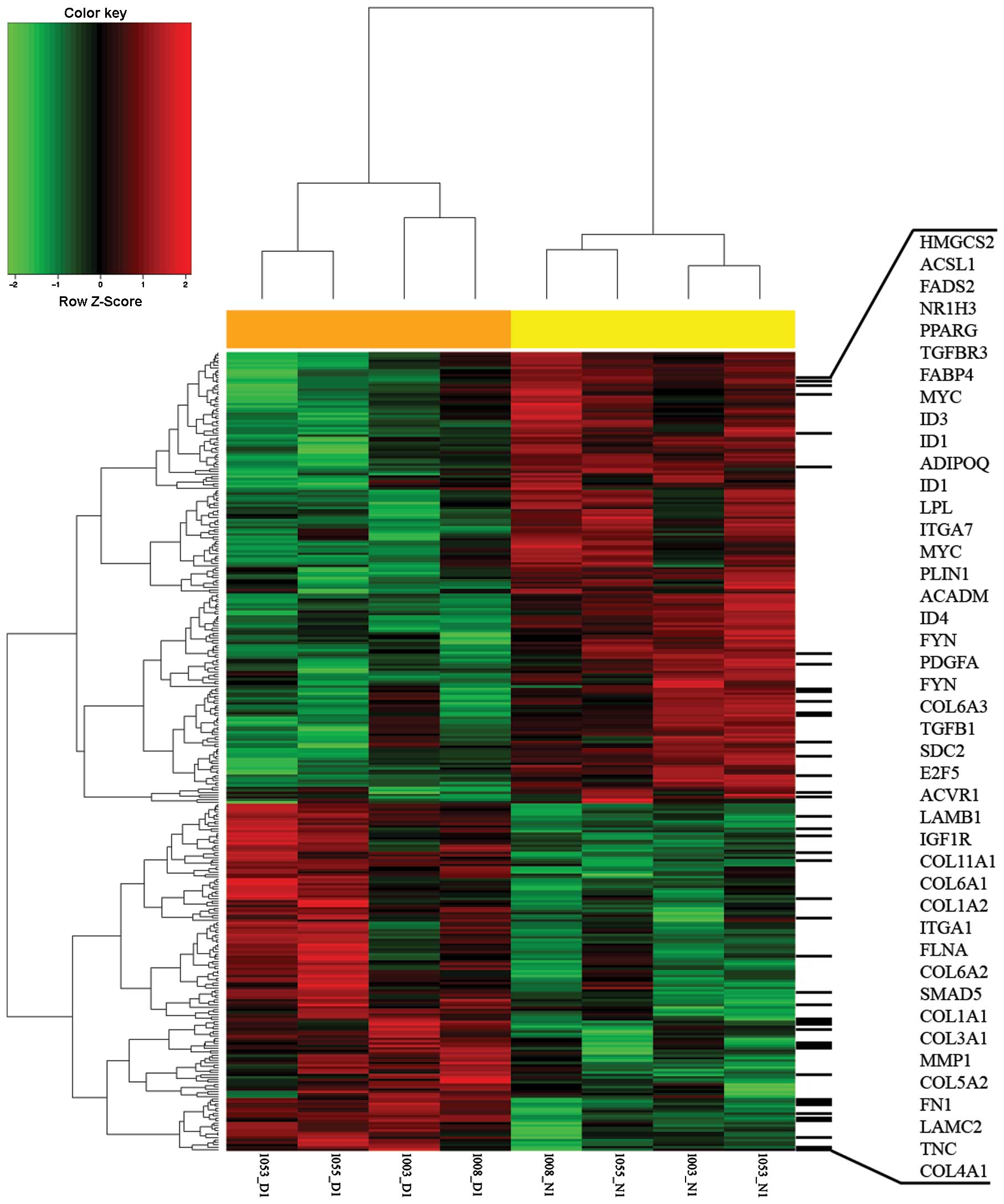
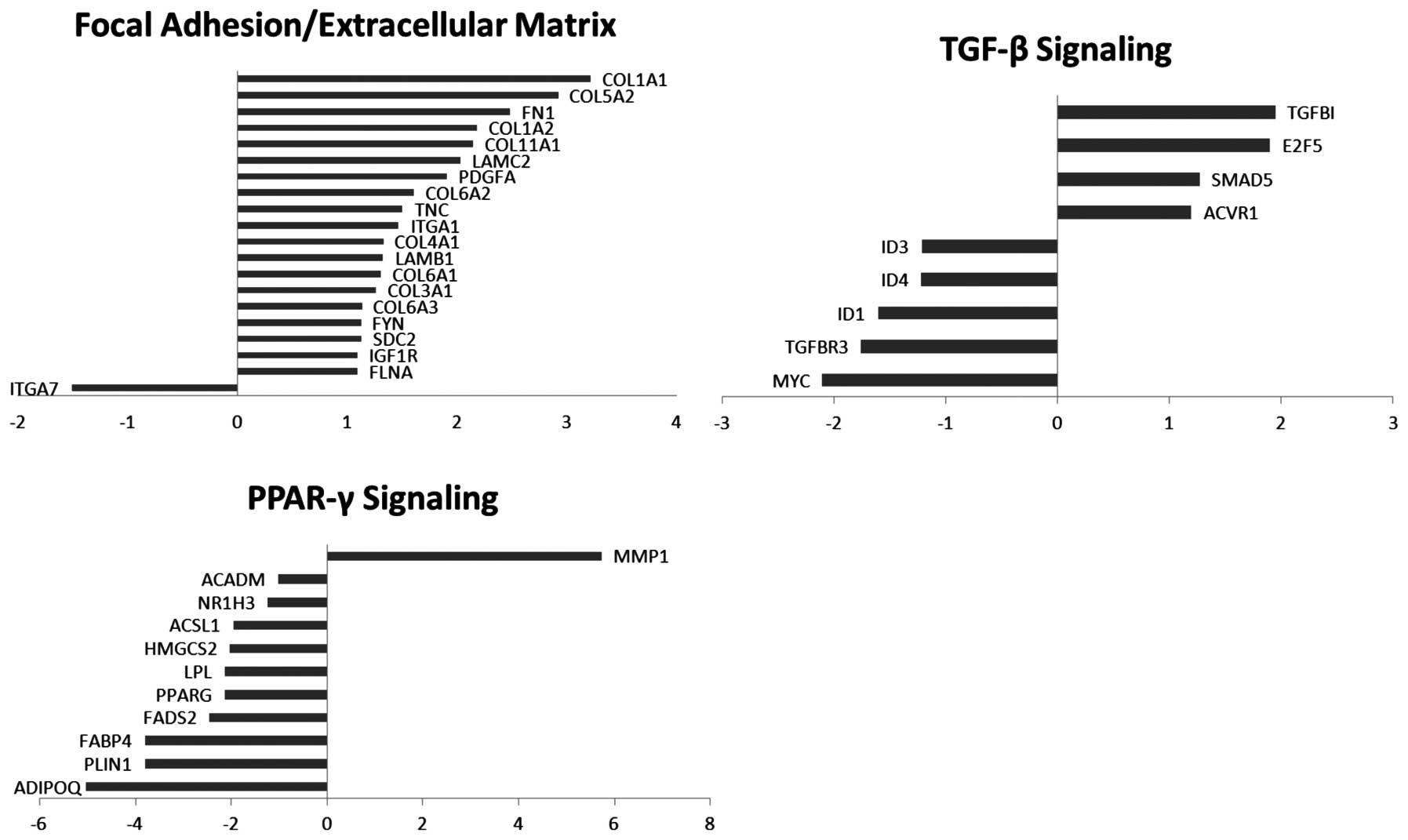
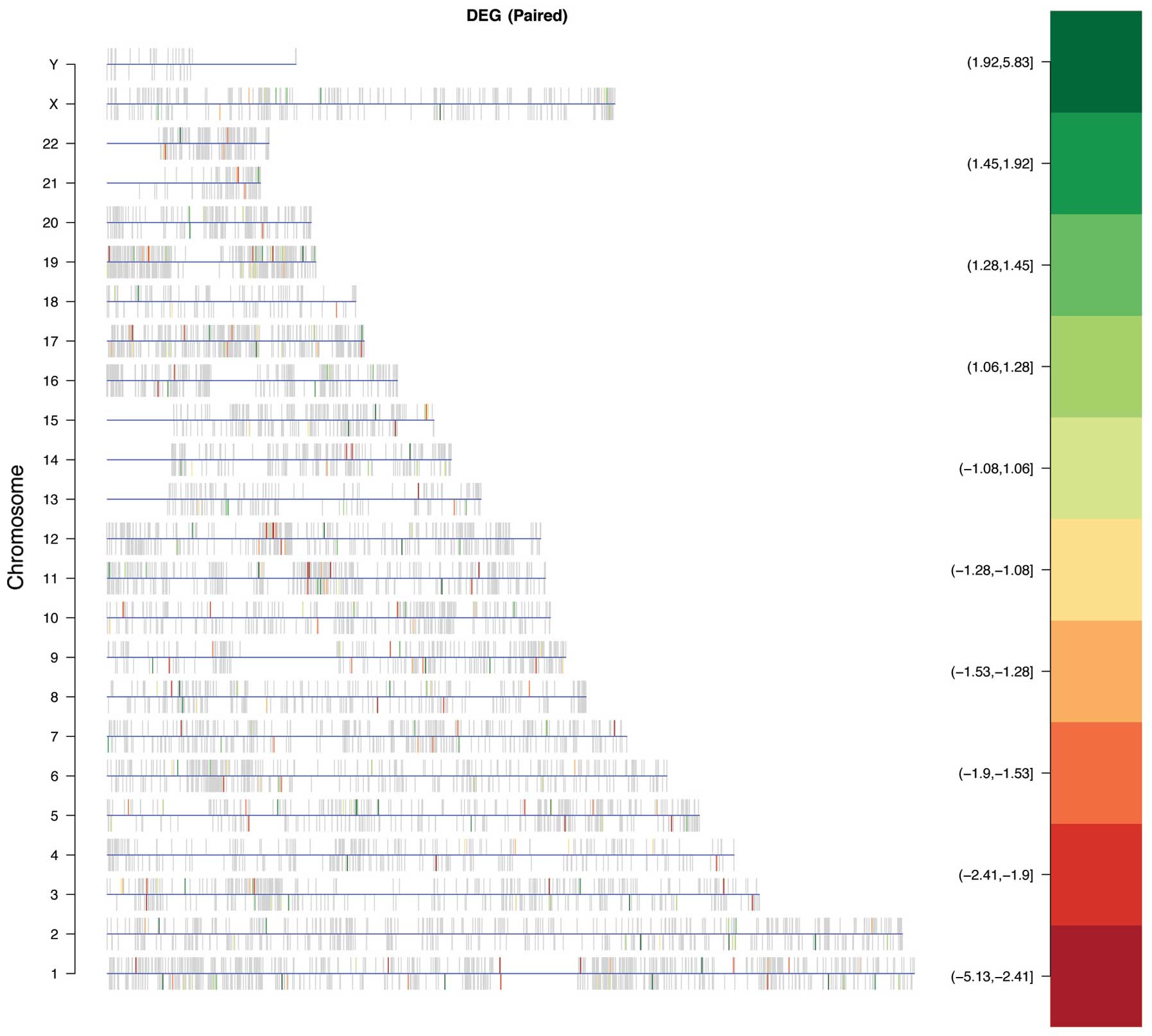
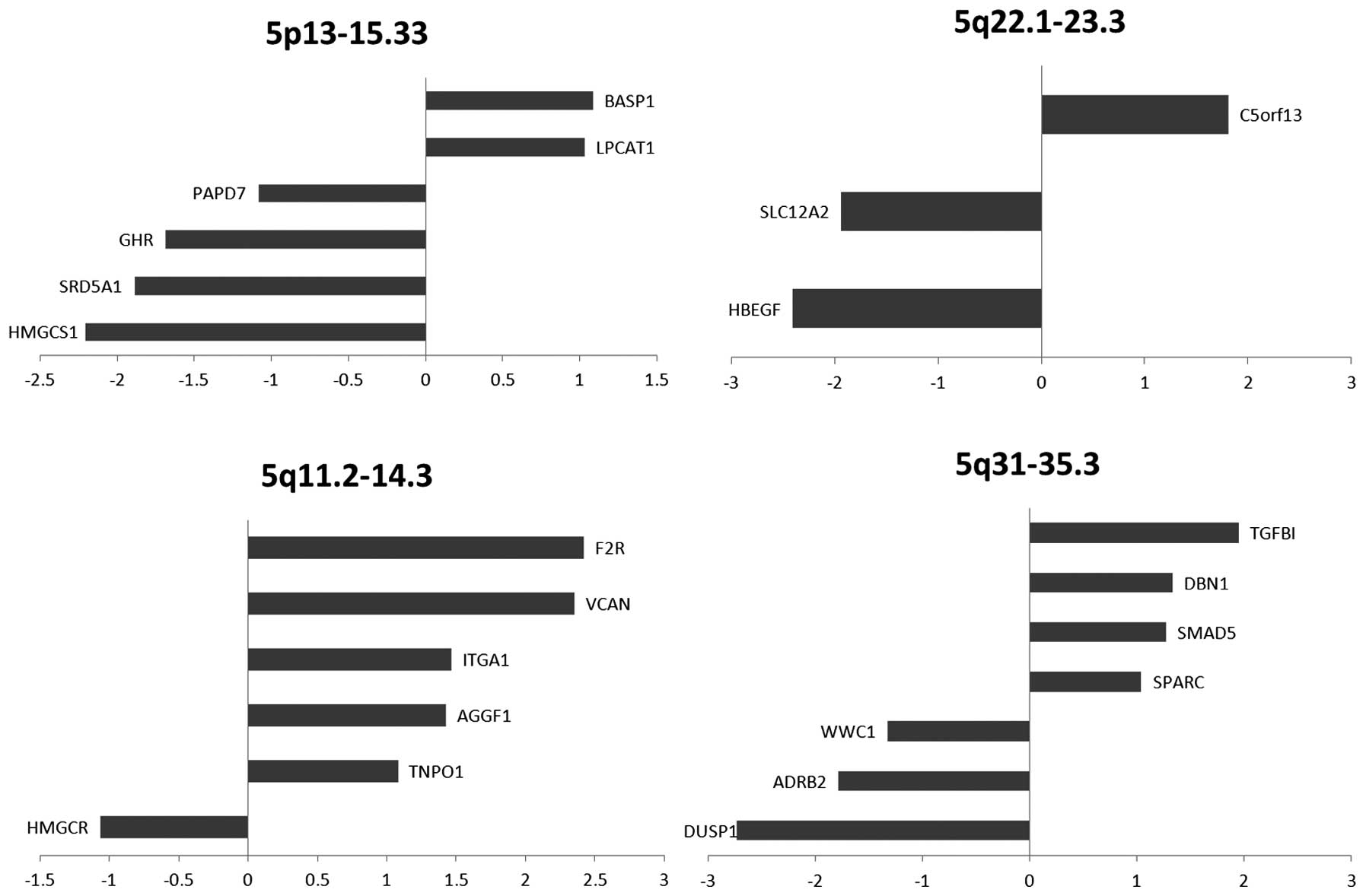
|
1.
|
Armstrong BK and Kricker A: Skin cancer.
Dermatol Clin. 13:583–594. 1995.
|
|
2.
|
Rubin AI, Chen EH and Ratner D: Basal-cell
carcinoma. N Engl J Med. 353:2262–2269. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Sexton M, Jones DB and Maloney ME:
