Introduction
Cervical cancer is the third most common cancer
among women worldwide (1) emerging
from pre-malignant precursor lesions that are gradually referred to
as cervical intraepithelial neoplasia (CIN), graded mild dysplasia
(CIN I), moderate dysplasia (CIN II) and severe dysplasia/carcinoma
in situ (CIN III). Cervical cancer shows a prevalence of
human papillomaviruses (HPV) of 99.7% (2). For humans, approximately 100
HPV-types have been described (3),
approximately 40 of which infect the anogenital tract (4), the most common carcinogenic types
being HPV16, 18, 31 and 45 (2,5,6). The
HPV life cycle requires infection of proliferating epithelial cells
and the ‘early’ proteins E5, E6 and E7 are known to foster
proliferation (6). During cervical
cancer development, the HPV genome may integrate into the host
genome, whereby parts of the HPV genome, including the E5 gene, are
deleted (7). The oncoproteins E6
and E7 can immortalize cells independently of each other but
transformation efficiency in the presence of both is increased
(8–11). Finally, during differentiation
‘late’ genes are expressed in the upper layers of the mucosa and
viral particles are produced and released (12). Despite this rather precise
understanding of the aetiology of carcinoma of the cervix, tumour
classification is often arduous to accomplish and there is still a
manifested need for better diagnostic and prognostic tools.
Tetraspanins represent a superfamily of
transmembrane proteins with 33 members in humans (13) that was first described in studies
of tumour associated proteins (14,15).
Tetraspanins are glycoproteins that consist of four transmembrane
domains, forming two hydrophilic extracellular domains as well as a
small intracellular loop. Both termini, carboxyl and amino, are
cytoplasmatic and the extracellular domains contain several
glycosylation sites (16–19). Tetraspanins are discriminated from
other integral 4-transmembrane proteins by virtue of a highly
conserved 4-amino acid motif within the large extracellular domain
and conserved cystein residues within each of the four
transmembrane domains (19). The
large extracellular domain consists of two subdomains that mediate
tetraspanin-tetraspanin homophilic binding or interactions with
other transmembrane proteins, respectively (20,21).
On their intracellular, membrane-proximal portion tetraspanins
contain cysteines that undergo palmitoylation, a feature deemed to
be important for protein-protein interactions (22). Tetraspanins are not only found at
the plasma membrane (PM), they also associate with membranes of
endo- and exosomes (23,24). Thus, via their large extracellular
domain and the intracellular palmitoylation moieties, tetraspanins
are able to bind and thereby form complexes with each other and
other transmembrane proteins, for instance members of the integrin
family (reviewed in ref. 25).
These tetraspanin network or tetraspanin enriched microdomains
(TEM) contribute to the organisation of cell surface proteins and
can influence signal transduction (26,27).
The role of tetraspanins in carcinogenesis and
cancer progression is probably multifaceted. For instance, levels
of the tetraspanin CD151 are elevated in several types of cancer
including high-grade breast cancer and poor-outcome endometrial
cancer and correlate with decreased survival (28–31).
Furthermore, CD151 is able to increase motility and invasiveness of
the breast cancer cell line MDA-MB-231 and depletion of CD151 leads
to reduced tumour progression in vivo (32). In contrast, tetraspanin CD9
inhibits cell motility and growth of the human lung adenocarcinoma
cell line MAC10 and the myeloma cell line ARH77 (33). An inverse correlation of CD9
expression versus metastasis was documented for breast cancer
(34) and reduction of CD9
expression correlates with poor prognosis in breast (35) or lung carcinoma (36). Beyond cancer, tetraspanins and TEMs
are involved in viral diseases. CD82 plays a role in hepatitis C
virus binding and infection (37).
Similarly the participation of TEMs containing CD9, CD63, CD81 and
CD82 has been reported in the context of HIV-1 infections (38). Recently, the involvement of TEMs
including CD63 and CD151 in the course of HPV16 infection was
documented (39).
The tetraspanin family member tetraspanin 1 (Tspan1,
NET-1) is upregulated in human hepatocellular carcinoma (42), gastric carcinoma (43), colorectal adenocarcinoma (44) and ovarian carcinomas (45), suggesting that tetraspanin 1 may be
involved in genesis and progression of various forms of neoplasias.
In line with this possibility tetraspanin 1 is important for
proliferation and invasion of various human carcinoma cell lines
(46,47). We have previously reported that
tetraspanin 1 expression is markedly increased at the RNA and
protein level in high-grade CIN and cervical cancer (40,41),
but its role in cervical cancer biology has remained obscure. In
the present study we investigated the function of tetraspanin 1 in
2 cervical cancer cell lines, looking both at functional effects
and the possible involvement of tetraspanin 1 in growth factor
signal transduction. Our data argue against a function of
tetraspanin 1 as a genuine signal transducing protein and rather
highlight an effect on cervical cancer cell invasion processes.
Materials and methods
Reagents and antibodies
The antibodies used for western blotting and
immunohistochemistry are listed in Table I. All antibodies except for Tspan1
were commercially available. For the generation of Tspan1
antibodies, the major extracellular domain of Tspan1 was expressed
as a histidine tagged fusion protein in E. coli (41). Gel purified protein was injected
intraperitoneally into Balb/c mice. The mice were boosted 2 and 4
weeks after immunization. Serum was collected 3 days after the last
boost to assess the immune response by indirect immune-fluorescence
on Tspan1 transfected Cos-7 cells. Mouse splenocytes were fused
with SP2/0 myeloma cells according to standard procedures.
Hybridomas producing antibodies with reactivity to Tspan1 were
selected by ELISA. Clone 4/12 was found to be suitable both for
immunocyto- and immunohistochemistry. Secondary peroxidase-coupled
antibodies were purchased from Dianova. EGF and poly-L-lysine were
purchased from Sigma-Aldrich and extracellular matrix proteins
(ECM)-Proteins fibronectin, laminin, collagen type I and III from
Millipore.
 | Table I.Antibodies used in the present
study. |
Table I.
Antibodies used in the present
study.
| Protein | Antibody | Species/type | Dilution | Company |
|---|
| EGF-receptor | α-EGFR-IgG |
Rabbit/polyclonal | 1:1,000 | Cell Signaling |
|
α-pEGFR(Y1068)-IgG |
Rabbit/polyclonal | 1:1,000 | Invitrogen |
| Erk1/2 | α-Erk1/2-IgG |
Rabbit/polyclonal | 1:1,000 | Cell Signaling |
|
α-pErk1/2(T202/Y204)-IgG |
Mouse/monoclonal | 1:1,000 | Cell Signaling |
| Focal | α-FAK-IgG |
Rabbit/polyclonal | 1:1,000 | Cell Signaling |
| Adhesion |
α-pFAK(Y397)-IgG |
Rabbit/polyclonal | 1:1,000 | Cell Signaling |
| Kinase |
α-pFAK(Y576/577)-IgG |
Rabbit/polyclonal | 1:1,000 | Cell Signaling |
|
α-pFAK(Y925)-IgG |
Rabbit/polyclonal | 1:1,000 | Cell Signaling |
| Paxillin | α-Paxillin-IgG |
Rabbit/polyclonal | 1:1,000 | Santa Cruz
Biotechnology |
|
α-pPaxillin(Y118)-IgG |
Rabbit/polyclonal | 1:1,000 | Cell Signaling |
| Ras | α-pan-Ras-IgG |
Mouse/monoclonal | 1:1,000 | Calbiochem |
| α-K-Ras-IgG |
Mouse/monoclonal | 1:1,000 | Santa Cruz
Biotechnology |
| α-N-Ras-IgG |
Mouse/monoclonal | 1:1,000 | Santa Cruz
Biotechnology |
| α-H-Ras-IgG |
Mouse/monoclonal | 1:1,000 | Santa Cruz
Biotechnology |
| Tetraspanin 1 | α-C4.8/NET-1-IgG
#4/12 |
Mouse/monoclonal | 1:1,000 | Jena University
Hospital
Department of Gynecology |
|
p16INK4a | α-
p16INK4a -IgG |
Mouse/monoclonal | Ready to use
CINtec®
Histology kit | MTM
Laboratories
Heidelberg |
| MMP14 | α-MMP14-IgG |
Rabbit/polyclonal | 1:1,000 | Abcam |
Cell culture
SiHa and HeLa cells (ATCC) were cultured in DMEM
(Gibco) supplemented with 10% fetal calf serum (FCS)
(Sigma-Aldrich) under 37°C, 95% humidity and 5% CO2
content. Experiments were done with cell passages 5–20.
Adenoviral transduction
Cells (4×105) seeded in 60 mm or
106 cells in 100 mm dishes were infected with adenovirus
particles coding for EGFP or tetraspanin 1, respectively, at a
multiplicity of infection of 10. Twenty-four hours later medium was
replaced and cells were cultured additional 24 h and processed for
further experiments. Transduction efficiency, as determined by
fluorescence microscopy (EGFP) and immunocytochemistry with the
monoclonal antibody clone 4/12 against tetraspanin 1, ranged
between 80 and 90%.
