Introduction
MicroRNAs (miRNAs) are small non-coding RNAs of
20–22 nucleotides, and function to suppress the expression of
target mRNAs by translation blockade and/or mRNA degradation
(1,2). They are involved in many biological
processes including cell proliferation, differentiation and
apoptosis, and their dysregulation can contribute to the
pathological state including cancer (1,3,4).
Several groups have documented miRNA expression profiling in
ovarian cancers using miRNA microarray and massive parallel
sequencing technology (5–10). miR-93, miR-141, miR-200 and miR-214
are frequently upregulated whereas miR-100, miR-143, miR-145 and
let-7 are downregulated in ovarian carcinomas compared with normal
counterparts (5–9). Abnormal miRNA expression is due to
DNA copy number amplification and deletion, epigenetic modification
and/or the dysregualtion of miRNA processing in cancer state
(7,11,12).
miR-214 upregulated in ovarian cancer can target PTEN tumor
suppressor gene whereas down-regulated let-7 can target the RAS
oncogene (8,13), suggesting that miRNAs may have a
role as novel class of oncogenes or tumor suppressor genes in
ovarian cancer (14).
Based on these findings, the clinical potential of
miRNAs as cancer biomarkers and/or therapeutic agents is widely
recognized and accepted (15). A
single miRNA can regulate multiple mRNA transcripts that
cooperatively work in cellular differentiation and function
(16–19). The use of miRNA mimics or
anti-miRNAs may represent powerful therapeutic tools to accomplish
regression and/or re-differentiation of cancer by effectively
targeting tumor suppressive or oncogenic genes with less toxicity
(15,20). Indeed, a number of pre-clinical
trials of miRNAs are currently in progress (21). In this study, we performed a gain
of function screen using miRNA mimics library containing 319 miRNAs
to identify miRNAs that can affect cell proliferation in A2780
ovary cancer cells. We found several anti-proliferative miRNAs
including miR-124, miR-192 and miR-193 in A2780, suggesting that
the potential of miRNA screens for discovering miRNAs as
therapeutic tools to treat ovarian cancer.
Materials and methods
Cell culture
Human ovarian cancer cell line A2780 was obtained
from Dr T. Tsuruo (22), and human
colorectal cancer cell line DLD-1 was obtained from the American
Type Culture Collection (ATCC, Manassas, VA, USA). A2780 and DLD-1
were cultured in RPMI-1640 (Gibco, Life Technologies, Carlsbad, CA,
USA) containing 50 IU/ml penicillin and 50 μg/ml
streptomycin (Gibco, Life Technologies), supplemented with 5%
(A2780) or 10% (DLD-1) fetal bovine serum (FBS, JRH Biosciences,
Lenexa, KS, USA) at 37°C in an atmosphere of 5% CO2.
miRNA library screening
A gain of function miRNA screen on cell viability
was performed using A2780 as previously described (23). A2780 was seeded at 2,500 cells per
well in 96-well plates the day before transfection. Synthetic miRNA
mimic library (human Pre-miR™ miRNA precursor library-ver.1,
Ambion, Applied Biosystems, Foster City, CA, USA) was screened
using 50 nM in a duplicate. The library contained 319 miRNAs
registered in miRBase ver. 7.1 (http://www.mirbase.org/). miRNA mimics were
transfected using Lipofectamine 2000 (Life Technologies) according
to the manufacturer’s protocols. Pre-miR miRNA precursor
molecule-negative control (13)
(Ambion, Applied Biosystems) was used as a negative control for
miRNA mimics. We confirmed transfection efficiency (>90%) using
siControl TOX transfection control (50 nM, Dharmacon, Lafayette,
CO, USA). After 3 days of transfection, the cell viability was
measured using the Cell Titer-Glo Luminescent Cell Viability Assay
(Promega, Madison, WI, USA) according to the manufacturer’s
instructions. Data were expressed as percentage of the negative
control. Several miRNA hits were selected to assess reproducibility
and dose-dependency (5, 25 and 50 nM).
BrdU incorporation assay
miRNA (25 nM)-transfected cells in 96-well format
were harvested for one day, and then were incubated with 10
μM of 5′-bromo-2-deoxy-uridine (BrdU) for 2–4 h. The cells
were fixed with cold ethanol/HCl, and the incorporated BrdU was
detected using BrdU labeling and detection kit III (Roche
Diagnostics GmbH, Mannheim, Germany) according to the
manufacturer’s instruction.
Caspase 3/7 activation assay
miRNA (25 nM)-transfected cells in 96-well format
were harvested for 2 days, and then used to measure caspase 3/7
activity using Caspase-Glo 3/7 assay (Promega) according to the
manufacturer’s instruction.
RNA isolation and whole genome
microarray
A2780 cells were transfected with miR-193a (Pre-miR
miRNA precursor molecules, hsa-miR-193a-3p, Ambion, Applied
Biosystems) or negative control miRNA (25 nM), and allowed to grow
in the medium (RPMI-1640) for 10 h before RNA isolation. Total RNA
was isolated using the RNeasy mini RNA isolation kit (Qiagen). The
integrity of the RNA was verified using an Agilent 2100 Bioanalyzer
(1.8–2.0: Agilent Technologies, Palo Alto, CA, USA). Transcriptome
microarray analysis was carried out using the 44K Whole Human
Genome Microarray chip (Agilent Technologies) according to the
manufacturer’s instructions. Scanning microarray chips and
processing data were done by Pharmafrontier Co., Ltd, Kyoto, Japan.
Differentially expressed probe sets were identified with a fold
change >1.5. Gene ontology (GO) pathway enrichment analysis was
performed among genes differentially expressed after miR-193a
transfection by SigTerm software (24). The downregulated genes with
miR-193a transfection were compared with predicted miR-193a target
genes searched by TargetScan (http://www.targetscan.org/). Over-representation of
predicted miR-193a target genes within downregulated gene sets was
assessed by SigTerm software.
Western blot analysis
miRNA or siRNA (25 nM)-transfected A2780 cells were
lysed in radio immunoprecipitation assay (RIPA) buffer [50 mM
Tris-HCl (pH 8.0), 150 mM sodium chloride, 1% NP-40, 0.5% sodium
deoxycholate, 0.1% sodium dodecyl sulfate] supplemented with 1% of
a protease inhibitor cocktail stock solution (set III, Roche
Diagnostics GmbH) after 1 or 2 days transfection. The following
pre-designed siRNA was used as a positive control: MCL1 siRNA
(Hs_MCL1_6 HP validated siRNA, SI02781205, Qiagen GmbH, Hilden,
Germany). Proteins (10 or 20 μg) were separated by SDS-PAGE.