Histologic pattern analysis of basal cell carcinoma. Study of a
series of 1039 consecutive neoplasms. J Am Acad Dermatol.
23:1118–1126. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Diepgen TL and Mahler V: The epidemiology
of skin cancer. Br J Dermatol. 146(Suppl. 61): S1–S6. 2002.
View Article : Google Scholar
|
|
5.
|
Reifenberger J, Wolter M, Knobbe CB, et
al: Somatic mutations in the PTCH, SMOH, SUFUH and TP53 genes in
sporadic basal cell carcinomas. Br J Dermatol. 152:43–51. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Benjamin CL, Melnikova VO and Ananthaswamy
HN: P53 protein and pathogenesis of melanoma and nonmelanoma skin
cancer. Adv Exp Med Biol. 624:265–282. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
De Zwaan SE and Haass NK: Genetics of
basal cell carcinoma. Australas J Dermatol. 51:81–93. 2010.
|
|
8.
|
Iwasaki JK, Srivastava D, Moy RL, Lin HJ
and Kouba DJ: The molecular genetics underlying basal cell
carcinoma pathogenesis and links to targeted therapeutics. J Am
Acad Dermatol. 66:E167–E168. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Lacour JP: Carcinogenesis of basal cell
carcinomas: genetics and molecular mechanisms. Br J Dermatol.
