Introduction
Development of cancers are affected by a number of
factors including environment and genetic background (1). Difference in incidence of certain
cancers between Asian and Caucasian populations is well-recognized
(2,3). Environmental and/or genetic factors
may contribute to these differences. For example, a high incidence
of gastric cancers have been reported in Asian countries including
Japan (3,4). However, the incidence of gastric
cancers in Japanese immigrants to Hawaii is much lower than the
frequency in Japanese in their homeland (4,5).
This high incidence of gastric cancer appears to be strongly
influenced by environmental factors. In contrast, Asian non-smoking
women have a high incidence of non-small cell lung cancer (NSCLC)
associated with an EGFR mutation irrespective of the country where
they live (6,7), suggesting a germline predisposition
for NSCLC with this mutation.
Chronic lymphocytic leukemia (CLL) is a common
hematological malignancy in Western countries and it is very rare
in Asian countries, including Japan (8–10).
Furthermore, Asians including Japanese immigrants to USA continue
to have a low incidence of CLL (11,12).
This difference in incidence of CLL between Asians
and Caucasians is not clear. One possible explanation for the
difference may be that CLL in these two distinct ethnic groups show
two distinctly different molecular signatures. Another possible
explanation may be that environmental factors, for example an
infectious disease, could cause the development of CLL and affect
the incidence of this disease in two geographically distinct
regions. This seems less likely because Asians in USA also have a
low incidence of CLL (11,12). A third possibility is germline
variation in the general population of these two ethnic groups
leads to the difference in the incidence of the development of
CLL.
To identify molecular signatures of Caucasian and
Asian CLL at the DNA level, we analyzed these cells using high
resolution single nucleotide polymorphism (SNP) genomic
oligonucleotide microarrays.
Materials and methods
Samples
Diagnosis of CLL was made based on the updated
National Cancer Institute-Working Group guidelines (13). All leukemic cells expressed CD5,
CD19 and CD20. Rearrangement of the immunoglobulin heavy chain gene
was confirmed by PCR as described previously (14). Detailed information of the Japanese
CLL has been reported previously (14). Leukemic cells were collected from
77 cases of Asian CLL (75 Japanese and 2 Chinese) and 55 Caucasian
CLL after their informed consents were obtained. Institutional
committees approved the experiments undertaken. DNA was extracted
using Qiagen DNA extraction kit according to the manufactuer’s
protocol.
SNP-chip analysis
The extracted DNAs were digested with Nsp-I
restriction enzyme, amplified and hybridization with
GeneChip250NspI chip from Affymetrix. Hybridized signals were
captured on GeneChip Scanner 3000 and the data were analyzed by
CNAG3.3 software (15,16). Significant differences were
analyzed using χ2 tests. P-values <0.05 were
considered statistically significant.
Results
Caucasian and Asian CLL shared the major
genomic copy number changes
We analyzed 55 cases of Caucasian CLL and 77 cases
of Asian CLL and found a number of genomic abnormalities in both
groups. The four well characterized genomic abnormalities of CLL,
including deletion of 13q14, trisomy 12, deletion/UPD of 17p and
deletion/UPD of 11q (ATM) (17,18),
were detected in both groups at a comparable frequency except for
deletion/UPD of 11q. Asian v/s Caucasian CLL had deletion of
13q14.3 (Asian: 39 cases, 51%; Caucasian: 26 cases, 48%), trisomy
12 (Asian: 15 cases, 20%; Caucasian: 16 cases, 30%), abnormalities
of 11q (Asian: 21 cases, 27%; Caucasian: 5 cases, 10%) and
abnormalities of 17p (Asian: 11 cases, 15%; Caucasian: 3 cases, 5%)
(Table I). Only the frequency of
11q was statistically different (P<0.01). Interestingly,
deletion of 13q14 and trisomy 12 was mutually exclusive in Asian
CLL cases (no cases with both trisomy 12 and 13q14 in the 77 Asian
CLL) and nearly same in Caucasian CLL (2 of the 55).
 | Table IFour common genomic abnormalities in
Asian and Caucasian CLL. |
Table I
Four common genomic abnormalities in
Asian and Caucasian CLL.
