Introduction
SIRT1 and SIRT2 are enzymes that belong to the
sirtuin family, also known as class III histone deacetylases. The
sirtuin prototype was found in yeast as a silent information
regulator 2 (Sir2). Seven mammalian sirtuins (SIRT1-SIRT7) have
been reported so far and they have roles in genome stability,
stress response, lifespan and tumorigenesis (1).
SIRT1, closest to yeast Sir2 in terms of sequence,
mediates heterochromatin formation through histone deacetylation.
Deacetylation of specific lysine residues in histone H1, H3 and H4
by SIRT1 plays an important role in chromatin regulation and
epigenetic modification. SIRT1 also has substrates which are
non-histone proteins such as p53, FOXO and Rb. Through
deacetylation of these substrates, SIRT1 is linked to a variety of
physiological functions (2). SIRT2
is mainly cytoplasmic and has been characterized for diverse
cellular functions. At first, SIRT2 was reported as a deacetylase
for α-tubulin, regulating the microtubule network (3). Furthermore, later, SIRT2 was also
found to deacetylate histone H4 during mitosis, implicating its
role as a mitotic checkpoint protein (4).
SIRT1 and SIRT2 seem to be involved in
tumorigenesis. Overexpression of SIRT1 was observed in several
tumors including leukemia, lymphoma, skin cancer, prostate cancer,
hepatocellular carcinoma, gastric carcinoma and colorectal cancer
(5). However, the function of
SIRT1 in tumorigenesis seems to be context-dependent, because there
are contradictory reports about whether SIRT1 acts as a tumor
suppressor or not. SIRT2 is involved in the regulation of cancer
cell motility with its tubulin deacetylation function.
Overexpression of SIRT2 increased cancer cell motility and
decreased sensitivity to paclitaxel treatment, a microtubule
inhibiting anticancer drug (6). In
addition, SIRT2 inhibition potentiated the effect of paclitaxel in
endothelial and tumor cells.
An anticancer effect by SIRT1/2 small molecule
inhibitors supports the role of SIRTs as tumor promoters. Several
small molecules have been discovered and proposed for cancer
therapy (2). The following are
reported as sirtuin inhibitors: EX-527 (7), splitomicin (8), sirtinol (9), cambinol (10), AGK2 (11), suramin (12), tenovins (13), salermide (14), JGB1741 (15), UVI5008 (16) and inauhzin (17). Out of these compounds, only four
compounds (tenovins, cambinol, UVI5008 and inauhzin) exhibited
in vivo antitumor effects in mouse models. The results from
sirtuin inhibitors reveal therapeutic potential for anticancer drug
development with the mechanism of SIRT inhibition.
In the search for small molecules inhibiting
SIRT1/2, toxoflavin, also known as xanthothricin, was identified
using biochemical enzymatic assay. Toxoflavin was originally known
as a toxin produced from bacteria with antibiotic function
(18). The SIRT1/2 inhibitory
action of toxoflavin was evaluated and cytotoxic activity against
cancer cells was investigated as well. With the characterization of
toxoflavin, novel and potent SIRT1/2 inhibitors with antitumor
property were presented in this study.
Materials and methods
In vitro SIRT1 assay
SIRT1 activity measurements were carried out in a
384-well black plate (7). The
enzymatic reaction was performed at room temperature in a buffer
solution containing 50 mM Tris-HCl pH 7.5, 100 mM NaCl, 7.5 mM
MgCl2, 3 mM KCl, 0.01% Tween-20, 0.1% BSA, 1 mM DTT, 125
nM biotinylated peptide with an acetylated FLAG sequence (Cisbio,
France), 130 μM NAD and the appropriate amount of SIRT1
enzyme (Biomol, Farmingdale, NY, USA). Compounds were serially
diluted ranging from 0.003 to 10 μM for SIRT1 inhibition
experiments. All reaction components except NAD were added to the
wells first and the reaction was started by adding NAD to the assay
mixture. After incubation for 3 h, the reaction was stopped by
adding the solution containing europium-cryptate conjugated
anti-FLAG antibody, streptavidin-XL665 (Cisbio) and SIRT1 inhibitor
nicotinamide. The plate was incubated further at room temperature
for 1 h. Time-resolved fluorescence resonance energy transfer
(TR-FRET) between europium cryptate (donor) and XL665 (acceptor)
was measured in EnVision multilabel reader (Perkin-Elmer, Waltham,
MA, USA). The signal ratio at 665/615 nm was used in all data
analyses.
In vitro SIRT2 assay
SIRT2 activity measurements were performed in a
96-well plate format. The substrate mixture containing biotin
labeled histone H4 peptide with an acetylated lysine 4 (Anaspec,
Fremont, CA, USA) and NAD was prepared in a buffer solution
containing 50 mM Tris-HCl (pH 8.0), 100 mM NaCl, 2 mM
MgCl2, 0.1% BSA and 1 mM DTT. The substrate mixture and
the appropriately diluted toxoflavin solution were distributed in a
96-well polystyrene plate for inhibition experiments. The enzyme
reaction was started by adding SIRT2 enzyme (SignalChem, Canada)
and processed at 37°C for 1 h with gentle shaking. Nicotinamide was
added to the wells to stop the reaction.
The resulting mixtures were transferred to the wells
of streptavidin-coated microplates (Pierce) and incubated for 1 h.
