depletion of
NEK2 expression results in growth suppression of breast and
colorectal cancers (29,30). PBK, a mitotic
serine/threonine kinase, is significantly upregulated in the
majority of breast cancers. siRNA-mediated knockdown of PBK
expression also results in significant suppression of cell growth
due to cytokinetic failure (31).
MELK, a member of the snf1/AMPK serine-threonine kinase
family, is involved in mammalian embryonic development and is also
frequently upregulated in breast cancers and brain tumors (33,41).
Suppression of MELK expression by siRNA significantly
inhibits the growth of human breast cancer cells (33). These findings strongly suggest that
these cancer-specific kinases, NEK2, PBK and MELK,
are promising therapeutic targets for TNBC.
Furthermore, we performed a gene-annotation
enrichment analysis using DAVID based on gene expression profiling
to elucidate the biological processes and pathways associated with
each gene cluster. We found that the vast majority of genes
upregulated in TNBC are functionally responsible for cell cycle
progression involved in nuclear division, microtubule organization,
kinetochore, and chromosome segregation, and that most inactivated
functions closely related to TNBC progression are involved in
cell-cell or cell-matrix interactions, which is consistent with
epithelial mesenchymal transition (EMT) features as a phenotype of
TNBC (42).
To further the development of novel anticancer drugs
with minimum adverse effects, we focused on the cancer-specific
cell-cycle associated genes ASPM and CEPNK as novel
molecular targets for TNBC therapy. ASPM has been reported
to play an essential role in nucleating microtubules at
centrosomes, to localize to the spindle poles during mitosis
(39) and to contribute to
glioblastoma cell growth (43),
but has not been associated with breast carcinogenesis, especially
TNBC. Here, we confirmed that ASPM is upregulated in
clinical samples and TNBC cell lines (Fig. 3) and that siRNA-mediated knockdown
of endogenous ASPM results in the loss of nucleating
microtubules through mitosis by impeding centrosome function,
resulting in G2/M cell cycle arrest and subsequent apoptosis. These
results suggest that aberrant ASPM expression might be
involved in the carcinogenesis of TNBC and that ASPM
targeting might be an attractive therapeutic option with less
adverse effects. CENPK is known to be a subunit of the
CENPH-I complex, and essential for proper kinetochore
assembly (39), but little is
known about the roles of CENPK in human cancer growth,
progression, and carcinogenesis. We also confirmed that
CENPK is upregulated in clinical samples and TNBC cell
lines, and that siRNA-mediated knockdown also causes cell growth
inhibition through G0/G1 cell cycle arrest due to a loss of correct
tubulin structures (Figs.
3–5). Interestingly, we
determined that other centromere or kinetochore-associated
proteins, CENPA, CENPF, CENPI, CENPM, NDC80 and
HJURP, were also significantly overexpressed in TNBC cases,
but not expressed in normal vital organs (Fig. 1C and Table IV). Human CENPA was first
identified based on autoantibodies found in patients suffering from
scleroderma (44) and is
over-expressed in colorectal cancers (45). CENPF is also reportedly
upregulated in head and neck squamous cell carcinomas and
pancreatic ductal carcinomas (46,47).
NDC80 and HJURP are reportedly overexpressed in
breast cancers and associated with tumor grade and poor prognosis
(48,49). These findings suggest that aberrant
regulation of kinetochore assembly and centromere function through
mitosis might contribute to the carcinogenesis of TNBC and that
destroying one component of the kinetochore, such as targeting
CENPK, might be a novel molecular target for TNBC
treatment.
TNBC is a heterogeneous subgroup of breast cancers;
therefore oncologists, pathologists, and geneticists had tried to
clarify TNBC by means of gene expression profiling and
immunohistochemical analyses. We also applied unsupervised
2-dimensional hierarchical clustering analysis to groups of genes
based on similarities in the expression pattern, but there is no
clustering for TNBC based on gene expression patterns, probably due
to the small sample size (data not shown). However, the information
provided in this study will facilitate the development of novel and
attractive molecular drug targets without adverse events.