| Asian CLL (77) | Caucasian CLL
(55) | P-value |
|---|
| 13q14 deletion | 39 (51%) | 26 (48%) | 0.60 |
| (sole 13q14
deletion) | 12 (16%) | 8 (15%) | 0.66 |
| 11q
deletion/UPD | 21 (27%) | 5 (10%) | <0.01a |
| 17p
deletion/UPD | 11 (15%) | 3 (5%) | 0.06 |
| Trisomy 12 | 15 (20%) | 16 (30%) | 0.09 |
Other genomic alterations involved four regions:
trisomy 3/duplication of 3q; trisomy 18/duplication of 18q;
deletion of 18p; and deletion of 8p (Table II). Asians more frequently had
either trisomy 3/duplication (dup) of 3q or trisomy 18/dup18q
compared to the Caucasian CLL samples (18 Asian v.s. 0 Caucasian
CLL sample) (Table II).
 | Table IIOther common genomic abnormalities
found in Asian and Caucasian CLL. |
Table II
Other common genomic abnormalities
found in Asian and Caucasian CLL.
| Asian CLL (77) | Caucasian CLL
(55) | P-value |
|---|
| Trisomy 3/Dup
3q | 6 (8%) | 0 (0%) | <0.01a |
| Trisomy 18/Dup
18q | 8 (11%) | 0 (0%) | <0.01a |
| Del 8p | 4 (7%) | 1 (2%) | 0.11 |
| Del 18p | 3 (4%) | 3 (5%) | 0.47 |
Novel candidate target genes associated
with development of CLL
In this study, we found novel tumor suppressor
candidate genes often deleted in CLL. Deletion of 11q is frequently
detected in CLL, involving the ATM gene (26/132 cases in total);
however, of interest, these 26 cases also had deletion of the
microRNA (miR) 34b/34c (Fig. 1A
and data not shown). A second novel finding was a homozygous
deletion of 11q22.3 involving caspase 1/4/5 genes in an Asian CLL
sample (Fig. 1A); and seven
additional cases had hemizygous deletion of these caspase genes
(Fig. 1A).
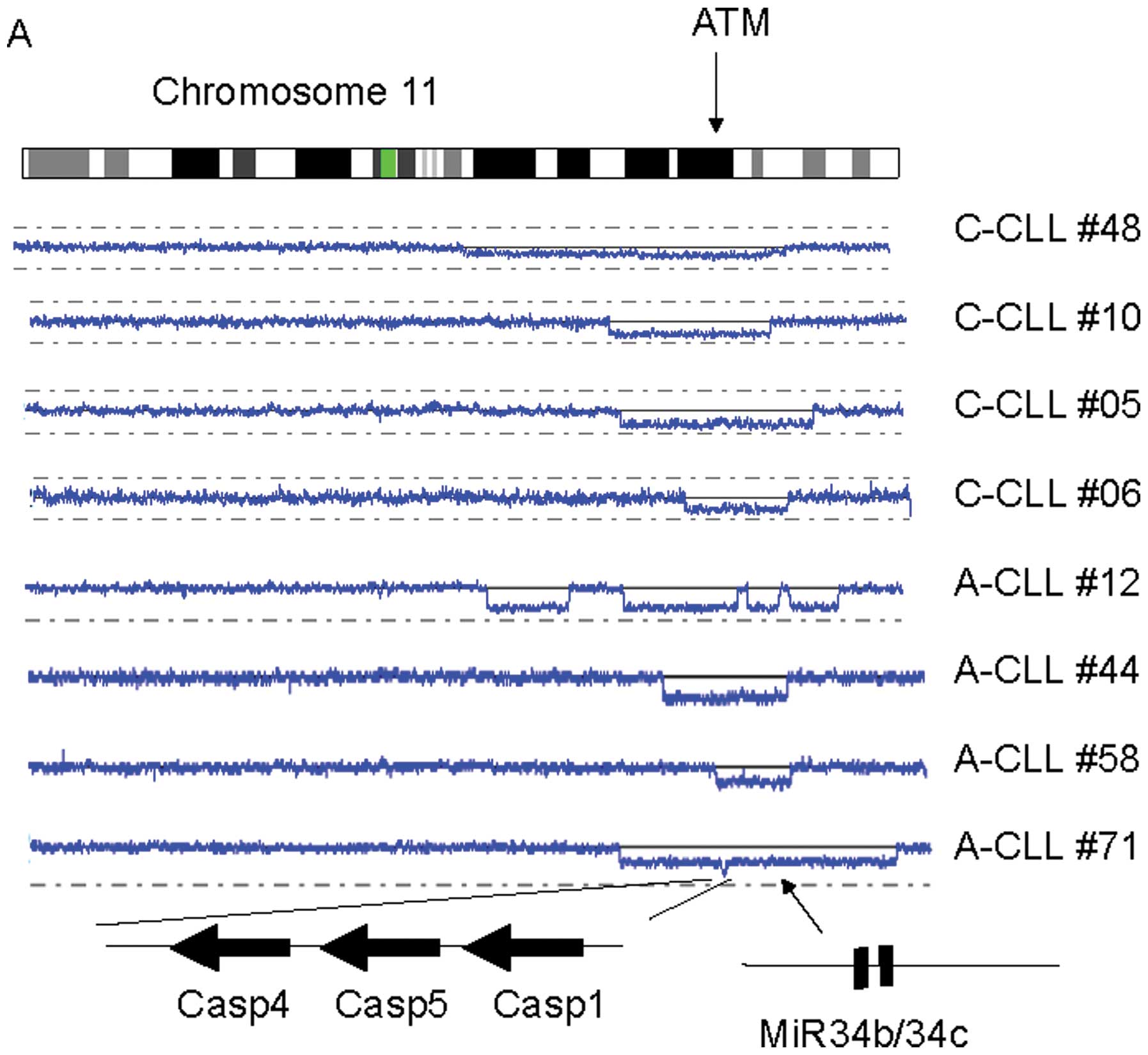 | Figure 1Novel common target candidate genes in
CLL. (A) Representative CLL cases with deletion of 11q. SNP-chip