After washing three times with the phosphate-buffered saline
containing 0.05% Tween-20 (PBST), the plates were incubated with
anti-acetyl-histone H4 (Lys16) antibody (Abcam 07-329, Cambridge,
MA) for 1 h. After removing the primary antibody and washing the
plates with PBST, horse-radish peroxidase (HRP)-conjugated
secondary antibody was added and incubated for 30 min. After
incubation, the wells were emptied and washed with PBST three
times. After the final wash, HRP substrates (Applied Biosystems,
Carlsbad, CA, USA) were added to the wells and incubated at room
temperature for 1 h without shaking. The extent of the reaction was
determined by measuring the luminescence signals from the
wells.
Synthesis of toxoflavin (1) and its derivatives (2–7)
Toxoflavin (1) and
its derivatives (2–7) were synthesized according to
literature procedures (19–21).
Cell culture and cytotoxicity assay
Cell lines were cultured under a humidified 5%
CO2 incubator at 37°C in DMEM (A549) or RPMI (DU-145,
SK-OV-3, MCF-7, MKN-45, PANC-1, HEL 92.1.7 and MV-4;11)
supplemented with 10% fetal bovine serum (FBS). Cells were plated
in 96-well plate (2,000 cells/well) and serial dilutions of
toxoflavin compound were added. At the end of the incubation period
(72 h), cell viability was measured using tetrazolium-based Ez
CyTox cell viability assay kit (Daeil, Korea). Growth inhibition
(50%) (GI50) was calculated by a non-linear regression
using Prism version 5.01 (Graphpad, La Jolla, CA, USA).
Immunoblot analysis
Samples of cell extracts prepared in SDS lysis
buffer (12 mM Tris-Cl, pH 6.8, 5% glycerol, 0.4% SDS) were resolved
by SDS-PAGE and transferred to PVDF membrane. The filters were
blocked in Tris-buffered saline (10 mM Tris-Cl, pH 7.4, 140 mM
NaCl) containing 0.1% Tween-20 and 5% non-fat dry milk and then
incubated with blocking solution containing the indicated
antibodies for 2 h. The filters were visualized with HRP-coupled
secondary antibodies and enhanced chemiluminiscence reagent (Thermo
Scientific Pierce, Rockford, IL, USA). The primary antibodies used
were SIRT1 (H-300), SIRT2 (H-95), p53 (Pab 1801) (Santa Cruz
Biotechnology Inc., Dallas, TX, USA), acetylated p53 (Lys382) (Cell
Signaling Technology, Danvers, MA, USA), β-actin (Sigma, St. Louis,
MO, USA). The secondary antibodies were purchased from Jackson
ImmunoReserach (West Grove, PA, USA).
Migration assay
For measurement of cell migration, cells were
incubated with various concentrations (0–10 μM) of
toxoflavin for 24 h. Cells (100,000/well) in DMEM with 0.1% FBS
were plated to the upper chamber of Transwell (Costar 3422,
Tweksbur, MA, USA) containing a membrane with pores of 8 μm.
The cells were allowed to migrate for 24 h into the lower chamber
containing DMEM with 10% FBS. Cells on top of the Transwell were
