Introduction
Prospective randomized clinical trials have shown
that tyrosine kinase inhibitors (TKI) gefitinib (1–3) and
erlotinib (4,5) as initial treatment for EGFR
mutation-positive advanced NSCLC improved outcomes compared with
chemotherapy. These molecules have thus been approved in many
countries worldwide. Therefore, routine analysis of pathological
specimens is mandatory in clinical practice to predict patient
response. The potential result is an increased likelihood that
patients will receive optimal therapy for their tumour and be
spared a course of therapy with no or significantly less benefit.
For that promise to be realized, a robust process, from patient
sampling to screening methods, has to be developed to be capable of
fast, reliable, sensitive and reproducible detection of the
mutations in patient tumor samples.
In current clinical practice, the samples available
for detection of somatic mutations are most of the time
formalin-fixed paraffin-embedded tissues of various tumor sites.
The samples are usually composed of mutant and wild-type DNA from
tumor cells and wild-type DNA from non-malignant cells (normal
epithelial cells, hematopoietic cells and stromal cells such as
fibroblasts). Therefore there is a need for a sensitive technique
and a complete reliable process. If standard dideoxy sequencing has
been the ‘gold standard’ for detecting mutations in constitutive
genetics, this robust method is however time-consuming, has only
moderate sensitivity and might suffer from a lack of robustness
when working on fragmented DNA extracted from formalin fixed
paraffin embedded tumors (6,7).
These limitations of direct sequencing for detecting somatic
mutations has led to the development of more sensitive, less
expensive, and faster methods. A number of alternative procedures
have therefore been developed to detect common cancer mutations,
such as HRM (8–10), allele-specific amplification
(11,12), primer extension (13), and pyrosequencing (14). In most cases, a better sensitivity
was obtained using targeted techniques as compared to direct
sequencing (15,16); reviewed in Ellison et al
(17).
We developed assays aiming at accurately detecting
EGFR mutations in patient tumor samples in routine screening. The
assays had to detect exon 19 deletions and the p.L858R (exon 21)
mutations, the two most common mutations in NSCLC that are clearly
associated with a clinical benefit. These assays, fragment analysis
(exon 19) and allele specific PCR (L858R) have been routinely used
for the last 3 years in our laboratory. Moreover, during this
period, we collected information on the patients (age, gender) and
the samples tested: histology, thyroid transcription factor-1
(TTF-1) expression, primary or metastatic lesion, type of specimen,
and tumor cell content. We undertook the analysis of the data
obtained. This allowed us to evaluate the impact of these
parameters on the frequency and spectrum of EGFR mutations in
Caucasian NSCLC patients. Here we report our experience testing for
EGFR mutations in a large number of samples using sensitive
techniques in a clinical setting.
Materials and methods
Patients
A total of 1,403 formalin-fixed paraffin-embedded
tumor samples from NSCLC patients were referred to our laboratory
for EGFR typing between January 2010 and June 2012. There were
1,243 adenocarcinomas, 49 squamous cell carcinomas and 111
non-small cell carcinomas, from 827 men and 576 women.
Sample processing and DNA extraction
Serial sections were cut from each paraffin block.
Tumor-rich areas were marked by the pathologist on a hematoxylin
and eosin 3 μm-thick stained section. To eliminate
non-malignant, stromal and contaminating inflammatory cells and to
enrich the analyzed specimen with tumor cells, these areas were
manually macro-dissected on 10 μm-thick sections using
single-use sterilized scalpels. DNA was then extracted after
paraffin removal (toluene 5 min, ethanol 3 min, ethanol 2 min)
using the Forensic kit and an iPrep system according to the
manufacturer’s recommendations (Invitrogen, Life Technologies SAS,
Villebon sur Yvette, France). DNA concentration was quantified by
spectro-photometry (NanoDrop ND-100 instrument, Thermo Fisher
Scientific, Waltham, MA) and normalized to 5 ng/μl.
Detection of the L858R mutation - allele
specific amplification
For codon 858 mutation, we designed two forward
primers with variations in their 3′ nucleotides such that each was
specific for the wild-type (858L; TCAAGATCACAGATTT TGGGCT) or the
mutated variant (858R; TCAAGATCACAG ATTTTGGGCG), and one common
reverse primer (AS; CATC CTCCCCTGCATGTGTTAAAC). The
sequence-specific forward and the reverse primer were then combined
in ‘Primer mix L’ (primers 858L and AS), and ‘Primer mix R’
(primers 858R and AS). The amplification conditions were optimized
for the RotorGene 3000 instrument (Qiagen, Courtaboeuf, France).
