Introduction
Hepatocellular carcinoma (HCC) is one of the most
common malignancies. Despite advances in the detection and
treatment of HCC, the mortality rate remains high because the
majority of HCC patients with local invasion and intrahepatic
metastasis for which most potentially curative therapies have
limited efficacy (1). However, it
may be an important therapeutic target for inhibition of invasion
and metastasis in HCC.
During recent years, various studies have described
cancer stem cell (CSC) hypothesis and these cells have
heterogeneous tumorigenic potential. Based on this hypothesis, only
a fraction of cells within a tumor is endowed with stem cell-like
features, including unlimited proliferative potential and
asymmetric cell division. This cellular subset, varying in size in
different tumors, can initiate and sustain tumor growth and is
believed to drive relapse (2).
According to the CSC hypothesis solely the CSC population is
responsible for early systemic dissemination and metastasis
formation (3). So it is important
to explore the potential mechanism of invasion and metastasis in
CSC for therapy of HCC. Prior reports have suggested that side
population (SP) cells separated from diverse cancer cells possess
stem cell-like properties (4–6). SP
cells refer to a unique cell population, first identified in bone
marrow cells by Goodell et al, which could efflux the
DNA-binding dye Hoechst 33342 (7).
Thereafter, SP cells have been defined in multiple species and
tissues and SP cells could be characterized as CSC in primary
tissues of cancer. SP cells were also shown in established tumor
cell lines with different origins, such as glioma (8), breast (9) and thyroid cancer monoclonal cell
lines (10). In a previous study,
we reported the identification and isolation of SP cell phenotype
in human HCC cell lines that demonstrate specific characteristics
of CSC when compared with MP cells, such as quiescence, elevated
chemoresistance, increased tumorigenicity, higher actin
polymerization ability and increased migration capacity towards the
chemokine CXCL12 (11). However,
it is unknown why SP cells have more migration and invasion
capabilities.
A recently identified class of non-coding small
RNAs, microRNAs (miRNAs), is a group of RNAs of 14–24 nucleotides
that can negatively regulate protein expression at the
post-transcriptional level by translational inhibition and/or mRNA
degradation (12). Dysregulated
expression of microRNAs has been identified in a variety of human
malignancies and is suggested to have a significant role in the
development or progression of tumors by inhibiting tumor suppressor
genes or activating oncogenes. Among them, microRNA-21 (miR-21) has
been widely suggested to be oncogenic in many tumors. It had
demonstrated that miR-21 can increase tumor cells migration and
invasion by directly targeting PTEN, RECK and PDCD4 (13–16).
Although miR-21 and its target have been widely explored as a
cancer-related target for tumors including HCC, there is no
information available concerning the relevance of the genes for
metastasis in CSC. The aims of this study were to examine miR-21 in
SP cells of HCC and to investigate the potential mechanism that
miR-21 participated in the migration and invasion of SP cells.
Materials and methods
Cell culture and reagents
The human liver non-tumor cell line (HL-7702) and
human HCC cell lines (SMMC-7721, MHCC97L and MHCC97H) were
cultivated in DMEM medium supplemented with 10% fetal calf serum
(Sigma Chemical Co., St. Louis, MO, USA). Primary antibodies
against PTEN, RECK, PDCD4 and GAPDH were purchased from Santa Cruz
Biotechnology (Santa Cruz, CA, USA). All secondary antibodies were
obtained from Pierce (Rockford, IL, USA). PTEN, RECK and PDCD4
small interfering RNA (siRNA) and siRNA controls were obtained from
Santa Cruz Biotechnology (Santa Cruz, CA, USA). Lipofectamine 2000
was purchased from Invitrogen (Carlsbad, CA, USA). All other
chemicals and solutions were purchased from Sigma-Aldrich unless
otherwise indicated.
Flow cytometry analysis
To identify and isolate the SP and MP fractions, we
used flow cytometry analysis described before (11).
Real-time PCR
Total RNA, including miRNAs, was isolated from
prepared liver samples or cells with TRIzol reagent (Invitrogen)
according to the manufacturer’s instructions. Expression of
hsa-miR-21 was analyzed with the miScript system (Qiagen, USA),
which consists of the miScript Reverse Transcription kit, miScript
Primer assays and miScript SYBR Green PCR kit, according to the
protocol provided by the company. Small nuclear RNA U6 was used for
normalization. For the analysis of BCRP1, AFP, CK19, PTEN, RECK and
PDCD4 expression, cDNA was synthesized using Moloney murine
leukaemia virus reverse transcriptase (Epicentre, Paris, France) as
described by the manufacturer. The housekeeping gene,
glyceraldehyde-3-phosphate dehydrogenase (GAPDH), was used for
normalization. Primers used are shown in Table I. Real-time PCR was run on the ABI
PRISM 7700 Sequence Detector (Applied Biosystems, USA). All of the
reactions were run in triplicate. The ΔΔCt method was
used for relative quantification of gene expression to determine
miR-21 and BCRP1, AFP, CK19, PTEN, RECK and PDCD4 mRNA expression
levels.
 | Table I.The primers used in the PCR
reaction. |
Table I.
The primers used in the PCR
reaction.
| Gene | Primer
sequences |
|---|
| BCRP1 | F:
5′-CAACCATTGCATCTTGGCTG-3′ |
| R:
5′-CAAGGCCACGTGATTCTTCC-3′ |
| AFP | F:
5′-CAGGAGGAAGAAAGGACAAAAAA-3′ |
| R:
5′-ATTCCTAAGGCATAGAAATCCCA-3′ |
| CK-19 | F:
5′-ATGGCCGAGCAGAACCGGAA-3′ |
| R:
5′-CCATGAGCCGCTGGTACTCC-3′ |
| PTEN | F:
5′-GCGTGCAGATAATGACAAGG-3′ |
| R:
5′-GGATTTGACGGCTCCTCTAC-3′ |
| RECK | F:
5′-TGCAAGCAGGCATCTTCAAA-3′ |
| R:
5′-ACCGAGCCCATTTCATTTCTG-3′ |
| PDCD4 | F:
5′-AGTGACGCCCTTAGAAGTGG-3′ |
| R:
5′-TCATATCCACCTCCTCCACA-3′ |
| GAPDH | F:
5′-CAAGGTCATCCATGACAACTTTG-3′ |
| R:
5′-GTCCACCACCCTGTTGCTGTAG-3′ |
Chemo-resistance assay
Sorted SP or MP cells were cultured with the
chemotherapeutic agent doxorubicin (DOX, 50 nM) or methotrexate
(MTX, 100 nM). After 72-h exposure, cell viability was examined by
an MTS-based Cell Titer 96 Aqueous One Solution Cell Proliferation
Assay (Promega).