removed by scraping and those on the bottom were stained with 1%
crystal violet solution and photographed.
Flow cytometry
A549 cells were treated with toxoflavin for 24 and
48 h, respectively. Cells were then fixed and stained with
propidium iodide (Sigma) and subjected to flow cytometry using
Accuri C6 Flow cytometer (BD Biosciences, Billerica, MA, USA). Data
were analyzed by CFlow Plus (BD Biosciences).
Results
Activity of toxoflavin on purified human
SIRT1 and SIRT2 protein
In the course of screening for SIRT1/2 inhibitors
with anticancer effects, toxoflavin was identified to affect both
SIRT1 and SIRT2 in vitro activity. Purified recombinant
protein for SIRT1 and SIRT2 were used for measurement of
deacetylase activity using TR-FRET and ELISA method, respectively.
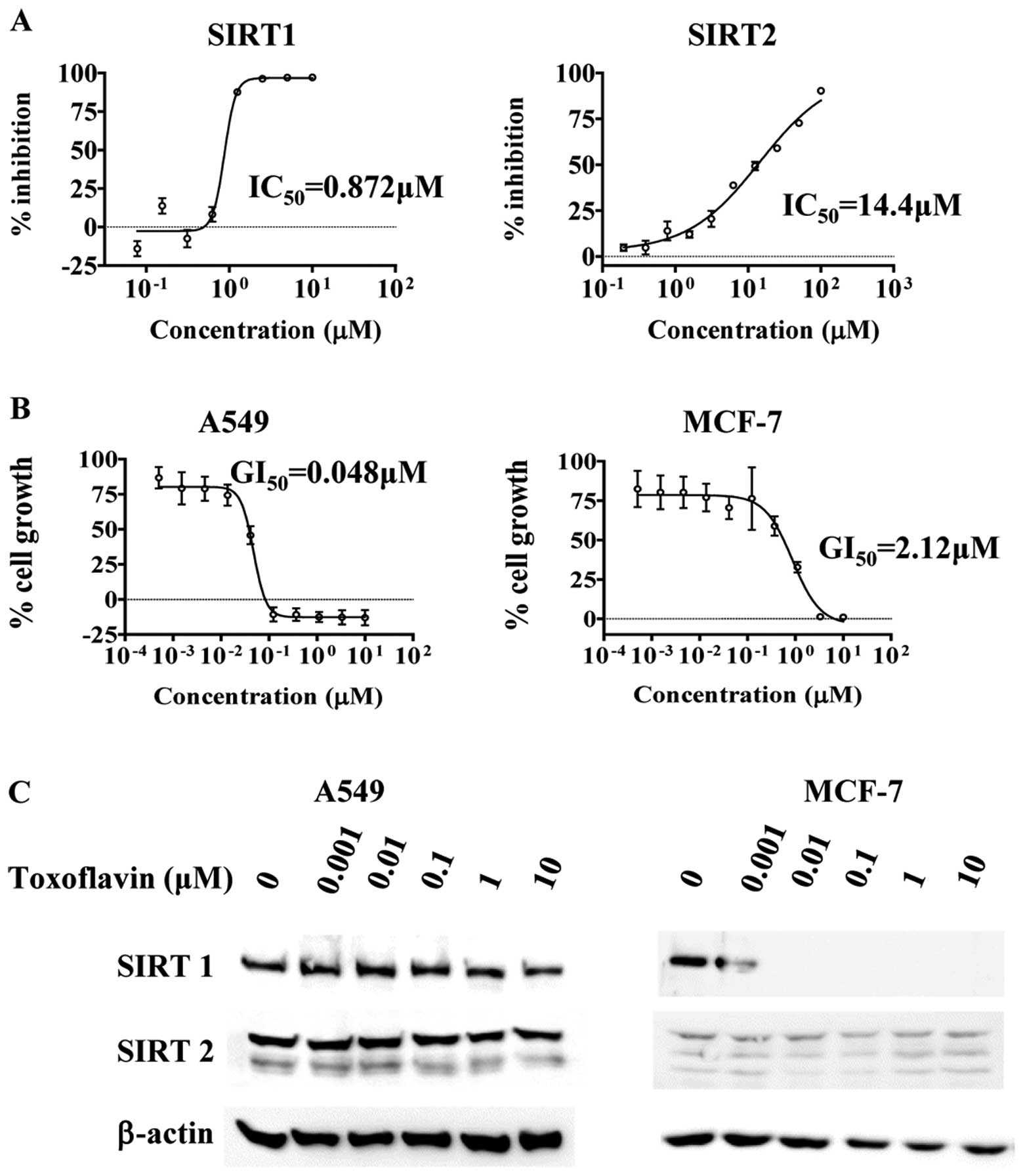
As shown in Fig. 1A, direct
inhibition of SIRT1 and SIRT2 activity by toxoflavin was observed.
Toxoflavin inhibited SIRT1 more potently than SIRT2
(IC50 0.872 μM for SIRT1 and IC50 14.4
μM for SIRT2). Potent small molecule SIRT1 inhibitor EX-527
(7), exhibited similar activity
with toxoflavin in SIRT1 inhibition (data not shown). Among SIRT1
inhibitors reported, EX-527 is one of the compounds with highest
inhibitory activity in in vitro recombinant enzyme assay
system (22). This great potency
of toxoflavin against the SIRT1 enzyme led us to further
characterize toxoflavin's anticancer effects with regard to SIRT1/2
inhibition.
Growth inhibition of various tumor cell
lines by toxoflavin treatment
Various cancer cell lines were treated with
toxoflavin, in order to investigate the effect of toxoflavin on the
growth of cancer cells (Table I).
Toxoflavin inhibited growth of A549 (lung cancer), MCF-7 (breast
cancer), SK-OV-3 (ovarian cancer), DU-145 (prostate cancer), MKN-45
(gastric cancer), PANC-1 (pancreatic cancer), U87MG (CNS cancer)
and HEL 91.1.7 (leukemia) and MV-4;11 (leukemia) cells. Non-small
cell lung cancer cell line A549 cell growth was affected by
toxoflavin with GI50 of a 48 nM and breast cancer cell
line MCF-7 cell growth was also inhibited by toxoflavin (Fig. 1B). However, the GI50 for
MCF-7 is 2.2 μM, which is 45-fold difference with A549
GI50, meaning toxoflavin is more sensitive in A549 cells
than MCF-7 cells. The expression of SIRT1 and SIRT2 proteins in
A549 and MCF-7 cells in the presence and absence of toxoflavin is
shown in Fig. 1C. In A549 cells,
SIRT1 and SIRT2 proteins are expressed and do not change with the
treatment of toxoflavin. However, SIRT1 expression in MCF-7 cells
is decreased with toxoflavin treatment dose-dependently. The
expression level of SIRT2 in MCF-7 cells is lower than that of A549
cells.
 | Table I.Growth inhibition of various tumor
cell lines by toxoflavin treatment.a |
Table I.