146(Suppl 61): S17–S19. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Muller PA, Vousden KH and Norman JC: p53
and its mutants in tumor cell migration and invasion. J Cell Biol.
192:209–218. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Gentleman RC, Carey VJ, Bates DM, et al:
Bioconductor: open software development for computational biology
and bioinformatics. Genome Biol. 5:R802004. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Wilson CL and Miller CJ: Simpleaffy: a
BioConductor package for Affymetrix Quality Control and data
analysis. Bioinformatics. 21:3683–3685. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Smyth GK: Linear models and empirical
bayes methods for assessing differential expression in microarray
experiments. Stat Appl Genet Mol Biol. 3:Article 3. 2004.PubMed/NCBI
|
|
14.
|
Benjamini Y: Controlling the false
discovery rate: a practical and powerful approach to multiple
testing. J R Stat Soc Series B. 57:289–300. 1995.
|
|
15.
|
Eisen MB, Spellman PT, Brown PO and
Botstein D: Cluster analysis and display of genome-wide expression
patterns. Proc Natl Acad Sci USA. 95:14863–14868. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Huang da W, Sherman BT, Tan Q, et al:
DAVID Bioinformatics Resources: expanded annotation database and
novel algorithms to better extract biology from large gene lists.
Nucleic Acids Res. 35:W169–W175. 2007.PubMed/NCBI
|
|
17.
|
Asplund A, Gry Bjorklund M, Sundquist C,
et al: Expression profiling of microdissected cell populations
selected from basal cells in normal epidermis and basal cell
carcinoma. Br J Dermatol. 158:527–538. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Howell BG, Solish N, Lu C, et al:
Microarray profiles of human basal cell carcinoma: insights into
tumor growth and behavior. J Dermatol Sci. 39:39–51. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
O’Driscoll L, McMorrow J, Doolan P, et al:
Investigation of the molecular profile of basal cell carcinoma
using whole genome microarrays. Mol Cancer. 5:742006.PubMed/NCBI
|
|
20.
|
Yu M, Zloty D, Cowan B, et al:
Superficial, nodular, and morpheiform basal-cell carcinomas exhibit
distinct gene expression profiles. J Invest Dermatol.
128:1797–1805. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Nijsten T, Geluyckens E, Colpaert C and
Lambert J: Peroxisome proliferator-activated receptors in squamous
cell carcinoma and its precursors. J Cutan Pathol. 32:340–347.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Tachibana K, Yamasaki D, Ishimoto K and
Doi T: The role of PPARs in cancer. PPAR Res. 2008:1027372008.
View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Reka AK, Goswami MT, Krishnapuram R,
Standiford TJ and Keshamouni VG: Molecular cross-regulation between
PPAR-gamma and other signaling pathways: implications for lung
cancer therapy. Lung Cancer. 72:154–159. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Sarraf P, Mueller E, Jones D, et al:
Differentiation and reversal of malignant changes in colon cancer
through PPARgamma. Nat Med. 4:1046–1052. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Guan YF, Zhang YH, Breyer RM, Davis L and
Breyer MD: Expression of peroxisome proliferator-activated receptor
gamma (PPARgamma) in human transitional bladder cancer and its role
in inducing cell death. Neoplasia. 1:330–339. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Mossner R, Schulz U, Kruger U, et al:
Agonists of peroxisome proliferator-activated receptor gamma
inhibit cell growth in malignant melanoma. J Invest Dermatol.
119:576–582. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Petta E, Sotiropoulou-Bonikou G, Kourelis
K, Melachrinou M and Bonikos DS: Differential expression and
cross-talk of peroxisome proliferator-activated receptor gamma and
retinoid-X receptor alpha in urothelial carcinomas of the bladder.