Migration and invasion assays
The cell migration was analyzed with
non-Matrigel-coated Transwell cell culture chambers (8-μm
pore size) (Millipore, Billerica, MA, USA). The cell invasion was
analyzed with Matrigel-coated Transwell cell culture chambers
(8-μm pore size) (Millipore). Briefly, cells
(5×104 cells/well) were serum starved for 24 h and
plated in the upper insert of a 24-well chamber in a serum-free
medium. A medium containing 10% serum or the chemokine CXCL12
(PeproTech) was added to the well. The cells were incubated for 24
h. Cells on the upper side of the filters were mechanically removed
by scrubbing with a cotton swab, after which the membrane was fixed
with 4% formaldehyde for 10 min at room temperature and stained
with 0.5% crystal violet for 10 min. Finally, invasive or migrated
cells were counted at magnification ×200 from 10 different fields
of each filter.
Actin polymerization
Sorted SP or MP cells were resuspended and kept at
37°C. Human CXCL12 (PeproTech) or PBS were added to cell
suspensions and aliquots were taken at the indicated times and
immediately fixed in 4% paraformaldehyde for 10 min. After washing,
samples were stained with FITCPhalloidin (Molecular Probes) stain
F-actin and analyzed by flow cytometry. The relative F-actin index
was determined as the ratio of the F-actin level of SP or MP cells
treated with CXCL12 to SP or MP cells treated with PBS.
Cell transfection
MiR-21 mimics, inhibitors (miR-21-AS) and their
respective negative controls (NC) were obtained from GenePharma Co.
(Shanghai, China). The day before transfection, SP cells were
seeded in antibiotic-free medium. Transfections were carried out
using Lipofectamine 2000 (Invitrogen) in accordance with the
manufacturer’s instructions. To monitor transfection efficiency,
fluorescein (FAM) siRNA (GenePharma) was used as control.
Successfully transfected cells were observed with a fluorescence
microscope. According to the protocol supplied with the
Lipofectamine 2000, SP cells were transfected with either siRNA or
control siRNA. siRNA-transfected cells were seeded into 6-well cell
culture plates at a density of 1×105 cells/well. The
cells were allowed to grow for an additional 24 h and were then
harvested for further analysis.
Plasmid construction and luciferase
reporter assay
The 3′-UTR of PTEN or RECK or PDCD4 containing the
PTEN or RECK or PDCD4-miR-21 response element was cloned into the
pIS0 control luciferase vector (Promega). A mutant 3′-UTR of PTEN
or RECK or PDCD4 was synthesised by PCR. SP cells were seeded in a
24-well plate (1×105 per well) and transiently
transfected with PTEN or RECK or PDCD4-UTR-pIS0/Mu-PTEN or RECK or
PDCD4-UTR-pIS0, Renilla luciferase control vector (20 ng) and
miR-21/NC. Luciferase activity was measured 48 h later using a dual
luciferase reporter assay system according to the manufacturer’s
protocol (Promega).
Protein extraction and western
blotting
The cells were lysed in lysis buffer [50 mmol/l Tris
(pH 7.5), 100 mmol/l NaCl, 1 mmol/l EDTA, 0.5% NP40, 0.5% Triton
X-100, 2.5 mmol/l sodium orthovanadate, 10 μl/ml protease
inhibitor cocktail and 1 mmol/l PMSF] by incubating for 20 min at
4°C. The protein concentration was determined using the Bio-Rad
assay system (Bio-Rad, Hercules, CA, USA). Total proteins were
fractionated using SDS-PAGE and transferred onto nitrocellulose
membranes. The membranes were blocked with 5% non-fat dried milk or
bovine serum albumin in 1X TBS buffer containing 0.1% Tween-20 and
then incubated with the appropriate primary antibodies. Horseradish
peroxidase-conjugated anti-rabbit or anti-mouse IgG was used as the
secondary antibody and the protein bands were detected using the
enhanced chemiluminescence detection system (Amersham Pharmacia
Biotech). Quantification of the western blots was performed using
laser densitometry and relative protein expression was then
normalized to GAPDH levels.
Statistical analysis
Each experiment was repeated at least three times.
All data are summarized and presented as means ± SDs. The
differences among means were statistically analyzed using a t-test.
All statistical analyses were performed using SPSS 13.0 software
(Chicago, IL, USA). P<0.05 was considered as statistically
significant.
Results
SP cells are an enriched source of
stem-like cells in HCC cell lines
Using flow cytometry, we were able to identify and
successfully isolate populations of SP and MP cells from the three
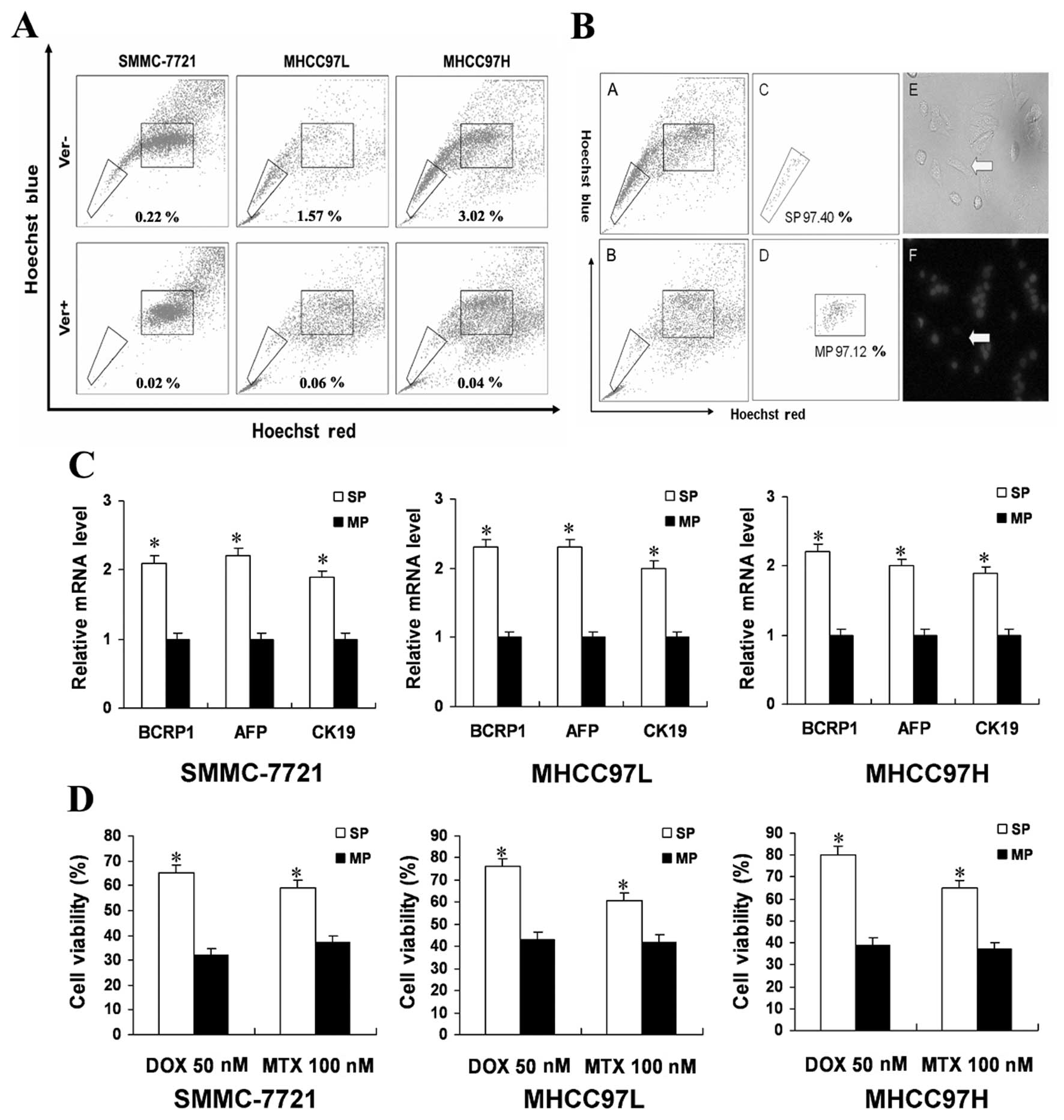
human HCC cell lines (Fig. 1A).