Growth inhibition of various tumor
cell lines by toxoflavin treatment.a
| Cell line | GI50
(μM) |
|---|
| U87MG (CNS
cancer) | 0.342 |
| SK-OV-3 (ovarian
cancer) | 0.145 |
| DU-145 (prostate
cancer) | 0.027 |
| MKN-45 (gastric
cancer) | 0.091 |
| PANC-1 (pancreatic
cancer) | 0.496 |
| HEL 92.1.7
(leukemia) | 1.042 |
| MV-4;11
(leukemia) | 0.593 |
Effects of toxoflavin on the acetylated
form of p53 and α-tubulin
After inhibition in vitro SIRT1/2 activity
and cancer cell growth by toxoflavin were confirmed, the toxoflavin
effect on the substrates of SIRT1 and SIRT2 in cell-based system
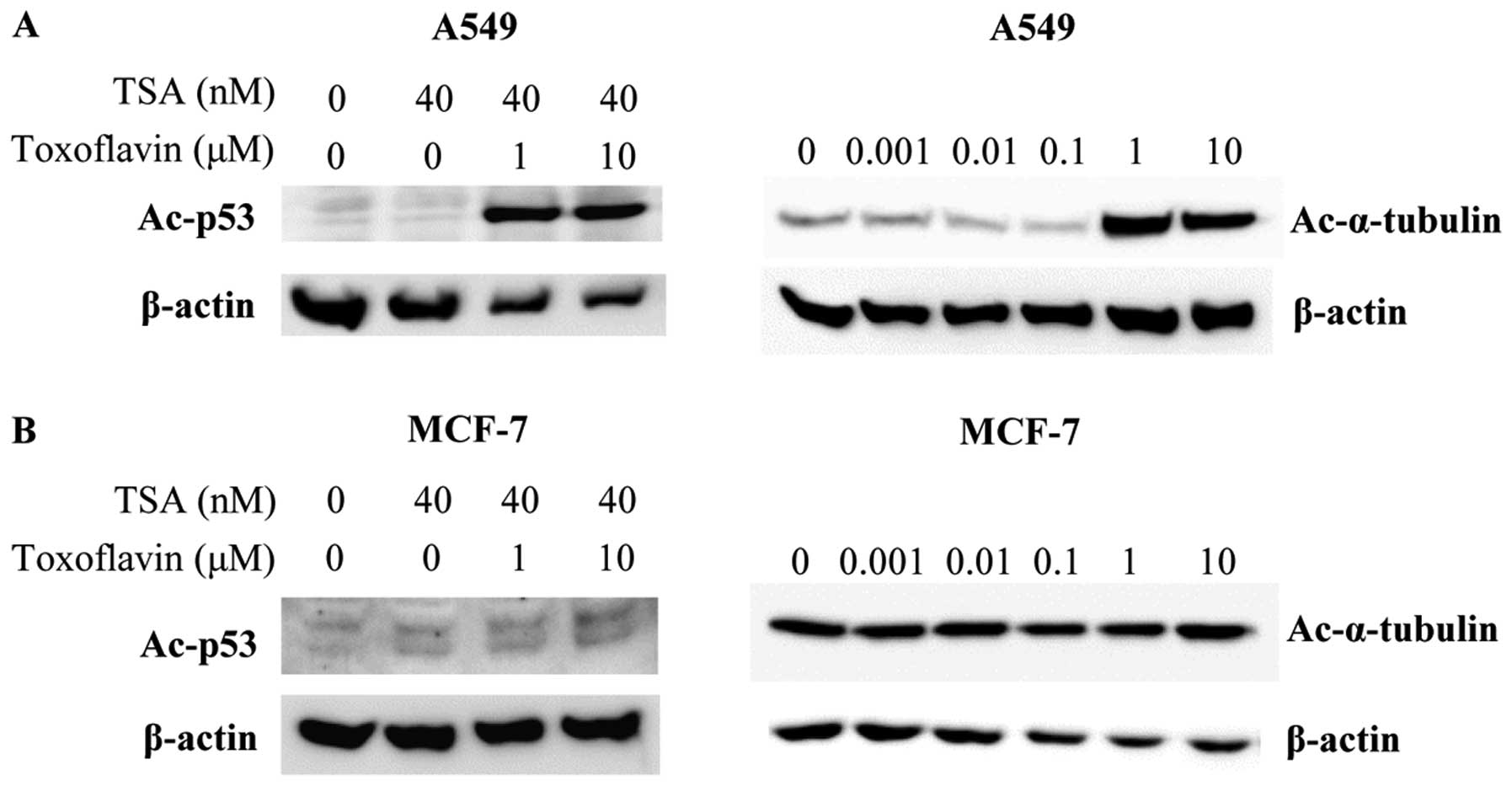
was examined. In A549 cells, toxoflavin and trichostatin A (TSA)
were treated for 6 h and acetylated p53 level was measured by
western blotting. p53 is deacetylated by SIRT1 (23,24).
Acetylated p53 level is increased by toxoflavin through inhibition
of SIRT1 deacetylase activity in the presence of TSA (Fig. 2). α-tubulin is deacetylated by
SIRT2 (4). Toxoflavin increased
the acetylated form of α-tubulin dose-dependently in A549 cells
through inhibition of SIRT2. In contrast to A549 cells, the
acetylation level of p53 and α-tubulin in MCF-7 cells was not
changed by toxoflavin treatment (Fig.
2). MCF-7 cells are not sensitive to toxoflavin treatment and
this may be related to the difference of cell growth inhibition
between A549 and MCF-7 cells.