analysis (blue lines) is shown with a scheme of the chromosome
(top), individual CLL samples and their altered genes (bottom). The
arrow on the chromosome panel indicates the position of the ATM
gene. At the bottom, the three large arrows show the direction of
transcription of the target genes; and the diagonal arrow
designates the position of the microRNAs, miR 34b/34c. The CLL case
numbers are shown on the right side (A, Asian; C, Caucasian). (B)
Representative CLL cases with deletion of 13q. The results of
SNP-chip analysis (blue lines) are shown together with the scheme
of the chromosome (top). Arrows at the bottom indicate the
positions of RB1 gene and the microRNAs, miR 15-a/16-1. The case
numbers are shown on the right side (A, Asian; C, Caucasian). (C)
Representative CLL cases with deletion of 8p. The result of
SNP-chip analysis (blue lines) is shown with the scheme of the
chromosome (top) and genes (bottom). The arrow indicates the
direction of transcription of the DLC1 gene. The case numbers are
shown on the right side (A, Asian; C, Caucasian). |
The third finding was near the well-known commonly
deleted region at 13q14.3 (65 cases in total) involving miR 15-a
and 16-1 (Table I and Fig. 1B). One case had two distinct
deletions in this region; one involved miR 15-a and 16-1; and the
other encompassed the Rb1 gene (Asian CLL #23 in Fig. 1B). On close inspection, 36% of
cases with deletion of 13q (23/65 cases), had a deletion of Rb1,
suggesting that Rb1 might be another target of deletion of 13q in
CLL.
Fourth, we found that deletion of 8p was common in
CLL (5 cases) (Table II and
Fig. 1C). One Asian-CLL had a very
small deletion only spanning the DLC1 gene (Asian-CLL #17, Fig. 1C). The DLC1 gene may be one of the
target genes in those samples with deletion of 8p in CLL.