Upon electroblotting to polyvinylidene fluoride (PVDF) membrane
(Immobilon-P, Millipore, Billerica, MA, USA), non-specific binding
sites were blocked by incubation in TBST (Tris-buffered
saline/0.05% Tween-20) containing 1% skim milk, and then incubated
with rabbit polyclonal anti-MCL1 (1:200, S-19, Santa Cruz
Biotechnology Inc., Santa Cruz, CA, USA), or mouse monoclonal
anti-α-tubulin (1:2,500, clone DM1A, Sigma, St. Louis, MO, USA) in
blocking solution. After washing with TBST, the membrane was
incubated with HRP-conjugated rabbit anti-mouse IgG secondary
antibody (P0161, Dako, Glostrup, Denmark) or HRP-conjugated swine
anti-rabbit IgG secondary antibody (P0217, Dako). Signals were
detected using enhanced chemiluminescence (ECL) or ECL-plus reagent
(Amersham™ GE Healthcare UK Ltd., Buckinghamshire, UK).
qRT-PCR
Total RNA was prepared from miRNA or siRNA (25
nM)-transfected cells 2 days after transfection using RNeasy mini
kit (Qiagen), and then first strand cDNA was synthesized using
SuperScript III (Life Technologies) according to the manufacturer’s
instruction. Real-time RT-PCR was performed using 7900 HT fast
real-time PCR system (Applied Biosystems Inc., Foster City, CA,
USA) with SYBR-Green as a reporter. The following primers were used
for detection: MCL1 (222 bp) forward: TCTAAGTGCTGACTGGCTACG,
reverse: CCTGGCACAGCTATCAAAAG; GAPDH (137 bp) forward:
ACTTTGTCAAGCTCATTTCCTG, reverse: CTCTCTTCCTCTTGGCTCTTG.
Luciferase miRNA target reporter
assay
3′-untranslated regions (UTRs) of MCL1 gene (1546
bp), containing predicted binding sites of miR-193a, were amplified
by PCR from A2780 cDNA, and inserted into the pGL3 control vector
(Promega) by using Xba-I site immediately downstream from the stop
codon of Firefly luciferase. The following primers were
used: MCL1 3′-UTR forward: CGGCTAGCGAAAAGCAAGTGGCAAGAGG, reverse:
CGGCTAGCAGGGAGGGTCACTCAGGTTT.
Deletion of the first 3 nucleotides corresponding
miR-193a seed-region complementary site was inserted in mutant
constructs using KOD-plus-Mutagenesis kit (Toyobo, Osaka, Japan),
according to the manufacturer’s protocol. The following primers
were used for generation of mutant constructs: MCL1-mutant-Primer
1: AGCCAGGCAAGTCATAGAATTGATT, MCL1-mutant-Primer 2:
GGCCACTTTCCTGTTCTCAACAAGG.
DLD-1 cells were cultured in 96-well formats and
co-transfected with 100 ng of pGL3 Firefly luciferase
reporter vector, 20 ng of pRL-TK Renilla luciferase control
vector (Promega) and 25 nM miRNA or negative control miRNA using
Lipofectamine 2000. Firefly and Renilla luciferase
activities were measured consecutively using the Dual-Luciferase
Reporter Assay System (Promega) 24 h after transfection. All the
experiments were done in triplicate and repeated at least twice on
different days.
Results
Effects of miRNA mimic library
transfection on cell proliferation of A2780 cell line
To identify miRNAs that affect cell proliferation of
ovarian cancer cells, we performed a gain of function screen using
synthetic miRNA mimic library (319 miRNAs) for human epithelial
ovary cancer cells (A2780). The library consists of miRNAs
registered in early version of miRBase (ver. 7.1 in October, 2005,
http://www.mirbase.org/), and many of them were
expressed in ovarian normal and cancer tissues and cell lines
(5). We detected cellular ATP to
assess cell viability in miRNA (50 nM)-transfected cells 3 days
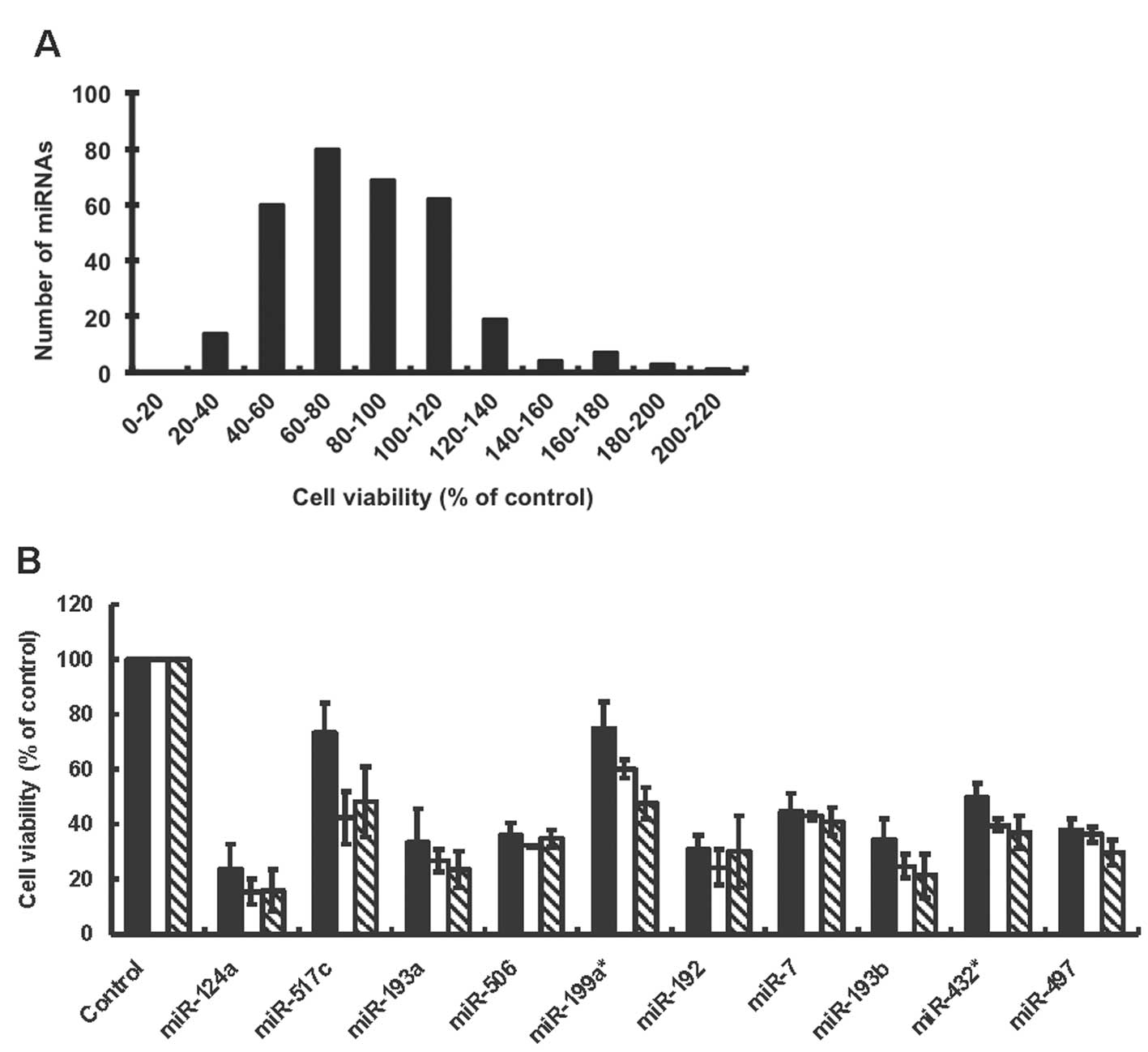
after transfection. Frequency distribution indicated that broad
ranges of miRNA mimic transfections affected the cell viability of
A2780 (Fig. 1A). A total of 46 out
of 319 miRNAs induced more than 50% changes in the cell viability
of A2780 after 3 days transfection. Table I shows top 10 miRNAs that increased
or decreased the cell viability of A2780. They included known
oncogenic miRNAs such as miR-372 (cell viability, 187%) and miR-373
(165%), and tumor suppressive miRNAs such as miR-124a (28.3%),
miR-7 (37.1%), miR-192 (36.6%) and miR-193a (29.7%) in several
different cancer types (18,25–27).