Effects of toxoflavin on tumor cell
death
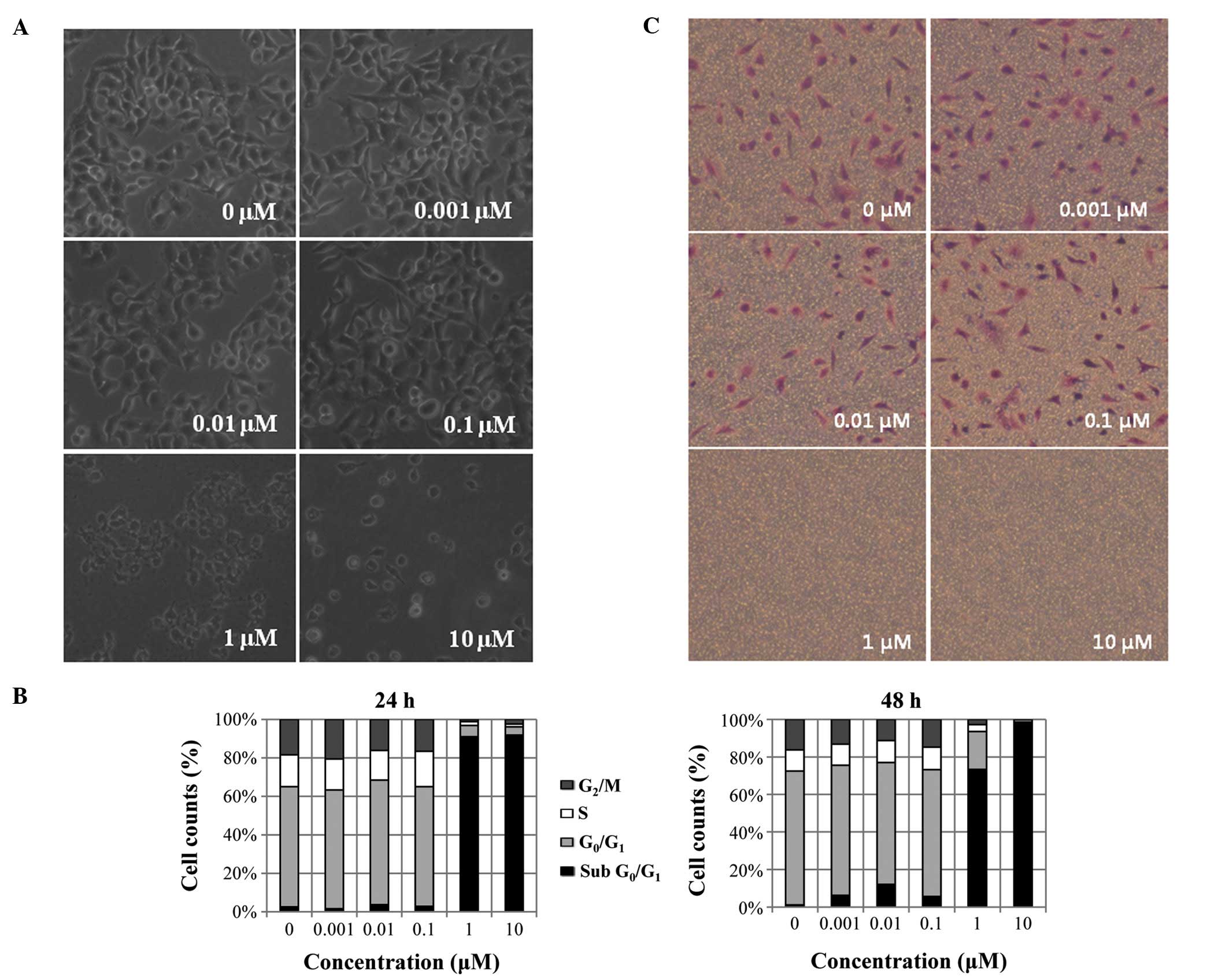
Toxoflavin induced cell death as shown in the phase
contrast image in Fig. 3A. After
72 h of toxoflavin treatment, complete cell death was observed in 1
and 10 μM of toxoflavin treated-A549 cells. Cell death was
also observed in flow cytometric results (Fig. 3B). After treatment with toxoflavin
for 24 and 48 h, A549 cells were subjected to cell cycle analysis.
The sub-G0/G1 population, indicating dead
cells, was increased by toxoflavin treatment dose-dependently.
Sub-G0/G1 population increased from 2.5 (DMSO) to 91%
(10 μM toxoflavin) with 24-h treatment and 1.1 (DMSO) to 98%
(10 μM toxoflavin) with 48-h treatment. Cell migration was
also reduced by toxoflavin. Using the Transwell assay, cell
migration was measured in the absence and presence of toxoflavin.
Migrated cells were stained with crystal violet. As shown in
Fig. 3C, dose-dependent inhibition
of cell migration was observed.
Structure-activity relationship of
toxoflavin derivatives
In order to examine the structure-activity
relationship of toxoflavin on the SIRT1/2 inhibitory activity,
several derivatives of toxoflavin were synthesized, as shown in
Table II. The inhibitory activity
of toxoflavin derivatives against SIRT1 and SIRT2 were measured
using in vitro deacetylase activity. Some of the derivatives
were more potent than toxoflavin (1) against SIRT1. However, all the
derivatives were inactive against SIRT2 in an enzymatic assay.
 | Table II.Structure-activity relationship of
toxoflavin (1)
derivatives.a |
Table II.
Structure-activity relationship of
toxoflavin (1)
derivatives.a

|
|---|
| Compound | R1 | R2 | X | SIRT1
IC50 (μM) | SIRT2
IC50 (μM) | SK-OV-3
GI50 (μM) |
|---|
| Toxoflavin
(1) | H | Me | N | 0.87 | 14.4 | 0.15 |
| 2 | H | Et | N | 0.35 | >100 | 0.55 |
| 3 | Bu | Me | N | 0.11 | >100 | 2.40 |
| 4 | p-tolyl | Me | N | 0.11 | >100 | 2.03 |
| 5 |
p-CF3-phenyl | Et | N | 0.50 | >100 | 3.20 |
| 6 |
4-bromothiophen-2-yl | Et | N | 0.92 | >100 | 17.5 |
| 7 | p-tolyl | Me | C | >100 | >100 | - |
Binding mode of the SIRT-toxoflavin
complex
To understand the interaction of the SIRT-toxoflavin
complex, a docking experiment was performed using the shape-based
docking algorithm, LigandFit in the Discovery Studio 3.5 (25). The crystal structure of SIRT2 used
was taken from the human sirtuin C-pocket (PDB entry; 1J8F)
(26). As the crystal structure of
human SIRT1 has not been resolved yet, a homology model obtained
through the hierarchical protein structure modeling approach based
on secondary-structure enhanced Profile-Profile threading Alignment
(PPA) and the iterative implementation of the Threading ASSEmbly
Refinement (TASSER) program (27).