The SP gate was defined as the region where cells were absent in
the presence of verapamil, an agent which blocks the efflux of
Hoechst 33342. The three cell lines, SMCC-7721, MHCC97L and
MHCC97H, contained 0.19±0.02, 1.34±0.06 and 2.95±0.24% SP cells,
respectively. Since MHCC97H contained the highest percentage of SP
cells further experiments were performed using these cells. The
isolated SP cells had high purity and can efflux the DNA-binding
dye Hoechst 33342 in MHCC97H (Fig.
1B). It showed similar results in MHCC97L and SMMC-7721 cells
(data not shown). Expression of the hepatocyte-specific marker AFP
and the cholangiocyte-specific marker CK19 has been associated with
subpopulations showing bipotential stem/progenitor properties in
HCC. BCRP1 has the capacity to efflux a broad range of cytotoxic
substances and is characteristic of the SP phenotype. These genes
were expressed in both SP and MP cells but upregulated in SP cells
relative to MP cells (Fig. 1C).
The chemo-resistance ability of SP cells has been reported to
depend mainly on ABC transporters (17). To determine whether SP cells are
able to resist ABC transporter-independent anticancer drugs more
than MP cells we tested DOX and MTX which are used for the
treatment of HCC (Fig. 1D). SP
cells were more resistant than MP cells, especially to DOX. Cells
were then stained with Hoechst 33342 and reanalyzed. A higher
percentage of SP cells were seen after 72 h of DOX or MTX
treatment. This suggested that SP cells might be more resistant to
anticancer drugs than MP cells. We also evaluated other data such
as repopulation capability and cell cycle analysis described before
(11). It showed similar results
to our previous study (11). From
these data, we can propose SP cells exhibit stem-like
characteristics.
SP cells show more migration and invasion
capabilities than MP cells
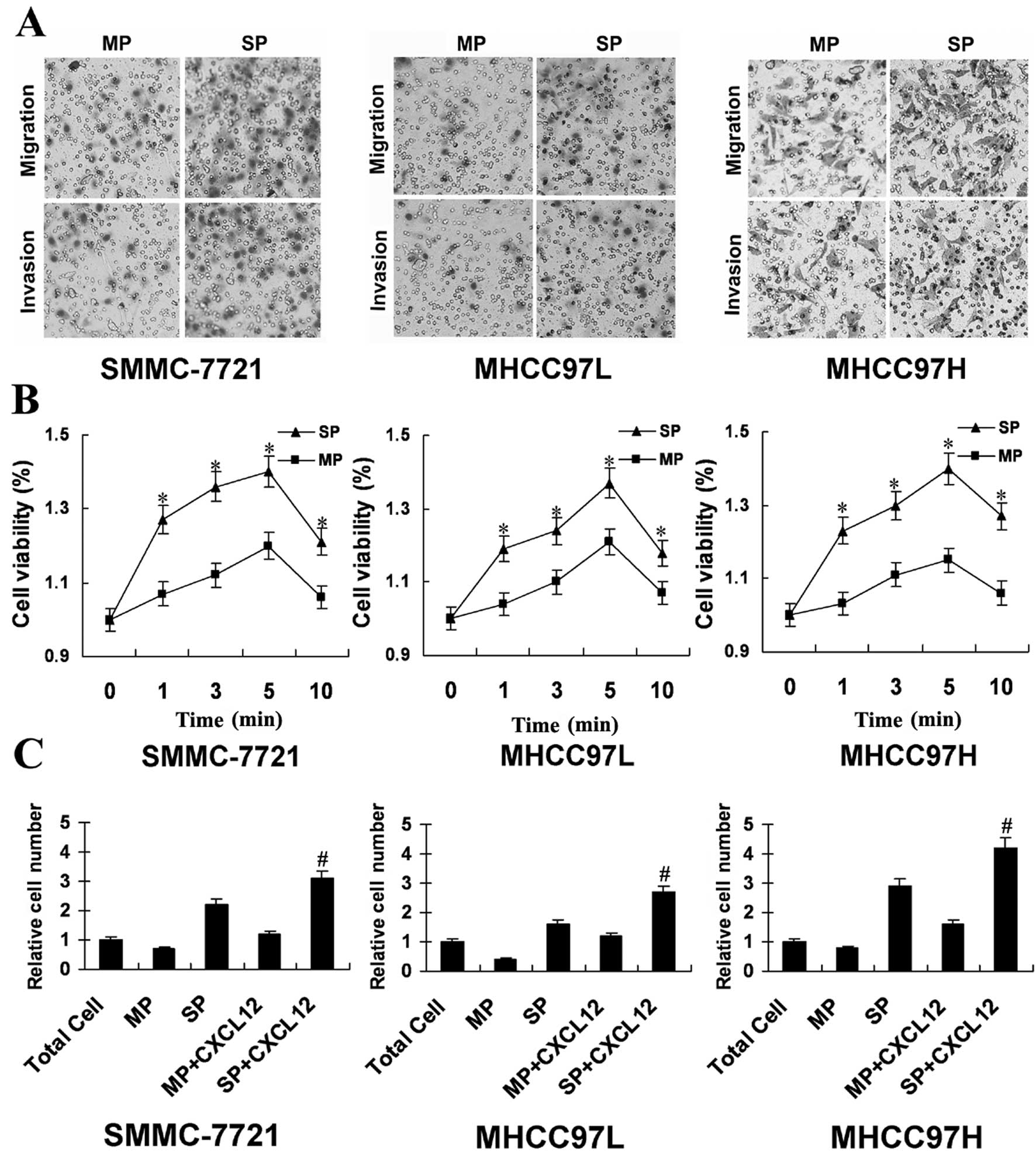
CSC in a tumor is proposed to mediate invasion and
metastasis and SP cells from tumor cells possess the properties
ascribed to CSC. Fig. 2A shows SP
cells with higher levels of penetration through Transwell cell
culture chambers versus the MP cells. Cell polarization requires
actin polymerization (18). As
shown in Fig. 2B, CXCL12 treatment
increased the extent of actin polymerization in SP cells more than
MP cells at the studied time-points. Following treatment with
CXCL12, the relative migrating index of SP cells was higher than MP
cells (Fig. 2C), suggested SP
cells show more potentiality of migration and invasion than MP
cells.