PCR amplifications were performed using the LC480 SYBR-Green mix
(Roche Diagnostics, Meylan, France). The reaction mixture contained
10 μl of the supplied 2X master mix, 0.5 μl of each
primer (10 μM each), and 9 μl of the template (45 ng
genomic DNA). The cycling conditions were as follows: denaturation
for 10 min at 95°C; amplification for 45 cycles, with denaturation
for 10 sec at 95°C, annealing for 15 sec at 65°C, and extension for
20 sec at 72°C. The specific 172-bp PCR products were amplified,
and the cycle threshold (Ct)-value was determined for a fixed
normalized fluorescence of 0.05. For each sample, the Ct-value was
determined for the 858L (control) PCR, and for the 858R (mutation
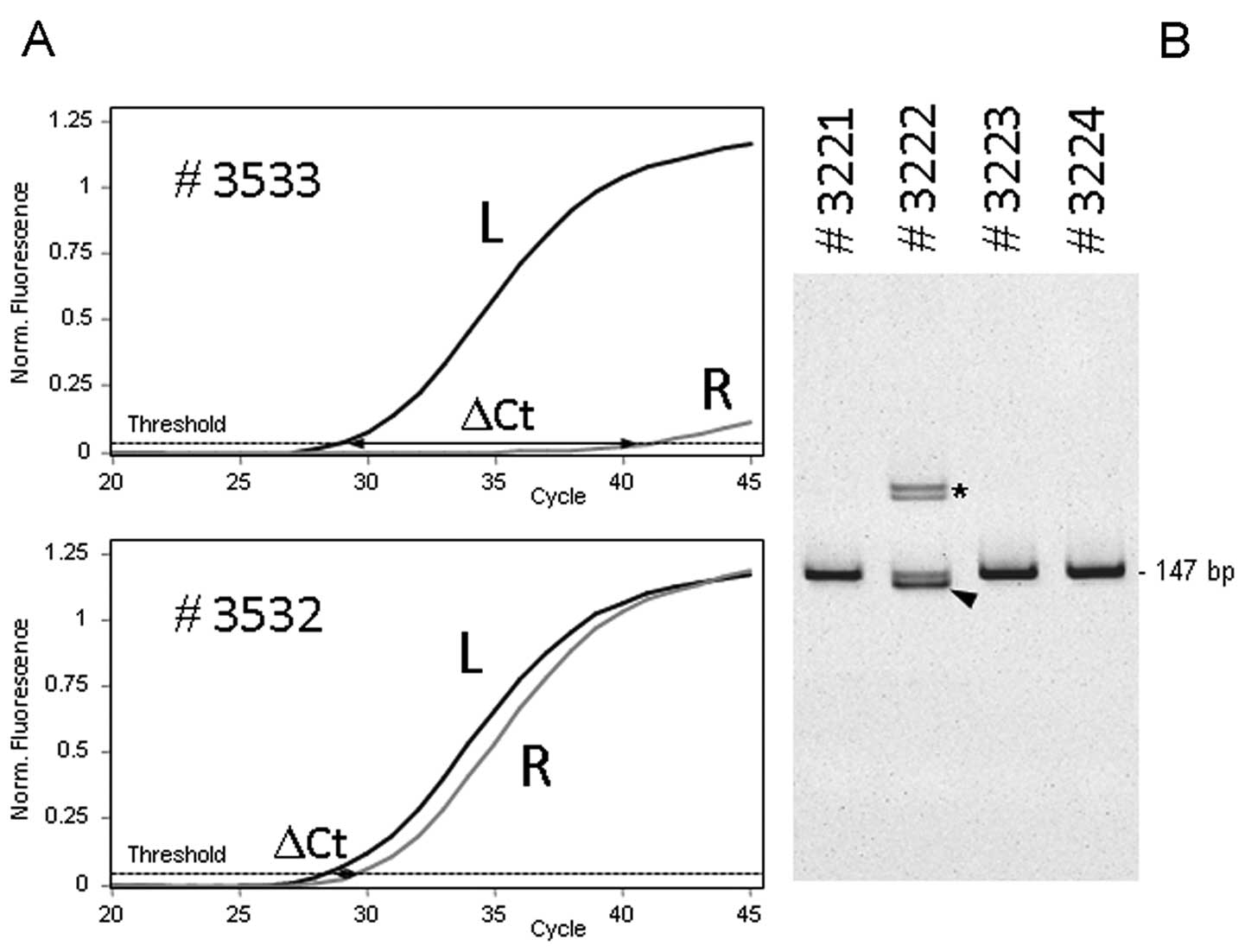
specific) PCR, and the difference was calculated (ΔCt, Fig. 1A). The lower the amount of mutated
DNA in the sample, the higher the ΔCt value. Samples with ΔCt<5
were considered as positive for the p.L858R mutation. Fig. 1A illustrates the assay performed on
formalin-fixed paraffin-embedded extracted DNAs containing a
p.L858R mutation (#3532, ΔCt=1.0) or wild-type for this allele
(#3533, ΔCt=12.6).