The seed family miRNAs that have the same sequences in seed region
(2nd to 8th nucleotide) of miRNAs showed similar effects on cell
viability in A2780 cells. For example,
miR-93/miR-302/miR-372/mir-373 seed family miRNAs (miR-93,
miR-302b, miR-302d, miR-372, miR-373) were pro-proliferative, while
miR-193 seed family miRNAs (miR-193a, miR-193b) were
anti-proliferative (Table I).
miR-200/miR-141 seed family miRNAs that are upregulated in ovarian
cancer (5,6,10)
had a little effect on the cell viability in A2780 cells (the cell
viability; 97.9, 113, 92.0 and 101% with miR-200a, miR-200b,
miR-200c and miR-141 transfection, respectively). miR-100, miR-143
and miR-145 that are down-regulated miRNAs in ovarian cancer
(5,6,8)
induced a 15–30% decrease in the cell viability of A2780 (the cell
viability; 84.1, 81.8 and 73.1 with miR-100, miR-143 and miR-145
transfection, respectively). We are interested in miRNA mimics that
decreased the cell viability of A2780 since these miRNA mimics
themselves could have therapeutic potential to treat ovarian
cancer. To further evaluate miRNA mimics on the inhibition of cell
proliferation in A2780, we selected top 10 anti-proliferative
miRNAs (miR-7, miR-124a, miR-192, miR-193a, miR-193b,
miR-199a*, miR-432*, miR-497, miR-506 and
miR-517c) from the first screen, and examined the cell viability in
A2780 cells transfected with different concentrations of miRNAs (5,
25, 50 nM). We confirmed results of our first screening at 50 nM,
and found that the transfection of miR-124a, miR-192, miR-193a and
miR-193b induced a large decrease in the cell viability of A2780
even at 5 nM (Fig. 1B), indicating
that these miRNAs had a profound anti-proliferative effect in A2780
cells. We examined whether miR-124a, miR-192, miR-193a and miR-193b
affected DNA synthesis to inhibit cell proliferation in A2780
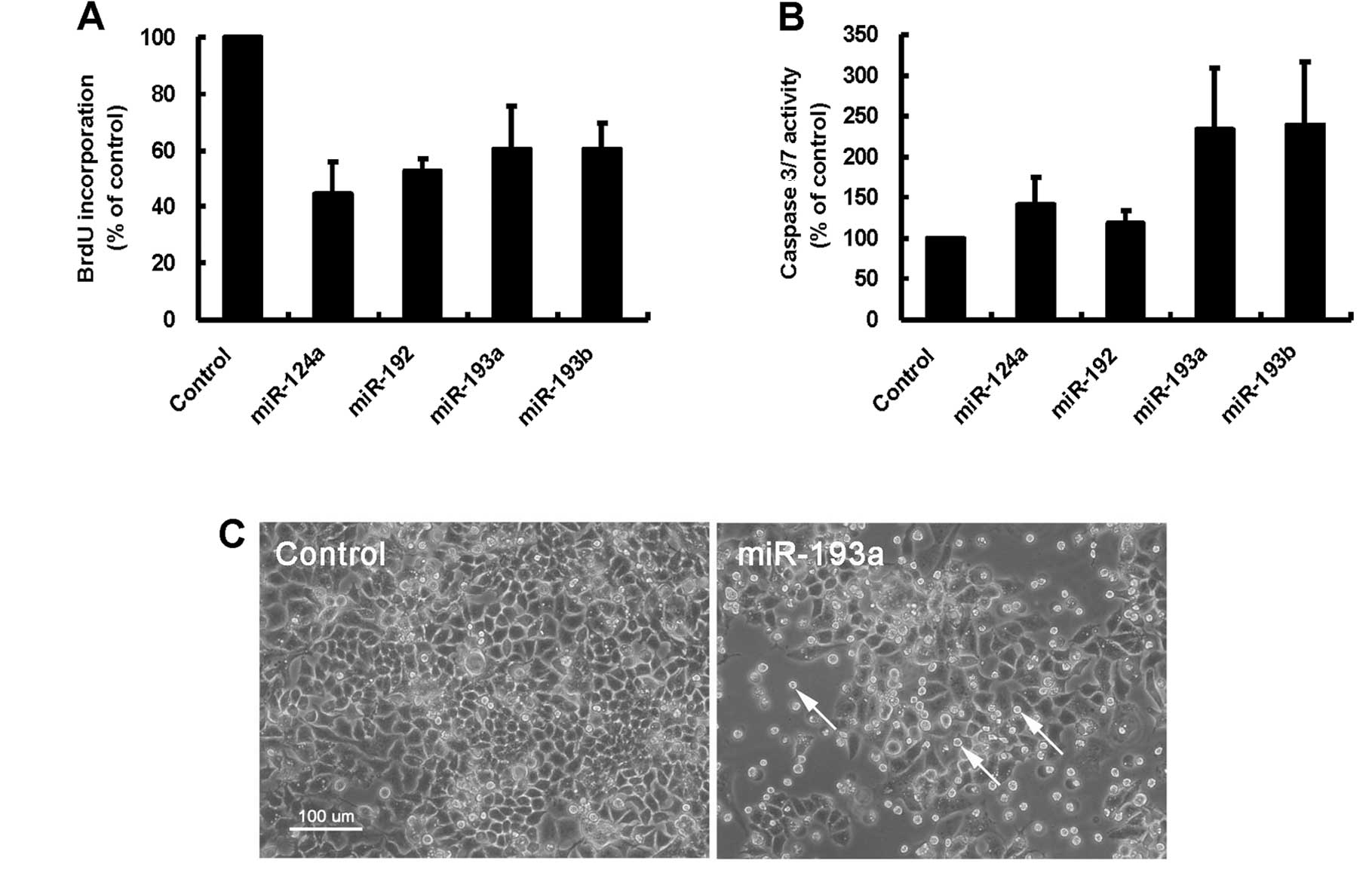
cells. One day after miRNA transfection, BrdU incorporation was
examined to evaluate DNA synthesis in transfected cells. As shown
in Fig. 2A, miR-124a, miR-192,
miR-193a and miR-193b decreased an incorporation of BrdU compared
with the negative control, indicating that these miRNAs induced the
inhibition of DNA synthesis in A2780 cells. We next examined
whether these miRNAs affected apoptotic pathway to inhibit cell
proliferation in A2780 cells. We found that miR-193a and miR-193b
but not miR-124a and miR-192 induced more than twofold increase in
an activity of caspase 3/7, the effector of apoptotic pathway, in
A2780 cells (Fig. 2B). The result
indicated that miR-193a and miR-193b could induce the apoptotic
cell death in A2780 cells. Actually, apoptotic cell debris was
frequently observed in miR-193a-transfected A2780 cells (Fig. 2C, arrows).