MiR-21 promotes SP cell migration and
invasion by targeting PTEN, RECK and PDCD4
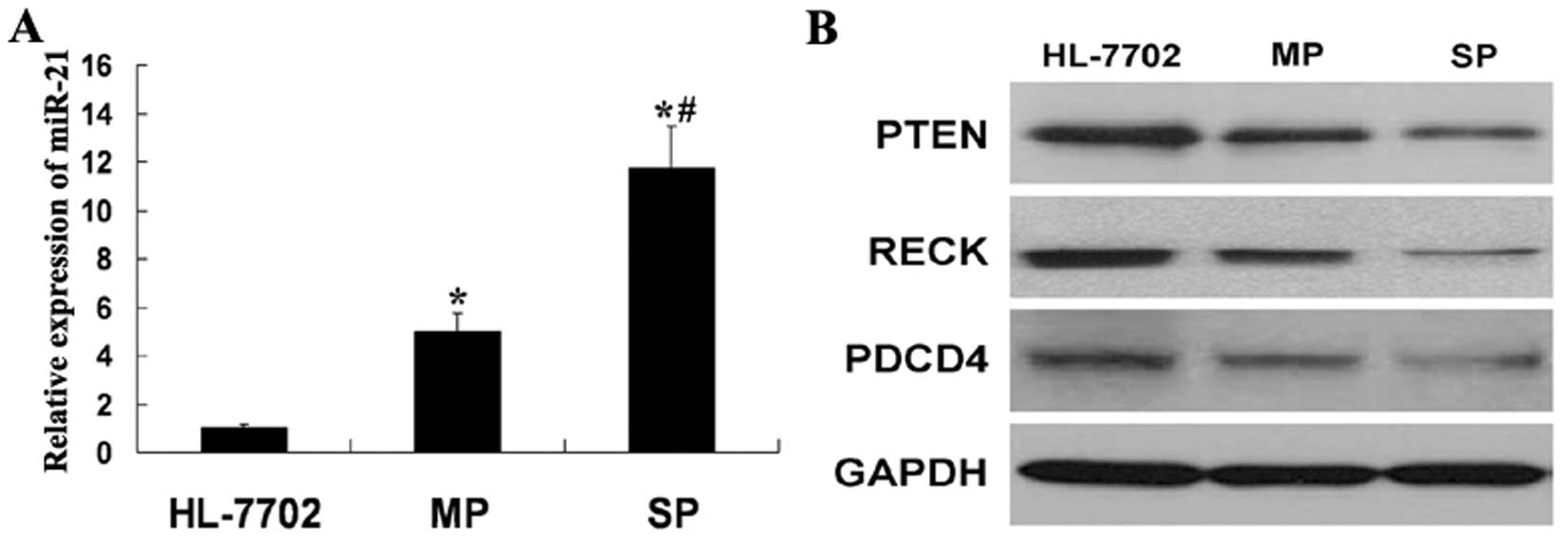
Increasing number of reports implicate miR-21
overexpression in carcinogenesis and metastasis. To assess the
relevance of miR-21 in SP cells, we first determined its expression
level comparing MP cells and liver non-tumor HL-7702 cells.
Interestingly, compared with HL-7702 cells, miR-21 expression was
found to be upregulated in SP and MP cells. These results indicated
miR-21 may be important in HCC. Furthermore, the higher expression
of miR-21 was observed in SP cells than MP cells (Fig. 3A). As a previous study described,
miR-21 can regulate HCC cells migration and invasion by targeting
PTEN, RECK and PDCD4 (13). We
also determined the protein level of PTEN, RECK and PDCD4 in
HL-7702, MP and SP cells. The lower expression of PTEN, RECK and
PDCD4, respectively, was observed in SP cells than MP and HL-7702
cells (Fig. 3B). There was an
inverse correlation between miR-21 and its target gene. These
results also indicated as in the cancer cells, miR-21 may play a
similar role in CSC.
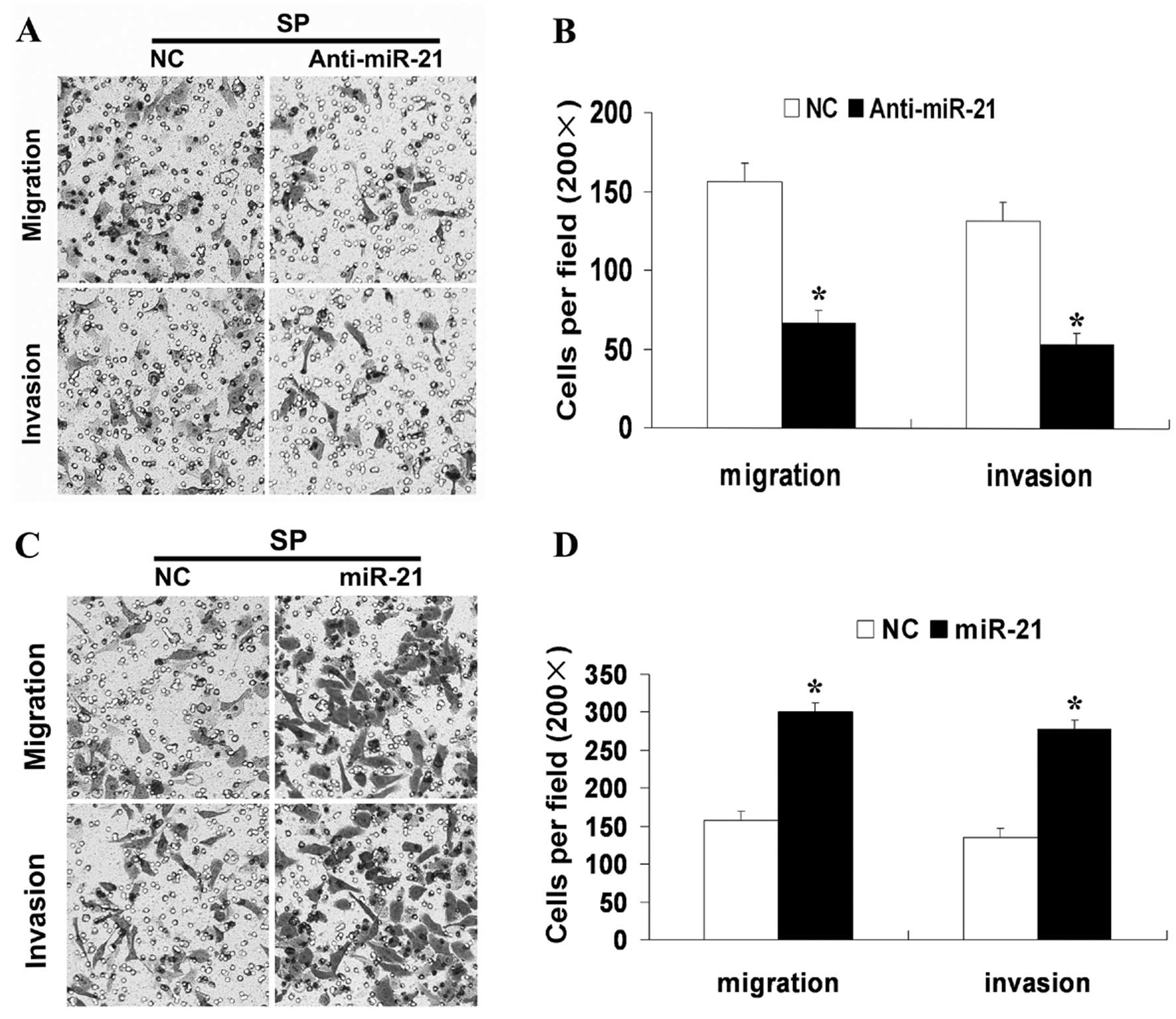
An important component of the invasive profile of a
cancer cell is its ability to be motile. High levels of miR-21 have
been associated with cell motility and metastasis of different
cancers. Having this background in mind, we first performed in
vitro loss-of-function analyses by silencing miR-21 with
miR-21-AS in SP cells. Silencing of miR-21 led to a more than 60%
reduction in the migration and invasion of these cells (Fig. 4A and B). Next, synthetic miR-21
mimics or NC were transfected into SP cells and overexpression of
miR-21 caused a 2-fold increase in cell migration and invasion
(Fig. 4C and D). These results
confirmed the involvement of miR-21 in the regulation of SP cell
motility and suggested a biological role for miR-21 in controlling
SP cell ability for migration and invasion.