Detection of exon 19 deletions - fragment
size analysis
PCR were performed as described above using primers
19S (GTCT TCCTTCTCTCTCTGTCATAG) and 19AS (CCACACAGCA
AAGCAGAAACTCAC). Following amplification, amplicons were analyzed
by polyacrylamide gel electrophoresis on 5-20% acrylamide gels
(Invitrogen, Life Technologies, Saint Aubin, France). Migration was
performed for 1 h at 100 V. The wild-type EGFR gene yielded a
147-bp amplicon. Deletions were identified as faster migrating
bands. Fig. 1B illustrates the
profiles obtained for exon 19 wild-type tumor (#3221, #3223 and
#3224) and a tumor presenting an exon 19 deletion (#3222). In case
of exon 19 deletions, a doublet (or in rare cases a single band)
was seen at higher molecular weight. We have previously described
that each deletion is characterized by specific additional bands
corresponding to heteroduplexes (18).
Statistical analysis
The χ2 test, the Fisher’s exact test and
the two-tailed non-parametric Mann-Whitney test were used to assess
the association between mutation status of EGFR and each of the
clinicopathological parameters. A p-value of 0.05 was considered
statistically significant.
Results
Assay performance
The sensitivity and specificity of our assay were
evaluated for the specified EGFR alteration in quality control
schemes.
First, serial dilutions of DNA extracted from cell
lines harboring a p.L858R mutation (NCI-H1975: p.L858R;
c.2573T>G) or an exon 19 deletion (NCI-H1650: p.E746- A750del;
c.2235-2249del) in wild-type DNA were analyzed. This revealed that
our techniques allowed us to detect an EGFR alteration when it is
present in at least 1% (p.L858R mutation) or 2% of cells (exon 19
deletions). However, DNA extracted from FFPE samples is of much
lower quality than DNA extracted from cell lines. Therefore, in
order to avoid false negative results, we considered that a minimum
of 10% of tumor cells should be present in the sample. If no
alteration was found in a sample presenting less than 10% of tumor
cells, we concluded that the test was not contributive.
To assess the specificity of our assays, we repeated
the analysis of a large number of samples using an approved kit
(Therascreen EGFR RGQ kit, Qiagen, Hilden, Germany). We tested 160
samples that did not present a mutation on exon 19 and 21, and 98
samples presenting an exon 19 deletion or the L858R point mutation.
Identical results were obtained for all these samples.
Finally, our laboratory has been involved in
external quality schemes organized in western France in 2010 and
2011. Twenty NSCLC samples were tested during this period and we
found concordant results with the 5 other centers in all these
cases. More recently we have also been involved in the first
external quality control scheme organized by the French National
Cancer Institute. Sections from 10 NSCLC specimens were sent to all
the laboratories performing EGFR testing in France. We obtained the
expected result for all these samples using our procedures.
In routine practice, FFPE sections were sent from
the department of pathology to the department of molecular biology
every wednesday, and the reports were sent to the oncologists the
next friday. The molecular biology laboratory turnaround time is
thus of 48 h.
