Introduction
Fusion genes are frequently observed in hematologic
malignancies and soft tissue sarcomas (1). These fusion genes are usually
associated with chromosome abnormalities such as translocations,
inversions, and deletions, but have also been identified in cryptic
chromosome abnormalities. Fusion genes have also been identified in
various solid tumors, including ETS-family fusion genes in prostate
cancer (2), ETV6-NTRK3 in
secretory breast cancer (3), and
ALK fusion genes in lung cancer (4). Many of these fusion genes create
in-frame fusion transcripts that result in the production of fusion
proteins, and some of which aid tumorigenesis. These fusion
proteins are often associated with disease phenotype and clinical
outcome, and act as markers for minimal residual disease and
indicators of therapeutic targets. However, several fusion genes
that do not create in-frame fusion transcripts have also been
identified. Oncogenic rearrangements of immunoglobulin (IG)
or T-cell receptor (TCR) genes are well-known fusion genes,
and some of these create fusion transcripts, such as
1GH-BACH2 by t(6;14)(q15;q32) in B-cell lymphoma/leukemia
(5), IGH-MMSET by
t(4;14)(p16.3;q32.3) (6) and
Cα-IRTA1 by t(1;14)(q21;q32) (7) in multiple myeloma, and
BCL11B-TRDC by inv(14)(q11.2q32.31) in T-cell acute
lymphoblastic leukemia (8).
Furthermore, chromosome abnormalities led to fusion transcripts in
the non-coding gene PVT1 such as PVT1-NBEA and
PVT1-WWOX in multiple myeloma (9), PVT1-CHD7 in small-cell lung
carcinoma (10), and
PVT1-MYC and PVT1-NDRG1 in medulloblastoma (11). The role of PVT1-fusions is
uncertain, but they may represent another type of fusion transcript
in cancer cells and possibly in normal cells.
In the present study, we unexpectedly identified
additional fusion genes involving 28S ribosomal DNA (RN28S1)
in hematologic malignancies.