CLL-associated genes are genomically
abnormal in CLL samples
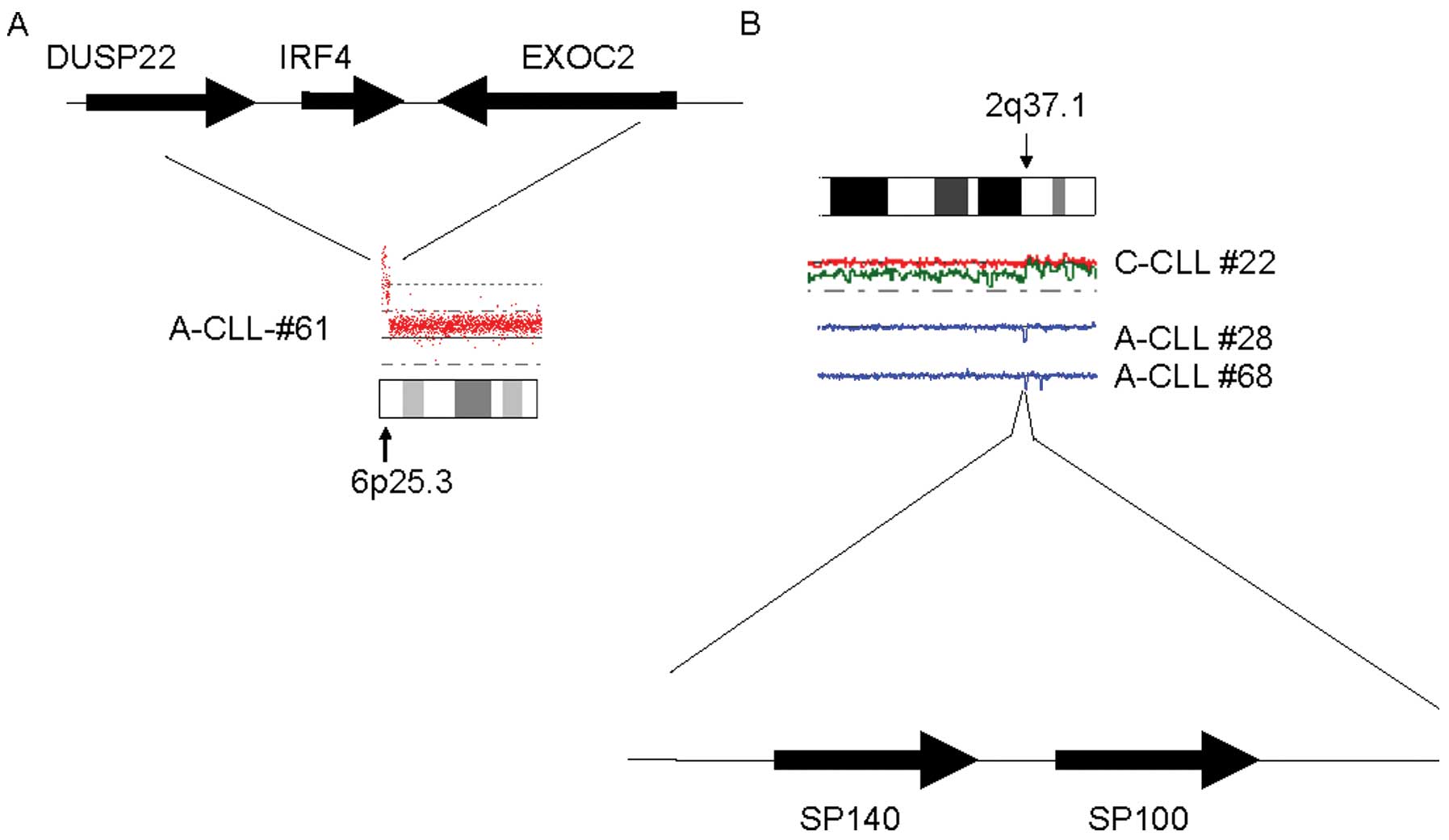
One Asian-CLL sample had high copy number
amplification of IRF4 (Fig. 2A).
Also, we found that SP140/SP100 genes were deleted in two Asian and
one Caucasian CLL samples (Fig.
2B).
A previous genome-wide association study (GWAS)
identifies SNP sites related to Caucasian CLL (19), including SNP-rs872071 (6p25.3)
located close to IRF4 gene and SNP-rs13397985 (2q37.1) located
close to the SP140/SP100 genes.
Discussion
CLL is extremely rare in Asians (10–12).