Acknowledgements
We thank Dr Tomoya Fukawa and Dr Le
Tan Dat for helpful and constructive discussions and Ms. Hitomi
Kawakami for technical assistance in microdissection. This work was
supported in part by a grant from Health Labour Research Grant
‘Third Term Comprehensive Control Research for Cancer
(H24-3rd-Gan-Ippan-006), and Kobayashi Foundation for cancer
Research (2009) (TK).
References
|
1.
|
Jemal A, Siegel R, Ward E, Hao Y, Xu J,
Murray T and Thun MJ: Cancer Statistics, 2008. CA Cancer J Clin.
58:71–96. 2008. View Article : Google Scholar
|
|
2.
|
Di Cosimo S and Baselga J: Management
breast cancer with targeted agents: importance of heterogeneity.
Nat Rev Clin Oncol. 7:139–147. 2010.PubMed/NCBI
|
|
3.
|
Rahman M, Pumphrey JG and Lipkowitz S: The
TRAIL to targeted therapy of breast cancer. Adv Cancer Res.
103:43–73. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Smith I, Procter M, Gelber RD, Guillaume
S, Feyereislova A, Dowsett M, Goldhirsch A, Untch M, Mariani G,
Baselga J, Kaufmann M, Cameron D, Bell R, Bergh J, Coleman R,
Wardley A, Harbeck N, Lopez RI, Mallmann P, Gelmon K, Wilcken N,
Wist E, Sánchez Rovira P and Piccart-Gebhart MJ: HERA study team:
2-year follow-up of trastuzumab after adjuvant chemotherapy in
HER2-positive breast cancer: a randomised controlled trial. Lancet.
369:29–36. 2007.
|
|
5.
|
Romond EH, Perez EA, Bryant J, Suman VJ,
Geyer CE Jr, Davidson NE, Tan-Chiu E, Martino S, Paik S, Kaufman
PA, Swain SM, Pisansky TM, Fehrenbacher L, Kutteh LA, Vogel VG,
Visscher DW, Yothers G, Jenkins RB, Brown AM, Dakhil SR, Mamounas
EP, Lingle WL, Klein PM, Ingle JN and Wolmark N: Trastuzumab plus
adjuvant chemotherapy for operable HER2-positive breast cancer. N
Engl J Med. 353:1673–1684. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Joensuu H, Kellokumpu-Lehtinen PL, Bono P,
Alanko T, Kataja V, Asola R, Utriainen T, Kokko R, Hemminki A,
Tarkkanen M, Turpeenniemi-Hujanen T, Jyrkkiö S, Flander M, Helle L,
Ingalsuo S, Johansson K, Jääskeläinen AS, Pajunen M, Rauhala M,
Kaleva-Kerola J, Salminen T, Leinonen M, Elomaa I and Isola J; for
the FinHer Study Investigators: Adjuvant docetaxel or vinorelbine
with or without trastuzumab for breast cancer. N Engl J Med.
354:809–820. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Foulkes WD, Smith IE and Reis-Filho JS:
Triple-negative breast cancer. N Engl J Med. 363:1938–1948. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Liedtke C, Mazouni C, Hess KR, André F,
Tordai A, Mejia JA, Symmans WF, Gonzalez-Angulo AM, Hennessy B,
Green M, Cristofanilli M, Hortobagyi GN and Pusztai L: Response to
neoadjuvant therapy and long-term survival in patients with
triple-negative breast cancer. J Clin Oncol. 26:1275–1281. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Petricoin EF III, Hackett JL, Lesko LJ,
Puri RK, Gutman SI, Chumakov K, Woodcock J, Feigal DW Jr, Zoon KC
and Sistare FD: Medical applications of microarray technologies: a
regulatory science perspective. Nat Genet. 32:474–479. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Huang DW, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
Bioinformatics Resources. Nat Protoc. 4:44–57. 2009.PubMed/NCBI
|
|
11.
|
Huang DW, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Hao JM, Chen JZ, Sui HM, Si-Ma XQ, Li GQ,
Liu C, Li JL, Ding YQ and Li JM: A five-gene signature as a
potential predictor of metastasis and survival in colorectal
cancer. J Pathol. 220:475–489. 2010.PubMed/NCBI
|
|
13.
|
Ueki T, Park JH, Nishidate T, Kijima K,
Hirata K, Nakamura Y and Katagiri T: Ubiquitination and
downregulation of BRCA1 by ubiquitin-conjugating enzyme E2T
overexpression in human breast cancer cells. Cancer Res.