Frequency and spectrum of EGFR
mutations
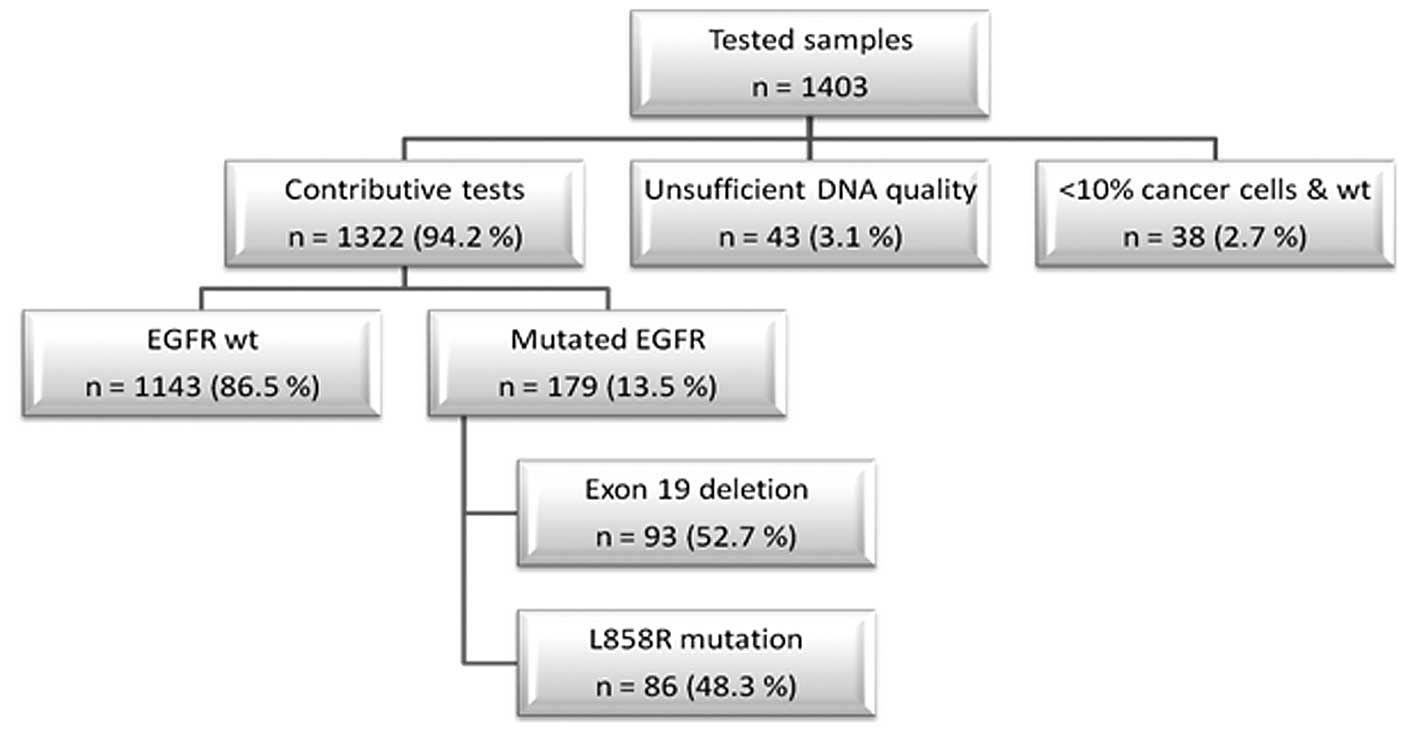
Our study population was composed of 1,403 NSCLC
patients (417 females and 725 males; median age 64). The tests
failed in 43 cases (3.1%), because of insufficient DNA quality
(Fig. 2). In 38 cases (2.7%) where
we were unable to detect an EGFR alteration, the content of tumor
cells in the sample was considered too low to conclude. The
remaining 1,322 tests performed (94.2%) were contributive. We were
able to identify an EGFR alteration in 179 patients (13.5%). These
included 93 exon 19 deletions (52.7% of the detected alterations),
and 86 L858R point mutations (48.3%) (Fig. 2).
The clinicopathologic features of patients with
EGFR- mutated or wild-type tumors in our cohort are summa rized in
Table I. Consistent with previous
observations, the mutation rate was significantly higher in women
than in men (23.0 vs 6.9%), and mutations were more frequent in
adenocarcinomas (168/1,144; 14.6%) than in other histological
types. Interestingly, EGFR-mutated patients were significantly
older than those with wild-type EGFR (median age 71 vs 63 years;
p<10−4).
 | Table I.Clinical and pathological
characteristics associated with EGFR mutational status in 1,322
french patients with lung tumors. |
Table I.