We have for the first time, analyzed a large number of Asian CLL
samples, using high resolution SNP-chips. Well-known common genomic
abnormalities (17,18) occured in Asian CLL at a comparable
frequency as Caucasian CLL except for deletion/UPD of 11q. Thus,
the classic genomic abnormalities of CLL in these two ethnic groups
are similar at the DNA level. However, the Asian CLL had trisomy
3/duplication of 3q and trisomy 18/duplication of 18q more
frequently than Caucasian CLL samples, suggesting that the
mechanism of development of CLL in Asians is slightly different
from Caucasian CLL.
The 18q region contains the BCL2 gene. BCL2 is
associated with an anti-apoptotic effect and the gene is often
overexpressed in CLL (20,21). CLL-related loss of 13q14, deletes
miR15-a/miR 16-1 (22–24). These two microRNAs target BCL2 and
their deletion causes high expression of BCL2 (24,25).
Trisomy 18/duplication of 18q may be an alternative mechanism to
overexpress BCL2. Also, the 3q region contains BCL6 (26,27),
whose protein antagonizes the TP53 tumor suppressor gene (27,28).
Seven cases had an 8p deletion (3/77 Asian CLL, 1/55
Caucasian CLL); and the minimally commonly deleted region involved
DLC1. In fact, one sample (Asian-CLL 17) had a deletion that only
involved the DLC1 gene. This gene encodes the GAP protein and plays
an important role in the RAS/MAP pathway in a manner similar to NF1
(29,30). This gene is frequently deleted in a
variety of cancers (30–33). Our data suggest that dysregulation
of RAS/MAP signal pathway occurs in CLL. Reagents targeting this
signaling pathway could be a promising treatment option for these
CLL cases.
Deletion of 8p, duplication of 3q and duplication of
18q are also frequently detected in other types of B-cell
malignancies including lymphomas (34,35).
Although a variety of B-cell lymphoid malignancies have their
distinct clinical features (36),
they may have dysregulation of common signaling pathways.
Identification of molecular signatures by SNP-chip may lead to
re-classification of B-cell malignancies and guide clinicians to
select optimal therapeutic options targeting the dysregulated
signaling pathways.
In this study, we also found novel candidate target
genes in altered chromosomal regions, which may be involved in
development of CLL. We found that Rb1 on the 13q chromo-some is
frequently deleted when 13q14 is deleted (Fig. 1B). This was especially evident in
the sample, A-CLL #23, which had two distinct deletions in 13q; one
targeting 13q14 (miR15-a/miR16-1) and the other targeting RB1.
Thus, Rb1 may be another tumor suppressor gene on 13q that is
altered in CLL. In addition, a homozygous deletion of three gene
family (Casp1/4/5) occurred at 11q in A-CLL #71. Both ATM and
miR34b/34c genes are on 11q and are frequently deleted in CLL. A
short distance away are these three caspase genes which can also be
target genes of this common deleted regions. Casp1/4/5 are
components of the inflamasome associated with inflamation and cell
growth (37,38). Deletion of these genes may cause an
abnormal response of leukemic cells to the immuno-surveillance
system, allowing proliferation of the leukemic cells.
GWAS co-relates presence or absence of a disease
with the patterns of SNPs in large populations, leading to
identification of high-risk alleles strongly associated with
development of the disease (39,40).
Interestingly, the genes close to these CLL-associated SNP-sites
(IRF4 and SP140/SP100) are moderately often mutated
(deleted/amplified) in CLL.
In conclusion, we found that: i) the fundamental
molecular signatures of CLL in the Caucasian and Asian populations
are fairly similar; ii) novel candidate target genes in commonly
altered genomic regions include Rb1, Casp1/4/5, DLC1, IRF4 and
SP140/SP100; iii) CLL-associated genes identified by the previous
GWAS were moderately often mutated in CLL. These findings will help
to re-classify CLL based on their molecular signatures and lead to
individualized treatment for CLL with reagents targeting signaling
pathways which are altered in each patient.
Acknowledgements
This study was supported by NIH grants
R01CA02603831 and U54CA143930 and A* STAR Investigator
Grant to H.P.K and the Tower Cancer Foundation and NIH grant
GM008243 to N.K. We thank Rocio Alvarez for technical
assistance.