69:8752–8760. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Loussouarn D, Campion L, Leclair F,
Campone M, Charbonnel C, Ricolleau G, Gouraud W, Bataille R and
Jézéquel P: Validation of UBE2C protein as a prognostic marker in
node-positive breast cancer. Br J Cancer. 101:166–173. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Arumugam T and Logsdon CD: S100P: a novel
therapeutic target for cancer. Amino Acids. 41:893–899. 2011.
View Article : Google Scholar
|
|
16.
|
Xiang T, Li L, Yin X, Yuan C, Tan C, Su X,
Xiong L, Putti TC, Oberst M, Kelly K, Ren G and Tao Q: The
ubiquitin peptidase UCHL1 induces G0/G1 cell cycle arrest and
apoptosis through stabilizing p53 and is frequently silenced in
breast cancer. PLoS One. 7:e297832012. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Yoon CH, Kim MJ, Lee H, Kim RK, Lim EJ,
Yoo KC, Lee GH, Cui YH, Oh YS, Gye MC, Lee YY, Park IC, An S, Hwang
SG, Park MJ, Suh Y and Lee SJ: PTTG1 oncogene promotes tumor
malignancy via epithelial to mesenchymal transition and expansion
of cancer stem cell population. J Biol Chem. 287:19516–19527. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Jin W, Liu Y, Xu SG, Yin WJ, Li JJ, Yang
JM and Shao ZM: UHRF1 inhibits MDR1 gene transcription and
sensitizes breast cancer cells to anticancer drugs. Breast Cancer
Res Treat. 124:39–48. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Ford HL, Landesman-Bollag E, Dacwag CS,
Stukenberg PT, Pardee AB and Seldin DC: Cell cycle-regulated
phosphorylation of the human SIX1 homeodomain protein. J Biol Chem.
275:22245–22254. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Shimo A, Nishidate T, Ohta T, Fukuda M,
Nakamura Y and Katagiri T: Elevated expression of protein regulator
of cytokinesis 1, involved in the growth of breast cancer cells.
Cancer Sci. 98:174–181. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Di Leo A and Isola J: Topoisomerase II
alpha as a marker predicting the efficacy of anthracyclines in
breast cancer: are we at the end of the beginning? Clin Breast
Cancer. 4:179–186. 2003.PubMed/NCBI
|
|
22.
|
Nakagawa M, Bando Y, Nagao T, Morimoto M,
Takai C, Ohnishi T, Honda J, Moriya T, Izumi K, Takahashi M, Sasa M
and Tangoku A: Expression of p53, Ki-67, E-cadherin, N-cadherin and
TOP2A in triple-negative breast cancer. Anticancer Res.
31:2389–2393. 2011.PubMed/NCBI
|
|
23.
|
Adélaïde J, Finetti P, Bekhouche I,
Repellini L, Geneix J, Sircoulomb F, Charafe-Jauffret E, Cervera N,
Desplans J, Parzy D, Schoenmakers E, Viens P, Jacquemier J,
Birnbaum D, Bertucci F and Chaffanet M: Integrated profiling of
basal and luminal breast cancers. Cancer Res. 67:11565–11575.
2007.PubMed/NCBI
|
|
24.
|
Liu RZ, Graham K, Glubrecht DD, Germain
DR, Mackey JR and Godbout R: Association of FABP5 expression with
poor survival in triple-negative breast cancer: implication for
retinoic acid therapy. Am J Pathol. 178:997–1008. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Kalashnikova EV, Revenko AS, Gemo AT,
Andrews NP, Tepper CG, Zou JX, Cardiff RD, Borowsky AD and Chen HW:
ANCCA/ATAD2 overexpression identifies breast cancer patients with
poor prognosis, acting to drive proliferation and survival of
triple-negative cells through control of B-Myb and EZH2. Cancer
Res. 70:9402–9412. 2010. View Article : Google Scholar
|
|
26.