 | Table IResults of miRNA library
screening. |
Table I
Results of miRNA library
screening.
| miRNAs that
increased cell viability | miRNAs that
decreased cell viability |
|---|
|
|
|---|
| miRNA | Cell viability
(%) | miRNA | Cell viability
(%) |
|---|
| miR-301 | 218 | miR-124a | 28.3 |
| miR-372 | 187 | miR-517c | 29.4 |
| miR-93 | 185 | miR-193a | 29.7 |
| miR-302b | 181 | miR-506 | 31.9 |
| miR-130a | 173 |
miR-199a* | 34.5 |
| miR-302d | 172 | miR-192 | 36.6 |
| miR-363 | 166 | miR-7 | 37.1 |
| miR-373 | 165 | miR-193b | 37.7 |
|
miR-9* | 162 |
miR-432* | 37.8 |
| miR-130b | 162 | miR-497 | 38.3 |
Transcriptome analysis to assess target
genes regulated by miR-193a
We further characterized the anti-proliferative
effect of miR-193a in A2780 cells. To examine target genes
regulated by miR-193a, we performed genome wide gene expression
analysis using miR-193a (25 nM)-transfected cells compared with the
negative control miRNA-transfected ones. We identified 518 genes
that were downregulated more than 1.5-fold by miR-193a transfection
after 10 h. To evaluate the potential functional significance of
the genes downregulated after miR-193a transfection, we subjected
the gene expression data to gene ontology (GO) pathway enrichment
analysis. The 20 most significantly over-represented pathways
listed in Table II include small
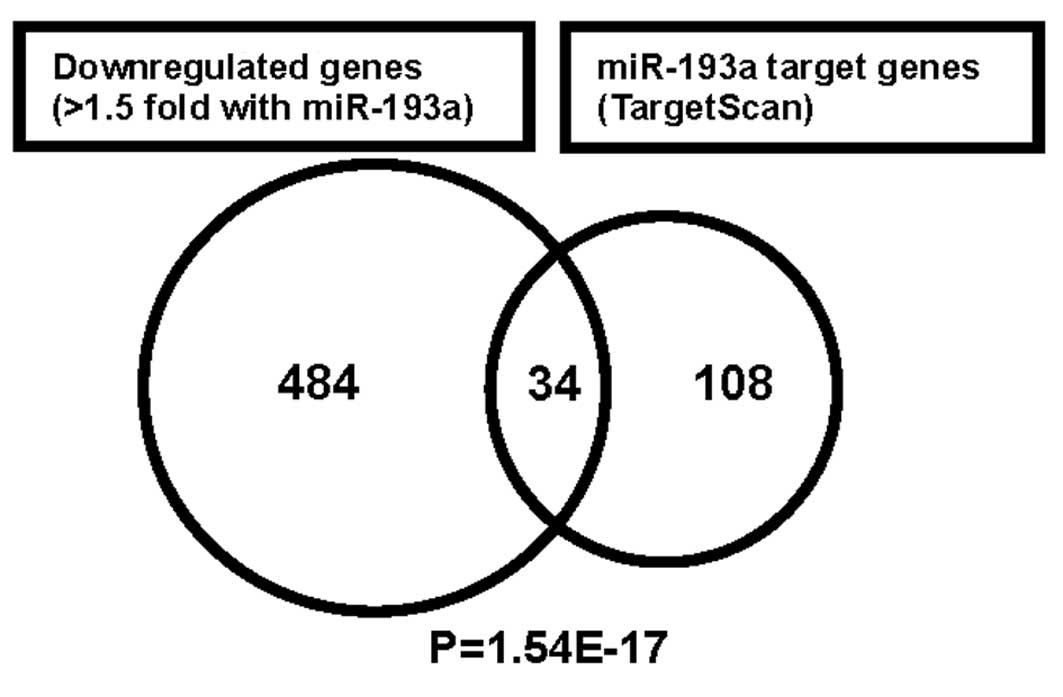
GTPase signaling and vesicular transport. We compared these
downregulated 518 genes with predicted miR-193a target genes (142
genes) obtained by TargetScan (Fig.