References
|
1.
|
DeVita VT, Lawrence TS, Rosenberg SA,
DePinho RA and Weinberg RA: DeVita, Hellman, and Rosenberg’s
Cancer: Principles and Practice of Oncology. 8th edition.
Lippncott: Williams & Wilkins; 2009
|
|
2.
|
Eley JW, Hill HA, Chen VW, et al: Racial
differences in survival from breast cancer. Results of the National
Cancer Institute Black/White Cancer Survival Study. JAMA. 272. pp.
947–954. 1994, View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
GLOBOCAN 2008/Cancer Incidence and
Mortality Worldwide in 2008. International Agency for Research on
Cancer. World Health Organization. http://globocan.iarc.fr/.
|
|
4.
|
Tsugane S and Sasazuki S: Diet and the
risk of gastric cancer: review of epidemiological evidence. Gastric
Cancer. 10:75–83. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Dunn JE: Cancer epidemiology in
populations of the United States - with emphasis on Hawaii and
California - and Japan. Cancer Res. 35:3240–3245. 1975.PubMed/NCBI
|
|
6.
|
Jänne PA, Engelman JA and Johnson BE:
Epidermal growth factor receptor mutations in non-small-cell lung
cancer: implications for treatment and tumor biology. J Clin Oncol.
23:3227–3234. 2005.PubMed/NCBI
|
|
7.
|
Bell DW, Lynch TJ, Haserlat SM, et al:
Epidermal growth factor receptor mutations and gene amplification
in non-small-cell lung cancer: molecular analysis of the
IDEAL/INTACT gefitinib trials. J Clin Oncol. 23:8081–8092. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Rai KR and Keating MJ: Chronic Lymphocytic
Leukemia Chapter 127 Cancer Medicine. 6th edition. BC Decker;
Hamilton, ON: 2000
|
|
9.
|
Redaelli A, Laskin BL, Stephens JM,
Botteman MF and Pashos CL: The clinical and epidemiological burden
of chronic lymphocytic leukaemia. Eur J Cancer Care. 13:279–287.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
SEER Stat Fact Sheets: Chronic Lymphocytic
Leukemia Surveillance epidemiology and end of results. National
Cancer Institute. http://seer.cancer.gov/statfacts/html/clyl.html.
|
|
11.
|
Gale RP, Cozen W, Goodman MT, Wang FF and
Bernstein L: Decreased chronic lymphocytic leukemia incidence in
Asians in Los Angeles County. Leuk Res. 24:665–669. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Pan JW, Cook LS, Schwartz SM and Weis NS:
Incidence of leukemia in Asian migrants to the United States and
their descendants. Cancer Causes Control. 13:791–795. 2002.
View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Hallek M, Cheson BD, Catovsky D, et al:
Guidelines for the diagnosis and treatment of chronic lymphocytic
leukemia: a report from the International Workshop on Chronic
Lymphocytic Leukemia updating the National Cancer Institute-Working
Group 1996 guidelines. Blood. 111:5446–5456. 2008. View Article : Google Scholar
|
|
14.
|
Nakahashi H, Tsukamoto N, Hashimoto Y, et
al: Characterization of immunoglobulin heavy and light chain gene
expression in chronic lymphocytic leukemia and related disorders.
Cancer Sci. 100:671–677. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Kawamata N, Ogawa S, Gueller S, et al:
Identified hidden genomic changes in mantle cell lymphoma using
high-resolution single nucleotide polymorphism genomic array. Exp
Hematol. 37:937–946. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Yamamoto G, Nannya Y, Kato M, et al:
Highly sensitive method for genomewide detection of allelic
composition in nonpaired, primary tumor specimens by use of
affymetrix single-nucleotide-polymorphism genotyping microarrays.
Am J Hum Genet. 81:114–126. 2007. View
Article : Google Scholar
|
|
17.
|
Lehmann S, Ogawa S, Raynaud SD, et al:
Molecular allelokaryo-typing of early-stage, untreated chronic
lymphocytic leukemia. Cancer. 112:1296–1305. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Caporaso N, Goldin L, Plass C, et al:
Chronic lymphocytic leukaemia genetics overview. Br J Haematol.
139:630–634. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Di Bernardo MC, Crowther-Swanepoel D,
Broderick P, et al: A genome-wide association study identifies six
susceptibility loci for chronic lymphocytic leukemia. Nat Genet.