J BUON. 15:740–745. 2010.
|
|
28.
|
Pancione M, Forte N, Sabatino L, et al:
Reduced beta-catenin and peroxisome proliferator-activated
receptor-gamma expression levels are associated with colorectal
cancer metastatic progression: correlation with tumor-associated
macrophages, cyclooxygenase 2, and patient outcome. Hum Pathol.
40:714–725. 2009. View Article : Google Scholar
|
|
29.
|
Mueller E, Sarraf P, Tontonoz P, et al:
Terminal differentiation of human breast cancer through PPAR gamma.
Mol Cell. 1:465–470. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Yao L, Liu F, Sun L, et al: Upregulation
of PPARgamma in tissue with gastric carcinoma. Hybridoma (Larchmt).
29:341–343. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Wang W, Wang R, Zhang Z, Li D and Yut Y:
Enhanced PPAR-gamma expression may correlate with the development
of Barrett’s esophagus and esophageal adenocarcinoma. Oncol Res.
19:141–147. 2011.PubMed/NCBI
|
|
32.
|
Li M, Lee TW, Mok TS, Warner TD, Yim AP
and Chen GG: Activation of peroxisome proliferator-activated
receptor-gamma by troglitazone (TGZ) inhibits human lung cell
growth. J Cell Biochem. 96:760–774. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Boiteux G, Lascombe I, Roche E, et al:
A-FABP, a candidate progression marker of human transitional cell
carcinoma of the bladder, is differentially regulated by PPAR in
urothelial cancer cells. Int J Cancer. 124:1820–1828. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Storch J and Thumser AE: Tissue-specific
functions in the fatty acid-binding protein family. J Biol Chem.
285:32679–32683. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Wetmore C: Sonic hedgehog in normal and
neoplastic proliferation: insight gained from human tumors and
animal models. Curr Opin Genet Dev. 13:34–42. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Green DR and Kroemer G: Cytoplasmic
functions of the tumour suppressor p53. Nature. 458:1127–1130.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Sun Y, Zeng XR, Wenger L, Firestein GS and
Cheung HS: P53 down-regulates matrix metalloproteinase-1 by
targeting the communications between AP-1 and the basal
transcription complex. J Cell Biochem. 92:258–269. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Jung YS, Qian Y and Chen X: Examination of
the expanding pathways for the regulation of p21 expression and
activity. Cell Signal. 22:1003–1012. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Tanaka K, Iwamoto Y, Ito Y, et al: Cyclic
AMP-regulated synthesis of the tissue inhibitors of
metalloproteinases suppresses the invasive potential of the human
fibrosarcoma cell line HT1080. Cancer Res. 55:2927–2935.
1995.PubMed/NCBI
|
|
40.
|
Tartaglia M and Gelb BD: Disorders of
dysregulated signal traffic through the RAS-MAPK pathway:
phenotypic spectrum and molecular mechanisms. Ann N Y Acad Sci.
1214:99–121. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Dunn KL, Espino PS, Drobic B, He S and
Davie JR: The Ras-MAPK signal transduction pathway, cancer and
chromatin remodeling. Biochem Cell Biol. 83:1–14. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
42.
|
Yang D, Tao J, Li L, et al: RasGRP3, a Ras
activator, contributes to signaling and the tumorigenic phenotype
in human melanoma. Oncogene. 30:4590–600. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
43.
|
Mathiasen DP, Egebjerg C, Andersen SH, et
al: Identification of a c-Jun N-terminal kinase-2-dependent signal
amplification cascade that regulates c-Myc levels in ras
transformation. Oncogene. 31:390–401. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
44.
|
Shi XH, Liang ZY, Ren XY and Liu TH:
Combined silencing of K-ras and Akt2 oncogenes achieves synergistic
effects in inhibiting pancreatic cancer cell growth in vitro and in
vivo. Cancer Gene Ther. 16:227–236. 2009.PubMed/NCBI
|
|
45.
|
Rauch J, Moran-Jones K, Albrecht V, et al:
c-Myc regulates RNA splicing of the A-Raf kinase and its activation
of the ERK pathway. Cancer Res. 71:4664–4674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46.
