Introduction
Colorectal carcinoma (CRC) is one of the most common
and well-studied malignancies in the Western world. The disease is
thought to originate in multi-potential stem cells located in the
intestinal crypts; (non-) polypoid precursor lesions initiate from
these cells, and metastatic CRC can develop (1,2). The
histological progression of colorectal carcinogenesis is
characterized by sequential genetic (3) and epigenetic alterations (4,5). To
date, intensive efforts have been made by scientists to find a
provocative factor of this major cancer; many epidemiologic studies
indicate that a Western-style diet is associated with a high
incidence of CRC (6,7).
The development of CRC is usually described as a
multistep model, in which the accumulation of genetic and
epigenetic events mediates the adenoma-carcinoma sequence (8). The accumulation of mutations is
driven through distinct pathways by different types of genomic
instability, chromosomal instability and microsatellite instability
(9). Additionally, an epigenetic
pathway has been proposed, in which tumor suppressor genes are
inactivated by promoter methylation and silencing of gene
transcription (10).
Runt-related transcription factor 3 (RUNX3) belongs
to the RUNX family of genes, which is important in mammalian
development and neoplasia (11–13).
RUNX3 cooperates with Sma and Mad related family 3 (Smad3)/Smad4 to
activate transforming growth factor-β (TGF-β)-dependent growth
inhibition and apoptosis by inducing p21 and Bim (14). The RUNX3 gene is localized to the
1p36 locus and is linked with gastric epithelial homeostasis and
gastric carcinogenesis. The 1p36 region is thought to harbor at
least one tumor suppressor gene, since this region exhibits
frequent loss of heterozygosity in colon, gastric, breast and
ovarian cancers (15).
Additionally, the introduction of a normal human 1p36 chromosome
fragment into colon cancer cells suppresses their tumorigenicity
(16). Interestingly, a
considerable proportion of gastric cancers do not express RUNX3 due
to hemizygous deletion and hypermethylation of the RUNX3 promoter
region (17). Hypermethylation of
the RUNX3 promoter occurs in 21% of colon cancer specimens and at
least 65% of colon cancer cell lines, suggesting that RUNX3 has a
tumor suppressive function in CRC (18).
The regulation of DNA methylation is not well
understood, but involves DNA methyltransferases (DNMTs), which
catalyze the transfer of methyl groups to the carbon-5 position of
cytosines in phosphodiester bond between the cytosine and the
guanine (CpG) islands. Three active DNMTs have been identified in
mammals, DNMT1, DNMT3A and DNMT3B. DNMT1 is largely responsible for
maintaining methylation, and contributes to de novo DNA
promoter methylation in cancer (19). Some models suggest that DNMT3B
cooperates with DNMT1 to maintain DNA methylation status (20). Epigenetic changes by DNMTs are
highly relevant to colon carcinogenesis; as such, they represent a
target for novel strategies that can prevent or treat cancer. DNA
methylation results in the local recruitment of histone
deacetylases to promoter regions, with co-localization of methyl
CpG binding protein 2. These eventually inhibit the binding of RNA
polymerase II, thereby repressing the expression of genes and their
products. Such observations led to the paradigm that DNA
methylation occurs upstream of histone acetylation in this pathway
of transcriptional repression. DNMTs produce methylation of CpG
repeats in the upstream promoter regions of genes, while
physiological upstream regulators of the activity and levels of
DNMTs have not yet been identified.
Recent studies suggest that DNA methylation in colon
cancer and NIH 3T3 cells may be regulated, in some circumstances by
extracellular signal-regulated kinase (ERK) activity (21). DNA methylation and DNMTs might be
modulated directly or indirectly by the extracellular milieu
through signal transduction pathways, which may include ERK or
other cellular kinases.
Recently, two well-characterized and
clinically-relevant DNMT inhibitors, 5-aza-cytidine (5-Aza) and its
metabolite 5-aza-2′deoxycytidine (5-Aza-2′dC), were identified as
nucleoside analogue mechanism-based inhibitors. They are
substituted for cytosine residues in the DNA strand during
replication, and inhibit DNMT activity by covalently binding to the
DNMT enzymes, resulting in reduced genomic DNA methylation
(22,23). The Food and Drug Administration has
approved these two inhibitors for the treatment of myelodysplastic
syndromes (24). Although
effective against certain hematopoietic disorders, these drugs show
some toxicity both in vitro and in vivo, and are
unstable in neutral solutions (25–27).
The inherent lack of specificity of 5-Aza and 5-Aza-2′dC for their
target genes allows for these undesirable effects. For example,
global demethylation by 5-Aza and 5-Aza-2′dC may result in the
expression of oncogenic loci and activation of transposable
elements, inducing the expression of multidrug resistance genes
(28–30). Hence, a non-toxic, highly stable,
and effective DNMT inhibitor would be an ideal epigenetic
therapeutic agent.
A number of saponins have been isolated from ginseng
and extensively investigated with regard to their possible
antitumor activity in various cancer cell lines (31–33).
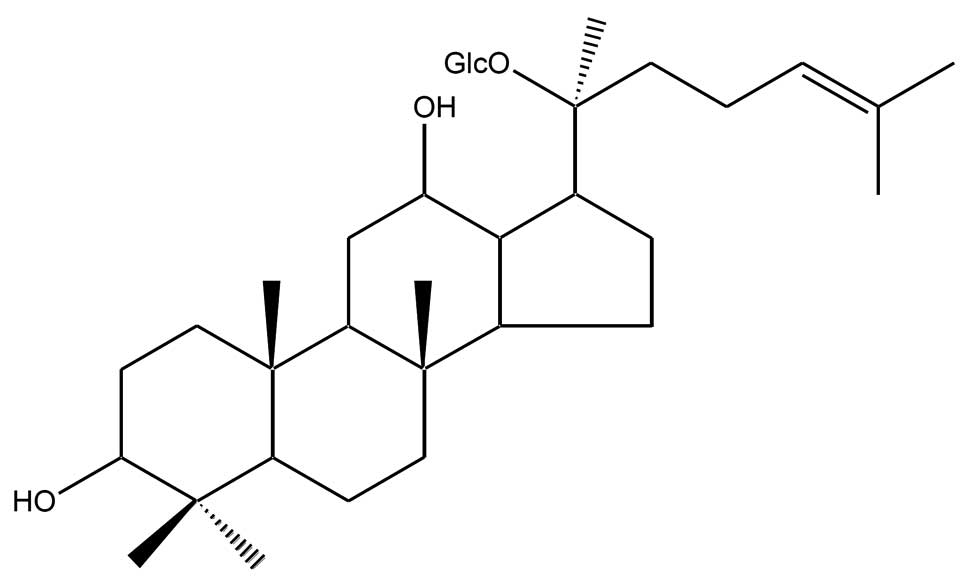
Among them, 20-O-(β-D-glucopyranosyl)-20(S)-protopanaxadiol
(compound K, Fig. 1) is the main
metabolite of protopanaxadiol-type ginsenoside, formed in the
intestine after oral administration (34–36).
We recently reported that compound K exhibited cytotoxicity by
inducing apoptosis, arrest of growth at the G1 phase of the cell
cycle, and inhibition of telomerase activity in human leukemia
cells (37,38). In addition, combined treatment with
compound K and γ-irradiation enhanced cell death in human lung
cancer cells (39). Finally,
compound K induced apoptosis in MCF-7 breast cancer cells by
modulating AMP-activated protein kinase (40).