Clinical and pathological
characteristics associated with EGFR mutational status in 1,322
french patients with lung tumors.
| Total | EGFR wt | EGFR mutated | P-value |
|---|
| Gender | | | | |
| Female | 543 | 418 (77.0%) | 125 (23.0%) | |
| Male | 779 | 725 (93.1%) | 54 (6.9%) | <0.0001 |
| Age, years | | | | |
| Median | | 63 | 71 | <0.0001 |
| Range | | 28–100 | 32–92 | |
| Histology | | | | |
|
Adenocarcinoma | 1,144 | 976 | 168 (14.7%) | |
| NSCLC-NOS | 101 | 97 | 4 (4.0%) | 0.004 |
| Squamous
cell | 45 | 42 | 3 (6.7%) | |
Recent data reported by Sun et al (19) showed a correlation between TTF-1
expression and EGFR mutation status. Thus we also analyzed TTF-1
expression in our series of patients. Our data also clearly
indicated that EGFR mutations are more likely to occur in TTF-1
positive tumors (145/675; 17.7%) than in TTF-1 negative
adenocarcinomas (3/218; 1.4%). Almost all (145/148; 98.0%) mutated
adenocarcinomas were TTF-1 positive (Table II).
 | Table II.Relationship between TTF-1
immunostaining and EGFR mutation frequency in adenocarcinomas. |
Table II.
Relationship between TTF-1
immunostaining and EGFR mutation frequency in adenocarcinomas.
| Total | EGFR wt | EGFR mutated | P-value |
|---|
| TTF-1 | | | | |
|
IHC+ | 820 | 675 (82.3%) | 145 (17.7%) | |
|
IHC− | 218 | 215 (98.6%) | 3 (1.4%) | <0.0001 |
We next addressed the influence of tumor site and
type of sampling (Table III). The
frequency of EGFR alteration was found to be slightly higher in
metastases than in primary tumors (16.2 vs 13.5%, respectively),
but it was not statistically different (p=0.27). Considering
primary tumors, we did not see a significant difference between
surgical specimen (35/294; 11.9%), bronchial biopsies (42/339;
12.4%) and transthoracic needle biopsies (37/258; 14.3%). When
considering metastases, we also observed similar mutation rate when
analyzing biopsies (23/183; 12.6%), surgical resection (20/135;
14.8%), and transbronchial needle aspirates (5/38; 13.2%). The
mutation rate seemed to be higher in pleural effusions (11/44;
25.0%), but it was not statistically significant.
 | Table III.Influence of tumor site and type of
sample on the EGFR mutation frequency. |
Table III.
Influence of tumor site and type of
sample on the EGFR mutation frequency.
| Total | EGFR wt | EGFR mutated |
|---|
 |
Finally, we analyzed our data according to the
percentage of cancer cells present in the samples tested. Similar
mutation rates were obtained, even with samples containing less
than 10% of cancer cells (Table
IV).
 | Table IV.Influence of the cellularity of
tested samples on the EGFR mutation frequency. |
Table IV.
Influence of the cellularity of
tested samples on the EGFR mutation frequency.
| Total | EGFR wt | EGFR mutated | |
|---|
| Tumor cell
content | | | | |
| >50% | 669 | 590 (88.2%) | 79 (11.8%) | |
| 25–50% | 477 | 407 (85.3%) | 70 (14.7%) | p=0.46, NS |
| 10–25% | 156 | 138 (85.5%) | 18 (11.5%) | |
| <10% | 45 | NC (84.4%)a | 7 (15.6%) | |
Discussion
EGFR is a target of the TKIs gefitinib and
erlotinib, which have been approved for advanced NSCLC treatment in
many countries. Various EGFR testing approaches have been used in
the different phase III trials that led to these approvals. The
Scorpion Amplification Refractory Mutation System (ARMS) was used
in the phase III Iressa Pan-Asia Study (IPASS) to determine
EGFR mutation status (1). A
variety of methods, including direct sequencing, PCR-invader,
PNA-LNA PCR clamp, fragment analysis, and cycleave PCR, were used
in the WJTOG3405 phase III study to select EGFR
mutation-positive patients (2),
and the PNA-LNA PCR clamp method was used in the NEJ002 study
(3). In the European EURTAC study,
tissue samples were analyzed with Sanger sequencing (exons 19 and
21), and EGFR mutations were confirmed with an independent
technique (deletions in exon 19 by length analysis and L858R
mutations in exon 21 were detected with a 5′ nuclease PCR assay)
(5). Finally, in the Optimal
trial, testing was done by PCR-based direct sequencing, and other
methods were applied for monitoring at the same time (gel
electrophoresis for EGFR exon 19 deletions and cycleave
real-time PCR for EGFR exon 21 L858R mutations) (4).