MiR-21 simultaneously regulates multiple programs
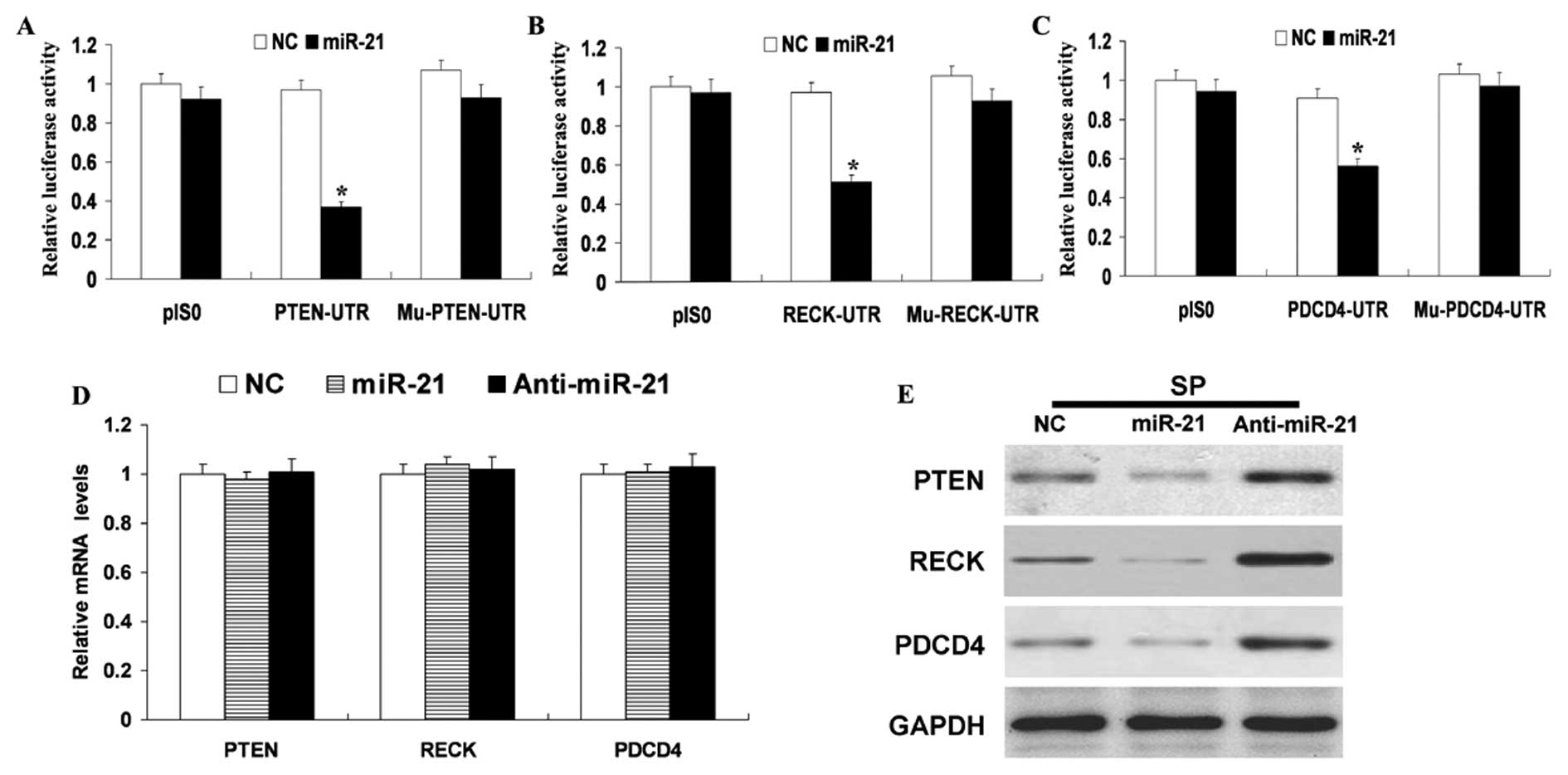
that enhance tumor invasiveness by targeting PTEN, PDCD4 and RECK
in HCC cells (13). As shown in
Fig. 5A–C, miR-21 overexpression
remarkably repressed the expression of luciferase containing a
wild-type miR-21 binding site (PTEN or RECK or PDCD4-UTR) compared
with NC. This suppression was restored by mutations in the seed
complementary sites of the 3′-UTR of PTEN or RECK or PDCD4
(Fig. 5A–C). To determine whether
miR-21 expression represses endogenous PTEN or RECK or PDCD4
expression through translational repression, synthetic miR-21
mimics and NC were transfected into SP cells. RT-PCR and western
blot analyses revealed that miR-21 overexpression did not cause
degradation of PTEN or RECK or PDCD4 mRNA (Fig. 5D) but did drastically inhibit its
protein expression (Fig. 5E).
Consistent with these results, silencing miR-21 with miR-10b-AS,
respectively, increased the level of PTEN, RECK or PDCD4 protein
(Fig. 5E) but did not alter PTEN,
RECK or PDCD4 mRNA levels (Fig.