The aim of this study was to determine whether
compound K reactivates tumor suppressor genes in human colorectal
cancer HT-29 cells by inhibiting DNMT. The results demonstrate that
compound K treatment results in demethylation of the silenced gene,
RUNX3, via inhibition of ERK and recruitment of RUNX3-mediated
proteins, such as Smad4 and Bim. These results show, for the first
time, that compound K alters DNA methylation patterns within the
RUNX3 gene promoter.
Materials and methods
Cell culture
Human colorectal cancer HT-29 cells were obtained
from the Korean Cell Line Bank (Seoul, Republic of Korea). Cells
were maintained in an incubator at 37°C with a humidified
atmosphere of 5% CO2. The cells were cultured in
RPMI-1640 medium containing 10% fetal calf serum, streptomycin (100
μg/ml) and penicillin (100 U/ml).
Cell proliferation assay
The effect of compound K on the proliferation of the
cells was determined using the
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT)
assay. Cells were seeded in a 96-well plate at a density of
1×105 cells/ml and treated with compound K. After
incubating for 48 h, 50 μl of the MTT stock solution (2
mg/ml) was added to each well to attain a total reaction volume of
250 μl. After 4 h incubation, the supernatants were
aspirated. The formazan crystals in each well were then dissolved
in 150 μl dimethylsulfoxide and absorbance at 540 nm was
read on a scanning multi-well spectrophotometer.
Reverse transcription polymerase chain
reaction (RT-PCR)
Total RNA was isolated from cells using TRIzol
reagent (Gibco-BRL, Grand Island, NY, USA). Complementary DNA (cDNA
1 μl) was amplified in a reverse-transcription reaction
containing primers, dNTPs, and 0.5 units of Taq DNA polymerase at a
final volume of 25 μl. The PCR conditions were: 5 min at
94°C for initial denaturation, followed by 35 cycles of 1 min at
94°C, 1 min at 55°C and 1 min at 72°C, and a final elongation
period of 7 min at 72°C. PCR amplification was carried out in a
programmable thermal cycler (Perkin-Elmer Cetus 9600, Roche
Molecular Systems Inc., Branchburg, NJ, USA). The primers used to
amplify the RUNX3 and DNMT1 cDNA (30,42)
were: RUNX3 sense, 5′-GGCAATGACGAGAA CTAC-3′ (located in exon 2),
antisense, 5′-GGAGAATGGGT TCAGTTC-3′ (located in exon 5); DNMT1
sense, 5′-CGCTGT ATCTAGCAAGGGTCA-3′, antisense, 5′-TCGAATCTCGCG
TAGTCTTG-3′; GAPDH sense, 5′-GTGGGCCGCCCTAGG CACCAGG-3′, antisense
5′-GGAGGAAGAGGATGCGGC AGTG-3′. The amplified products were resolved
on 1% agarose gels, stained with ethidium bromide and photographed
under ultraviolet light, using Image Quant™ TL analysis software
(Amersham Bioscience, Uppsala, Sweden).
Western blot analysis
Cells were harvested and lysed on ice in 1 ml of
lysis buffer (10 mM Tris-HCl, pH 7.9, 10 mM NaCl, 3 mM
MgCl2 and 1% NP-40) for 4 min. After centrifugation for
10 min at 3,000 × g, the pellets were re-suspended in 50 μl
of extraction buffer (20 mM HEPES, pH 7.9, 20% glycerol, 1.5 mM
MgCl2, 0.2 mM EDTA, 1 mM DTT and 1 mM PMSF). This was
incubated on ice for 30 min and centrifuged at 13,000 × g for 5
min. Following measurement of the protein concentration,
supernatants were stored at −70°C. Aliquots of the lysates (40
μg of protein) were boiled for 5 min and electrophoresed on
a 10% SDS-polyacrylamide gel. Proteins were transferred onto
nitrocellulose membranes, which were subsequently incubated with
primary antibodies. The membranes were further incubated with
secondary immunoglobulin-G-horseradish peroxidase conjugates
(Pierce, Rockford, IL, USA). Protein bands were detected using an
enhanced chemiluminescence western blot analysis detection kit
(Amersham, Little Chalfont, UK). The protein bands were visualized
using a luminescent image analyzer.
Methylation-specific (MS)-PCR
The bisulfite modification of DNA was conducted
using a Methylamp™ DNA modification kit (Epigentek, Pittsburgh, PA,
USA) according to the manufacturer’s instructions. For analysis of
DNA methylation of RUNX3, MS-PCR was conducted using an Epitect MSP
kit (Qiagen, Valencia, CA, USA). PCR products were separated on 6%
non-denaturing polyacrylamide gels, stained with ethidium bromide,
and visualized under UV light. The methylated or unmethylated RUNX3
primer sets were (17):
unmethylated RUNX3 sense, 5′-TTATGAGGGGTGGTTG TATGTGGG-3′,
antisense, 5′-AAAACAACCAACACAAAC ACCTCC-3′; methylated RUNX3 sense,
5′-TTACGAGGGG CGGTCGTACGCGGG-3′, antisense, 5′-AAAACGACCGAC
GCGAACGCCTCC-3′.
Immunocytochemistry
Cells plated on coverslips were fixed with 4%
paraformaldehyde for 30 min and permeabilized with 0.1% Triton
X-100 in PBS for 2.5 min. Cells were treated with blocking medium
(3% bovine serum albumin in PBS) for 1 h and incubated for 2 h with
the RUNX3 antibody diluted in blocking medium. The primary RUNX3
antibody was detected by a FITC-conjugated secondary antibody
(1:500; Santa Cruz Biotechnology, Santa Cruz, CA, USA) for 1 h.
After washing with PBS, stained cells were mounted onto microscope
slides in mounting medium with DAPI (Vector, Burlingame, CA, USA),
and imaged using the LSM 510 program on a Zeiss confocal
microscope.
DNMT activity
Nuclear extracts were prepared using a nuclear
protein extraction kit (Cayman Chemical, Ann Arbor, MI, USA). After
measuring the protein concentration, the nuclear fractions were
stored at −70°C. DNMT activity was detected using a EpiQuik DNA
methyltransferase activity assay kit (Epigentek). In this assay,
the cytosine-rich DNA substrate is stably coated on the strip
wells. DNMT transfers a methyl group from S-adenosylmethionine to a
cytosine in the DNA substrate. Methylated DNA is recognized using a
5-methylcytosine antibody. The levels of methylated DNA, which are
proportional to enzymatic activity, are then quantified
colorimetrically using an ELISA-like reaction. The results are
expressed as absorbance units at 450 nm and represent percentage
activity.
Immunoprecipitation
Smad4 was immunoprecipitated from the nuclear
extracts using the Smad4 antibody. Immune complexes were collected
with protein G beads and washed with immunoprecipitation buffer.
Equal amounts of the precipitates were run on an SDS-polyacrylamide
gel followed by western blot analysis with antibodies specific for
RUNX3.
Chromatin immunoprecipitation (ChIP)
assay
The ChIP assay was performed using a Simple ChIP™
enzymatic chromatin IP kit (Cell Signaling Technology, Danvers, MA,
USA) according to the manufacturer’s protocol with slight
modifications. Briefly, cells were treated with 20 μg/ml of
compound K for 48 h and then cross-linked by addition of 1%
formaldehyde. Chromatin was prepared and digested with nuclease for
12 min at 37°C. ChIP was performed with the RUNX3 antibody (Abcam,
Cambridge, MA, USA) and normal mouse IgG. Antibodies were added to
the chromatin digests and incubated with constant rotation
overnight at 4°C. ChIP-grade protein G magnetic beads were then
added to capture the immune complexes. The beads were washed and
the immunoprecipitates eluted with ChIP elution buffer. The
cross-links were reversed by incubation at 65°C for 30 min.