Materials and methods
Clinical sample and cell lines
Leukemic cells from a 15-year-old boy with
mixed-lineage (T/myeloid) acute leukemia having t(6;14)(q25;q32)
were analyzed after obtaining informed consent from the patient’s
parents. In addition, 14 B-cell non-Hodgkin’s lymphoma, 11 multiple
myeloma, 4 B-cell precursor acute lymphoblastic leukemia cell
lines, and 3 EB virus-transformed B-cell lines from normal healthy
volunteers were analyzed; the cell lines were as described
previously (5). The Institutional
Review Board of Kyoto Prefectural University of Medicine approved
this study.
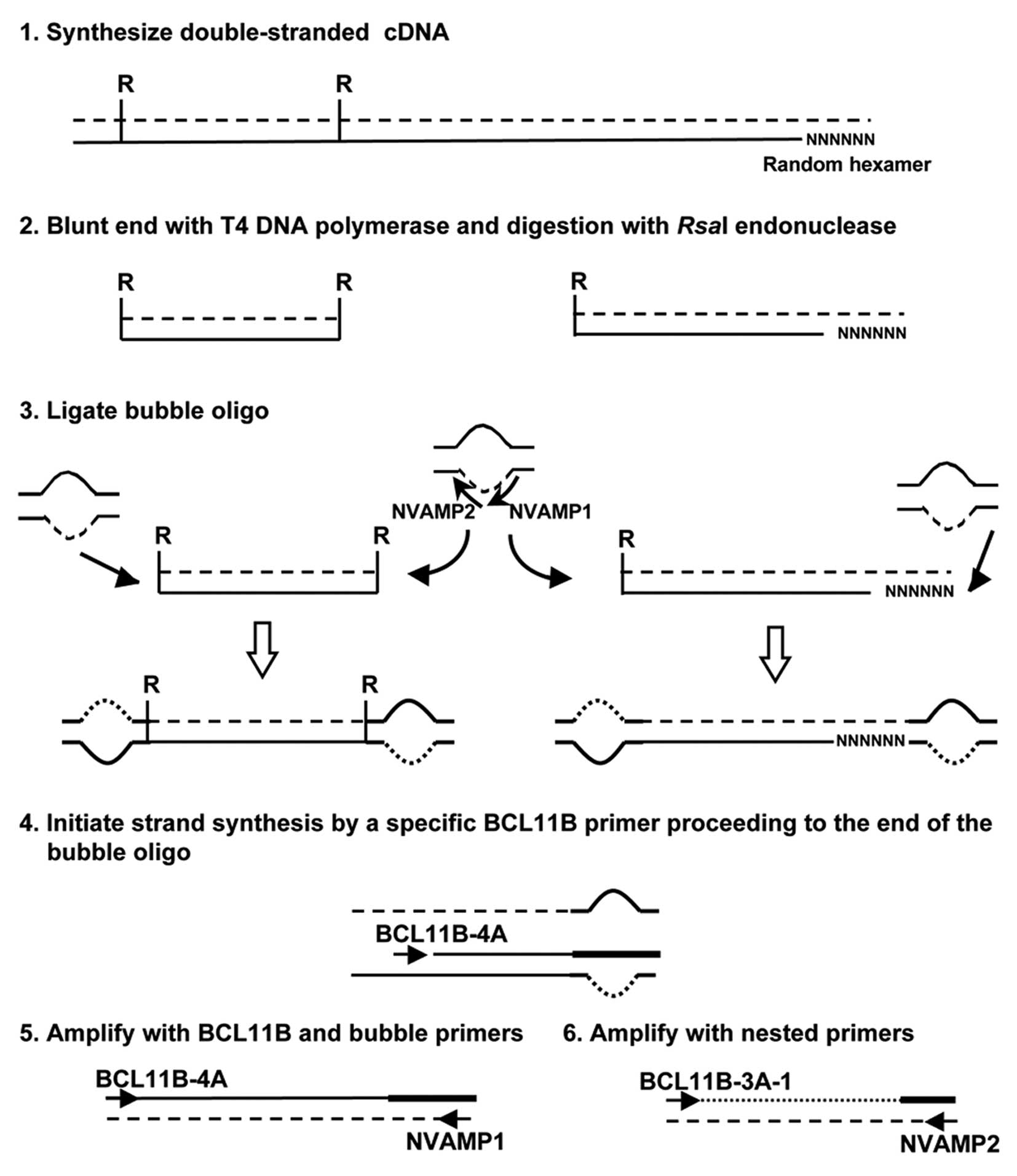
cDNA bubble PCR
cDNA bubble PCR was used to detect the fusion
partner of BCL11B, as described previously (Fig. 1) (12). Total RNA was used to generate
double-stranded cDNA. Primers used were: NVAMP-1 and BCL11B-4A for
first-round PCR, and NVAMP-2 and BCL11B-3A-1 for second-round PCR
(Fig. 1 and Table I).
 | Table I.Primers used for PCR. |
Table I.
Primers used for PCR.
| Primer | Sequence 5′-3′ |
|---|
| BCL11B-3A-1 |
ACGCAGAGGTGAAGTGATCAC |
| BCL11B-3A-2 |
GACAACTGACACTGGCATCC |
| BCL11B-4A |
ACCACGCGCTGTTGAAGGG |
| RN28S1-GA1 |
CCTTAGCGGATTCCGACTTCCAT |
| RN28S1-GA2 |
GTCCTGCTGTCTATATCAACCAACAC |
| IGKV3-20-2F |
GGCTCCTCATCTATGGTGCATC |
| COG1-11F |
AACAGCAACCTTCATCGCCTG |
Reverse transcription (RT)-PCR
RT-PCR analysis was performed as described
previously (5). Primers used for
the detection of BCL11B-RN28S1 fusion transcripts were
BCL11B-3A-2 and RN28S1-GA1 for first-round PCR, and BCL11B-3A-1 and
RN28S1-GA2 for second-round PCR; those for IGKV3-20-RN28S1
were IGKV3-20-2F and RN28S1-GA1; and those for COG1-RN28S1
were COG1-11F and RN28S1-GA1 (Table
I).
Nucleotide sequencing
Nucleotide sequences of PCR products and, if
necessary, subcloned PCR products were analyzed as previously
described (5).