Matrix invasion
Cell invasion was determined using Matrigel coated
cell culture inserts (8-μm pore size for 24-well cell
culture plates) (BD Biosciences, BD Falcon™) following the
manufacturer’s instructions. Briefly, Matrigel was diluted 1:3 in
serum-free DMEM and 30 μl diluted Matrigel were poured into
each insert and allowed to gelatinise for 1 h at 37°C. The upper
chamber was filled with 105 of untransduced, EGFP or
tetraspanin 1 expressing SiHa cells in 200 μl growth medium
(containing 10% FCS) while the lower compartment contained 500
μl growth medium. After 72 h cells at the lower membrane
side were rinsed with double distilled water (ddH2O) and
fixed with ice-cold 80% ethanol for 20 min. Wells were washed with
ddH2O and matrix material and remaining cells inside the
insert were thoroughly removed with a cotton tip. The fixed cells
were stained with 0.1% crystal violet for 5 min and washed once
with ddH2O. Stained cells were decolourised with 10%
acetic acid and absorbance was measured at 630 nm (48). Four independent experiments with
4-fold determinations were carried out.
Proliferation assay
Untransduced, EGFP or tetraspanin 1 expressing SiHa
cells (2×104) were seeded on a 24-well cell culture
plate. Cells were cultured for 24 h and deprived of serum for 6 h.
Medium was changed to DMEM supplemented with 10% FCS and
proliferation was determined 6, 24, 48 and 72 h later by using the
CellTiter 96® Non-Radioactive Cell Proliferation assay
(Promega) following the manufacturer’s protocol. For precise
quantification, four independent experiments with duplicates for
SiHa and five independent experiments with triplicates and
duplicates for EGFP and tetraspanin 1, respectively, were performed
at 6 h after serum re-addition.
Colony formation assay
Agarose (0.5%) (5 ml) in DMEM supplemented with FCS
were added to each well of a 6-well cell culture plate and allowed
to gelatinise for 3 h at room temperature. Agarose (0.3%) (3 ml) in
DMEM supplemented with 10% FCS and containing 5×104
untransduced, EGFP or tetraspanin 1 expressing SiHa cells was
poured on top of the agarose layer. Cells were cultured over 3
weeks and foci were scored manually under a microscope. Four
independent experiments with triplicates were done.
Cell adhesion
Cell culture plates (96-well) (BD Falcon™, BD
Biosciences) were coated with 100 μl/well of 150
μg/ml Poly-L-lysine or 10 μg/ml fibronectin, laminin,
collagen type I and III (Millipore), respectively. Proteins were
diluted in sterile PBS and coated overnight at room temperature
followed by blocking with 100 μl/well 2% bovine serum
albumin (BSA) in PBS for 2 h at room temperature and 2 washes with
200 μl/well PBS. Untransduced, EGFP or tetraspanin 1
expressing SiHa cells (2×104) were added to each well
and incubated for 2 h. Wells were washed twice with PBS and cells
were stained with 0.1% crystal violet for 5 min followed by 2
washes with PBS. Stained cells were de-stained with 10% acetic acid
and absorbance was measured at 630 nm. Four independent experiments
with 4-fold determination were done.
Immunohistochemistry and
-cytochemistry
For immunohistochemistry, tissue arrays with
paraffin embedded samples of normal cervical tissue, low- and
high-grade cervical lesions and cervical cancer tissues were
stained with a monoclonal antibody, clone 4/12, against tetraspanin
1 and antibodies against p16INK4a. Signal detection was
performed using EnVison™ Detection System (Dako). For
immunocytochemistry, adenovirally transduced SiHa cells were
spotted on a glass slide and fixed with 4% paraformaldehyde in
TBS-Tween (50 mM Tris, 150 mM NaCl, 0.1% Tween-20). Fixed cells
were incubated with 0.6% hydrogen peroxide for 7 min followed by 20
min blocking with goat serum, 1:5 diluted in TBS-Tween. Cells were
incubated with monoclonal antibody against tetraspanin 1 for 1 h
and bound antibodies were detected with the EnVison Detection
System (Dako). Between every incubation step a 5 min wash step with
TBS-Tween was implemented. All steps were done at room
temperature.
Cell stimulation and western
blotting
Growth factor stimulation: 4×105 cells
seeded on 60 mm cell culture dishes were subjected to adenoviral
transduction as described above and deprived of serum overnight
prior to stimulation with 100 ng/ml EGF. Cells were lysed in 500
μl ice-cold lysis buffer: 50 mM Tris, 50 mM NaCl, 5 mM
MgCl2, 1 mM EGTA, 1% NP-40, 10% glycerol, protease
inhibitor mix HP (Serva) and phosphatase inhibitors. Extracts were
cleared by centrifugation and the supernatants were processed by
SDS-PAGE separation and western blotting using the antibodies
listed in Table I.
Integrin activation: 60-mm cell culture dishes were
coated with matrix proteins as described before. Adenovirally
transduced and untransduced cells were starved for 16 h in
serum-free DMEM, detached by trypsinization and resuspended in
serum-free DMEM supplemented with 10 μg/ml trypsin inhibitor
(Gibco, Invitrogen) and 2% BSA. Cells were rotated for 1 h at 37°C
and 3×105 cells in 1 ml were added to each dish. Cells
were cultivated for 2 h and lysed with 100 μl ice-cold lysis
solution: 50 mM Tris; 150 mM NaCl, 5 mM MgCl2, 1 mM
EGTA, 1% NP-40, protease inhibitor Mix HP (Serva) and phosphatase
inhibitors. Lysates were cleared by centrifugation and processed
for SDS-PAGE and western blotting.
Expression of matrix metalloproteinases
(MMPs)
Expression of MMPs was analysed: i) by
semiquantitative RT-PCR for one sample set (control, EGFP and
tetraspanin 1 expressing SiHa and HeLa cells, respectively), ii) by
western blotting of two sample sets, and iii) via
immunohistochemistry with tissue arrays. Methods were described in
detail previously (49).
Ethic statement
This study was approved by the ethics committee of
the Friedrich-Schiller University Jena (reference number
175-02/00).
Statistical analysis
Statistical significance was determined by
non-parametric, independent homogeneity tests Kruskal-Wallis test
for more than two groups and Mann-Whitney test for two groups,
using analysis software SPSS Statistics 19 (IBM).
Results
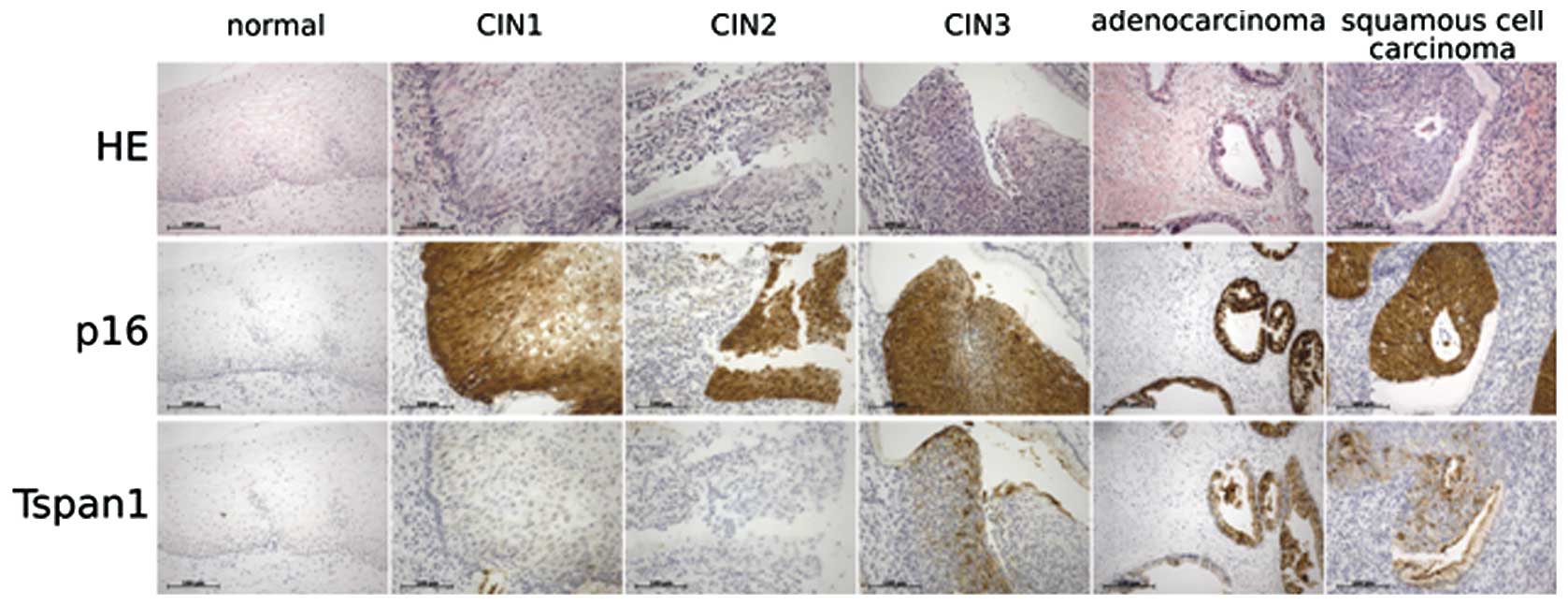
Tetraspanin 1 is upregulated at the
growth front of CIN III and cervical cancer
We have previously reported that tetraspanin 1
expression becomes detectable earliest in high-grade cervical
neoplasia and continues in cervical cancer (40,41),
but its precise role in cancer development remains unclear. To
determine more accurately the expression pattern of tetraspanin 1
in cervical cancer, tissue arrays were analysed by
immunohistochemistry using the newly generated monoclonal antibody
#4/12 (see Materials and methods). The arrays contained normal
cervical tissues, low- and high-grade cervical lesions and cervical
carcinoma tissue. As expected from our previous analyses using
polyclonal antibodies, tetraspanin 1 expression was not detectable
in normal cervical tissue and low-grade cervical intraepithelial
lesions (CIN) I and II (Fig. 1 and
data not shown). Expression of tetraspanin 1 was, however,
extensive in high-grade CIN III and cervical cancer tissue.