3). This resulted in the match of 34 candidate miR-193a target
genes, and they were significantly over-represented in the
downregulated gene sets by using the SigTerm software (24). Table
III showed 34 candidate miR-193a target genes obtained by our
transcriptome analysis. The candidate genes include ARHGAP19
(RhoGAP19), CCND1 (cyclin D1), ERBB4, KRAS and MCL1 that function
in cell signaling, cell cycle and apoptotic pathway.
 | Table IITwenty most significantly enriched
(P<0.05) gene ontology (GO) pathways among downregulated genes
after miR-193a transfection into A2780 cells. |
Table II
Twenty most significantly enriched
(P<0.05) gene ontology (GO) pathways among downregulated genes
after miR-193a transfection into A2780 cells.
| Term | P-value |
|---|
| Small GTPase
regulator activity | 0.0022 |
| Ras
guanyl-nucleotide exchange factor activity | 0.0030 |
| Rho
guanyl-nucleotide exchange factor activity | 0.0052 |
| Regulation of Rho
protein siganal transduction | 0.0060 |
| Blood vessel
development | 0.0069 |
| Cytoplasmic vesicle
part | 0.0072 |
| Vasculature
development | 0.0080 |
| Post-Golgi
vesicle-mediated transport | 0.0080 |
| Protein
localization | 0.0149 |
| Phospholipid
transporter activity | 0.0155 |
| Regulation of small
GTPase mediated signal transduction | 0.0156 |
| Guanyl-nucleotide
exchange factor activity | 0.0168 |
| GTPase regulator
activity | 0.0181 |
| Macromolecule
localization | 0.0187 |
| Insulin receptor
signaling pathway | 0.0193 |
| Guanylate kinase
activity | 0.0196 |
| Early endosome | 0.0213 |
| Neuron
projection | 0.0217 |
| Intracellular
signaling cascade | 0.0225 |
| One-carbon compound
metabolic process | 0.0229 |
 | Table IIICandidate miR-193a target genes
downregulated in miR-193a-transfectants. |
Table III
Candidate miR-193a target genes
downregulated in miR-193a-transfectants.
| Entrez gene ID | Symbol | Fold change |
|---|
| 23119 | HIC2 | −6.20 |
| 10152 | ABI2 | −3.34 |
| 595 | CCND1 | −3.30 |
| 54756 | IL17RD | −3.20 |
| 10238 | WDR68 | −3.15 |
| 3925 | STMN1 | −2.97 |
| 5324 | PLAG1 | −2.97 |
| 3845 | KRAS | −2.76 |
| 4076 | CAPRIN1 | −2.66 |
| 2066 | ERBB4 | −2.58 |
| 57704 | GBA2 | −2.45 |
| 84986 | ARHGAP19 | −2.23 |
| 10620 | ARID3B | −2.17 |
| 7342 | UBP1 | −2.09 |
| 27242 | TNFRSF21 | −2.07 |
| 4170 | MCL1 | −2.00 |
| 56262 | LRRC8A | −1.93 |
| 10160 | FARP1 | −1.91 |
| 57472 | CNOT6 | −1.91 |
| 23179 | RGL1 | −1.79 |
| 23341 | DNAJC16 | −1.79 |
| 88455 | ANKRD13A | −1.70 |
| 4215 | MAP3K3 | −1.67 |
| 114991 | ZNF618 | −1.66 |
| 23492 | CBX7 | −1.64 |
| 23365 | ARHGEF12 | −1.64 |
| 22883 | CLSTN1 | −1.61 |
| 9939 | RBM8A | −1.60 |
| 54890 | ALKBH5 | −1.59 |
| 115 | ADCY9 | −1.57 |
| 4189 | DNAJB9 | −1.51 |
| 1173 | AP2M1 | −1.51 |
| 9962 | SLC23A2 | −1.51 |
| 23384 | SPECC1L | −1.50 |
MCL1 is a direct target gene of miR-193a
in A2780 cells
From our results of transcriptome analysis, we
focused on MCL1 gene as miR-193a targets, since MCL1 was an
anti-apoptotic gene of BCL2 family (28), and therefore might contribute to
miR-193a-induced cell death in A2780 cells. MCL1 3′UTR contains one
potential target site of miR-193a and the site is conserved between
human and mouse. To examine the regulation of miR-193a on MCL1
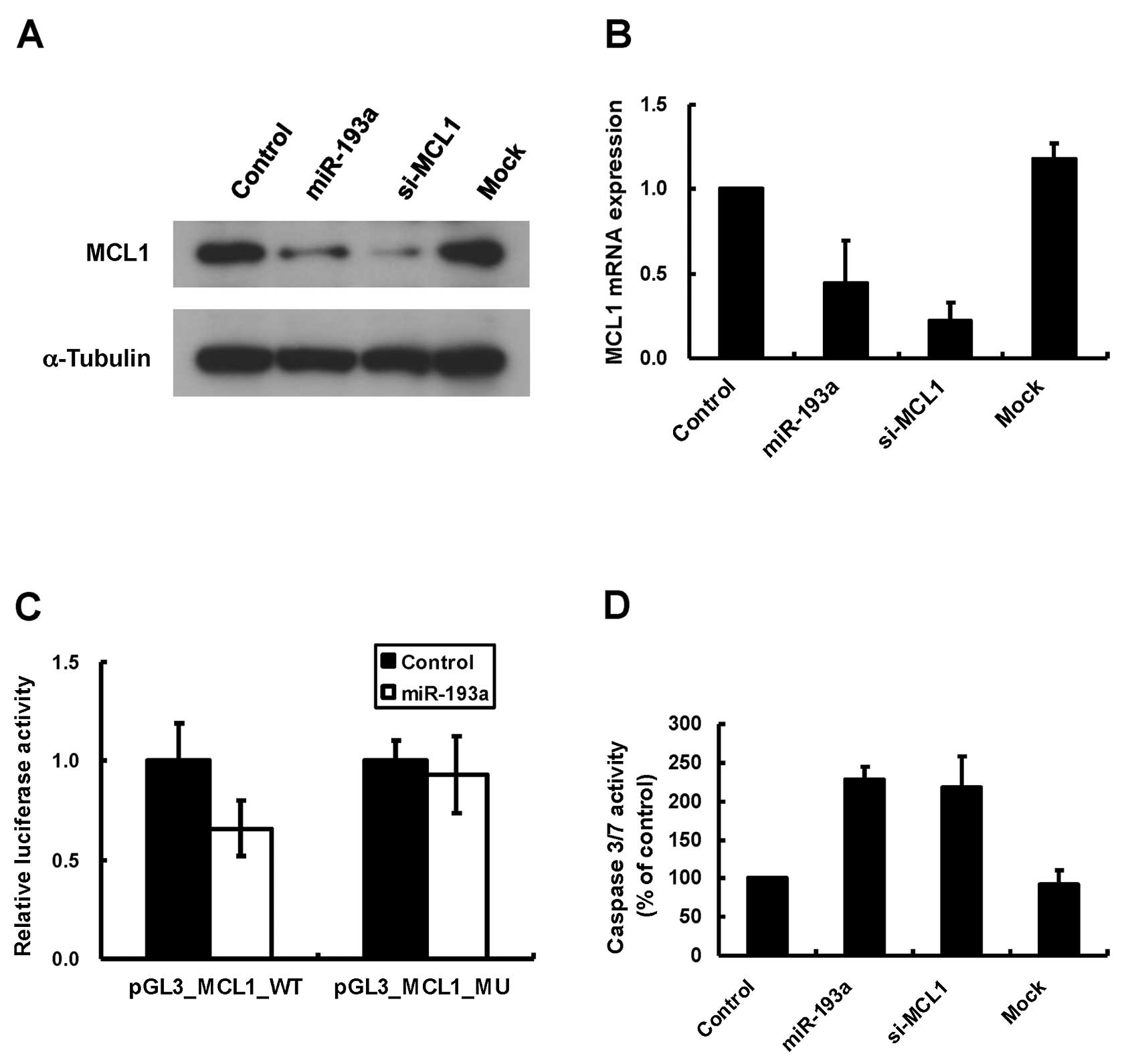
protein expression, we performed western blot analysis with
miR-193a-transfected A2780 cells. Transfection of positive control
MCL1 siRNA induced the decrease in endogenous MCL1 proteins in
A2780 cells (Fig. 4A). We
demonstrated that overexpression of miR-193a decreased MCL1
proteins in A2780 cells (Fig. 4A).