The proteins were prepared in Discovery Studio 3.5, with the
default parameters. The starting conformation was a low-energy
conformer generated using modified CHARMm force field-based 3D
structure minimization implemented in Discovery Studio's ‘prepare
ligands’ protocol (28). Results
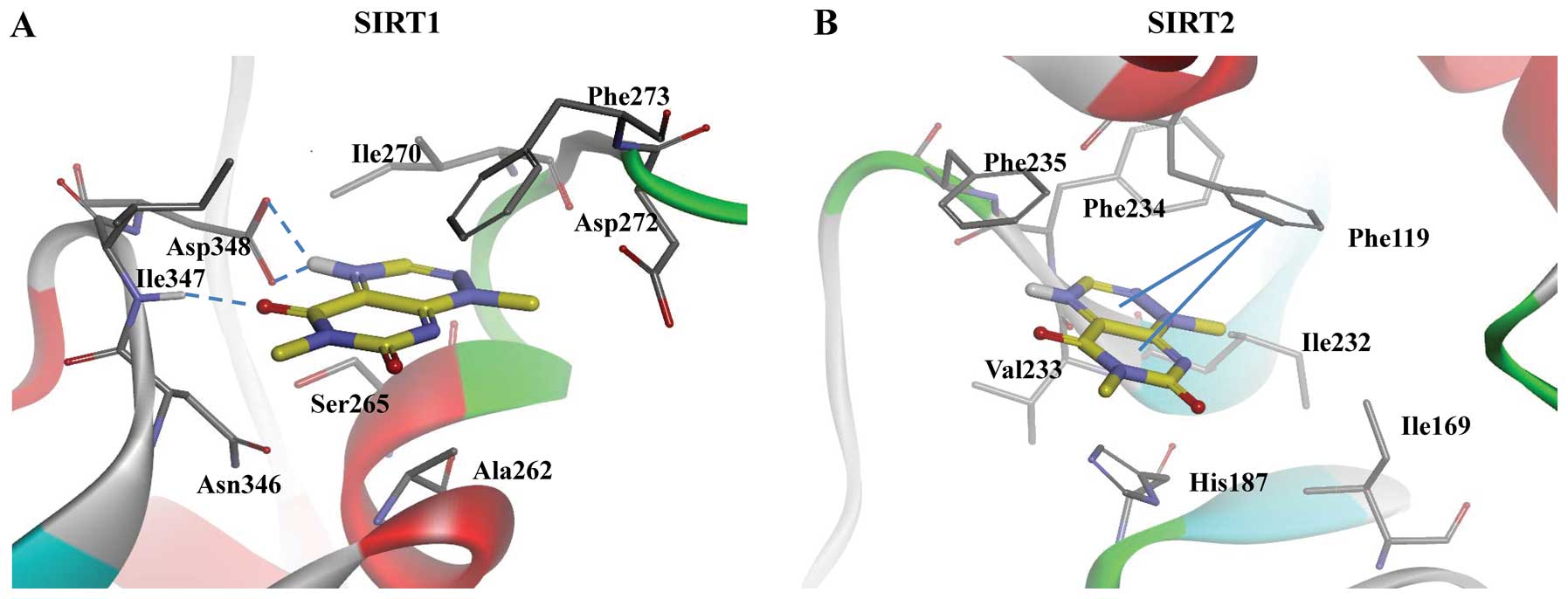
of the docking studies suggested that toxoflavin would dock
strongly into the binding site of the ribose and nicotinamide
moieties of NAD+ in SIRT1 and SIRT2. The strong
interaction of toxoflavin with SIRT1 is attributed to the H-bonding
interaction of both side chains of Asp348 and backbone - NH of
Ile347 residues, as shown in Fig.
4. While the interaction of toxoflavin with SIRT2 forms a
π-stacking interaction with Phe119, H-bonding interaction is not
present. The binding energy of SIRT1- and SIRT2-toxoflavin
complexes were −181.20 and −121.81 kcal/mol, respectively,
suggesting toxoflavin could bind to SIRT1 with an improved affinity
compared with that of SIRT2.
Discussion
Sirtuins have received tremendous attention in the
research fields of cancer, metabolic diseases, neurodegeneration
and aging (2). Thus, small
molecule sirtuin modulators of sirtuins have been explored
targeting these diseases. SIRT1 activators are pursued for
metabolic/neurodegenerative diseases and SIRT inhibitors are
investigated in the oncology area. Controversy exists on the role
of SIRT1 in tumorigenesis. Some evidence supports the role of SIRT1
as a tumor suppressor and some reports show that SIRT1 has an
oncogenic function (1,29). Thus, the identification of SIRT
inhibitors will be important to elucidate the role of SIRT in
tumorigenesis. Identified SIRT inhibitors can be useful tools for
proving the concept of SIRT as a therapeutic target for cancer.