|
Su E, Han X and Jiang G: The transforming
growth factor beta 1/SMAD signaling pathway involved in human
chronic myeloid leukemia. Tumori. 96:659–666. 2010.PubMed/NCBI
|
|
47.
|
Chen CR, Kang Y, Siegel PM and Massague J:
E2F4/5 and p107 as Smad cofactors linking the TGFbeta receptor to
c-myc repression. Cell. 110:19–32. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
48.
|
Jiang Y, Yim SH, Xu HD, et al: A potential
oncogenic role of the commonly observed E2F5 overexpression in
hepatocellular carcinoma. World J Gastroenterol. 17:470–477. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
49.
|
Polanowska J, Le Cam L, Orsetti B, et al:
Human E2F5 gene is oncogenic in primary rodent cells and is
amplified in human breast tumors. Genes Chromosomes Cancer.
28:126–130. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
50.
|
Van Haren R, Feldman D and Sinha AA:
Systematic comparison of nonmelanoma skin cancer microarray
datasets reveals lack of consensus genes. Br J Dermatol.
161:1278–1287. 2009.PubMed/NCBI
|
|
51.
|
Tanaka K, Iyama K, Kitaoka M, et al:
Differential expression of alpha 1(IV), alpha 2(IV), alpha 5(IV)
and alpha 6(IV) collagen chains in the basement membrane of basal
cell carcinoma. Histochem J. 29:563–570. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
52.
|
Bishop DT, Demenais F, Iles MM, et al:
Genome-wide association study identifies three loci associated with
melanoma risk. Nat Genet. 41:920–925. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
53.
|
Gerstenblith MR, Shi J and Landi MT:
Genome-wide association studies of pigmentation and skin cancer: a
review and meta-analysis. Pigment Cell Melanoma Res. 23:587–606.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
54.
|
Gudbjartsson DF, Sulem P, Stacey SN, et
al: ASIP and TYR pigmentation variants associate with cutaneous
melanoma and basal cell carcinoma. Nat Genet. 40:886–891. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
55.
|
Han J, Kraft P, Nan H, et al: A
genome-wide association study identifies novel alleles associated
with hair color and skin pigmentation. PLoS Genet. 4:e10000742008.
View Article : Google Scholar : PubMed/NCBI
|
|
56.
|
Han J, Qureshi AA, Prescott J, et al: A
prospective study of telomere length and the risk of skin cancer. J
Invest Dermatol. 129:415–421. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
57.
|
Liboutet M, Portela M, Delestaing G, et
al: MC1R and PTCH gene polymorphism in French patients with basal
cell carcinomas. J Invest Dermatol. 126:1510–1517. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
58.
|
Nan H, Kraft P, Hunter DJ and Han J:
Genetic variants in pigmentation genes, pigmentary phenotypes, and
risk of skin cancer in Caucasians. Int J Cancer. 125:909–917. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
59.
|
Nan H, Xu M, Kraft P, et al: Genome-wide
association study identifies novel alleles associated with risk of
cutaneous basal cell carcinoma and squamous cell carcinoma. Hum Mol
Genet. 20:3718–3724. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
60.
|
Rafnar T, Sulem P, Stacey SN, et al:
Sequence variants at the TERT-CLPTM1L locus associate with many
cancer types. Nat Genet. 41:221–227. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
61.
|
Stacey SN, Gudbjartsson DF, Sulem P, et
al: Common variants on 1p36 and 1q42 are associated with cutaneous
basal cell carcinoma but not with melanoma or pigmentation traits.
Nat Genet. 40:1313–1318. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
62.
|
Stacey SN, Sulem P, Masson G, et al: New
common variants affecting susceptibility to basal cell carcinoma.
Nat Genet. 41:909–914. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
63.
|
Wu C, Hu Z, He Z, et al: Genome-wide
association study identifies three new susceptibility loci for
esophageal squamous-cell carcinoma in Chinese populations. Nat
Genet. 43:679–684. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
64.
|
Christensen GB, Baffoe-Bonnie AB, George
A, et al: Genome-wide linkage analysis of 1,233 prostate cancer
pedigrees from the International Consortium for Prostate Cancer
Genetics using novel sumLINK and sumLOD analyses. Prostate.