|
Parris TZ, Danielsson A, Nemes S, Kovács
A, Delle U, Fallenius G, Möllerström E, Karlsson P and Helou K:
Clinical implications of gene dosage and gene expression patterns
in diploid breast carcinoma. Clin Cancer Res. 16:3860–3874. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Ai L, Tao Q, Zhong S, Fields CR, Kim WJ,
Lee MW, Cui Y, Brown KD and Robertson KD: Inactivation of Wnt
inhibitory factor-1 (WIF1) expression by epigenetic silencing is a
common event in breast cancer. Carcinogenesis. 27:1341–1348. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Cheng CJ, Lin YC, Tsai MT, Chen CS, Hsieh
MC, Chen CL and Yang RB: SCUBE2 suppresses breast tumor cell
proliferation and confers a favorable prognosis in invasive breast
cancer. Cancer Res. 69:3634–3641. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Tsunoda N, Kokuryo T, Oda K, Senga T,
Yokoyama Y, Nagino M, Nimura Y and Hamaguchi M: Nek2 as a novel
molecular target for the treatment of breast carcinoma. Cancer Sci.
100:111–116. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Suzuki K, Kokuryo T, Senga T, Yokoyama Y,
Nagino M and Hamaguchi M: Novel combination treatment for
colorectal cancer using Nek2 siRNA and cisplatin. Cancer Sci.
101:1163–1169. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Park JH, Lin ML, Nishidate T, Nakamura Y
and Katagiri T: PDZ-binding kinase/T-LAK cell-originated protein
kinase, a putative cancer/testis antigen with an oncogenic activity
in breast cancer. Cancer Res. 66:9186–9195. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Ueki T, Nishidate T, Park JH, Lin ML,
Shimo A, Hirata K, Nakamura Y and Katagiri T: Involvement of
elevated expression of multiple cell-cycle regulator, DTL/RAMP
(denticleless/RA-regulated nuclear matrix associated protein), in
the growth of breast cancer cells. Oncogene. 27:5672–5683. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Lin ML, Park JH, Nishidate T, Nakamura Y
and Katagiri T: Involvement of maternal embryonic leucine zipper
kinase (MELK) in mammary carcinogenesis through interaction with
Bcl-G, a pro-apoptotic member of the Bcl-2 family. Breast Cancer
Res. 9:R172007. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Shimo A, Tanikawa C, Nishidate T, Lin ML,
Matsuda K, Park JH, Ueki T, Ohta T, Hirata K, Fukuda M, Nakamura Y
and Katagiri T: Involvement of kinesin family member 2C/mitotic
centromere-associated kinesin overexpression in mammary
carcinogenesis. Cancer Sci. 99:62–70. 2008.PubMed/NCBI
|
|
35.
|
Chan SH, Yee Ko JM, Chan KW, Chan YP, Tao
Q, Hyytiainen M, Keski-Oja J, Law S, Srivastava G, Tang J, Tsao SW,
Chen H, Stanbridge EJ and Lung ML: The ECM protein LTBP-2 is a
suppressor of esophageal squamous cell carcinoma tumor formation
but higher tumor expression associates with poor patient outcome.
Int J Cancer. 129:565–573. 2011. View Article : Google Scholar
|
|
36.
|
Sathyanarayana UG, Maruyama R, Padar A,
Suzuki M, Bondaruk J, Sagalowsky A, Minna JD, Frenkel EP, Grossman
HB, Czerniak B and Gazdar AF: Molecular detection of noninvasive
and invasive bladder tumor tissues and exfoliated cells by aberrant
promoter methylation of laminin-5 encoding genes. Cancer Res.
64:1425–1430. 2004. View Article : Google Scholar
|
|
37.
|
Senchenko VN, Krasnov GS, Dmitriev AA,
Kudryavtseva AV, Anedchenko EA, Braga EA, Pronina IV, Kondratieva
TT, Ivanov SV, Zabarovsky ER and Lerman MI: Differential expression
of CHL1 gene during development of major human cancers. PLoS One.