Proteinase K was added and incubated at 65°C for 2 h. The
immunoprecipitated DNA fragments were then purified using spin
columns. DNA recovered from the immune-precipitated complex was
subjected to 35 cycles of PCR. The primers for the Bim (RUNX3
binding site) gene promoter were: sense, 5′-GGCAATGACGAGAACTAC-3′;
antisense, 5′-GGAG AATGGGTTCAGTTC-3′. The PCR products were
separated on 2% agarose gels, and DNA bands visualized using the
Image program (NIH, Bethesda, MD, USA).
Pyrosequencing analysis
The promoter regions of all genes were amplified
using a (biotinylated) forward primer and a (biotinylated) reverse
primer, and a sequencing primer designed by PSQ Assay Design
(Qiagen); a (biotinylated) sense, 5′-(biotinylated)
GGYGGGAGGYGGYGGTAGYGGTATAGTT-3′ (Y; T or C); a (biotinylated)
antisense, 5′-(biotinylated) RCAACCTACCCR ACTAATCCC-3′ (R; A or G);
sequencing primer, 5′-RCAACC TACCCR ACTAAT-3′. In addition, genomic
DNA (30 ng) was modified by sodium bisulfite using the EZ DNA
methylation kit (Zymo Research Co., Irvine, CA, USA) according to
the manufacturer’s instructions. Bisulfite-modified DNA was
amplified in a 50 μl reaction using biotinylated primer sets
and 5 units of Taq polymerase (Enzynomics Co., Daejeon, Republic of
Korea). The samples were heated at 94°C for 5 min and then
amplified for 45 cycles consisting of 94°C for 30 sec, 55°C for 30
sec and 72°C for 30 sec. All reactions were then incubated at 72°C
for 5 min and cooled to 4°C. The PCR products were visualized on a
2% agarose gel by ethidium bromide staining. Pyrosequencing
reactions were conducted with sequencing primers on the PyroMark ID
(Qiagen) according to the manufacturer’s specifications.
Unmethylated Cytosine (C) is measured according to the relative T
content at the CpG site, and methylated Cytosine (mC) is measured
according to the relative C content at the CpG site. The
methylation index of each gene promoter and of each sample was
calculated as the average value of mC/(mC+C) for all examined CpGs
in the target region.
Transient transfection of small RNA
interference (siRNA)
Cells were seeded at 1.5×105 cells/well
in 24-well plates and allowed to reach approximately 50% confluence
on the day of transfection. The siRNA constructs used were: a
mismatched siRNA control (siControl; Santa Cruz Biotechnology); a
siRNA against ERK, and an siRNA against DNMT1 (Santa Cruz
Biotechnology). Cells were transfected with 10–50 nM siRNA using
lipofectamine RNAiMax (Invitrogen, Carlsbad, CA, USA) based on the
manufacturer’s instructions. At 24 h after transfection, the cells
were treated with compound K for 48 h and examined by either
western blot analysis or MTT assay.
Statistical analysis
All measurements were performed in triplicate and
values expressed as the mean ± the standard error of the mean
(SEM). The results were examined using analysis of variance (ANOVA)
and Tukey’s test to determine pairwise differences. p<0.05 was
considered significant.
Results
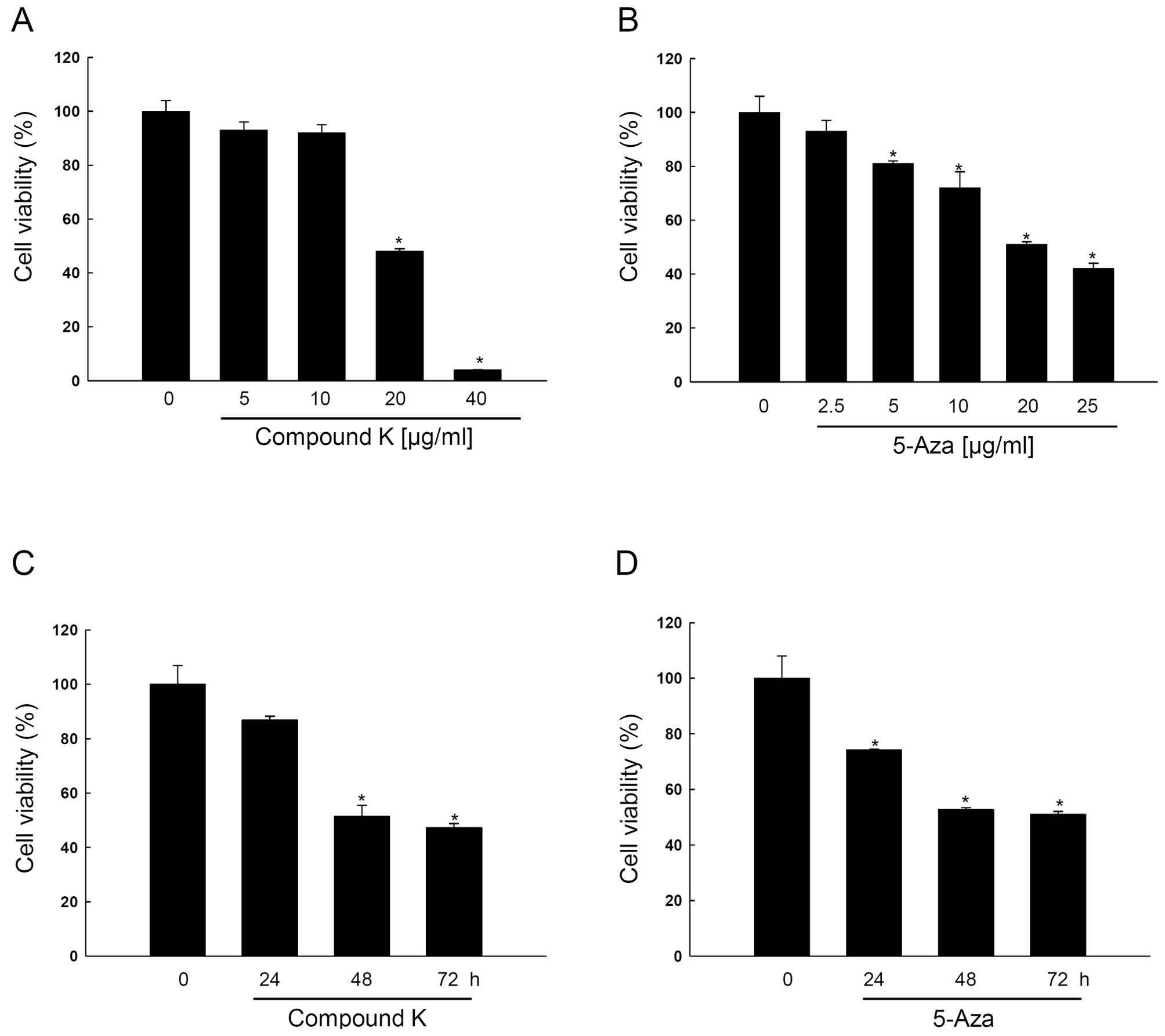
Compound K inhibits HT-29 cell growth in
a dose- and time-dependent manner
RUNX3 in HT-29 cells is epigenetically silenced
(41–45) and the present study used HT-29
cells to elucidate the epigenetic mechanisms of RUNX3. As shown in
Fig. 2A and B, compound K and
5-Aza inhibited HT-29 cell growth in a dose-dependent manner at 10,
20, 30, 40 μg/ml and 2.5, 5, 10, 20, 25 μg/ml,
respectively, and the concentration that yielded 50% growth
inhibition (IC50) was for compound K 20±1.0
μg/ml, and for 5-Aza 19±0.6 μg/ml, and both inhibited
cell growth in a time-dependent manner (Fig. 2C and D).