70:735–744. 2010.
|
|
65.
|
Xu J, Dimitrov L, Chang BL, et al: A
combined genomewide linkage scan of 1,233 families for prostate
cancer-susceptibility genes conducted by the international
consortium for prostate cancer genetics. Am J Hum Genet.
77:219–229. 2005. View
Article : Google Scholar
|
|
66.
|
Sun LD, Xiao FL, Li Y, et al: Genome-wide
association study identifies two new susceptibility loci for atopic
dermatitis in the Chinese Han population. Nat Genet. 43:690–694.
2011. View
Article : Google Scholar : PubMed/NCBI
|
|
67.
|
Beyer K, Nickel R, Freidhoff L, et al:
Association and linkage of atopic dermatitis with chromosome
13q12–14 and 5q31–33 markers. J Invest Dermatol. 115:906–908.
2000.
|
|
68.
|
Duffy DL, Zhao ZZ, Sturm RA, Hayward NK,
Martin NG and Montgomery GW: Multiple pigmentation gene
polymorphisms account for a substantial proportion of risk of
cutaneous malignant melanoma. J Invest Dermatol. 130:520–528. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
69.
|
Han J, Colditz GA, Samson LD and Hunter
DJ: Polymorphisms in DNA double-strand break repair genes and skin
cancer risk. Cancer Res. 64:3009–3013. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
70.
|
Sulem P, Gudbjartsson DF, Stacey SN, et
al: Two newly identified genetic determinants of pigmentation in
Europeans. Nat Genet. 40:835–837. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
71.
|
Sulem P, Gudbjartsson DF, Stacey SN, et
al: Genetic determinants of hair, eye and skin pigmentation in
Europeans. Nat Genet. 39:1443–1452. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
72.
|
Han J, Hankinson SE, Colditz GA and Hunter
DJ: Genetic variation in XRCC1, sun exposure, and risk of skin
cancer. Br J Cancer. 91:1604–1609. 2004.PubMed/NCBI
|
|
73.
|
Kang SY, Lee KG, Lee W, et al:
Polymorphisms in the DNA repair gene XRCC1 associated with basal
cell carcinoma and squamous cell carcinoma of the skin in a Korean
population. Cancer Sci. 98:716–720. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
74.
|
Brown KM, Macgregor S, Montgomery GW, et
al: Common sequence variants on 20q11.22 confer melanoma
susceptibility. Nat Genet. 40:838–840. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
75.
|
Smyth I, Narang MA, Evans T, et al:
Isolation and characterization of human patched 2 (PTCH2), a
putative tumour suppressor gene inbasal cell carcinoma and
medulloblastoma on chromosome 1p32. Hum Mol Genet. 8:291–297. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
76.
|
Fan Z, Li J, Du J, et al: A missense
mutation in PTCH2 underlies dominantly inherited NBCCS in a Chinese
family. J Med Genet. 45:303–308. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
77.
|
Grachtchouk M, Mo R, Yu S, et al: Basal
cell carcinomas in mice overexpressing Gli2 in skin. Nat Genet.
24:216–217. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
78.
|
Friedman E, Gejman PV, Martin GA and
McCormick F: Non-sense mutations in the C-terminal SH2 region of
the GTPase activating protein (GAP) gene in human tumours. Nat
Genet. 5:242–247. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
79.
|
Hahn H, Wicking C, Zaphiropoulous PG, et
al: Mutations of the human homolog of Drosophila patched in the
nevoid basal cell carcinoma syndrome. Cell. 85:841–851. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
80.
|
Aszterbaum M, Epstein J, Oro A, et al:
Ultraviolet and ionizing radiation enhance the growth of BCCs and
trichoblastomas in patched heterozygous knockout mice. Nat Med.
5:1285–1291. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
81.
|
Svard J, Heby-Henricson K, Persson-Lek M,
et al: Genetic elimination of Suppressor of fused reveals an
essential repressor function in the mammalian Hedgehog signaling
pathway. Dev Cell. 10:187–197. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
82.
|
Ling G, Ahmadian A, Persson A, et al:
PATCHED and p53 gene alterations in sporadic and hereditary basal
cell cancer. Oncogene. 20:7770–7778. 2001. View Article : Google Scholar : PubMed/NCBI
|