Overall, tetraspanin 1 expression was rather heterogeneous but
there was a marked, consistent accumulation of tetraspanin 1 at
growth fronts of CIN III and cancer tissues (Fig. 1). In these images dysplastic tumour
areas are highlighted by the presence of p16INK4a, which
accumulates as a consequence of HPV E7 protein action. These data
point to a potential participation of tetraspanin 1 in cell
proliferation or invasive growth of dysplastic cells in
vivo.
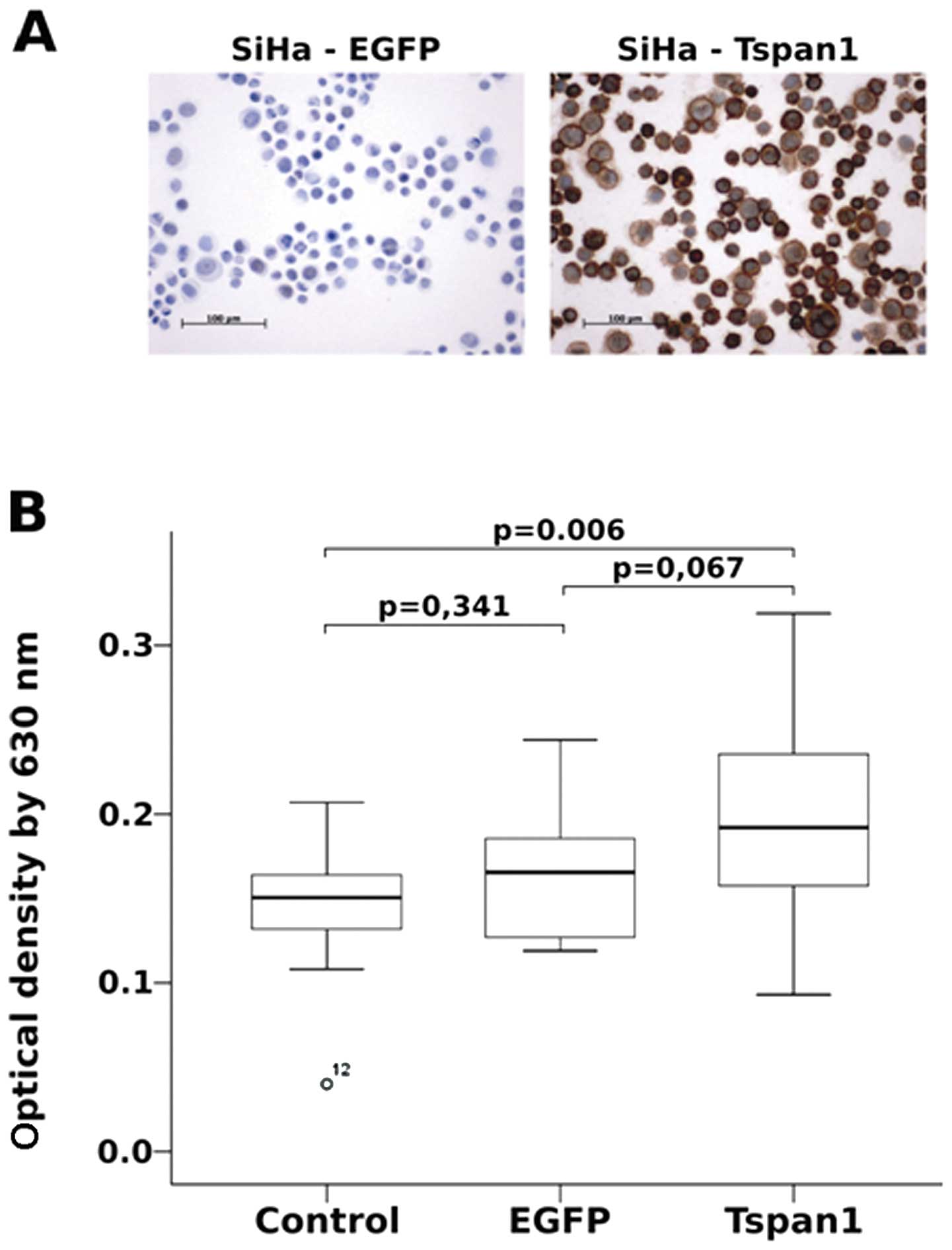
Tetraspanin 1 enhances cervical cancer
cell invasion
The expression of tetraspanin 1 at the growth front
of high-grade cervical neoplasias and cervical cancer suggested
that tetraspanin 1 might play a role in tissue invasion. To test
this hypothesis we analysed the invasive potential of 2 cervical
carcinoma cell lines in the presence and absence of tetraspanin 1.
The cervical carcinoma cell lines SiHa and HeLa, which are devoid
of tetraspanin 1 (Fig. 2A and data
not shown), were transduced with an adenovirus driving tetraspanin
1 expression (Fig. 2A). As shown
in Fig. 2B tetraspanin 1
significantly increased the invasion of SiHa cells through matrigel
as compared to untransduced or EGFP-transduced SiHa cells.
Virtually the same effect was observed in HeLa cells (data not
shown). These findings supported the notion that tetraspanin 1 was
involved in the modulation of cell invasion of cervical cancer
cells.
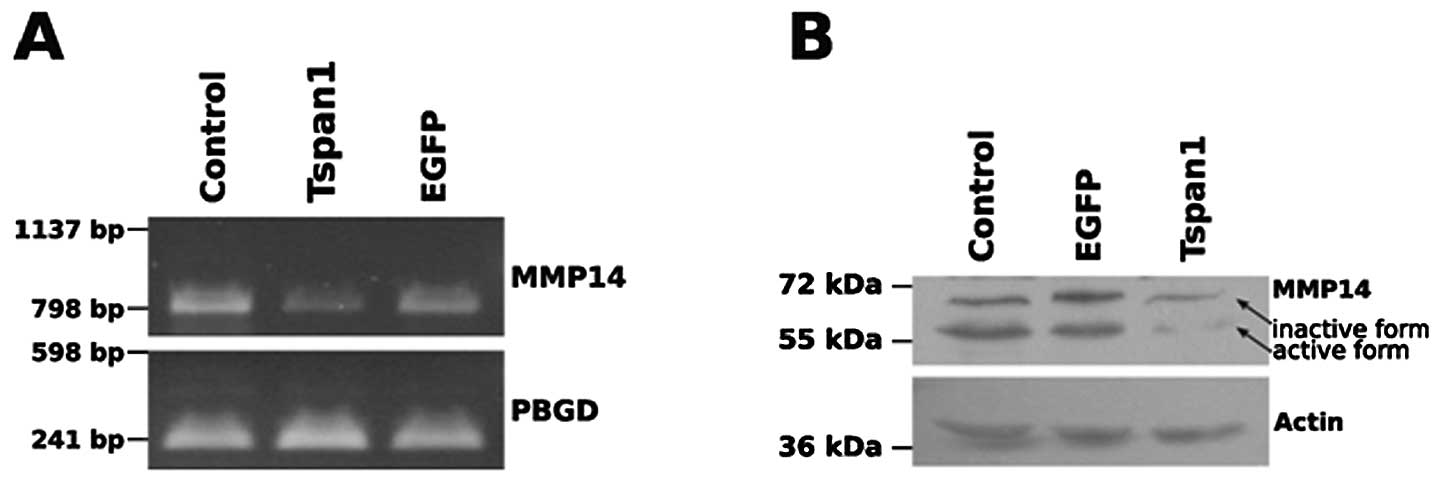
Control of MMP expression by tetraspanin
1
Tissue invasion by carcinoma cells involves the
aberrant expression of enzymes in the remodelling of the
extracellular space. MMPs are often upregulated in transformed
cells concomitantly with an increase in their invasive properties
(reviewed in ref. 50). To learn
whether or not the effect of tetraspanin 1 on cell invasion was due
to altered MMP expression we monitored the expression of all 23
known human MMP isoforms in SiHa cells in the presence/absence of
tetraspanin 1 by conventional PCR and at the protein level via
western blot analysis of cell extracts (49) (data not shown). The expression
screen at the mRNA level revealed a decreased expression of MMP14
[or membrane-type 1 MMP (MT1-MMP)] in SiHa cells expressing
tetraspanin 1 (Fig. 3A). Reduced
MMP14 expression in response to tetraspanin 1 expression was also
detected at the protein level (Fig.