Several years ago, we developed a sensitive
procedure for routine analysis of NSCLC tumors, and the aim of our
study was to evaluate its performance through analysis of the
results obtained during a long period. Between January 2010 and
June 2012, we tested 1,403 tumors in a routine clinical setting.
The tests failed in only 3.1% of the samples tested because of
insufficient quality of DNA. These not contributive tests were not
associated with the type or the size of the samples tested. But
this rate was significantly higher in samples processed in 2 of the
pathology centers that sent samples to our platform (not shown). We
are at present trying to identify the cause(s) of the lower quality
of DNA in some tissues processed in these centers. When excluding
these centers from the analysis, the failure rate decreased down to
1% (12/1,194 samples).
We found that 179 of the tumors tested harbored an
exon 19 deletion or an L858R point mutation. This corresponded to
13.5% of the 1,322 contributive samples. These findings are in
keeping with previous reports on Caucasian patients. For instance,
molecular testing of 755 patients from UK revealed a mutation
prevalence of 13% (20), and 13.1%
of Portuguese patients were found to present an EGFR mutation
(21). A large analysis of Spanish
patients revealed a slightly higher mutation rate (16.6%), but the
authors hypothesized that the participating centers included more
samples from women and patients who had never smoked (22).
In 2004, several groups correlated responses to
gefitinib or erlotinib with the presence of somatic mutations
clustered around the ATP-binding pocket of the tyrosine kinase
domain (23,24). Subsequent reports indicated that
the somatic mutational status of EGFR correlated with female sex
(25,26), smoking history (25,27,28),
adenocarcinoma histology (26,29)
and Asian origin (26).
We were not able to collect the smoking history of
the patients tested during this period in our platform, but the
mutation rate was clearly associated with female sex and
adenocarcinoma histology. Moreover, patients with a mutation were
clearly older than wild-type patients, as previously described for
patients from Japan (30) and
Korea (31). In addition, we
clearly demonstrated that the overwhelming majority of EGFR mutated
tumors expressed the TTF-1 antigen.
In a recent report, Girard et al collected
clinical and pathological data on a large number of patients with
NSCLC who had their tumors genotyped for EGFR mutations at
different institutions. Variables of interest (smoking history,
histological subtype, sex, stage of disease and age) were
integrated in a multivariate logistic regression model, and they
could thus build a model-based nomogram to allow for prediction of
the presence of EGFR mutations in NSCLC (32). Unfortunately, TTF-1 expression data
were lacking, and they could not include this covariate in the
final model.
In our series, the EGFR mutation prevalence was
clearly higher for women with a TTF-1 positive adenocarcinoma
(28.3%; 101/357). The mutation prevalence increased to 46.7%
(64/137) when selecting women older than 70.
Our analysis of this large series of patients
indicated that the mutation rate was not significantly different
between primary tumors and metastases. More importantly, we
demonstrated that there is no impact of the type of specimen tested
on the mutation prevalence. Similar levels of contributive results
(not shown) and of EGFR mutation rates were obtained using
different type of samples, including pleural fluids and
trans-bronchial needle aspirates.
Finally, we also found, using a sensitive approach,
that the mutation rate was not dependent on the sample cellularity.
Indeed, 23 patients for whom the tested samples contained less than
25% of cancer cells (<10 and 10–25%) presented an EGFR mutation
(23/170; 13.5%). Using a less sensitive technique such as direct
sequencing, we would have most likely missed these mutations.
Consequently, these patients would have been treated by
chemotherapy, and not benefited from TKI. That is the reason why,
in routine diagnosis, we do not reject samples containing less than
10% tumor cells. In a recent report Warth et al described an
algorithm for Sanger sequencing-based EGFR mutation analyses
(33). They demonstrated that
sequencing generated 80% reliable analysis of biopsy specimens, but
that in 20% of cases rebiopsy had to be recommended. A more
sensitive technique such as the one we use on a routine practice
avoids performing additional biopsies.
The clinical impact of detecting mutations present
in a low percentage of tumor cells has been assessed by Zhou et
al (34). They determined
whether abundance of EGFR mutations in tumors predicts
benefit from treatment with EGFR-TKIs for advanced NSCLC. They
detected EGFR mutations in lung cancer samples using both direct
DNA sequencing and the sensitive ARMS technique. Mutation-positive
tumors by both methods carried high abundance of EGFR mutations,
and tumors that were mutation positive by ARMS but mutation
negative by direct DNA sequencing harbored low abundance of EGFR
mutations. Median progression free survival of patients with low
abundance of EGFR mutations was significantly longer than for those
with wild-type tumors, and the difference between patients with
high and low abundance of EGFR mutations was not significant
regarding overall response rate and overall survival. Thus, it is
worth using a sensitive technique to detect patients with a low
abundance of mutations.