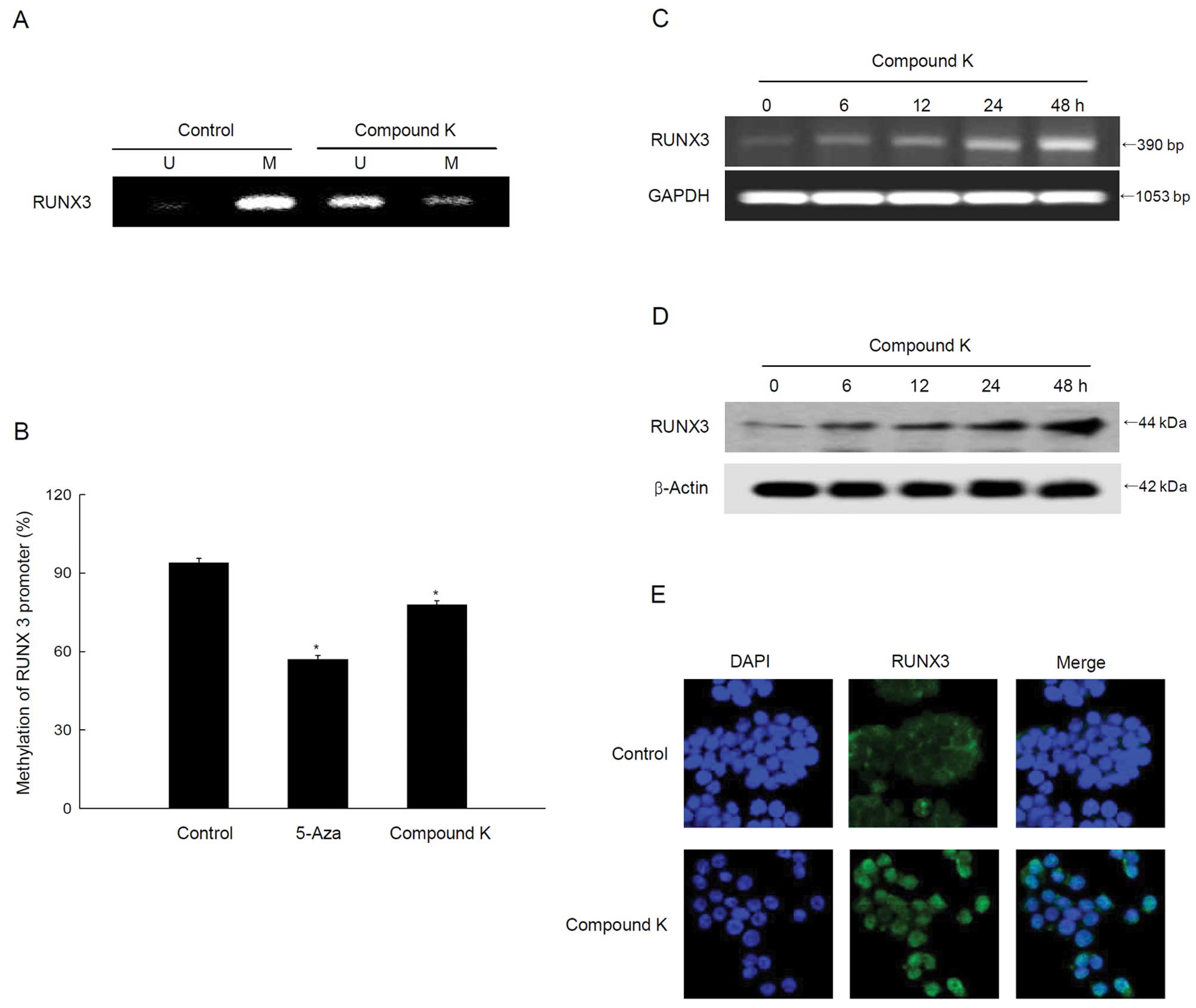
Compound K reverses methylation and
reactivates the tumor suppressor gene RUNX3
Methylation-specific PCR data revealed specific
unmethylation of a RUNX3 promoter in compound K-treated cells
(Fig. 3A) and the quantitative
pyrosequencing data showed a decrease in RUNX3 methylation
(Fig. 3B). The unmethylation of
RUNX3 in compound K-treated cells resulted in the re-expression of
RUNX3 mRNA and protein (Fig. 3C and
D). Mislocalization of nuclear RUNX3 protein to the cytoplasm
is initially observed in various cancers, including colon cancer
(47,48). However, microscopic data showed
that compound K-treated cells increased the RUNX3 protein
localization in the nucleus (Fig.
3E).
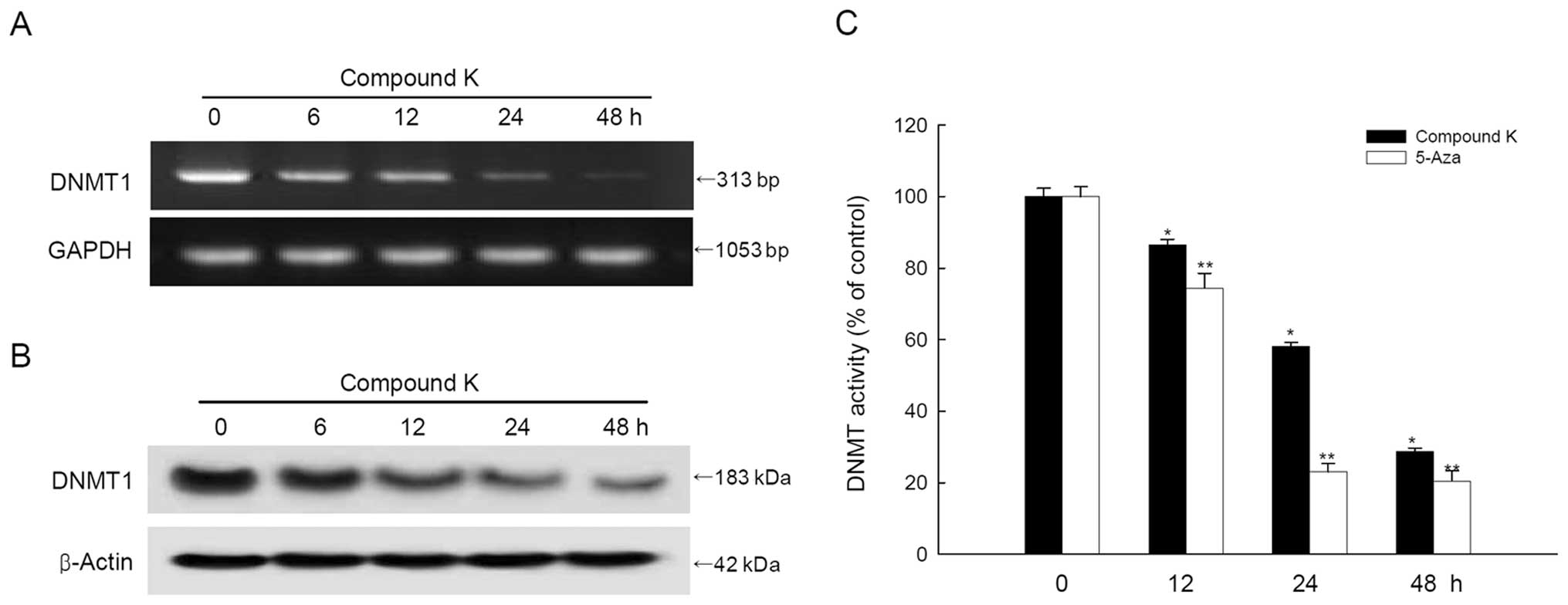
Compound K inhibits the expression and
activity of DNMT1
Compound K treatment decreased DNMT1 mRNA and
protein expression in a time-dependent manner (Fig. 4A and B). DNMT activity in the
nuclear extract of compound K-treated cells decreased in a
time-dependent manner, similar to the mRNA and protein pattern of
DNMT1 by 5-Aza (Fig. 4C).