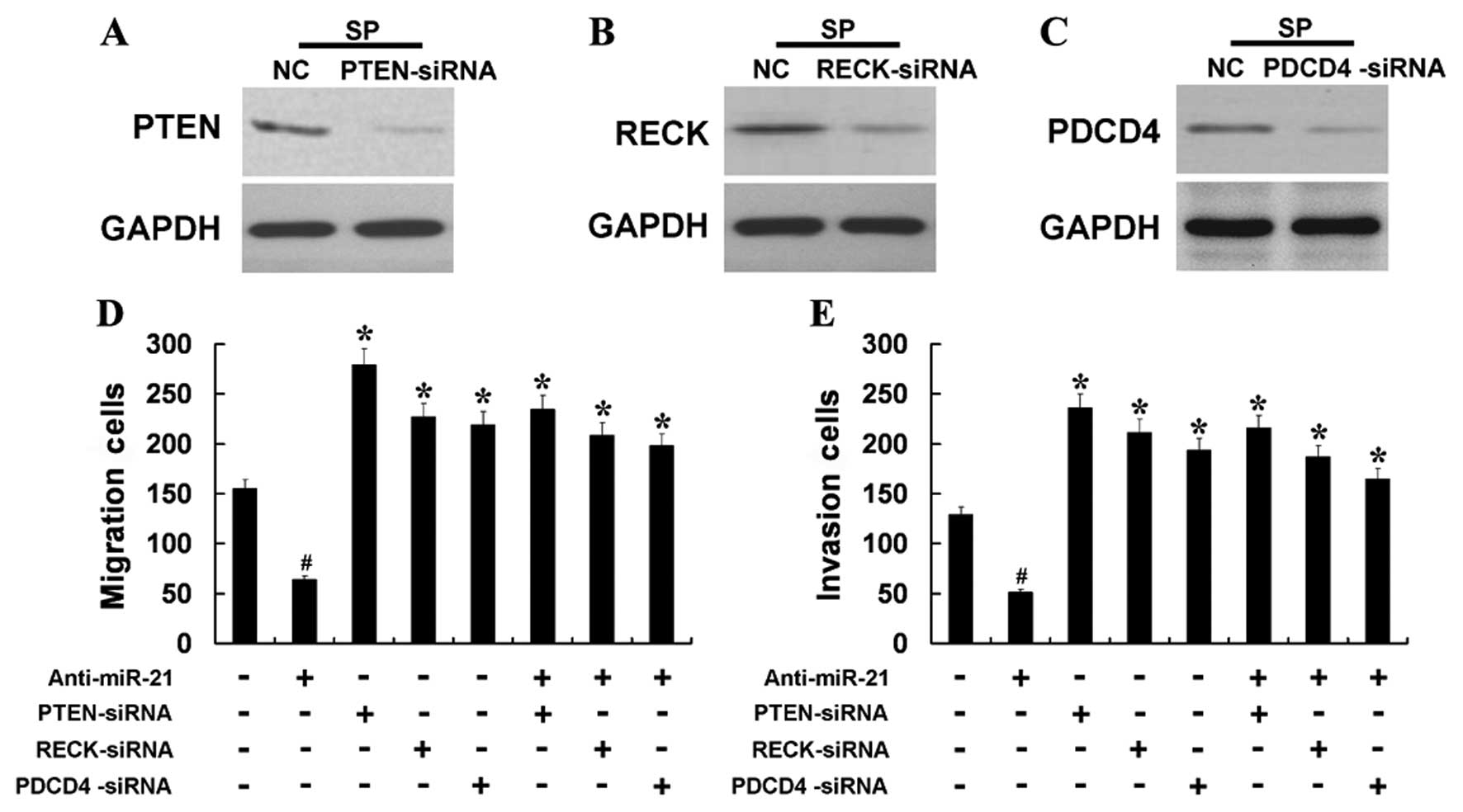
5D). We examined whether a reduction in the expression of PTEN
or RECK or PDCD4 might mediate the induction of cell migration and
invasion observed following miR-21 overexpression. Silencing of
PTEN, RECK or PDCD4 with siRNA in SP cells led to increased cell
migration and invasion (Fig. 6D and
E), demonstrating a negative role for PTEN, RECK or PDCD4 in
the migration and invasion of SP cells. The knockdown efficiency of
PTEN, RECK or PDCD4 was verified by western blot analysis (Fig. 6A–C). Further, we co-transfected SP
cells with siRNA for PTEN, RECK and PDCD4 mRNA, respectively, and
miR-21-AS and found that the effect of miR-21-AS was partially
attenuated by silencing of PTEN, RECK or PDCD4 mRNA (Fig. 6D and E). These results indicated
that the prometastatic effect of miR-21 is partly mediated through
regulation of PTEN, RECK or PDCD4 expression.
Discussion
Hepatocellular carcinoma (HCC) is the most common
malignancy of the liver and surgical resection remains the major
treatment. However, long-term results after resection of HCC are
still unsatisfactory. The main cause for the poor prognosis is the
high recurrence rate and a risk of vascular invasion, and
metastasis even after curative resection of HCC. Therefore, further
improvement of long-term survival after hepatic resection may
depend on prevention and treatment of the recurrent tumor, which
has attracted attention of researchers in recent years. Although
studies have focused on HCC invasion and metastasis, the underlying
mechanisms are still not fully understood (19).
There is increasing evidence indicating that the
maintenance and spreading of a variety of tumors is sustained by a
small subset of cancer cells, termed CSC. These cells possess the
ability for self-renewal, unlimited proliferation potential and
capacity to generate differentiated cells which constitute the
major tumor population (20).
Currently, human CSC have been identified in a variety of tumor
types (5,21–23).
It has been reported that CSCs are resistant to chemotherapy and
targeted therapy, which resulted in cancer relapse and metastasis.
An important criterion is that tumors with high percentages of
cancer stem cells will be more aggressive (17). On the other hand, in the analysis
of hematopoietic stem cells, a sub-population that effluxes the
DNA-binding dye Hoechst 33342 out of the cell membrane through an
ATP-binding cassette (ABC) transporter was recognised as a stem
cell population (7). This cell
population expressing the ABC transporter was defined as side
population (SP) cells, which were distinguished from cells of the
other population (main population; MP). Moreover, recent work has
led to the detection of the SP cells in a variety of tumor types
(8,10,24).
SP cells from tumor cells possess the properties ascribed to cancer
stem cells and have been proposed to play a critical role in
metastasis progression (20). Our
present results showed that SP cells had more migration and
invasion capabilities than MP cells, therefore, this may be one of
the potential mechanisms of HCC cell invasion and metastasis.
MiRNAs have a broad impact on gene expression
through translational repression or post-transcriptional
suppression (25). MiRNAs play
important roles in multiple biological processes such as
development, differentiation and cellular stress response. Previous
studies have linked deregulation of miRNAs to various diseases
including cancer (26,27). A single miRNA can potentially bind
to hundreds of mRNA targets, thereby having an important role in
various biological processes (28). Furthermore, it has been
demonstrated that miRNAs can induce tumorigenesis, for example, by
downregulating tumor suppressor genes. Among such oncogenic miRNAs,
miR-21 has been shown to be overexpressed in a variety of
malignancies (29–31). Therefore, miR-21 has been
recognized as an oncomir. Elevated miR-21 expression has been
causally linked to cell metastasis (32,33).
In our data, expression of miR-21 was more significantly increased
in SP cells compared to MP cells and changes of miR-21 can regulate
migration and invasion of SP cells. The link between miR-21 and SP
cell metastasis suggests the presence of metastatic pathways and
raised the question regarding the mechanisms of how miR-21 may
impact the metastatic potential of SP cells.
The metastatic process is complex and often
associated with alterations in the different metastasis-related
protein of the tumor cells. PTEN is a phosphoinositide phosphatase
which was originally identified as a multifunctional tumor
suppressor frequently lost in various human cancers (34,35).