We next performed qRT-PCR with miR-193a-transfected cells to
examine whether miR-193a affected MCL1 mRNA expression. We found
that miR-193a induced about 50% decrease in MCL1 mRNA expression in
A2780 cells (Fig. 4B). These
results indicated that miR-193a affected MCL1 expression at both
protein and mRNA levels. To validate whether miR-193a can directly
regulate the translation of MCL1 mRNAs, we constructed a luciferase
reporter plasmid that inserted MCL1 3′UTR (around 1.5 kb) at the
downstream of Firefly luciferase gene, and tested the
luciferase activity. As shown in Fig.
4C, co-transfection of miR-193a and MCL1 3′UTR reporter vector
induced around 40% reduction of the luciferase activity compared
with co-transfection of the negative control miRNA and the reporter
vector. The decrease of the luciferase activity was attenuated by
using the mutant reporter vector deleting miR-193a seed region
complementary sites in MCL1 3′UTR (Fig. 4C, MCL1-3′UTR-MU). These results
indicated that MCL1 would be a direct target of miR-193a. We
further examined whether the downregulation of endogenous MCL1
could induce apoptosis in A2780 cells. As shown in Fig. 4D, the transfection of MCL1 siRNA
(25 nM) induced caspase 3/7 activation comparable with miR-193a
transfection in A2780 cells (Fig.
4D), indicating that the downregulation of MCL1 by miR-193a
could contribute to miR-193a-induced apoptosis in A2780 cells.
Discussion
Several studies reveal that global miRNA expression
is dysregulated in ovarian cancer (5–10),
and miRNAs may represent new targets for detection, diagnosis and
therapy in ovarian cancer (14).
However, functions of many miRNAs in ovarian cancer remain to be
elucidated. In this study, we performed a gain-of-function screen
using a miRNA mimic library (319 miRNA species) to identify those
affecting cell proliferation in epithelial ovarian cancer cells
(A2780). The library consists of miRNAs registered in early version
of miRBase (ver. 7.1 in October, 2005, http://www.mirbase.org/), and many of them were
expressed in ovarian normal and cancer tissues and cell lines
(5). We discovered
pro-proliferative miRNAs (miR-9*, miR-93, miR-130a,
miR-130b, miR-301, miR-302b, miR-302d, miR-363, miR-372, miR-373),
and anti-proliferative miRNAs (miR-7, miR-124a, miR-192, miR-193a,
miR-193b, miR-199a*, miR-432*, miR-497,
miR-506, miR-517c) in A2780 cells. By the same miRNA mimics library
screening, we found that miR-93/miR-372/miR-373 and miR-124a were
pro-proliferative and anti-proliferative, respectively, in DLD-1
colorectal cancer cells (23),
suggesting consistent roles of these miRNAs on cell proliferation
in ovary and colorectal cancer cells. The base-pairing between
target mRNAs and the seed region (2nd to 8th nucleotides) of miRNA
is important for miRNAs to function to regulate their target genes
(2). The seed family miRNAs
induced similar cellular phenotypes on cell proliferation in this
study (ex. pro-proliferative miR-93, miR-302b, miR-302d, miR-372,
miR-373 and anti-proliferative miR-193a, miR-193b), supporting the
importance of the seed region of miRNA on its function. Our miRNA
hits did not always correspond to dysregulated miRNAs reported in
ovarian cancer (5–10), but included pro-proliferative
miR-93 that was upregulated in primary ovarian carcinomas (6), supporting an oncogenic role of this
miRNA in ovarian cancer. Our miRNA hits also included tumor
suppressive miR-7, miR-124a, miR-192 and miR-193a in several cancer
types (18,25–27),
suggesting that these miRNAs could be tumor suppressive in ovarian
cancer. Among our miRNA hits, we further characterized miR-124a,
miR-192, miR-193a and miR-193b that induced a large decrease in the
cell viability of A2780 cells. miR-124a and miR-192 induced a
decrease in BrdU incorporation, indicating that these miRNAs
affected cell cycle resulting in inhibition of DNA synthesis in
A2780 cells. Inhibitory effects of miR-124a and miR-192 on cell
cycle gene pathway are reported in several cancer cell lines.
miR-124a targets cyclin dependent kinase 6 (CDK6), and thereby
inhibits the phosphorylation of retinoblastoma (Rb) in HCT116 cells
(29). miR-192 is upregulated by
genotoxic stress in HCT116, A549 and U2OS cell lines bearing
wild-type p53, and induces the cell cycle arrest by enhancing
CDKN1A/p21 expression (18,30).
We showed that miR-193a and miR-193b inhibited BrdU
incorporation and induced caspase 3/7 activation in A2780 cells,
indicating that these miRNAs could affect cell cycle and apoptotic
gene pathways. Our transcriptome analysis with miR-193a-transfected
A2780 cells identified ARHGAP19, CCND1, ERBB4, KRAS, MCL1 as
potential miR-193a target genes. We demonstrated that the
translation of MCL1 proteins was suppressed by miR-193a, suggesting
that anti-apoptotic MCL1 would be one of the target genes for
miR-193a-induced cell death in A2780. Anti-proliferative and
pro-apoptotic functions of miR-193 are reported in several cancer
cell lines including MDA-MB-453 (breast cancer), Malme-3M,
SKMEL-28, SKMEL-5 (melanoma), HO-1-N-1, HSC-2 (oral squamous cell
carcinoma), 22Rv1 (prostate cancer), SK-Hep-1 (hepato-cellular
carcinoma) and Kasumi-1 (acute myeloid leukemia) (26,31–36).