Compound K induces RUNX3-mediated
expression of Smad4 and Bim
RUNX3 cooperates with Smad3/Smad4 to activate
TGF-β-dependent growth inhibition and induces apoptosis by inducing
p21 and Bim (14,42). Compound K treatment enhanced Smad4
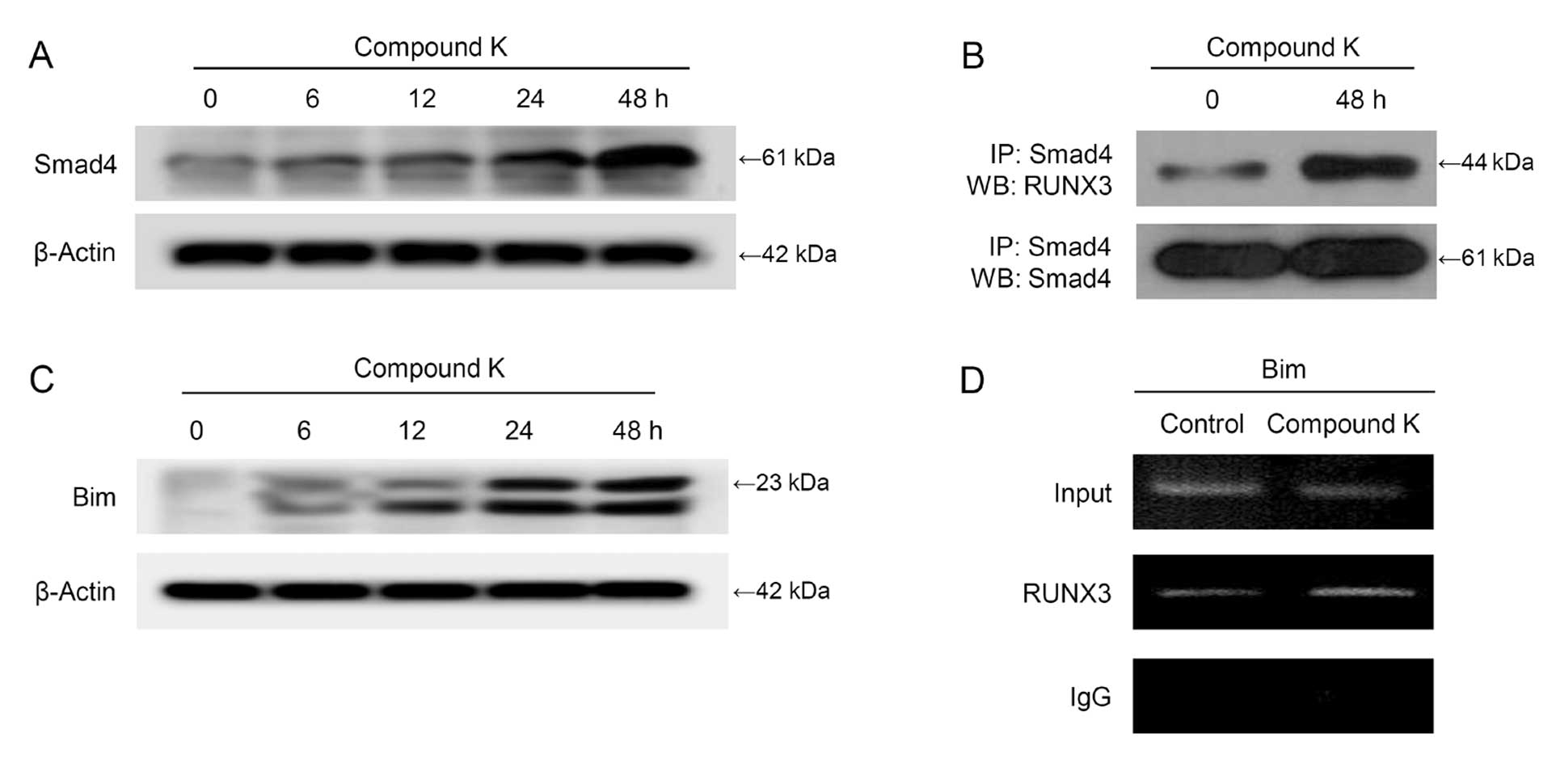
expression in a time-dependent manner (Fig. 5A). Additionally,
immunoprecipitation data showed that RUNX3 interacted with Smad4
(Fig. 5B); this complex induced
Bim expression via binding to the Bim promoter region (Fig. 5C and D).
Compound K suppresses DNMT1 activity by
inhibiting the ERK pathway
Lu et al (21) reported that the ERK pathway might
regulate DNA methylation in colon cancer cells. In the present
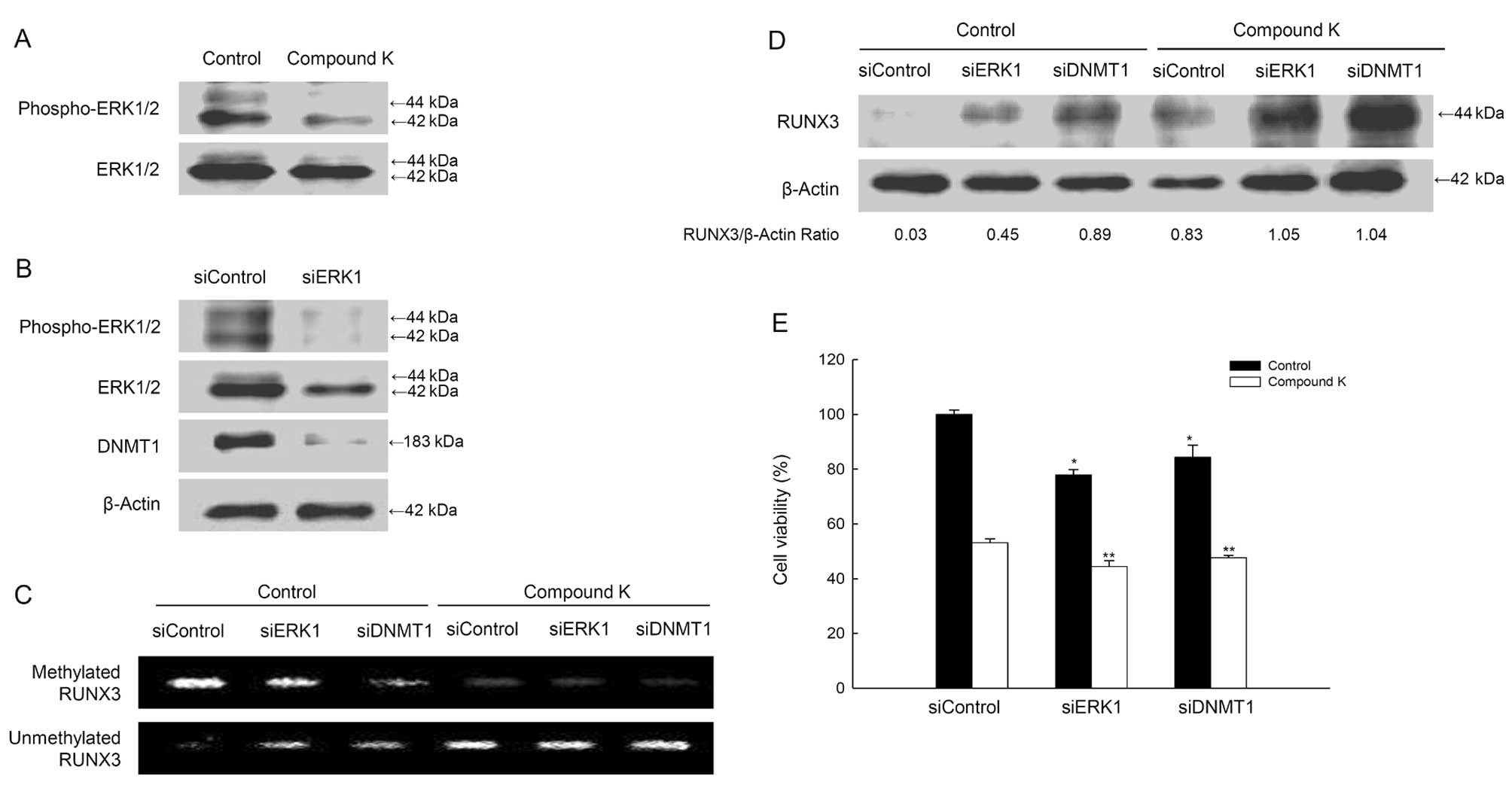
study, compound K-treated cells showed the decreased levels of
phospho-ERK (active form) (Fig.