3B) and affected in particular the proteolytically processed,
activate form of MMP14 (Fig. 3B).
These findings revealed an inhibitory effect of tetraspanin 1 on
the expression and activation levels of MMP14. At first sight these
results appear counter-intuitive with regard to the enhanced
invasiveness of tetraspanin 1 expressing SiHa cells. Indeed,
immunohistochemistry of cervical cancer samples revealed no
differences in MMP14 expression between tetraspanin 1-positive and
-negative areas (data not shown), suggesting that the
down-modulation of MMPs observed in SiHa cells did not reflect the
situation in the native tumour and was therefore probably not
relevant to the physiological function of tetraspanin 1 in
carcinoma of the cervix. Taken together, we concluded that the
increased ability of SiHa cells to invade Matrigel following the
forced expression of tetraspanin 1 is unlikely to result from an
altered MMP expression pattern in cervical cancer cells.
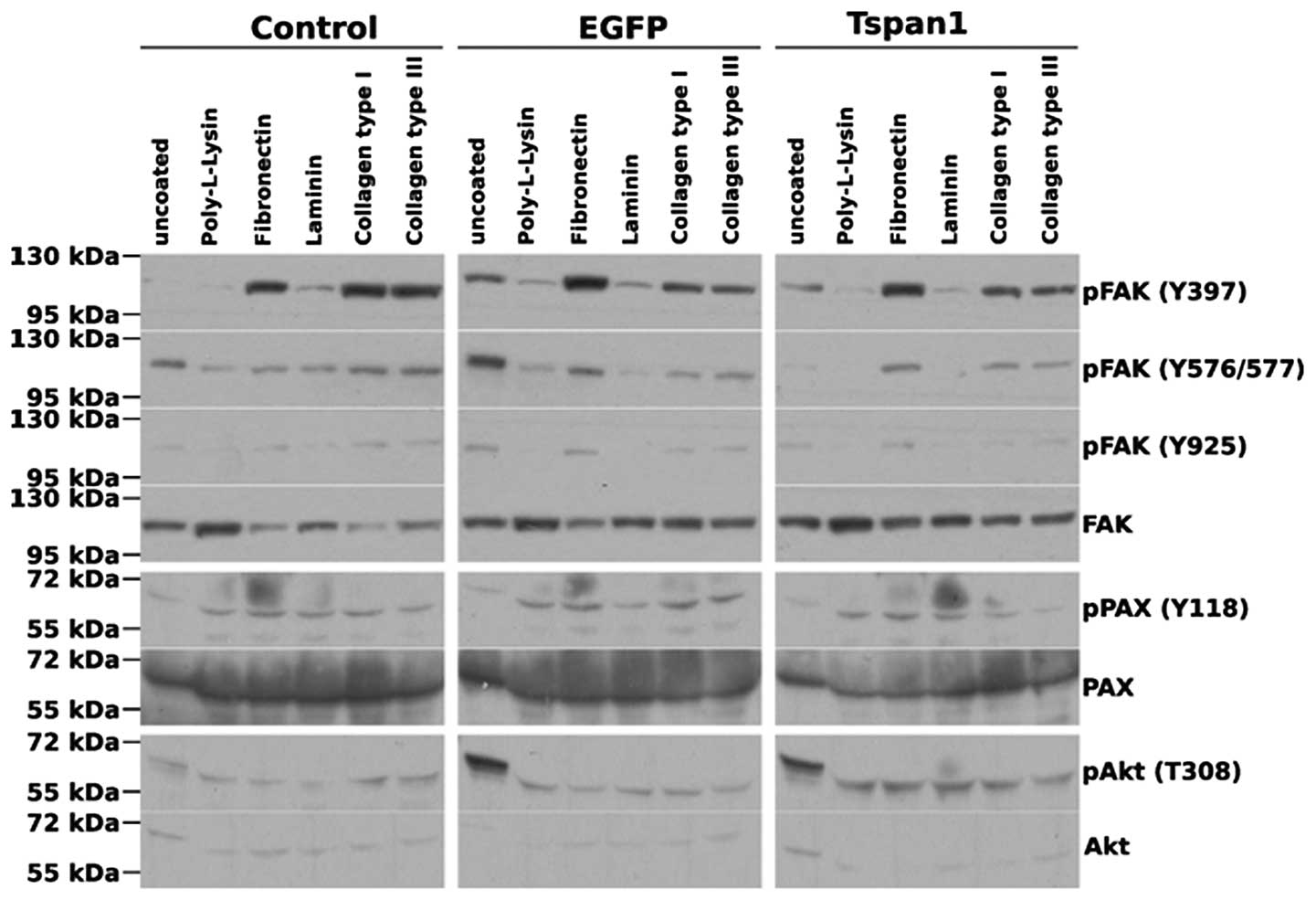
Tetraspanin 1 does not mediate outside-in
integrin signalling to focal adhesion kinase
Beyond the release of MMP activity, cell
invasiveness requires the concerted regulation of numerous cell
biological processes including the spatiotemporal modulation of
integrin-dependent signalling and adhesion. Since other members of
the tetraspanin family have been linked to the control of integrin
activity in the past (reviewed in ref. 25), we moved on to investigate the
effect of tetraspanin 1 overexpression on signalling events
downstream of integrins. Focal adhesion kinase (FAK) is a major,
pivotal transducer of integrin outside-in signals. Adhesion of SiHa
cells to fibronectin or collagen type I or III, but not laminin or
the inert, charged polymer poly-L-lysine, lead to an activation of
FAK, as illustrated by the increase in the phosphorylation of
various key tyrosine residues (Fig.
4). Intriguingly, FAK activation/phosphorylation did not
translate into paxillin phosphorylation at Tyr118, a bona
fide phosphorylation site of FAK (51), suggesting that in SiHa cells
paxillin phosphorylation is under control of factors other than the
activation status of FAK. Importantly, overexpression of
tetraspanin 1 did not markedly affect the phosphorylation pattern
of FAK in adhered cells (Fig. 4),
strongly indicating that tetraspanin 1 did not regulate cell
invasiveness by modulating or transmitting integrin sparked
signals.
Effects of tetraspanin 1 on cell
adhesion
Since tetraspanin 1 did not affect FAK
phosphorylation in response to matrix adhesion we hypothesized that
cell adhesion itself would not be grossly affected by tetraspanin
1. To test this assert and since aberrant cell adhesion is a
hallmark of invasive transformed cells, we measured cell adhesion
of SiHa cells to the extracellular matrix (ECM) proteins
fibronectin, laminin and collagen type I and III in dependency of
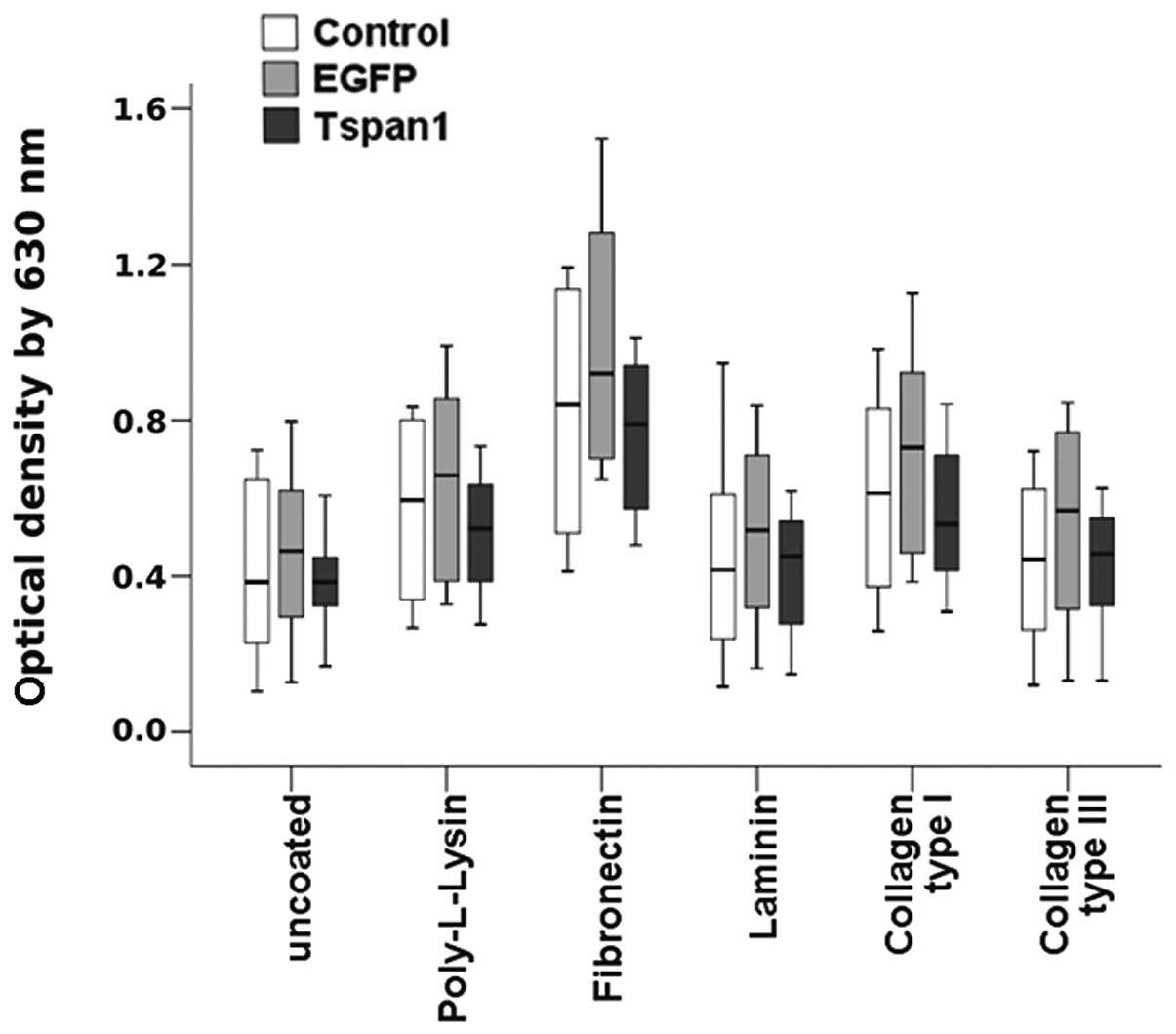
tetraspanin 1 expression. No significant differences in adhesion
strength to any of the tested ECM proteins, or to uncoated and
poly-L-lysine coated dishes, were detected between tetraspanin 1
expressing and untransduced SiHa cells (Fig. 5). These data confirmed the findings
shown above and argued against a role of tetraspanin 1 in the
control of carcinoma of the cervix cell adhesion to ECM
proteins.