Consistent with our results, CCND1, KRAS and MCL1 are identified as
miR-193 target genes (26,32,33,37).
miR-193a gene locus (chromosomal region 17q11.2) has CpG islands
that are hyper-methylated in oral cancer (26) and acute myeloid leukemia (36) compared with normal tissues and
cells. miR-193a is downregulated in epithelial ovary cancer
compared with normal counterparts (10), but study is needed on whether the
miR-193a gene locus is hyper-methylated in ovary cancer.
Exogenous expression of a single miRNA mimic can
coordinately regulate gene expression on cellular function, which
encourages the therapeutic use of miRNA to direct cancer cell death
and/or re-differentiation without undesirable side-effects
(15,20). One of the challenges to the
therapeutic use of miRNA is to predict precisely molecular
consequences induced by modulating cellular miRNAs. Transcriptome
analysis by microarray has been widely used for miRNA target
identification at the transcription level. Protein-profiling
techniques have been applied to miRNA-transfected cells for the
identification of miRNA targets at the translational level
(38–41).
In summary, we performed a gain-of-function miRNA
screen and discovered several miRNAs affecting cell proliferation
and death in A2780 ovary cancer cells. Among them, we identified
miR-193a as strong anti-proliferative miRNAs in A2780 cells.
miR-193a induced the inhibition of DNA synthesis and apoptosis by
targeting genes including ARHGAP19, CCND1, ERBB4, KRAS, MCL1,
indicating a tumor suppressive role of this miRNA in epithelial
ovarian cancer cells. Our study suggests the potential of miRNA
screens to discover miRNAs as therapeutic tools to treat ovarian
cancer.
Acknowledgements
We thank Ms. K. Hayama, Ms. I. Taki
and Ms. M. Kamigaki for their technical assistance. This research
was partly supported by a grant from the New Energy and Industrial
Technology Development Organization (NEDO).
References
|
1
|
He L and Hannon GJ: MicroRNAs: small RNAs
with a big role in gene regulation. Nat Rev Genet. 5:522–531. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fabian MR, Sonenberg N and Filipowicz W:
Regulation of mRNA translation and stability by microRNAs. Annu Rev
Biochem. 79:351–379. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Garzon R, Calin GA and Croce CM: MicroRNAs
in cancer. Annu Rev Med. 60:167–179. 2009. View Article : Google Scholar
|
|
5
|
Iorio MV, Visone R, Di Leva G, Donati V,
Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H,
Calin GA, Menard S and Croce CM: MicroRNA signatures in human
ovarian cancer. Cancer Res. 67:8699–8707. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nam EJ, Yoon H, Kim SW, Kim H, Kim YT, Kim
JH, Kim JW and Kim S: MicroRNA expression profiles in serous
ovarian carcinoma. Clin Cancer Res. 14:2690–2695. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang L, Volinia S, Bonome T, Calin GA,
Greshock J, Yang N, Liu CG, Giannakakis A, Alexiou P, Hasegawa K,
Johnstone CN, Megraw MS, Adams S, Lassus H, Huang J, Kaur S, Liang
S, Sethupathy P, Leminen A, Simossis VA, Sandaltzopoulos R, Naomoto
Y, Katsaros D, Gimotty PA, DeMichele A, Huang Q, Butzow R, Rustgi
AK, Weber BL, Birrer MJ, Hatzigeorgiou AG, Croce CM and Coukos G:
Genomic and epigenetic alterations deregulate microRNA expression
in human epithelial ovarian cancer. Proc Natl Acad Sci USA.
105:7004–7009. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yang H, Kong W, He L, Zhao JJ, O’Donnell
JD, Wang J, Wenham RM, Coppola D, Kruk PA, Nicosia SV and Cheng JQ:
MicroRNA expression profiling in human ovarian cancer: miR-214
induces cell survival and cisplatin resistance by targeting PTEN.
Cancer Res. 68:425–433. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Dahiya N, Sherman-Baust CA, Wang TL,
Davidson B, Shih IeM, Zhang Y, Wood W III, Becker KG and Morin PJ:
MicroRNA expression and identification of putative miRNA targets in
ovarian cancer. PLoS One. 3:e24362008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wyman SK, Parkin RK, Mitchell PS, Fritz
BR, O’Briant K, Godwin AK, Urban N, Drescher CW, Knudsen BS and
Tewari M: Repertoire of microRNAs in epithelial ovarian cancer as
determined by next generation sequencing of small RNA cDNA
libraries. PLoS One. 4:e53112009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang L, Huang J, Yang N, Greshock J,
Megraw MS, Giannakakis A, Liang S, Naylor TL, Barchetti A, Ward MR,
Yao G, Medina A, O’brien-Jenkins A, Katsaros D, Hatzigeorgiou A,
Gimotty PA, Weber BL and Coukos G: microRNAs exhibit high frequency
genomic alterations in human cancer. Proc Natl Acad Sci USA.
103:9136–9141. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Merritt WM, Lin YG, Han LY, Kamat AA,
Spannuth WA, Schmandt R, Urbauer D, Pennacchio LA, Cheng JF, Nick
AM, Deavers MT, Mourad-Zeidan A, Wang H, Mueller P, Lenburg ME,
Gray JW, Mok S, Birrer MJ, Lopez-Berestein G, Coleman RL, Bar-Eli M
and Sood AK: Dicer, Drosha, and outcomes in patients with ovarian
cancer. N Engl J Med. 359:2641–2650. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Johnson SM, Grosshans H, Shingara J, Byrom
M, Jarvis R, Cheng A, Labourier E, Reinert KL, Brown D and Slack
FJ: RAS is regulated by the let-7 microRNA family. Cell.
120:635–647. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Dahiya N and Morin PJ: MicroRNAs in
ovarian carcinomas. Endocr Relat Cancer. 17:F77–89. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Garofalo M and Croce CM: microRNAs: Master
regulators as potential therapeutics in cancer. Annu Rev Pharmacol
Toxicol. 51:25–43. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Linsley PS, Schelter J, Burchard J,
Kibukawa M, Martin MM, Bartz SR, Johnson JM, Cummins JM, Raymond
CK, Dai H, Chau N, Cleary M, Jackson AL, Carleton M and Lim L:
Transcripts targeted by the microRNA-16 family cooperatively
regulate cell cycle progression. Mol Cell Biol. 27:2240–2252. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
He L, He X, Lim LP, de Stanchina E, Xuan
Z, Liang Y, Xue W, Zender L, Magnus J, Ridzon D, Jackson AL,
Linsley PS, Chen C, Lowe SW, Cleary MA and Hannon GJ: A microRNA
component of the p53 tumour suppressor network. Nature.