6A), and the decreased ERK activity by ERK1 siRNA led to
reduced DNMT1 protein levels (Fig.
6B). However, siDNMT1 did not inhibit ERK expression (data not
shown), indicating that ERK acts upstream of DNMT1. The siERK1 or
siDNMT1 results showed that the methylation status in RUNX3
promoter decreased and the unmethylation status increased, which
was enhanced by compound K (Fig.
6C). These data are consistently RUNX3 protein expression in
siERK1 or siDNMT1-transfected cells with compound K treatment
(Fig. 6D). The combination of
compound K treatment with siERK1 or siDNMT1-transfection decreased
cell viability (Fig. 6E).
Discussion
This study shows that the ginseng saponin
metabolite, compound K, actively regulates cell growth in HT-29
cells. Most importantly, the results show that compound K can
reactivate a silenced tumor suppression gene by inhibiting DNMT1
protein expression and activity. Interestingly, the effects of
compound K were similar to those achieved using the clinically-used
compound, 5-Aza (49,50), a potent but toxic synthetic DNMT
inhibitor. Moreover, these findings are comparable with those
observed for non-toxic nutriceuticals, such as tea catechins, soy
bioflavonoids, and apple polyphenols, each of which can modulate
DNMT1 protein levels (51–53). Compound K is an active metabolite
of ginsenosides that exhibits antitumor effects against various
types of cancer cells, including HT-29 colorectal cancer cells
(34–40,46).
DNMTs may be regulated in a cell cycle-dependent
pattern in some cell types (54);
for example, the level of DNMT expression is lower during the G0/G1
phase. However, cancer cells maintain higher methylation levels
compared with normal cells, even during the G0/G1 phase. In our
system, HT-29 cells showed methylation and silencing of RUNX3. Some
reports demonstrated that lack of expression of RUNX3 in HT-29
cells can lead to inactivation of the TGF-β-mediated apoptosis
pathway (55,56). Re-expression of RUNX3 after
compound K treatment might be the direct result of its
unmethylation. Alternatively, there might be an additional
transcriptional component, contributed by compound K, once the
silencing methylation is reversed. Also, our results showed
compound K-treatment increased the localization of RUNX3 from
cytosol into nucleus. Not only is epigenetic silencing of RUNX3 a
frequent occurrence in cancer, cytoplasmic sequestration of RUNX3
was reported in a significant proportion of various cancer cases,
especially, 15% of colorectal cancer (14,18,47,48).
These findings indicate that nuclear RUNX3 and its downstream
transcriptional targets are crucial for tumor suppression.
Moreover, activation of TGF-β pathway triggered nuclear
translocation of endogenous RUNX3 in SNU16 cells and this
subsequently led to growth inhibition (48). With the TGF-β pathway frequently
impaired in cancer tissues, this would mean frequent cytoplasmic
sequestration and, thus, functional inactivation of RUNX3. It is
not known how TGF-β elicits nuclear translocation of RUNX3.
However, because SMAD and RUNX3 proteins interact (14), it is possible that perturbation of
Smad nuclear import/export may promote cytoplasmic accumulation
(14,57). Therefore, aberrant signals from
oncogenic pathways may mediate post-translational modification and
subsequent cytoplasmic mislocalization of RUNX3 in cancer
cells.
RUNX3 is now known to have a function in
TGF-β-induced growth inhibition. After stimulation by TGF-β
cytokines, RUNX3 cooperates with SMAD proteins to directly
upregulate transcription of the proapoptotic gene Bim and increase
expression of the cyclin-dependent kinase inhibitor p21 in gastric
cancer cells (43). Moreover, the
ability of RUNX3 to target SMADs to distinct nuclear foci on
stimulation by TGF-β further strengthens the case for RUNX3 as a
regulator of the TGF-β signaling pathway (57). In this study, compound K induced
expression of Smad4 and Bim, which is associated with RUNX3
reaction. Our results showed that compound K downregulated DNMT1
protein and mRNA levels in a time-dependent manner. Therefore, it
seems to that compound K regulated DNMT1 expression in HT-29 cells
at transcriptional level. This possibility is currently being
explored. The repression of DNMT1 levels by compound K is the
likely mechanism by which silencing of these tumor suppressor genes
is reversed; the downregulation of DNMT1 by siRNA is sufficient to
unmethylate RUNX3 and allow re-expression. Compound K treatment
also inhibited the regulation of ERK. ERK1-specific siRNA treatment
downregulated DNMT1 resulting in the unmethylation of RUNX3. These
results are consistent with a model in which ERK acts an upstream
regulator of DNMT1 (21). In our
system, suppression of DNMT1 did not inhibit ERK regulation,
whereas ERK inhibition downregulated DNMT1. Downregulation of DNMT1
resulted in unmethylation of RUNX3 and expression of the
RUNX3-related proteins, Smad4 and Bim, which caused cell growth
inhibition.