PTEN expression is associated with tumor invasion and
tumor-node-metastasis (TNM) stage (36,37).
As a tumor suppressor in multiple cancers including HCC, PTEN can
affects the Akt and ERK signaling pathways (38). These pathways are linked to cell
survival, proliferation, differentiation, cell migration and
invasion. Previous studies showed the modulation of expression of
PTEN in HCC can impact on the activity of critical downstream
mediators of tumor progression and metastases (14). RECK, a membrane-anchored
glycoprotein, is able to suppress tumor invasion and metastasis and
is known to inhibit both catalytic and processing steps of
pro-MMP-2 activity and associated with the suppression of MMP-2,
MMP-9 and MMP-14 secretion (39,40).
In tumors, RECK is absent or diminished and MMPs are highly active,
facilitating tumor promotion and progression. The expression of
RECK is often decreased during cancer progression (41,42)
and is a molecular marker for cancer prognosis and controller of
cellular metastatic capacity (40). RECK expression levels are
predictive in determining prognoses in a number of common cancers
and low levels of RECK are often associated with increased
invasiveness and poor prognosis (43,44).
Programmed cell death 4 (PDCD4) is a tumor suppressor gene that
inhibits metastasis in human cancer cells (45). PDCD4 is a downstream component of
the Akt pathway and PDCD4 has been shown to inhibit neoplastic
transformation, tumor development and malignant progression
(46,47). Studies investigating cellular
functions of PDCD4 demonstrated that it suppresses the expression
and/or activity of the invasion-related proteins such as AKT
(48), MAP4K1 (49) and increases the release of
metastasis suppressor proteins such as E-cadherin (50) and TIMP2 (51). In hepatocellular carcinoma cells,
PDCD4 has recently been shown to correlate inversely with
metastatic capacity (52). Thus,
change of these proteins can be involved in migrating and invading
of tumor cells. Our studies showed that the protein of PTEN, RECK
or PCCD4 decreased in SP cells. The results indicated these
proteins may participate in migration and invasion of SP cells. A
few targets of miR-21 have been experimentally validated, including
some metastasis-related protein such as PTEN (14), PDCD4 (15), RECK (16). Recent studies had showed that
miR-21 simultaneously regulates multiple programs that enhance
tumor invasiveness by targeting PTEN, PDCD4 and RECK in HCC cells
(13). We hypothesized that miR-21
may play an important role in regulating migration and invasion of
cancer stem-like cells by targeting PTEN, PDCD4 and RECK.
Although PTEN, PDCD4 and RECK are direct targets of
miR-21 and their expression is inhibited by miR-21 in HCC cells, we
still question whether miR-21 could regulate these proteins in
cancer stem-like cells. One of the best ways to understand miRNA
function is via the elucidation of functional targets, which
usually involves analysis of changes in target proteins following
either a gain- or loss-of function of the specific miRNA. In our
present study, we also found that miR-21 overexpression drastically
inhibits PTEN, RECK or PDCD4 protein expression and silencing
miR-21 increased the level of PTEN, RECK or PDCD4 protein. On the
other hand, we also observed that SP cell migration and
invasiveness weakened by anti-miR-21 was ‘rescued’ by knockdown of
PTEN, PDCD4 or RECK. Taken together the biological effects of
miR-21 on SP cell invasion are probably due to the simultaneous
repression of migration suppressive proteins such as PTEN, RECK or
PDCD4.
In conclusion, the results of this study revealed
the aberrant expression of miR-21 in SP cells and showed that
miR-21 regulates the expression of multiple target proteins that
are associated with tumor dissemination, many of which are
implicated in SP cell biology. Importantly, repression of miR-21
inhibited SP cell migration and invasion in vitro, possibly
due to downregulation of the tumor suppressor PTEN, RECK or PDCD4.
These compelling data provide preliminary evidence that miR-21 is a
pro-metastatic miRNA in SP cells and raise the possibility that
therapy of HCC may be improved by pharmaceutical strategies
directed towards miR-21.
Acknowledgements
We are grateful to Fu-Gin Zhang for
technical help. This study was supported by grants from the
National Natural Science Foundation of China (grants no.
81101619/H1607).
References
|
1.
|
Lau WY and Lai EC: Hepatocellular
carcinoma: current management and recent advances. Hepatobiliary
Pancreat Dis Int. 7:237–257. 2008.PubMed/NCBI
|
|
2.
|
Visvader JE and Lindeman GJ: Cancer stem
cells in solid tumours: accumulating evidence and unresolved
questions. Nat Rev Cancer. 8:755–768. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Simeone DM: Pancreatic cancer stem cells:
implications for the treatment of pancreatic cancer. Clin Cancer
Res. 14:5646–5648. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Ho MM, Ng AV, Lam S and Hung JY: Side
population in human lung cancer cell lines and tumors is enriched
with stem-like cancer cells. Cancer Res. 67:4827–4833. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Prince ME, Sivanandan R, Kaczorowski A, et
al: Identification of a subpopulation of cells with cancer stem
cell properties in head and neck squamous cell carcinoma. Proc Natl
Acad Sci USA. 104:973–978. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Wang J, Guo LP, Chen LZ, Zeng YX and Lu
SH: Identification of cancer stem cell-like side population cells
in human nasopharyngeal carcinoma cell line. Cancer Res.
67:3716–3724. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Goodell MA, Brose K, Paradis G, Conner AS
and Mulligan RC: Isolation and functional properties of murine
hematopoietic stem cells that are replicating in vivo. J Exp Med.
183:1797–1806. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Kondo T, Setoguchi T and Taga T:
Persistence of a small subpopulation of cancer stem-like cells in
the C6 glioma cell line. Proc Natl Acad Sci USA. 101:781–786. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Kruger M, Schwarz A and Blumich B:
Investigations of silicone breast implants with the NMR-MOUSE. Magn
Reson Imaging. 25:215–218. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Mitsutake N, Iwao A, Nagai K, et al:
Characterization of side population in thyroid cancer cell lines:
cancer stem-like cells are enriched partly but not exclusively.
Endocrinology. 148:1797–1803. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Zhang N, Li R, Tao KS, et al:
Characterization of a stem-like population in hepatocellular
carcinoma MHCC97 cells. Oncol Rep. 23:827–831. 2010.PubMed/NCBI
|
|
12.
|
Ambros V: MicroRNA pathways in flies and
worms: growth, death, fat, stress and timing. Cell. 113:673–676.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Liu C, Yu J, Yu S, et al: MicroRNA-21 acts
as an oncomir through multiple targets in human hepatocellular
carcinoma. J Hepatol. 53:98–107. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Meng F, Henson R, Wehbe-Janek H, Ghoshal
K, Jacob ST and Patel T: MicroRNA-21 regulates expression of the
PTEN tumor suppressor gene in human hepatocellular cancer.
Gastroenterology. 133:647–658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Asangani IA, Rasheed SA, Nikolova DA, et
al: MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor
suppressor Pdcd4 and stimulates invasion, intravasation and
metastasis in colorectal cancer. Oncogene. 27:2128–2136. 2008.