Tetraspanin 1 does not affect cell
proliferation in culture or in semi-solid media
Previous studies have documented the involvement of
tetraspanins in the control of cell proliferation (15). Since proliferation and
motility/invasion are intimately intertwined processes we wished to
analyse the influence of tetraspanin 1 on tumour cell-specific cell
proliferation. To this end, we first assessed growth of SiHa cells
in a semi-solid medium, as an indicator of aberrant growth
properties. Next, EGFP or tetraspanin 1 transduced SiHa cells and
untransduced cells were seeded in semi-solid media and colony
numbers and size distribution were assessed 3 days later. As
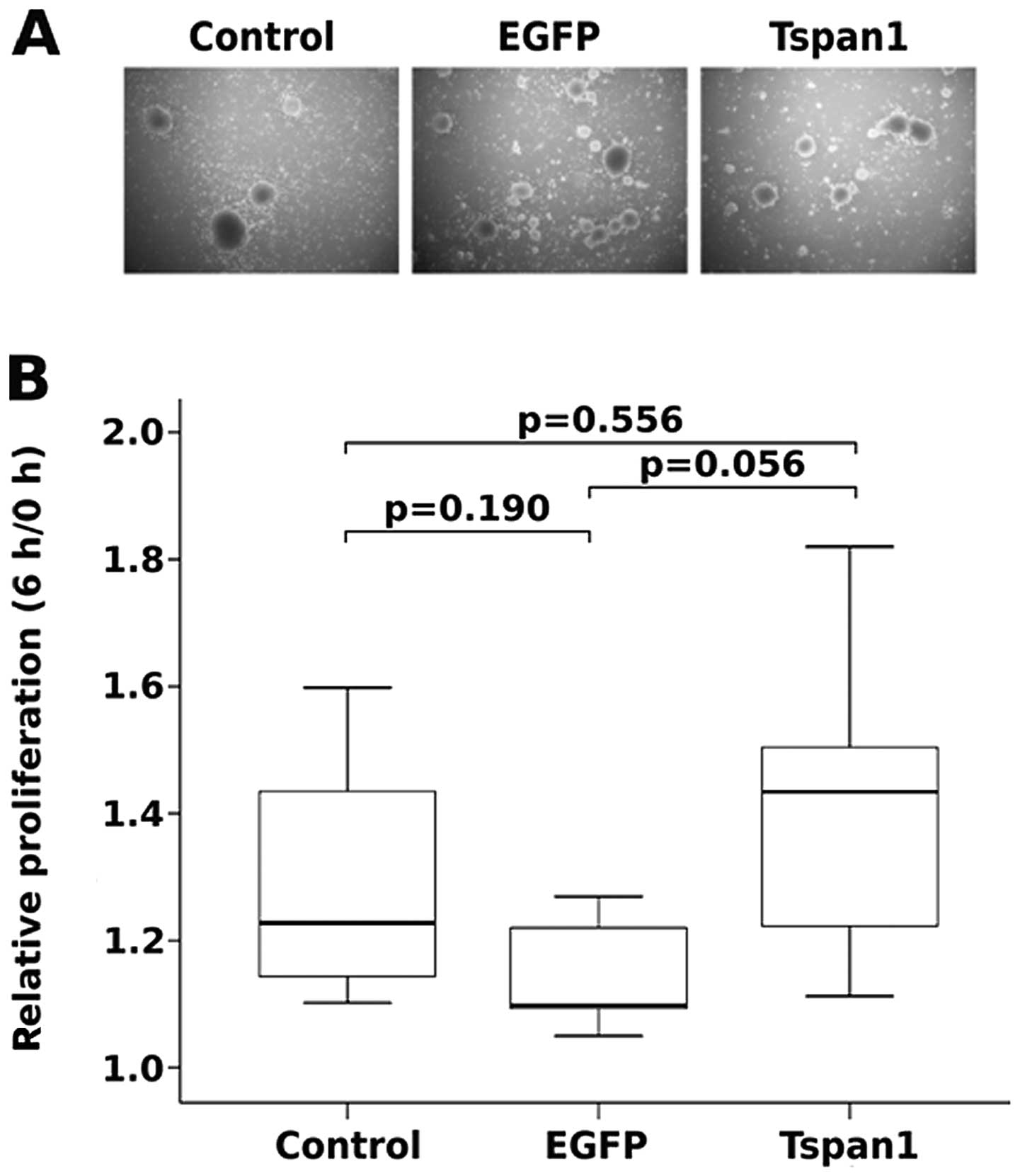
illustrated in Fig. 6A no
significant differences in colony numbers or foci size were
observed between native, EGFP or tetraspanin 1 expressing SiHa
cells indicating that tetraspanin 1 did not enhance
anchorage-independent proliferation of SiHa cells. To test whether
or not tetraspanin 1 affected proliferation of cervical cancer
cells in standard cell culture, native, EGFP or tetraspanin 1
expressing SiHa cells were subjected to a proliferation assay for
72 h (data not shown) and for detailed analysis 6 h (Fig. 6B). Again, no differences in
proliferation among the various cell populations could be
discerned. These data suggested that tetraspanin 1 does not affect
proliferation, either anchorage-dependent or -independent, of
cervical cancer cells.
Involvement of tetraspanin 1 in growth
factor signalling
Since tetraspanin 1 did not upregulate cervical
cancer cell proliferation, we tested whether or not tetraspanin 1
mediated signalling by EGF, a major mitogen of cervical carcinoma
cell lines (52–54). EGF induces a very strong activation
of Ras and its downstream effector Erk1/2 in these cells (55,56).
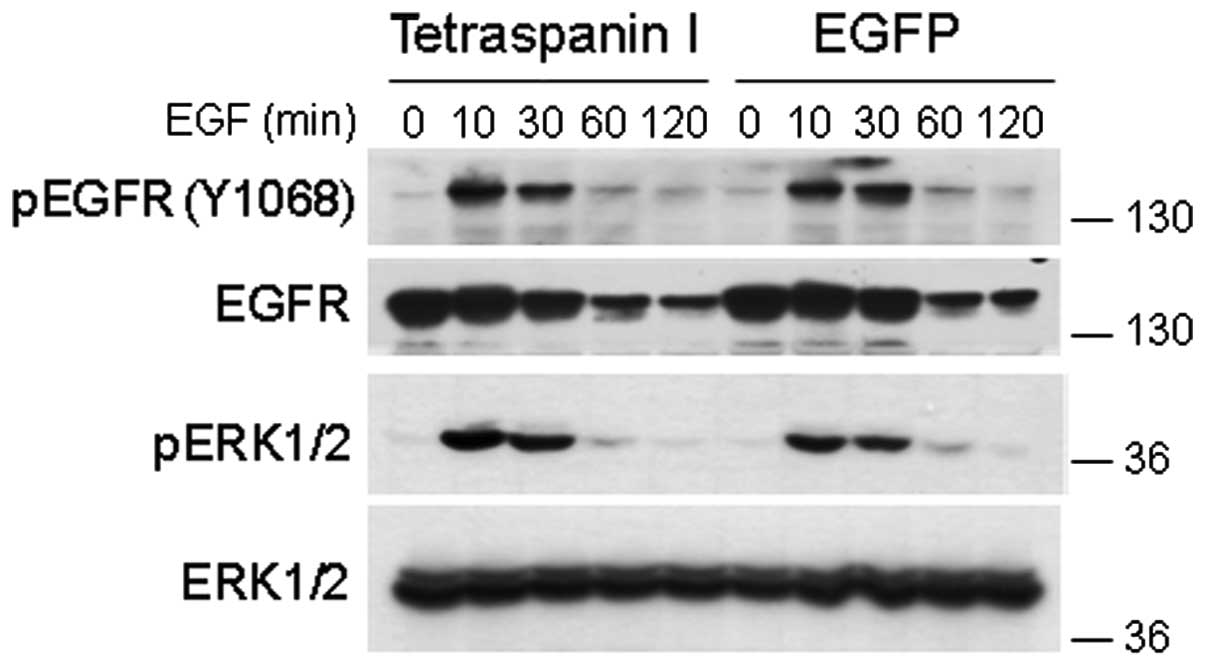
EGF-triggered activation of Erk1/2 in SiHa (Fig. 7) and HeLa cells (data not shown),
monitored by assessing phosphorylation of Erk, was not altered in
the presence of tetraspanin 1. Consistently, tetraspanin 1
expression did not affect the extent of agonist-induced
EGF-receptor (EGFR) autophosphorylation, nor did it affect EGFR
internalization and degradation, as highlighted by the virtually
identical time-dependent reduction of the EGFR signal (Fig. 7). An absence of tetraspanin 1
effects on EGF-signalling was also observed upstream of Erk1/2, at
the level of Ras-GTP formation (data not shown). In conclusion,
these findings documented that tetraspanin 1 was not involved in
receptor-proximal signalling events downstream of the epidermal
growth factor receptor.