40:1204–1210. 2008.
|
|
20.
|
Kitada S, Andersen J, Akar S, et al:
Expression of apoptosis-regulating proteins in chronic lymphocytic
leukemia: correlations with in vitro and in vivo chemoresponses.
Blood. 91:3379–3389. 1998.PubMed/NCBI
|
|
21.
|
Majid A, Tsoulakis O, Walewska R, et al:
BCL2 expression in chronic lymphocytic leukemia: lack of
association with the BCL2 938A>C promoter single nucleotide
polymorphism. Blood. 111:874–877. 1998.PubMed/NCBI
|
|
22.
|
Calin GA, Dumitru CD, Shimizu M, et al:
Frequent deletions and down-regulation of micro- RNA genes miR15
and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad
Sci USA. 99:15524–15529. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Calin GA, Ferracin M, Cimmino A, et al: A
MicroRNA signature associated with prognosis and progression in
chronic lymphocytic leukemia. N Engl J Med. 353:1793–1801. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Calin GA and Croce CM: Chronic lymphocytic
leukemia: interplay between noncoding RNAs and protein-coding
genes. Blood. 114:4761–4770. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Cimmino A, Calin GA, Fabbri M, et al:
miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl
Acad Sci USA. 102:13944–13949. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Abramson JS and Shipp MA: Advances in the
biology and therapy of diffuse large B-cell lymphoma: moving toward
a molecularly targeted approach. Blood. 106:1164–1174. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Basso K and Dalla-Favera R: BCL6: master
regulator of the germinal center reaction and key oncogene in B
cell lymphoma-genesis. Adv Immunol. 105:193–210. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Phan RT and Dalla-Favera R: The BCL6
proto-oncogene suppresses p53 expression in germinal-centre B
cells. Nature. 432:635–639. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Ching YP, Wong CM, Chan SF, et al: Deleted
in liver cancer (DLC) 2 encodes a RhoGAP protein with growth
suppressor function and is underexpressed in hepatocellular
carcinoma. J Biol Chem. 278:10824–10830. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Lahoz A and Hall A: DLC1: a significant
GAP in the cancer genome. Genes Dev. 22:1724–1730. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Yin XL, Pang JC and Ng HK: Identification
of a region of homozygous deletion on 8p22-23.1 in medulloblastoma.
Oncogene. 21:1461–1468. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Yuan BZ, Jefferson AM, Baldwin KT,
Thorgeirsson SS, Popescu NC and Reynolds SH: DLC-1 operates as a
tumor suppressor gene in human non-small cell lung carcinomas.
Oncogene. 23:1405–1411. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Zender L, Xue W, Zuber J, et al: An
oncogenomics-based in vivo RNAi screen identifies tumor suppressors
in liver cancer. Cell. 135:852–864. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Rinaldi A, Mian M, Chigrinova E, et al:
Genome-wide DNA profiling of marginal zone lymphomas identifies
subtype-specific lesions with an impact on the clinical outcome.
Blood. 117:1595–1604. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Tagawa H, Suguro M, Tsuzuki S, et al:
Comparison of genome profiles for identification of distinct
subgroups of diffuse large B-cell lymphoma. Blood. 106:1770–1777.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Swerdlow SH, Campo E, Harris NL, et al:
WHO Classification of Tumours of Haematopoietic and Lymphoid
Tissues. 4th edition. World Health Organization; 2008
|
|
37.
|
Martinon F and Tschopp J: Inflammatory
caspases and inflammasomes: master switches of inflammation. Cell
Death Differ. 14:10–22. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Davis BK, Wen H and Ting JP: The
inflammasome NLRs in immunity, inflammation, and associated
diseases. Annu Rev Immunol. 29:707–735. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Altshuler D, Daly MJ and Lander ES:
Genetic mapping in human disease. Science. 322:881–888. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Clifford RJ, Edmonson MN, Nguyen C,
Scherpbier T, Hu Y and Buetow KH: Bioinformatics tools for single
nucleotide polymorphism discovery and analysis. Ann NY Acad Sci.
1020:101–109. 2004. View Article : Google Scholar : PubMed/NCBI
|