447:1130–1134. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Georges SA, Biery MC, Kim SY, Schelter JM,
Guo J, Chang AN, Jackson AL, Carleton MO, Linsley PS, Cleary MA and
Chau BN: Coordinated regulation of cell cycle transcripts by
p53-inducible microRNAs, miR-192 and miR-215. Cancer Res.
68:10105–10112. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lim LP, Lau NC, Garrett-Engele P, Grimson
A, Schelter JM, Castle J, Bartel DP, Linsley PS and Johnson JM:
Microarray analysis shows that some microRNAs downregulate large
numbers of target mRNAs. Nature. 433:769–773. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mishra PJ and Merlino G: MicroRNA
reexpression as differentiation therapy in cancer. J Clin Invest.
119:2119–2123. 2009.PubMed/NCBI
|
|
21
|
Wahid F, Shehzad A, Khan T and Kim YY:
MicroRNAs: synthesis, mechanism, function, and recent clinical
trials. Biochim Biophys Acta. 1803:1231–1243. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Tsuruo T, Hamilton TC, Louie KG, Behrens
BC, Young RC and Ozols RF: Collateral susceptibility of
adriamycin-, melphalanand cisplatin-resistant human ovarian tumor
cells to bleomycin. Jpn J Cancer Res. 77:941–945. 1986.PubMed/NCBI
|
|
23
|
Nakano H, Miyazawa T, Kinoshita K, Yamada
Y and Yoshida T: Functional screening identifies a microRNA,
miR-491 that induces apoptosis by targeting Bcl-X(L) in colorectal
cancer cells. Int J Cancer. 127:1072–1080. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Creighton CJ, Nagaraja AK, Hanash SM,
Matzuk MM and Gunaratne PH: A bioinformatics tool for linking gene
expression profiling results with public databases of microRNA
target predictions. RNA. 14:2290–2296. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kefas B, Godlewski J, Comeau L, Li Y,
Abounader R, Hawkinson M, Lee J, Fine H, Chiocca EA, Lawler S and
Purow B: microRNA-7 inhibits the epidermal growth factor receptor
and the Akt pathway and is down-regulated in glioblastoma. Cancer
Res. 68:3566–3572. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kozaki K, Imoto I, Mogi S, Omura K and
Inazawa J: Exploration of tumor-suppressive microRNAs silenced by
DNA hypermethylation in oral cancer. Cancer Res. 68:2094–2105.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Voorhoeve PM, le Sage C, Schrier M, Gillis
AJ, Stoop H, Nagel R, Liu YP, van Duijse J, Drost J, Griekspoor A,
Zlotorynski E, Yabuta N, De Vita G, Nojima H, Looijenga LH and
Agami R: A genetic screen implicates miRNA-372 and miRNA-373 as
oncogenes in testicular germ cell tumors. Adv Exp Med Biol.
604:17–46. 2007. View Article : Google Scholar
|
|
28
|
Nijhawan D, Fang M, Traer E, Zhong Q, Gao
W, Du F and Wang X: Elimination of Mcl-1 is required for the
initiation of apoptosis following ultraviolet irradiation. Genes
Dev. 17:1475–1486. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lujambio A, Ropero S, Ballestar E, Fraga
MF, Cerrato C, Setien F, Casado S, Suarez-Gauthier A,
Sanchez-Cespedes M, Git A, Spiteri I, Das PP, Caldas C, Miska E and
Esteller M: Genetic unmasking of an epigenetically silenced
microRNA in human cancer cells. Cancer Res. 67:1424–1429. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Braun CJ, Zhang X, Savelyeva I, Wolff S,
Moll UM, Schepeler T, Orntoft TF, Andersen CL and Dobbelstein M:
p53-responsive micrornas 192 and 215 are capable of inducing cell
cycle arrest. Cancer Res. 68:10094–10104. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ovcharenko D, Kelnar K, Johnson C, Leng N
and Brown D: Genome-scale microRNA and small interfering RNA
screens identify small RNA modulators of TRAIL-induced apoptosis
pathway. Cancer Res. 67:10782–10788. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Iliopoulos D, Rotem A and Struhl K:
Inhibition of miR-193a expression by Max and RXRalpha activates
K-Ras and PLAU to mediate distinct aspects of cellular
transformation. Cancer Res. 71:5144–5153. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Chen J, Zhang X, Lentz C, Abi-Daoud M,
Pare GC, Yang X, Feilotter HE and Tron VA: miR-193b Regulates Mcl-1
in Melanoma. Am J Pathol. 179:2162–2168. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Xu C, Liu S, Fu H, Li S, Tie Y, Zhu J,
Xing R, Jin Y, Sun Z and Zheng X: MicroRNA-193b regulates
proliferation, migration and invasion in human hepatocellular
carcinoma cells. Eur J Cancer. 46:2828–2836. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Rauhala HE, Jalava SE, Isotalo J, Bracken
H, Lehmusvaara S, Tammela TL, Oja H and Visakorpi T: miR-193b is an
epigenetically regulated putative tumor suppressor in prostate
cancer. Int J Cancer. 127:1363–1372. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gao XN, Lin J, Gao L, Li YH, Wang LL and
Yu L: MicroRNA-193b regulates c-Kit proto-oncogene and represses
cell proliferation in acute myeloid leukemia. Leuk Res.
35:1226–1232. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Chen J, Feilotter HE, Pare GC, Zhang X,
Pemberton JG, Garady C, Lai D, Yang X and Tron VA: MicroRNA-193b
represses cell proliferation and regulates cyclin D1 in melanoma.
Am J Pathol. 176:2520–2529. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Baek D, Villen J, Shin C, Camargo FD, Gygi
SP and Bartel DP: The impact of microRNAs on protein output.
Nature. 455:64–71. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Selbach M, Schwanhausser B, Thierfelder N,
Fang Z, Khanin R and Rajewsky N: Widespread changes in protein
synthesis induced by microRNAs. Nature. 455:58–63. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Leivonen SK, Makela R, Ostling P, Kohonen
P, Haapa-Paananen S, Kleivi K, Enerly E, Aakula A, Hellstrom K,
Sahlberg N, Kristensen VN, Borresen-Dale AL, Saviranta P, Perala M
and Kallioniemi O: Protein lysate microarray analysis to identify
microRNAs regulating estrogen receptor signaling in breast cancer
cell lines. Oncogene. 28:3926–3936. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Leivonen SK, Rokka A, Ostling P, Kohonen
P, Corthals GL, Kallioniemi O and Perala M: Identification of
miR-193b targets in breast cancer cells and systems biological
analysis of their functional impact. Mol Cell Proteomics.
10:M110.0053222011. View Article : Google Scholar : PubMed/NCBI
|