View Article : Google Scholar
|
|
16.
|
Gabriely G, Wurdinger T, Kesari S, et al:
MicroRNA 21 promotes glioma invasion by targeting matrix
metalloproteinase regulators. Mol Cell Biol. 28:5369–5380. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Dean M, Fojo T and Bates S: Tumour stem
cells and drug resistance. Nat Rev Cancer. 5:275–284. 2005.
View Article : Google Scholar
|
|
18.
|
Devreotes P and Janetopoulos C: Eukaryotic
chemotaxis: distinctions between directional sensing and
polarization. J Biol Chem. 278:20445–20448. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Tang DJ, Dong SS, Ma NF, et al:
Overexpression of eukaryotic initiation factor 5A2 enhances cell
motility and promotes tumor metastasis in hepatocellular carcinoma.
Hepatology. 51:1255–1263. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Reya T, Morrison SJ, Clarke MF and
Weissman IL: Stem cells, cancer and cancer stem cells. Nature.
414:105–111. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Al-Hajj M, Wicha MS, Benito-Hernandez A,
Morrison SJ and Clarke MF: Prospective identification of
tumorigenic breast cancer cells. Proc Natl Acad Sci USA.
100:3983–3988. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Matsui W, Huff CA, Wang Q, et al:
Characterization of clonogenic multiple myeloma cells. Blood.
103:2332–2336. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Singh SK, Hawkins C, Clarke ID, et al:
Identification of human brain tumour initiating cells. Nature.
432:396–401. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Szotek PP, Pieretti-Vanmarcke R, Masiakos
PT, et al: Ovarian cancer side population defines cells with stem
cell-like characteristics and Mullerian Inhibiting Substance
responsiveness. Proc Natl Acad Sci USA. 103:11154–11159. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Lu J, Getz G, Miska EA, et al: MicroRNA
expression profiles classify human cancers. Nature. 435:834–838.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Iorio MV, Visone R, Di Leva G, et al:
MicroRNA signatures in human ovarian cancer. Cancer Res.
67:8699–8707. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Bartel DP: MicroRNAs: genomics,
biogenesis, mechanism and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Bloomston M, Frankel WL, Petrocca F, et
al: MicroRNA expression patterns to differentiate pancreatic
adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA.
297:1901–1908. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Feber A, Xi L, Luketich JD, et al:
MicroRNA expression profiles of esophageal cancer. J Thorac
Cardiovasc Surg. 135:255–260. 2008. View Article : Google Scholar
|
|
31.
|
Yanaihara N, Caplen N, Bowman E, et al:
Unique microRNA molecular profiles in lung cancer diagnosis and
prognosis. Cancer Cell. 9:189–198. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Krichevsky AM and Gabriely G: miR-21: a
small multi-faceted RNA. J Cell Mol Med. 13:39–53. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Grunder E, D’Ambrosio R, Fiaschetti G, et
al: MicroRNA-21 suppression impedes medulloblastoma cell migration.
Eur J Cancer. 47:2479–2490. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Li J, Yen C, Liaw D, et al: PTEN, a
putative protein tyrosine phosphatase gene mutated in human brain,
breast and prostate cancer. Science. 275:1943–1947. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Salmena L, Carracedo A and Pandolfi PP:
Tenets of PTEN tumor suppression. Cell. 133:403–414. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Bedolla R, Prihoda TJ, Kreisberg JI, et
al: Determining risk of biochemical recurrence in prostate cancer
by immunohistochemical detection of PTEN expression and Akt
activation. Clin Cancer Res. 13:3860–3867. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Yoshimoto M, Cunha IW, Coudry RA, et al:
FISH analysis of 107 prostate cancers shows that PTEN genomic
deletion is associated with poor clinical outcome. Br J Cancer.
97:678–685. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38.
|
Yu J, Zhang SS, Saito K, et al: PTEN
regulation by Akt-EGR1-ARF-PTEN axis. EMBO J. 28:21–33. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Noda M, Oh J, Takahashi R, Kondo S,
Kitayama H and Takahashi C: RECK: a novel suppressor of malignancy
linking oncogenic signaling to extracellular matrix remodeling.
Cancer Metastasis Rev. 22:167–175. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Takahashi C, Sheng Z, Horan TP, et al:
Regulation of matrix metalloproteinase-9 and inhibition of tumor
invasion by the membrane-anchored glycoprotein RECK. Proc Natl Acad
Sci USA. 95:13221–13226. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Clark JC, Thomas DM, Choong PF and Dass
CR: RECK - a newly discovered inhibitor of metastasis with
prognostic significance in multiple forms of cancer. Cancer
Metastasis Rev. 26:675–683. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42.
|
Noda M and Takahashi C: Recklessness as a
hallmark of aggressive cancer. Cancer Sci. 98:1659–1665. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
43.
|
Kotzsch M, Farthmann J, Meye A, et al:
Prognostic relevance of uPAR-del4/5 and TIMP-3 mRNA expression
levels in breast cancer. Eur J Cancer. 41:2760–2768. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
44.
|
Takenaka K, Ishikawa S, Kawano Y, et al:
Expression of a novel matrix metalloproteinase regulator, RECK and
its clinical significance in resected non-small cell lung cancer.
Eur J Cancer. 40:1617–1623. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
45.
|
Allgayer H: Pdcd4, a colon cancer
prognostic that is regulated by a microRNA. Crit Rev Oncol Hematol.
73:185–191. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
46.
|
Cmarik JL, Min H, Hegamyer G, et al:
Differentially expressed protein Pdcd4 inhibits tumor
promoter-induced neoplastic transformation. Proc Natl Acad Sci USA.
96:14037–14042. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
47.
|
Jansen AP, Camalier CE and Colburn NH:
Epidermal expression of the translation inhibitor programmed cell
death 4 suppresses tumorigenesis. Cancer Res. 65:6034–6041. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
48.
|
Lankat-Buttgereit B and Goke R: The tumour
suppressor Pdcd4: recent advances in the elucidation of function
and regulation. Biol Cell. 101:309–317. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49.
|
Yang HS, Matthews CP, Clair T, et al:
Tumorigenesis suppressor Pdcd4 down-regulates mitogen-activated
protein kinase kinase kinase kinase 1 expression to suppress colon
carcinoma cell invasion. Mol Cell Biol. 26:1297–1306. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
50.
|
Wang Q, Sun Z and Yang HS: Downregulation
of tumor suppressor Pdcd4 promotes invasion and activates both
beta-catenin/Tcf and AP-1-dependent transcription in colon
carcinoma cells. Oncogene. 27:1527–1535. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
51.
|
Nieves-Alicea R, Colburn NH, Simeone AM
and Tari AM: Programmed cell death 4 inhibits breast cancer cell
invasion by increasing tissue inhibitor of metalloproteinase-2
expression. Breast Cancer Res Treat. 114:203–209. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
52.
|
Zhang S, Li J, Jiang Y, Xu Y and Qin C:
Programmed cell death 4 (PDCD4) suppresses metastastic potential of
human hepatocellular carcinoma cells. J Exp Clin Cancer Res.
28:712009. View Article : Google Scholar : PubMed/NCBI
|