In conclusion, we demonstrated that performing EGFR
testing in routine diagnostic and clinical practice using sensitive
approaches can be successful. In our French population, the EGFR
mutation prevalence is higher in older patients, women,
adenocarcinomas and TTF-1 expressing adenocarcinomas.
Acknowledgements
The authors gratefully acknowledge all
the people involved in the different steps of the process from
tissue processing (technicians and secretaries from the Department
of Pathology) to molecular analysis (Réjane Lapied and Séverine
Huet), and all the Pathologists that provided tumors for EGFR
testing. The Departments of Biochemistry and Pathology are labelled
and funded by the French National Cancer Institute (INCa) to
perform EGFR testing of lung cancer patients.
References
|
1.
|
Mok TS, Wu YL, Thongprasert S, et al:
Gefitinib or carboplatinpaclitaxel in pulmonary adenocarcinoma. N
Engl J Med. 361:947–957. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Mitsudomi T, Morita S, Yatabe Y, et al:
Gefitinib versus cisplatin plus docetaxel in patients with
non-small-cell lung cancer harbouring mutations of the epidermal
growth factor receptor (WJTOG3405): an open label, randomised phase
3 trial. Lancet Oncol. 11:121–128. 2010. View Article : Google Scholar
|
|
3.
|
Maemondo M, Inoue A, Kobayashi K, et al:
Gefitinib or chemotherapy for non-small-cell lung cancer with
mutated EGFR. N Engl J Med. 362:2380–2388. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Zhou C, Wu Y-L, Chen G, et al: Erlotinib
versus chemotherapy as first-line treatment for patients with
advanced EGFR mutation-positive non-small-cell lung cancer
(OPTIMAL, CTONG-0802): a multicentre, open-label, randomised, phase
3 study. Lancet Oncol. 12:735–742. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Rosell R, Carcereny E, Gervais R, et al:
Erlotinib versus standard chemotherapy as first-line treatment for
European patients with advanced EGFR mutation-positive
non-small-cell lung cancer (EURTAC): a multicentre, open-label,
randomised phase 3 trial. Lancet Oncol. 13:239–246. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Querings S, Altmuller J, Ansen S, et al:
Benchmarking of mutation diagnostics in clinical lung cancer
specimens. PLoS One. 6:e196012011. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Marchetti A, Felicioni L and Buttitta F:
Assessing EGFR mutations. N Engl J Med. 354:526–528. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Takano T, Ohe Y, Tsuta K, et al: Epidermal
growth factor receptor mutation detection using high-resolution
melting analysis predicts outcomes in patients with advanced non
small cell lung cancer treated with gefitinib. Clin Cancer Res.
13:5385–5390. 2007. View Article : Google Scholar
|
|
9.
|
Do H, Krypuy M, Mitchell P, Fox S and
Dobrovic A: High resolution melting analysis for rapid and
sensitive EGFR and KRAS mutation detection in formalin fixed
paraffin embedded biopsies. BMC Cancer. 8:1422008. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Fukui T, Ohe Y, Tsuta K, et al:
Prospective study of the accuracy of EGFR mutational analysis by
high-resolution melting analysis in small samples obtained from
patients with non-small cell lung cancer. Clin Cancer Res.
14:4751–4757. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Didelot A, Le Corre D, Luscan A, et al:
Competitive allele specific TaqMan PCR for KRAS, BRAF and EGFR
mutation detection in clinical formalin fixed paraffin embedded
samples. Exp Mol Pathol. 92:275–280. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Kimura H, Kasahara K, Kawaishi M, et al:
Detection of epidermal growth factor receptor mutations in serum as
a predictor of the response to gefitinib in patients with
non-small-cell lung cancer. Clin Cancer Res. 12:3915–3921. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Lin CH, Yeh KT, Chang YS, Hsu N and Chang
JG: Rapid detection of epidermal growth factor receptor mutations
with multiplex PCR and primer extension in lung cancer. J Biomed
Sci. 17:372010. View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Dufort S, Richard M-J, Lantuejoul S and de
Fraipont F: Pyrosequencing, a method approved to detect the two
major EGFR mutations for anti EGFR therapy in NSCLC. J Exp Clin
Cancer Res. 30:572011. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Ellison G, Donald E, McWalter G, et al: A
comparison of ARMS and DNA sequencing for mutation analysis in
clinical biopsy samples. J Exp Clin Cancer Res. 29:132–139. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Goto K, Satouchi M, Ishii G, et al: An
evaluation study of EGFR mutation tests utilized for non-small-cell
lung cancer in the diagnostic setting. Ann Oncol. 23:2914–2919.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Ellison G, Zhu G, Moulis A, Dearden S,
Speake G and McCormack R: EGFR mutation testing in lung cancer: a
review of available methods and their use for analysis of tumour
tissue and cytology samples. J Clin Pathol. 66:79–89. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Denis MG, Theoleyre S, Chaplais C, et al:
Efficient detection of EGFR alterations in lung adenocarcinomas.