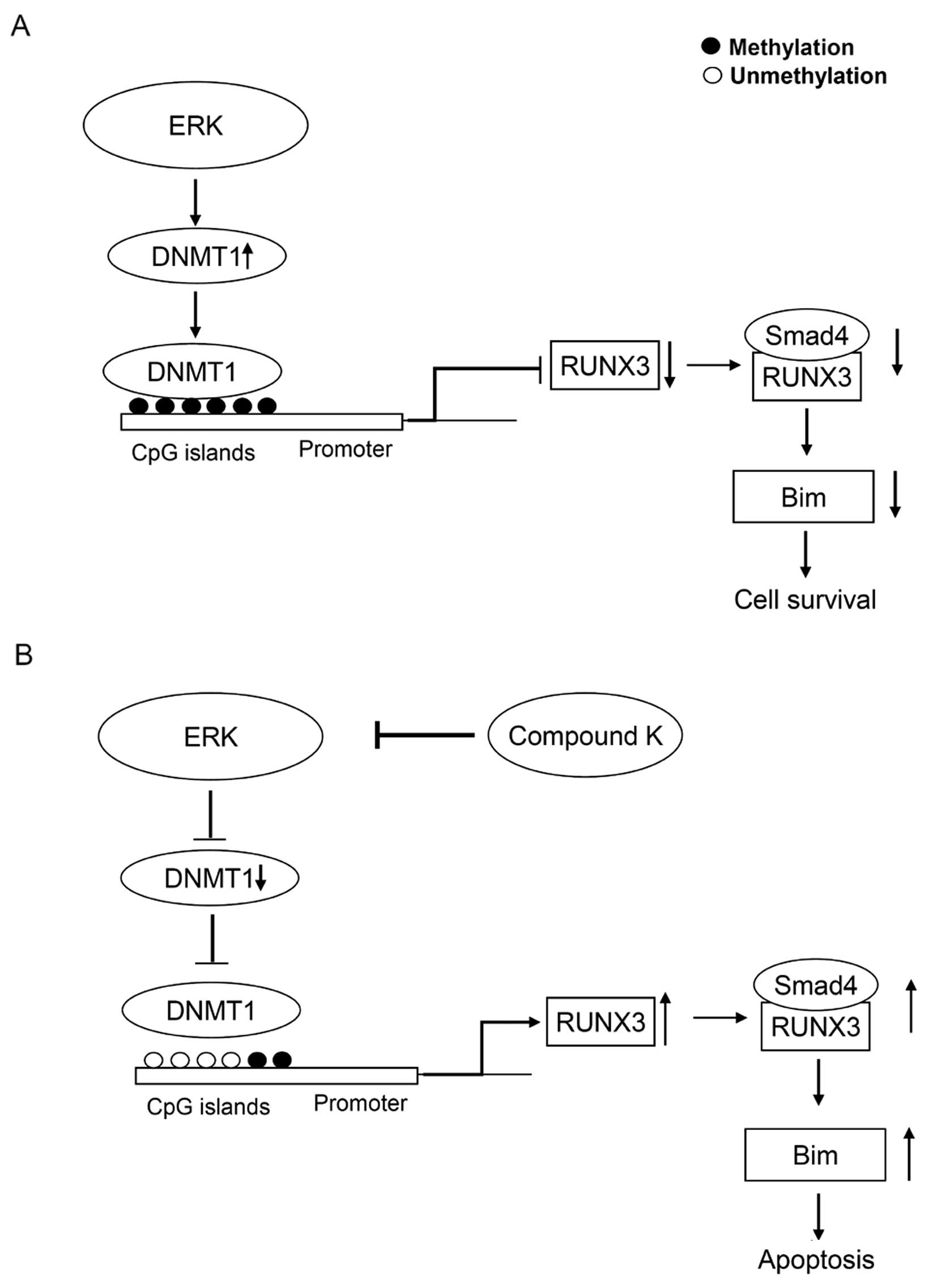
Although it remains unknown how ERK modulates DNMT1
protein levels, we propose a model in which compound K influences
the methylation status of RUNX3 (Fig.
7). In conclusion, compound K has potent demethylating
activity, mediated via inhibition of ERK-DNMT signaling. The lack
of toxicity associated with this ginseng saponin compound makes it
an excellent and novel candidate DNMT1 inhibitor for the
chemo-prevention or treatment of CRC.
Acknowledgements
This study was supported by a grant
from the National R&D Program for Cancer Control, Ministry for
Health and Welfare, Republic of Korea (1120340).
References
|
1.
|
Kudo S, Lambert R, Allen JI, et al:
Nonpolypoid neoplastic lesions of the colorectal mucosa.
Gastrointest Endosc. 68:S3–S47. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Smith JJ, Deane NG, Dhawan P and Beauchamp
RD: Regulation of metastasis in colorectal adenocarcinoma: A
collision between development and tumor biology. Surgery.
144:353–366. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Kinzler KW and Vogelstein B: Lessons from
hereditary colorectal cancer. Cell. 87:159–170. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Grady WM and Carethers JM: Genomic and
epigenetic instability in colorectal cancer pathogenesis.
Gastroenterology. 135:1079–1099. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Wong JJ, Hawkins NJ and Ward RL:
Colorectal cancer: a model for epigenetic tumorigenesis. Gut.
56:140–148. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Matos E and Brandani A: Review on meat
consumption and cancer in South America. Mutat Res.
506–507:243–249. 2002.PubMed/NCBI
|
|
7.
|
Nkondjock A and Ghadirian P: Associated
nutritional risk of breast and colon cancers: a population-based
case control study in Montreal, Canada. Cancer Lett. 223:85–91.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Fearon ER and Vogelstein B: A genetic
model for colorectal tumorigenesis. Cell. 61:759–767. 1990.
View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Grady WM: Genomic instability and colon
cancer. Cancer Metastasis Rev. 23:11–27. 2004. View Article : Google Scholar
|
|
10.
|
Herman JG and Baylin SB: Gene silencing in
cancer in association with promoter hypermethylation. N Engl J Med.
349:2042–2054. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Levanon D, Brenner O, Negreanu V, et al:
Spatial and temporal expression pattern of Runx3 (Aml2) and Runx1
(Aml1) indicates non-redundant functions during mouse
embryogenesis. Mech Dev. 109:413–417. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Woolf E, Xiao C, Fainaru O, et al: Runx3
and Runx1 are required for CD8 T cell development during
thymopoiesis. Proc Natl Acad Sci USA. 100:7731–7736. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Li QL, Kim HR, Kim WJ, et al:
Transcriptional silencing of the RUNX3 gene by CpG hypermethylation
is associated with lung cancer. Biochem Biophys Res Commun.
314:223–228. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Chuang LS and Ito Y: RUNX3 is
multifunctional in carcinogenesis of multiple solid tumors.
Oncogene. 29:2605–2615. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Ragnarsson G, Eiriksdottir G,
Johannsdottir JT, Jonasson JG, Egilsson V and Ingvarsson S: Loss of
heterozygosity at chromosome 1p in different solid human tumours:
association with survival. Br J Cancer. 79:1468–1474. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Tanaka K, Yanoshita R, Konishi M, et al:
Suppression of tumourigenicity in human colon carcinoma cells by
introduction of normal chromosome 1p36 region. Oncogene.
8:2253–2258. 1993.PubMed/NCBI
|
|
17.
|
Li QL, Ito K, Sakakura C, et al: Causal
relationship between the loss of RUNX3 expression and gastric
cancer. Cell. 109:113–124. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Goel A, Arnold CN, Tassone P, et al:
Epigenetic inactivation of RUNX3 in microsatellite unstable
sporadic colon cancers. Int J Cancer. 112:754–759. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Jair KW, Bachman KE, Suzuki H, et al: De
novo CpG island methylation in human cancer cells. Cancer Res.
66:682–692. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Rhee I, Bachman KE, Park BH, et al: DNMT1
and DNMT3b cooperate to silence genes in human cancer cells.
Nature. 416:552–556. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Lu R, Wang X, Chen ZF, Sun DF, Tian XQ and
Fang JY: Inhibition of the extracellular signal-regulated
kinase/mitogenactivated protein kinase pathway decreases DNA
methylation in colon cancer cells. J Biol Chem. 282:12249–12259.
2007. View Article : Google Scholar
|
|
22.
|
Santi DV, Norment A and Garrett CE:
Covalent bond formation between a DNA-cytosine methyltransferase
and DNA containing 5-azacytosine. Proc Natl Acad Sci USA.
81:6993–6997. 1984. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Gabbara S and Bhagwat AS: The mechanism of
inhibition of DNA (cytosine-5-)-methyltransferases by 5-azacytosine
is likely to involve methyl transfer to the inhibitor. Biochem J.
307:87–92. 1995.PubMed/NCBI
|
|
24.
|
Mack GS: Epigenetic cancer therapy makes
headway. J Natl Cancer Inst. 98:1443–1444. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Beisler JA: Isolation, characterization,
and properties of a labile hydrolysis product of the antitumor
nucleoside, 5-azacytidine. J Med Chem. 21:204–208. 1978. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Constantinides PG, Jones PA and Gevers W:
Functional striated muscle cells from non-myoblast precursors
following 5-azacytidine treatment. Nature. 267:364–366. 1977.