Discussion
Tetraspanins are integral components of distinct
multiprotein membrane microdomains where they interact with other
tetraspanins and other membrane proteins such as major
histo-compatibility complex (MHC) (57) or integrins (26,58–60)
depending on cellular context, tetraspanin isoform and other
parameters. These tetraspanin enriched microdomains (TEMs)
reportedly play a role in the control of diverse cell biological
phenomena such as cell proliferation (15), cell-cell adhesion (61), cell motility and migration
(58,62) or signal transduction (63,64).
In the present study we investigated the role of tetraspanin 1 in
cervical cancer cells. Previously, we have reported a prominent
overexpression of tetraspanin 1 in high-grade cervical
intraepithelial neoplasia (CIN) and cervical cancer when compared
to normal cervical tissue and low-grade CIN. In particular,
tetraspanin 1 expression was associated with neoplastic cell
proliferation and undifferentiated squamous cell cancers (40,41).
In an attempt to determine the role of tetraspanin 1 in cervical
cancer, we have used the cervical cancer cell lines SiHa and HeLa,
both of which lack detectable tetraspanin 1 expression and studied
the consequences of forcing tetraspanin 1 expression for a number
of cell biological read-outs. Our data document a role for
tetraspanin 1 as a positive regulator of cervical cell motility and
invasion, a function that could underlie the observed accumulation
of tetraspanin 1 at the growth border of late stage cancers
(Fig. 1). This notion is not
unprecedented since siRNA-mediated knockdown of tetraspanin 1
expression has previously been observed to severely affect
migration and invasion in the A431 and HCT-8 carcinoma lines
(46,47), suggesting that tetraspanin 1 might
be a global regulator of cell motility in numerous types of cancer.
Thus, beyond pointing to a potentially relevant function of
tetraspanin 1, these experiments lend support to the possibility of
using tetraspanin 1 as a potential marker for disease progression.
However, it is likely that tetraspanin 1 may not be the only
tetraspanin variant involved in the regulation of cell motility and
invasion. Overexpression of the tetraspanin CD151 stimulated
migration of HeLa cells (65) and
promoted migration and invasion in prostate cancer cell lines LNCaP
and PC3 (66). Another tetraspanin
family member, CD9, is expressed in normal cervical epithelium and
downregulated in cervical cancer, but re-expressed in invasive
regions of the tumour (67).
Considering all data together, it is tempting to speculate that
distinct combinations of tetraspanins could act in concert in
distinct cell types to regulate and govern various aspects related
to cancer cell motility, adhesion and migration.
In order to understand the mechanism by which
tetraspanin 1 regulates the invasive properties of SiHa and HeLa
cells we focused on the expression of MMPs. While we did observe a
downregulation of matrix metalloproteinase 14 (MMP14, MT1-MMP) at
RNA and protein level in tetraspanin 1 expressing SiHa cells
(Fig. 3), immunohistochemical
staining of cervical cancer samples did not show any tetraspanin
1-dependent changes in MMP14 levels (data not shown), suggesting
that changes in MMP expression levels did probably not mediate the
effect of tetraspanin 1 on cell motility. A previous report linked
the downregulation of MMP14 following the knockdown of tetraspanins
CD9, CD81 and TSPAN2 in the breast cancer cell line MCF-7 to a
reduced invasiveness and colony formation in fibronectin and
collagen gels (68), suggesting
that the role of MMPs in the control of tetraspanin-dependent
cellular process varies depending on the cell type. Interestingly,
the downregulation or inhibition of MMP14 is not only expected to
dictate the extent of ECM component degradation, but has also been
shown to affect, by unknown means, the phosphorylation/activation
status of intracellular focal adhesion components FAK and Akt
(69). Spatiotemporal modulation
of integrin-dependent signalling and adhesion is indeed of
paramount importance for correct cell motility and invasion. Our
analysis of ECM induced activation of FAK, paxillin and Akt, all
critical mediators of outside-in integrin signalling, evidenced no
significant differences between cervical carcinoma cells lacking or
harbouring tetraspanin 1. Again, this scenario differs from reports
in the literature where the knockdown of other tetraspanins, such
as CD151 in MDA-MB-231 cells, lead to a reduction of global FAK
phosphorylation and to reduced activity of other downstream
mediators of integrin signalling like Rac (32). Thus, we hypothesize that the
particular mode of action of distinct tetraspanins is likely to
vary markedly depending on the tetraspanin species and the cell
type under investigation.
For proper cell migration and tissue invasion, cells
need to interact with the extracellular matrix via dynamic adhesion
complexes and integrin-tetraspanin complexes are components of
these structures (70). However,
we observed no tetraspanin 1-dependent alteration in adhesion to
the matrix polypeptides tested (Fig.
5), which lead us to conclude that the control of cell
motility/invasion by tetraspanin 1 was not a consequence of changes
in the adhesive properties of the cells. We are currently
investigating whether or not tetraspanin 1 affects the GDP/GTP
loading status of small GTPases known to be involved in the
regulation of cell polarity, cell shape and actin cytoskeleton
dynamics. It is worth to note at this point that, as before, the
reported effects of tetraspanins on cellular adhesion are rather
conflictive and hard to combine in a unifying model framework. For
instance, CD9 enhances integrin-dependent adhesion of the colon
carcinoma cell line Colo320 (71)
and the tetraspanin CD82 stimulates cell adhesion of ovarian cancer
cell lines (72). By contrast
other carcinoma lines exhibit decreased adhesion to laminin and
fibronectin following forced expression of the CD82 (73,74).
Since tetraspanins have also been implicated in the
direct or indirect control of cell proliferation (71,75,76)
we wished to understand to what extent tetraspanin 1 played a role
in anchorage-dependent or independent cell proliferation of
cervical cancer cells. The results shown in Fig. 6A argued clearly against a role of
tetraspanin 1 as a driver of aberrant cell proliferation in
carcinoma of the cervix. Interestingly, other carcinoma cell lines
appear to rely heavily on tetraspanin 1 for proliferation (46,47),
evidencing once more that tetraspanin 1 is likely to fulfil
different functions in different tissues. In line with the lack of
an obvious effect on cell growth, tetraspanin 1 expression did not
affect signal transduction pathways engaged by EGF (a known mitogen
of SiHa and HeLa cells), corroborating that tetraspanin 1 is not a
constituent of mitogenic signalling pathways.
In conclusion, in the present study we document an
increased invasiveness of cervical cancer cell lines driven by the
expression of tetraspanin 1. At the same time our data argue that
tetraspanin 1 is not a genuine signalling mediator of adhesive or
mitogenic pathways in cervical carcinoma cells. While the precise
molecular mechanisms underlying the control of the cellular
motility machinery by tetraspanin 1 remain obscure, our present
observations confirm previous findings (40,41)
and identify tetraspanin 1 as a potentially useful marker for
discriminating neoplasias with low versus high progression
potency.
Acknowledgements
We thank Ute Wittig for excellent
technical assistance. We greatly appreciate the advice and support
given by Hans-Walter Zentgraf for generating monoclonal
antibodies.
References
|
1.
|
GLOBOCAN 2008 (IARC), Section of Cancer
Information. International Agency for Research on Cancer 2008;
(accessed 13/12/2012).
|
|
2.
|
Walboomers J, Jacobs M, Manos M, et al:
Human papilloma-virus is a necessary cause of invasive cervical
cancer worldwide. J Pathol. 189:12–19. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
de Villiers E, Fauquet C, Broker T,
Bernard H and zur Hausen H: Classification of papillomaviruses.