ASCO Meeting abstracts. 28:105562010.
|
|
19.
|
Sun P-L, Seol H, Lee HJ, et al: High
incidence of EGFR mutations in Korean men smokers with no
intratumoral heterogeneity of lung adenocarcinomas: correlation
with histologic subtypes, EGFR/TTF-1 expressions, and clinical
features. J Thorac Oncol. 7:323–330. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Leary AF, Castro DGd, Nicholson AG, et al:
Establishing an EGFR mutation screening service for non-small cell
lung cancer - sample quality criteria and candidate histological
predictors. Eur J Cancer. 48:61–67. 2012. View Article : Google Scholar
|
|
21.
|
Castro AS, Parente B, Goncalves I, et al:
Epidermal growth factor recetor mutation study for 5 years, in a
population of patients with non-small cell lung cancer. Rev Port
Pneumol. 19:7–12. 2013.PubMed/NCBI
|
|
22.
|
Rosell R, Moran T, Queralt C, et al:
Screening for epidermal growth factor receptor mutations in lung
cancer. N Engl J Med. 361:958–967. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Lynch TJ, Bell DW, Sordella R, et al:
Activating mutations in the epidermal growth factor receptor
underlying responsiveness of non small-cell lung cancer to
gefitinib. N Engl J Med. 350:2129–2139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Paez JG, Janne PA, Lee JC, et al: EGFR
mutations in lung cancer: correlation with clinical response to
gefitinib therapy. Science. 304:1497–1500. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Matsuo K, Ito H, Yatabe Y, et al: Risk
factors differ for non-small-cell lung cancers with and without
EGFR mutation: assessment of smoking and sex by a case-control
study in Japanese. Cancer Sci. 98:96–101. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Shigematsu H, Lin L, Takahashi T, et al:
Clinical and biological features associated with epidermal growth
factor receptor gene mutations in lung cancers. J Natl Cancer Inst.
97:339–346. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Kosaka T, Yatabe Y, Endoh H, Kuwano H,
Takahashi T and Mitsudomi T: Mutations of the epidermal growth
factor receptor gene in lung cancer: biological and clinical
implications. Cancer Res. 64:8919–8923. 2004. View Article : Google Scholar
|
|
28.
|
Yatabe Y, Kosaka T, Takahashi T and
Mitsudomi T: EGFR mutation is specific for terminal respiratory
unit type adenocarcinoma. Am J Surg Pathol. 29:633–639. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Yatabe Y and Mitsudomi T: Epidermal growth
factor receptor mutations in lung cancers. Pathol Int. 57:233–244.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
30.
|
Ueno T, Toyooka S, Suda K, et al: Impact
of age on epidermal growth factor receptor mutation in lung cancer.
Lung Cancer. 78:207–211. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31.
|
Choi YH, Lee JK, Kang HJ, et al:
Association between age at diagnosis and the presence of EGFR
mutations in female patients with resected non-small cell lung
cancer. J Thorac Oncol. 5:1949–1952. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Girard N, Sima CS, Jackman DM, et al:
Nomogram to predict the presence of EGFR activating mutation in
lung adenocarcinoma. Eur Respir J. 39:366–372. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33.
|
Warth A, Penzel R, Brandt R, et al:
Optimized algorithm for Sanger sequencing-based EGFR mutation
analyses in NSCLC biopsies. Virchows Arch. 460:407–414. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Zhou Q, Zhang X-C, Chen Z-H, et al:
Relative abundance of EGFR mutations predicts benefit from
Gefitinib treatment for advanced non small-cell lung cancer. J Clin
Oncol. 29:3316–3321. 2011. View Article : Google Scholar : PubMed/NCBI
|