View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Lyko F and Brown R: DNA methyltransferase
inhibitors and the development of epigenetic cancer therapies. J
Natl Cancer Inst. 97:1498–1506. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Gaudet F, Hodgson JG, Eden A, et al:
Induction of tumors in mice by genomic hypomethylation. Science.
300:489–492. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Howard G, Eiges R, Gaudet F, Jaenisch R
and Eden A: Activation and transposition of endogenous retroviral
elements in hypomethylation induced tumors in mice. Oncogene.
27:404–408. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Deng T and Zhang Y: 5-Aza-2′-deoxycytidine
reactivates expression of RUNX3 by deletion of DNA
methyltransferases leading to caspase independent apoptosis in
colorectal cancer Lovo cells. Biomed Pharmacother. 63:492–500.
2009.
|
|
31.
|
Hong SY, Cho JY and Seo DW: Ginsenoside
Rp1 inhibits proliferation and migration of human lung cancer
cells. Biomol Ther. 19:411–418. 2011. View Article : Google Scholar
|
|
32.
|
Park JW, Lee JC, Ann S, et al: A fermented
ginseng extract, BST204, inhibits proliferation and motility of
human colon cancer cells. Biomol Ther. 19:211–217. 2011. View Article : Google Scholar
|
|
33.
|
Seo EY and Kim WK: Red ginseng extract
reduced metastasis of colon cancer cells in vitro and in vivo. J
Ginseng Res. 35:315–324. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Akao T, Kanaoka M and Kobashi K:
Appearance of compound K, a major metabolite of ginsenoside Rb1 by
intestinal bacteria, in rat plasma after oral
administration-measurement of compound K by enzyme immunoassay.
Biol Pharm Bull. 21:245–249. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Hasegawa H, Sung JH and Huh JH: Ginseng
intestinal bacterial metabolite IH901 as a new anti-metastatic
agent. Arch Pharm Res. 20:539–544. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Quan LH, Cheng LQ, Kim HB, et al:
Bioconversion of ginsenoside Rd into compound K by Lactobacillus
pentosus DC101 isolated from Kimchi. J Gins Res. 34:288–295.
2010. View Article : Google Scholar
|
|
37.
|
Kang KA, Kim YW, Kim SU, et al: G1 phase
arrest of the cell cycle by a ginseng metabolite, compound K, in
U937 human monocytic leukamia cells. Arch Pharm Res. 28:685–690.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Kang KA, Lim HK, Kim SU, et al: Induction
of apoptosis by ginseng saponin metabolite in U937 human monocytic
leukemia cells. J Food Biochem. 29:27–40. 2005. View Article : Google Scholar
|
|
39.
|
Chae S, Kang KA, Chang WY, et al: Effect
of compound K, a metabolite of ginseng saponin, combined with
gamma-ray radiation in human lung cancer cells in vitro and in
vivo. J Agric Food Chem. 57:5777–5782. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Kim AD, Kang KA, Zhang R, et al: Ginseng
saponin metabolite induces apoptosis in MCF-7 breast cancer cells
through the modulation of AMP-activated protein kinase. Environ
Toxicol Pharmacol. 30:134–140. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Ito K, Lim AC, Salto-Tellez M, et al:
RUNX3 attenuates beta-catenin/T cell factors in intestinal
tumorigenesis. Cancer Cell. 14:226–237. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
42.
|
Ku JL, Kang SB, Shin YK, et al: Promoter
hypermethylation downregulates RUNX3 gene expression in colorectal
cancer cell lines. Oncogene. 23:6736–6742. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
43.
|
Yano T, Ito K, Fukamachi H, et al: The
RUNX3 tumor suppressor upregulates Bim in gastric epithelial cells
undergoing transforming growth factor beta-induced apoptosis. Mol
Cell Biol. 26:4474–4488. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
44.
|
Tong DD, Jiang Y, Li M, et al: RUNX3
inhibits cell proliferation and induces apoptosis by
TGF-beta-dependent and -independent mechanisms in human colon
carcinoma cells. Pathobiology. 76:163–169. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45.
|
Kodach LL, Jacobs RJ, Heijmans J, et al:
The role of EZH2 and DNA methylation in the silencing of the tumour
suppressor RUNX3 in colorectal cancer. Carcinogenesis.
31:1567–1575. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46.
|
Lee IK, Kang KA, Lim CM, et al: Compound
K, a metabolite of ginseng saponin, induces mitochondria-dependent
and caspase-dependent apoptosis via the generation of reactive
oxygen species in human colon cancer cells. Int J Mol Sci.
11:4916–4931. 2010. View Article : Google Scholar
|
|
47.
|
Subramaniam MM, Chan JY, Soong R, et al:
RUNX3 inactivation by frequent promoter hypermethylation and
protein mislocalization constitute an early event in breast cancer
progression. Breast Cancer Res Treat. 113:113–121. 2009. View Article : Google Scholar
|
|
48.
|
Ito K, Liu Q, Salto-Tellez M, et al:
RUNX3, a novel tumor suppressor, is frequently inactivated in
gastric cancer by protein mislocalization. Cancer Res.
65:7743–7750. 2005.PubMed/NCBI
|
|
49.
|
Mani S and Herceg Z: DNA demethylating
agents and epigenetic therapy of cancer. Adv Genet. 70:327–240.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
50.
|
Howell PM, Liu Z and Khong HT:
Demethylating agents in the treatment of cancer. Pharmaceuticals.
3:2022–2044. 2010. View Article : Google Scholar
|
|
51.
|
Fang MZ, Wang Y, Ai N, et al: Tea
polyphenol (-)-epigallocatechin-3-gallate inhibits DNA
methyltransferase and reactivates methylation-silenced genes in
cancer cell lines. Cancer Res. 63:7563–7570. 2003.PubMed/NCBI
|
|
52.
|
Fang MZ, Chen D, Sun Y, Jin Z, Christman
JK and Yang CS: Reversal of hypermethylation and reactivation of
p16INK4a, RARbeta, and MGMT genes by genistein and other
isoflavones from soy. Clin Cancer Res. 11:7033–7041. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
53.
|
Fini L, Selgrad M, Fogliano V, et al:
Annurca apple polyphenols have potent demethylating activity and
can reactivate silenced tumor suppressor genes in colorectal cancer
cells. J Nutr. 137:2622–2628. 2007.PubMed/NCBI
|
|
54.
|
Kishikawa S, Murata T, Ugai H, Yamazaki T
and Yokoyama KK: Control elements of Dnmt1 gene are regulated in
cell-cycle dependent manner. Nucleic Acids Res (Suppl). 3:307–308.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
55.
|
Torquati A, O’rear L, Longobardi L, et al:
RUNX3 inhibits cell proliferation and induces apoptosis by
reinstating transforming growth factor beta responsiveness in
esophageal adenocarcinoma cells. Surgery. 136:310–316. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
56.
|
Yamamura Y, Lee WL, Inoue K, Ida H and Ito
Y: RUNX3 cooperates with FoxO3a to induce apoptosis in gastric
cancer cells. J Biol Chem. 281:5267–5276. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
57.
|
Zaidi SK, Sullivan AJ, van Wijnen AJ,
Stein JL, Stein GS and Lian JB: Integration of Runx and Smad
regulatory signals at transcriptionally active subnuclear sites.
Proc Natl Acad Sci USA. 99:8048–8053. 2002. View Article : Google Scholar : PubMed/NCBI
|