6:e156122011. View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
do Carmo Avides M and Glover DM: Abnormal
spindle protein, Asp, and the integrity of mitotic centrosomal
microtubule organizing centers. Science. 283:1733–1735.
1999.PubMed/NCBI
|
|
39.
|
Cheeseman IM, Hori T, Fukagawa T and Desai
A: KNL1 and the CENP-H/I/K complex coordinately direct kinetochore
assembly in vertebrates. Mol Biol Cell. 19:587–594. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Hayward DG and Fry AM: Nek2 kinase in
chromosome instability and cancer. Cancer Lett. 237:155–166. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Nakano I, Masterman-Smith M, Saigusa K,
Paucar AA, Horvath S, Shoemaker L, Watanabe M, Negro A, Bajpai R,
Howes A, Lelievre V, Waschek JA, Lazareff JA, Freije WA, Liau LM,
Gilbertson RJ, Cloughesy TF, Geschwind DH, Nelson SF, Mischel PS,
Terskikh AV and Kornblum HI: Maternal embryonic leucine zipper
kinase is a key regulator of the proliferation of malignant brain
tumors, including brain tumor stem cells. J Neurosci Res. 86:48–60.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
42.
|
Jeong H, Ryu YJ, An J, Lee Y and Kim A:
Epithelial-mesenchymal transition in breast cancer correlates with
high histological grade and triple-negative phenotype.
Histopathology. 60:E87–E95. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
43.
|
Horvath S, Zhang B, Carlson M, Lu KV, Zhu
S, Felciano RM, Laurance MF, Zhao W, Qi S, Chen Z, Lee Y, Scheck
AC, Liau LM, Wu H, Geschwind DH, Febbo PG, Kornblum HI, Cloughesy
TF, Nelson SF and Mischel PS: Analysis of oncogenic signaling
networks in glioblastoma identifies ASPM as a molecular target.
Proc Natl Acad Sci USA. 103:17402–17407. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
44.
|
Moroi Y, Peebles C, Fritzler MJ,
Steigerwald J and Tan EM: Autoantibody to centromere (kinetochore)
in scleroderma sera. Proc Natl Acad Sci USA. 77:1627–1631. 1980.
View Article : Google Scholar : PubMed/NCBI
|
|
45.
|
Tomonaga T, Matsushita K, Yamaguchi S,
Oohashi T, Shimada H, Ochiai T, Yoda K and Nomura F: Overexpression
and mistargeting of centromere protein-A in human primary
colorectal cancer. Cancer Res. 63:3511–3516. 2003.PubMed/NCBI
|
|
46.
|
de la Guardia C, Casiano CA,
Trinidad-Pinedo J and Báez A: CENP-F gene amplification and
overexpression in head and neck squamous cell carcinomas. Head
Neck. 23:104–112. 2001.PubMed/NCBI
|
|
47.
|
Grützmann R, Pilarsky C, Ammerpohl O,
Lüttges J, Böhme A, Sipos B, Foerder M, Alldinger I, Jahnke B,
Schackert HK, Kalthoff H, Kremer B, Klöppel G and Saeger HD: Gene
expression profiling of microdissected pancreatic ductal carcinomas
using high-density DNA microarrays. Neoplasia. 6:611–622.
2004.PubMed/NCBI
|
|
48.
|
Bièche I, Vacher S, Lallemand F,
Tozlu-Kara S, Bennani H, Beuzelin M, Driouch K, Rouleau E,
Lerebours F, Ripoche H, Cizeron-Clairac G, Spyratos F and Lidereau
R: Expression analysis of mitotic spindle checkpoint genes in
breast carcinoma: role of NDC80/HEC1 in early breast
tumorigenicity, and a two-gene signature for aneuploidy. Mol
Cancer. 10:232011.PubMed/NCBI
|
|
49.
|
Hu Z, Huang G, Sadanandam A, Gu S, Lenburg
ME, Pai M, Bayani N, Blakely EA, Gray JW and Mao JH: The expression
level of HJURP has an independent prognostic impact and predicts
the sensitivity to radiotherapy in breast cancer. Breast Cancer
Res. 12:R182010. View Article : Google Scholar : PubMed/NCBI
|