Toxoflavin was isolated 80 years ago from
Pseudomonas (30) and its
toxicity and antibiotic function are presumed to originate from
inhibition of the respiratory chain (31). Toxoflavin derivatives were
identified as actives in several high-throughput screening
campaigns. Screening for Polo-like kinase 1, Akt and NIMA-related
kinase 2 resulted in a series of compounds containing toxoflavin
core structure (32–34). However, detailed characterization
regarding the kinase inhibition was not performed. In this study,
the screening of SIRT1 inhibitory activity identified toxoflavin as
a potent in vitro SIRT1 modulator. EX-527 (also known as
SEN0014196 and selisistat) is currently the strongest SIRT1
inhibitor in terms of in vitro activity (5). However, SIRT1-inhibitory activity is
not observed in cell-based assay (data not shown). Toxoflavin has
an in vitro activity similar to EX-527 and exhibits strong
SIRT1 inhibition in cell-based assay (Fig. 2). Thus toxoflavin can be a better
chemical probe than EX527 in the sirtuin research field.
In the inhibition of in vitro deacetylase
activity, toxoflavin showed higher selectivity toward SIRT1
(IC50 0.872 μM) than SIRT2 (IC50 14.4
μM) (Fig. 1A). In western
blotting for acetylated substrates of SIRT1 and SIRT2, however,
toxoflavin exhibited similar activity toward both SIRT1 and SIRT2
(Fig. 2). The acetylated form of
p53 and α-tubulin were increased by toxoflavin from 1 μM
concentration. The inconsistency between the in vitro
activity and cell-based activity may be due to the complex sirtuin
biological system and remains to be elucidated.
Toxoflavin did not affect the acetylated level of
p53 and α-tubulin in MCF-7 cells, while levels in A549 cells were
increased to a great degree (Fig.
2). The α-tubulin acetylation level change toxoflavin was also
examined in various other cell lines (data not shown). There are
increases in SK-OV-3, DU-145, MKN-45 and U87MG cells. In addtion,
in PANC-1 and MCF-7 cells, change in α-tubulin acetylation level
was not observed up to 10 μM of toxoflavin treatment. There
are various factors affecting these differences, such as SIRT2
expression level and basal α-tubulin acetylation level in cell
lines. It will be important to establish the mechanism of
toxoflavin sensitivity in order to find biomarkers for SIRT
inhibitors.
The preliminary study of structure-activity
relationship for toxoflavin revealed that the triazine ring is
critical to inhibitory potencies against SIRT1/2. Interestingly,
substitution of the N-methyl group with N-ethyl group and
replacement of hydrogen in the triazine ring of toxoflavin with
alkyl and aryl groups dramatically reduced SIRT2 inhibitory
potency. Moreover, replacement of triazine with diazine abolished
the inhibitory effect of toxoflavin derivatives against both SIRT1
and SIRT2.
SIRT1/2 are emerging as therapeutic targets for
cancer and identification of SIRT1/2 inhibitors will expedite the
development of an anticancer agent with a SIRT1/2 inhibitory
mechanism. Here, toxoflavin is described as a potent and direct
SIRT1/2 inhibitor with cytotoxic activity. Although clinical
development of toxoflavin is limited by its toxicity, toxoflavin
and its derivatives will serve as great chemical probes for SIRT1
and in the anticancer agent research area.
Acknowledgements
This study was supported by a grant of
the NRF (2011-0010374) funded by the government of Korea (MEST), a
grant from the Ministry of Trade, Industry and Energy (MOTIE),
Korea Institute for Advancement of Technology (KIAT) through
inter-ER cooperation projects (A004500005) and a grant
(12182MFDS666) from Ministry of Food and Drug Safety.
References
|
1.
|
Bosch-Presegue L and Vaquero A: The dual
role of sirtuins in cancer. Genes Cancer. 2:648–662. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Carafa V, Nebbioso A and Altucci L:
Sirtuins and disease: the road ahead. Front Pharmacol. 3:42012.
View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
North BJ, Marshall BL, Borra MT, Denu JM
and Verdin E: The human Sir2 ortholog, SIRT2, is an
NAD+-dependent tubulin deacetylase. Mol Cell.
11:437–444. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Vaquero A, Scher MB, Lee DH, et al: SirT2
is a histone deacetylase with preference for histone H4 Lys 16
during mitosis. Genes Dev. 20:1256–1261. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Stunkel W and Campbell RM: Sirtuin 1
(SIRT1): the misunderstood HDAC. J Biomol Screen. 16:1153–1169.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Bonezzi K, Belotti D, North BJ, et al:
Inhibition of SIRT2 potentiates the anti-motility activity of
taxanes: implications for antineoplastic combination therapies.
Neoplasia. 14:846–854. 2012.PubMed/NCBI
|
|
7.
|
Napper AD, Hixon J, McDonagh T, et al:
Discovery of indoles as potent and selective inhibitors of the
deacetylase SIRT1. J Med Chem. 48:8045–8054. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
8.
|
Bedalov A, Gatbonton T, Irvine WP,
Gottschling DE and Simon JA: Identification of a small molecule
inhibitor of Sir2p. Proc Natl Acad Sci USA. 98:15113–15118. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Mai A, Massa S, Lavu S, et al: Design,
synthesis, and biological evaluation of sirtinol analogues as class
III histone/protein deacetylase (Sirtuin) inhibitors. J Med Chem.