Virology. 324:17–27. 2004.
|
|
4.
|
zur Hausen H: Papillomaviruses causing
cancer: evasion from host-cell control in early events in
carcinogenesis. J Natl Cancer Inst. 92:690–698. 2000.PubMed/NCBI
|
|
5.
|
Munoz N, Bosch F, de Sanjose S, et al:
Epidemiologic classification of human papillomavirus types
associated with cervical cancer. N Engl J Med. 348:518–527. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Munoz N, Castellsague X, de Gonzalez AB
and Gissmann L: Chapter 1: HPV in the etiology of human cancer.
Vaccine. 24(Suppl 3): S3/1–10. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Schwarz E, Freese U, Gissmann L, Mayer W,
Roggenbuck B, Stremlau A and Hausen H: Structure and transcription
of human papillomavirus sequences in cervical-carcinoma cells.
Nature. 314:111–114. 1985. View
Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Phelps W, Yee C, Munger K and Howley P:
The human papillomavirus type-16 E7 gene encodes transactivation
and transformation functions similar to those of adenovirus-E1a.
Cell. 53:539–547. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Munger K, Phelps W, Bubb V, Howley P and
Schlegel R: The E6-gene and E7-gene of the human papillomavirus
type-16 together are necessary and sufficient for transformation of
primary human keratinocytes. J Virol. 63:4417–4421. 1989.PubMed/NCBI
|
|
10.
|
Hawleynelson P, Vousden KH, Hubbert NL,
Lowy DR and Schiller JT: HPV16 E6-proteins and E7-proteins
cooperate to immortalize human foreskin keratinocytes. EMBO J.
8:3905–3910. 1989.PubMed/NCBI
|
|
11.
|
Halbert C, Demers G and Galloway D: The E7
gene of human papillomavirus type-16 is sufficient for
immortalization of human epithelial cells. J Virol. 65:473–478.
1991.PubMed/NCBI
|
|
12.
|
zur Hausen H: Papillomaviruses and cancer:
from basic studies to clinical application. Nat Rev Cancer.
2:342–350. 2002.PubMed/NCBI
|
|
13.
|
Huang S, Yuan S, Dong M, et al: The
phylogenetic analysis of tetraspanins projects the evolution of
cell-cell interactions from unicellular to multicellular organisms.
Genomics. 86:674–684. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Hotta H, Ross A, Huebner K, et al:
Molecular-cloning and characterization of an antigen associated
with early stages of melanoma tumor progression. Cancer Res.
48:2955–2962. 1988.PubMed/NCBI
|
|
15.
|
Oren R, Takahashi S, Doss C, Levy R and
Levy S: Tapa-1, the target of an antiproliferative antibody,
defines a new family of transmembrane proteins. Mol Cell Biol.
10:4007–4015. 1990.PubMed/NCBI
|
|
16.
|
Boucheix C, Benoit P, Frachet P, Billard
M, Worthington R, Gagnon J and Uzan G: Molecular-cloning of the Cd9
antigen - a new family of cell-surface proteins. J Biol Chem.
266:117–122. 1991.PubMed/NCBI
|
|
17.
|
Levy S, Nguyen VQ, Andria ML and Takahashi
S: Structure and membrane topology of tapa-1. J Biol Chem.
266:14597–14602. 1991.PubMed/NCBI
|
|
18.
|
Wright MD and Tomlinson MG: The ins and
outs of the trans-membrane-4 superfamily. Immunol Today.
15:588–594. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Maecker HT, Todd SC and Levy S: The
tetraspanin superfamily: Molecular facilitators. FASEB J.
11:428–442. 1997.PubMed/NCBI
|
|
20.
|
Kitadokoro K, Bordo D, Galli G, et al:
CD81 extracellular domain 3D structure: Insight into the
tetraspanin superfamily structural motifs. EMBO J. 20:12–18. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Seigneuret M, Delaguillaumie A,
Lagaudriere-Gesbert C and Conjeaud H: Structure of the tetraspanin
main extracellular domain - a partially conserved fold with a
structurally variable domain insertion. J Biol Chem.
276:40055–40064. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Charrin S, Manie S, Oualid M, Billard M,
Boucheix C and Rubinstein E: Differential stability of
tetraspanin/tetraspanin interactions: Role of palmitoylation. FEBS
Lett. 516:139–144. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Escola JM, Kleijmeer MJ, Stoorvogel W,
Griffith JM, Yoshie O and Geuze HJ: Selective enrichment of
tetraspan proteins on the internal vesicles of multivesicular
endosomes and on exosomes secreted by human B-lymphocytes. J Biol
Chem. 273:20121–20127. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Wubbolts R, Leckie RS, Veenhuizen PTM, et
al: Proteomic and biochemical analyses of human B cell-derived
exosomes - potential implications for their function and
multivesicular body formation. J Biol Chem. 278:10963–10972. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Boucheix C and Rubinstein E: Tetraspanins.
Cell Mol Life Sci. 58:1189–1205. 2001. View Article : Google Scholar
|
|
26.
|
Rubinstein E, Le Naour F,
Lagaudrière-Gesbert C, Billard M, Conjeaud H and Boucheix C: CD9,
CD63, CD81 and CD82 are components of a surface tetraspan network
connected to HLA-DR and VLA integrins. Eur J Immunol. 26:2657–2665.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Berditchevski F, Odintsova E, Sawada S and
Gilbert E: Expression of the palmitoylation-deficient CD151 weakens
the association of alpha(3)beta(1) integrin with the
tetraspanin-enriched micro-domains and affects integrin-dependent
signaling. J Biol Chem. 277:36991–37000. 2002. View Article : Google Scholar
|
|
28.
|
Tokuhara T, Hasegawa H, Hattori N, et al:
Clinical significance of CD151 gene expression in non-small cell
lung cancer. Clin Cancer Res. 7:4109–4114. 2001.PubMed/NCBI
|
|
29.
|
Ang J, Lijovic M, Ashman LK, Kan K and
Frauman AG: CD151 protein expression predicts the clinical outcome
of low-grade primary prostate cancer better than histologic
grading: a new prognostic indicator? Cancer Epidemiol Biomarkers
Prev. 13:1717–1721. 2004.PubMed/NCBI
|
|
30.
|
Sadej R, Romanska H, Baldwin G, et al:
CD151 regulates tumori-genesis by modulating the communication
between tumor cells and endothelium. Mol Cancer Res. 7:787–798.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Voss MA, Gordon N, Maloney S, et al:
Tetraspanin CD151 is a novel prognostic marker in poor outcome
endometrial cancer. Br J Cancer. 104:1611–1618. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Yang XH, Richardson AL, Torres-Arzayus MI,
et al: CD151 accelerates breast cancer by regulating alpha(6)
integrin function, signaling and molecular organization. Cancer
Res. 68:3204–3213. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Ikeyama S, Koyama M, Yamaoko M, Sasada R
and Miyake M: Suppression of cell motility and metastasis by
transfection with human motility-related protein (mrp-1/cd9) dna. J
Exp Med. 177:1231–1237. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Miyake M, Nakano K, Ieki Y, et al:
Motility related protein-1 (mrp-1/cd9) expression - inverse
correlation with metastases in breast cancer. Cancer Res.
55:4127–4131. 1995.PubMed/NCBI
|
|
35.
|
Miyake M, Nakano K, Itoi S, Koh T and Taki
T: Motility-related protein-1 (MRP-1/CD9) reduction as a factor of
poor prognosis in breast cancer. Cancer Res. 56:1244–1249.
1996.PubMed/NCBI
|
|
36.
|
Higashiyama M, Taki T, Ieki Y, et al:
Reduced motility related protein-1 (mrp-1/cd9) gene-expression as a
factor of poor-prognosis in non-small-cell lung cancer. Cancer Res.
55:6040–6044. 1995.PubMed/NCBI
|
|
37.
|
Pileri P, Uematsu Y, Campagnoli S, et al:
Binding of hepatitis C virus to CD81. Science. 282:938–941. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Nydegger S, Khurana S, Krementsov DN, Foti
M and Thali M: Mapping of tetraspanin-enriched microdomains that
can function as gateways for HIV-1. J Cell Biol. 173:795–807. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Spoden G, Freitag K, Husmann M, Boller K,
Sapp M, Lambert C and Florin L: Clathrin- and caveolin-independent
entry of human papillomavirus type 16-involvement of
tetraspanin-enriched microdomains (TEMs). Plos One. 3:e33132008.
View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Nees M, van Wijngaarden E, Bakos E,
Schneider A and Durst M: Identification of novel molecular markers
which correlate with HPV-induced tumor progression. Oncogene.
16:2447–2458. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Wollscheid V, Kuhne-Heid R, Stein I,
Jansen L, Kollner S, Schneider A and Durst M: Identification of a
new proliferation-associated protein NET-1/C4.8 characteristic for
a subset of high-grade cervical intraepithelial neoplasia and
cervical carcinomas. Int J Cancer. 99:771–775. 2002. View Article : Google Scholar
|
|
42.
|
Chen L, Wang Z, Zhan X, Li D, Zhu Y and
Zhu J: Association of NET-1 gene expression with human
hepatocellular carcinoma. Int J Surg Pathol. 15:346–353. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
43.
|
Chen L, Li X, Wang G, Wang Y, Zhu Y and
Zhu J: Clinicopathological significance of overexpression of
TSPAN1, K167 and CD34 in gastric carcinoma. Tumori. 94:531–538.