48:7789–7795. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Heltweg B, Gatbonton T, Schuler AD, et al:
Antitumor activity of a small-molecule inhibitor of human silent
information regulator 2 enzymes. Cancer Res. 66:4368–4377. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Outeiro TF, Kontopoulos E, Altmann SM, et
al: Sirtuin 2 inhibitors rescue alpha-synuclein-mediated toxicity
in models of Parkinson's disease. Science. 317:516–519.
2007.PubMed/NCBI
|
|
12.
|
Trapp J, Meier R, Hongwiset D, Kassack MU,
Sippl W and Jung M: Structure-activity studies on suramin analogues
as inhibitors of NAD+-dependent histone deacetylases
(sirtuins). ChemMedChem. 2:1419–1431. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Lain S, Hollick JJ, Campbell J, et al:
Discovery, in vivo activity, and mechanism of action of a
small-molecule p53 activator. Cancer Cell. 13:454–463. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Lara E, Mai A, Calvanese V, et al:
Salermide, a Sirtuin inhibitor with a strong cancer-specific
proapoptotic effect. Oncogene. 28:781–791. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15.
|
Kalle AM, Mallika A, Badiger J, Alinakhi,
Talukdar P and Sachchidanand: Inhibition of SIRT1 by a small
molecule induces apoptosis in breast cancer cells. Biochem Biophys
Res Commun. 401:13–19. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Nebbioso A, Pereira R, Khanwalkar H, et
al: Death receptor pathway activation and increase of ROS
production by the triple epigenetic inhibitor UVI5008. Mol Cancer
Ther. 10:2394–2404. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17.
|
Zhang Q, Zeng SX, Zhang Y, et al: A small
molecule Inauhzin inhibits SIRT1 activity and suppresses tumour
growth through activation of p53. EMBO Mol Med. 4:298–312. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Machlowitz RA, Fisher WP, Betsey McKay S,
Tytell AA and Charney J: Xanthothricin, a new antibiotic. Antibiot
Chemother. 4:259–261. 1954.PubMed/NCBI
|
|
19.
|
Black TH: An improved, large-scale
synthesis of xanthothricin and reumycin. J Heterocyclic Chem.
24:1373–1375. 1987. View Article : Google Scholar
|
|
20.
|
Spinks D, Shanks EJ, Cleghorn LA, et al:
Investigation of trypanothione reductase as a drug target in
Trypanosoma brucei. ChemMedChem. 4:2060–2069. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Todorovic N, Giacomelli A, Hassell JA,
Frampton CS and Capretta A: Microwave-assisted synthesis of
3-arylpyrimido[5,4-e][1,2,4]triazine-5,7(1H,6H)-dione libraries:
derivatives of toxoflavin. Tetrahedron Lett. 51:6037–6040.
2010.
|
|
22.
|
Peck B, Chen CY, Ho KK, et al: SIRT
inhibitors induce cell death and p53 acetylation through targeting
both SIRT1 and SIRT2. Mol Cancer Ther. 9:844–855. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Lu X, Magrane G, Yin C, Louis DN, Gray J
and Van Dyke T: Selective inactivation of p53 facilitates mouse
epithelial tumor progression without chromosomal instability. Mol
Cell Biol. 21:6017–6030. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Vaziri H, Dessain SK, Ng Eaton E, et al:
hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell.
107:149–159. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
25.
|
Venkatachalam CM, Jiang X, Oldfield T and
Waldman M: LigandFit: a novel method for the shape-directed rapid
docking of ligands to protein active sites. J Mol Graph Model.
21:289–307. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Finnin MS, Donigian JR and Pavletich NP:
Structure of the histone deacetylase SIRT2. Nat Struct Biol.
8:621–625. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Wu S, Skolnick J and Zhang Y: Ab initio
modeling of small proteins by iterative TASSER simulations. BMC
Biol. 5:172007. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Brooks BR, Bruccoleri RE, Olafson BD,
States DJ, Swaminathan S and Karplus M: CHARMM: A program for
macromolecular energy, minimization, and dynamics calculations. J
Comput Chem. 4:187–217. 1983. View Article : Google Scholar
|
|
29.
|
Song S-H, Lee M-O, Lee J-S, Oh J-S, Cho
S-U and Cha H-J: Sirt1 promotes DNA damage repair and cellular
survival. Biomol Ther. 19:282–287. 2011. View Article : Google Scholar
|
|
30.
|
van Veen AG and Mertens WK: Das
Toxoflavin, der gelbe Giftstoff der Bongkrek. Rec Trav Chim.
53:398–404. 1934.PubMed/NCBI
|
|
31.
|
Latuasan HE and Berends W: On the origin
of the toxicity of toxoflavin. Biochim Biophys Acta. 52:502–508.
1961. View Article : Google Scholar : PubMed/NCBI
|
|
32.
|
Goh KC, Wang H, Yu N, et al: PLK1 as a
potential drug target in cancer therapy. Drug Dev Res. 62:349–361.
2004. View Article : Google Scholar
|
|
33.
|
Hayward DG, Newbatt Y, Pickard L, et al:
Identification by high-throughput screening of viridin analogs as
biochemical and cell-based inhibitors of the cell cycle-regulated
nek2 kinase. J Biomol Screen. 15:918–927. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
34.
|
Burns S, Travers J, Collins I, et al:
Identification of small-molecule inhibitors of protein kinase B
(PKB/AKT) in an AlphaScreen™ high-throughput screen. J Biomol
Screen. 11:822–827. 2006.PubMed/NCBI
|