2008.PubMed/NCBI
|
|
44.
|
Chen L, Zhu Y, Zhang X, et al: TSPAN1
protein expression: a significant prognostic indicator for patients
with colorectal adenocarcinoma. World J Gastroenterol.
15:2270–2276. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45.
|
Scholz C, Kurzeder C, Koretz K, Windisch
J, Kreienberg R, Sauer G and Deissler H: Tspan-1 is a tetraspanin
preferentially expressed by mucinous and endometrioid subtypes of
human ovarian carcinomas. Cancer Lett. 275:198–203. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
46.
|
Chen L, Yuan D, Zhao R, Li H and Zhu J:
Suppression of TSPAN1 by RNA interference inhibits proliferation
and invasion of colon cancer cells in vitro. Tumori. 96:744–750.
2010.PubMed/NCBI
|
|
47.
|
Chen L, Zhu Y, Li H, et al: Knockdown of
TSPAN1 by RNA silencing and antisense technique inhibits
proliferation and infiltration of human skin squamous carcinoma
cells. Tumori. 96:289–295. 2010.PubMed/NCBI
|
|
48.
|
Saito K, Oku T, Ata N, Miyashiro H,
Hattori M and Saiki I: A modified and convenient method for
assessing tumor cell invasion and migration and its application to
screening for inhibitors. Biol Pharm Bull. 20:345–348. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
49.
|
Koehrmann A, Kammerer U, Kapp M, Dietl J
and Anacker J: Expression of matrix metalloproteinases (MMPs) in
primary human breast cancer and breast cancer cell lines: New
findings and review of the literature. BMC Cancer. 9:1882009.
View Article : Google Scholar : PubMed/NCBI
|
|
50.
|
Stamenkovic I: Matrix metalloproteinases
in tumor invasion and metastasis. Semin Cancer Biol. 10:415–433.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
51.
|
Bellis SL, Miller JT and Turner CE:
Characterization of tyrosine phosphorylation of paxillin in vitro
by focal adhesion kinase. J Biol Chem. 270:17437–17441. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
52.
|
Ueda M, Ueki M, Terai Y, Morimoto A, Fujii
H, Yoshizawa K and Yanagihara T: Stimulatory effects of EGF and
TGF-alpha on invasive activity and 5′-deoxy-5-fluorouridine
sensitivity in uterine cervical-carcinoma SKG-IIIb cells. Int J
Cancer. 72:1027–1033. 1997.PubMed/NCBI
|
|
53.
|
Ueda M, Fujii H, Yoshizawa K, Terai Y,
Kumagai K, Ueki K and Ueki M: Effects of EGF and TGF-alpha on
invasion and proteinase expression of uterine cervical
adenocarcinoma OMC-4 cells. Invasion Metastasis. 18:176–183. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
54.
|
Narayanan R, Kim HN, Narayanan NK, Nargi D
and Narayanan B: Epidermal growth factor-stimulated human cervical
cancer cell growth is associated with EGFR and cyclin D1
activation, independent of COX-2 expression levels. Int J Oncol.
40:13–20. 2012.
|
|
55.
|
Beeser A, Jaffer ZM, Hofmann C and
Chernoff J: Role of group A p21-activated kinases in activation of
extracellular-regulated kinase by growth factors. J Biol Chem.
280:36609–36615. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
56.
|
Antonyak MA, Li B, Regan AD, Feng Q,
Dusaban SS and Cerione RA: Tissue transglutaminase is an essential
participant in the epidermal growth factor-stimulated signaling
pathway leading to cancer cell migration and invasion. J Biol Chem.
284:17914–17925. 2009. View Article : Google Scholar
|
|
57.
|
Angelisova P, Hilgert I and Horejsi V:
Association of four antigens of the tetraspans family (CD37, CD53,
TAPA-1, and R2/C33) with MHC class II glycoproteins.
Immunogenetics. 39:249–256. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
58.
|
Rubinstein E, Lenaour F, Billard M,
Prenant M and Boucheix C: Cd9 antigen is an accessory subunit of
the vla integrin complexes. Eur J Immunol. 24:3005–3013. 1994.
View Article : Google Scholar : PubMed/NCBI
|
|
59.
|
Berditchevski F, Bazzoni G and Hemler ME:
Specific association of Cd63 with the vla-3 and vla-6 integrins. J
Biol Chem. 270:17784–17790. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
60.
|
Berditchevski F, Zutter MM and Hemler ME:
Characterization of novel complexes on the cell surface between
integrins and proteins with 4 transmembrane domains (TM4 proteins).
Mol Biol Cell. 7:193–207. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
61.
|
Masellissmith A, Jensen GS, Seehafer JG,
Slupsky JR and Shaw ARE: Anti-Cd9 monoclonal-antibodies induce
homotypic adhesion of pre-B cell-lines by a novel mechanism. J
Immunol. 144:1607–1613. 1990.PubMed/NCBI
|
|
62.
|
Miyake M, Koyama M, Seno M and Ikeyama S:
Identification of the motility-related protein (mrp-1), recognized
by monoclonal-antibody M31-15, which inhibits cell motility. J Exp
Med. 174:1347–1354. 1991. View Article : Google Scholar : PubMed/NCBI
|
|
63.
|
Olweus J, Lundjohansen F and Horejsi V:
Cd53, a protein with 4 membrane-spanning domains, mediates
signal-transduction in human monocytes and B-cells. J Immunol.
151:707–716. 1993.PubMed/NCBI
|
|
64.
|
Schick MR, Nguyen VQ and Levy S:
Anti-tapa-1 antibodies induce protein-tyrosine phosphorylation that
is prevented by increasing intracellular thiol levels. J Immunol.
151:1918–1925. 1993.
|
|
65.
|
Testa JE, Brooks PC, Lin JM and Quigley
JP: Eukaryotic expression cloning with an antimetastatic monoclonal
antibody identifies a tetraspanin (PETA-3/CD151) as an effector of
human tumor cell migration and metastasis. Cancer Res.
59:3812–3820. 1999.
|
|
66.
|
Ang J, Fang B, Ashman LK and Frauman AG:
The migration and invasion of human prostate cancer cell lines
involves CD151 expression. Oncol Rep. 24:1593–1597. 2010.PubMed/NCBI
|
|
67.
|
Sauer G, Windisch J, Kurzeder C, Heilmann
V, Kreienberg R and Deissler H: Progression of cervical carcinomas
is associated with down-regulation of CD9 but strong local
re-expression at sites of transendothelial invasion. Clin Cancer
Res. 9:6426–6431. 2003.PubMed/NCBI
|
|
68.
|
Lafleur MA, Xu D and Hemler ME:
Tetraspanin proteins regulate membrane type-1 matrix
metalloproteinase-dependent pericellular proteolysis. Mol Biol
Cell. 20:2030–2040. 2009. View Article : Google Scholar
|
|
69.
|
Takino T, Watanabe Y, Matsui M, Miyamori
H, Kudo T, Seiki M and Sato H: Membrane-type 1 matrix
metalloproteinase modulates focal adhesion stability and cell
migration. Exp Cell Res. 312:1381–1389. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
70.
|
Berditchevski F and Odintsova E:
Characterization of integrintetraspanin adhesion complexes: Role of
tetraspanins in integrin signaling. J Cell Biol. 146:477–492. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
71.
|
Ovalle S, Gutierrez-Lopez MD, Olma N, et
al: The tetraspanin CD9 inhibits the proliferation and
tumorigenicity of human colon carcinoma cells. Int J Cancer.
121:2140–2152. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
72.
|
Ruseva Z, Geiger PXC, Hutzler P, et al:
Tumor suppressor KAI1 affects integrin alphavbeta3-mediated ovarian
cancer cell adhesion, motility, and proliferation. Exp Cell Res.
315:1759–1771. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
73.
|
He B, Liu L, Cook GA, Grgurevich S,
Jennings LK and Zhang XA: Tetraspanin CD82 attenuates cellular
morphogenesis through down-regulating integrin alpha 6-mediated
cell adhesion. J Biol Chem. 280:3346–3354. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
74.
|
Lee H, Park I, Byun H, Jeoung D, Kim Y and
Lee H: Metastasis suppressor KAI1/CD82 attenuates the matrix
adhesion of human prostate cancer cells by suppressing fibronectin
expression and beta(1) integrin activation. Cell Physiol Biochem.
27:575–586. 2011. View Article : Google Scholar
|
|
75.
|
Murayama Y, Miyagawa JI, Oritani K, et al:
CD9-mediated activation of the p46 shc isoform leads to apoptosis
in cancer cells. J Cell Sci. 117:3379–3388. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
76.
|
Ko E, Lee IY, Cheon IS, et al: Monoclonal
antibody to CD9 inhibits platelet-induced human endothelial cell
proliferation. Mol Cells. 22:70–77. 2006.PubMed/NCBI
|