Results
Identification of RN28S1-BCL11B fusion
transcript by cDNA bubble PCR method
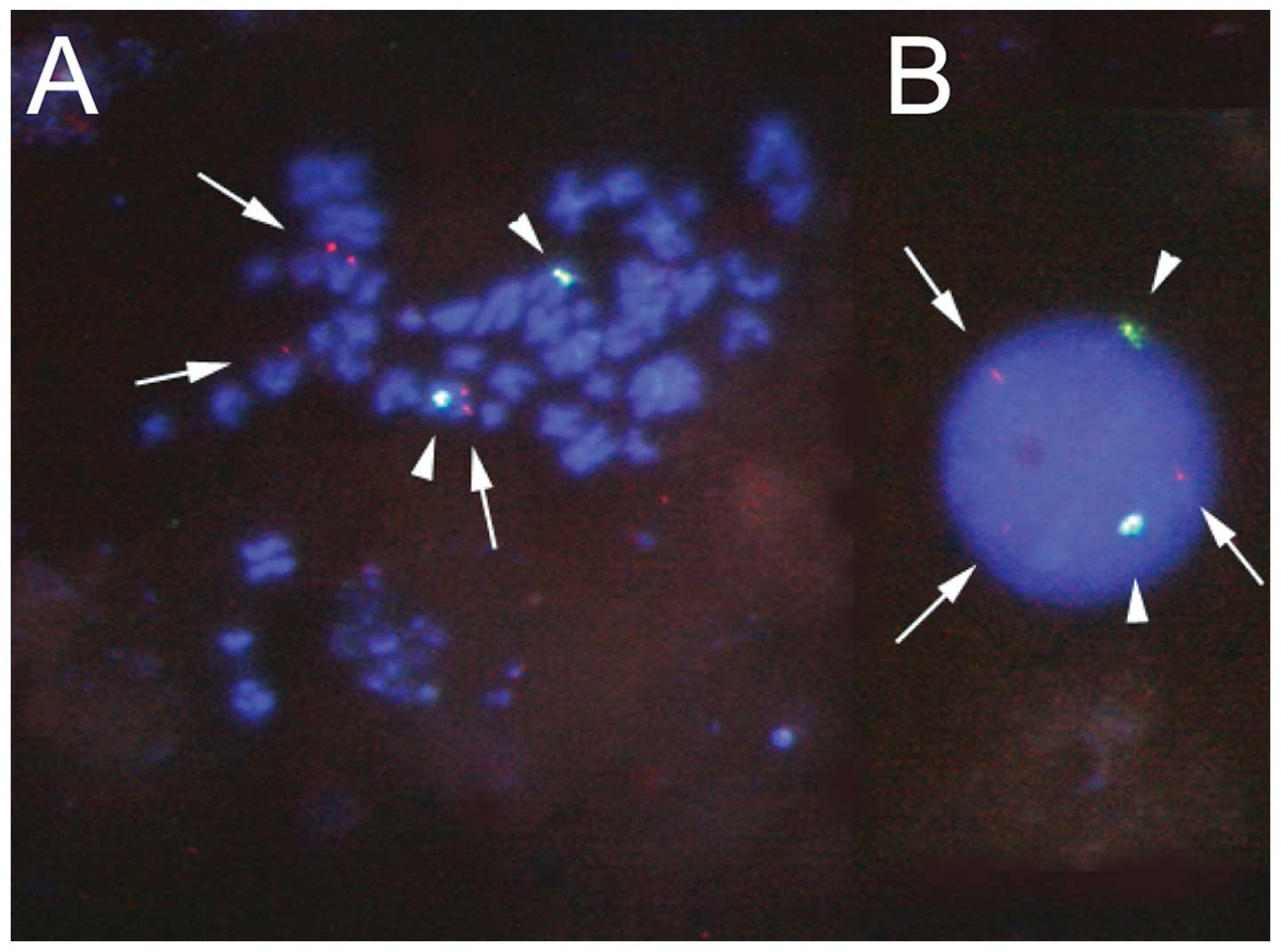
Leukemic cells from a patient with mixed-lineage
acute leukemia having t(6;14)(q25;q32) were analyzed using
fluorescence in situ hybridization analysis, and
rearrangement of the B-cell leukemia/lymphoma 11B (BCL11B)
gene was suggested (Fig. 2). Thus,
we performed cDNA bubble PCR to identify the gene on chromosome
6q25 that was fused to the BCL11B gene on 14q32. Sequence
analysis of multiple products amplified by second-round PCR of
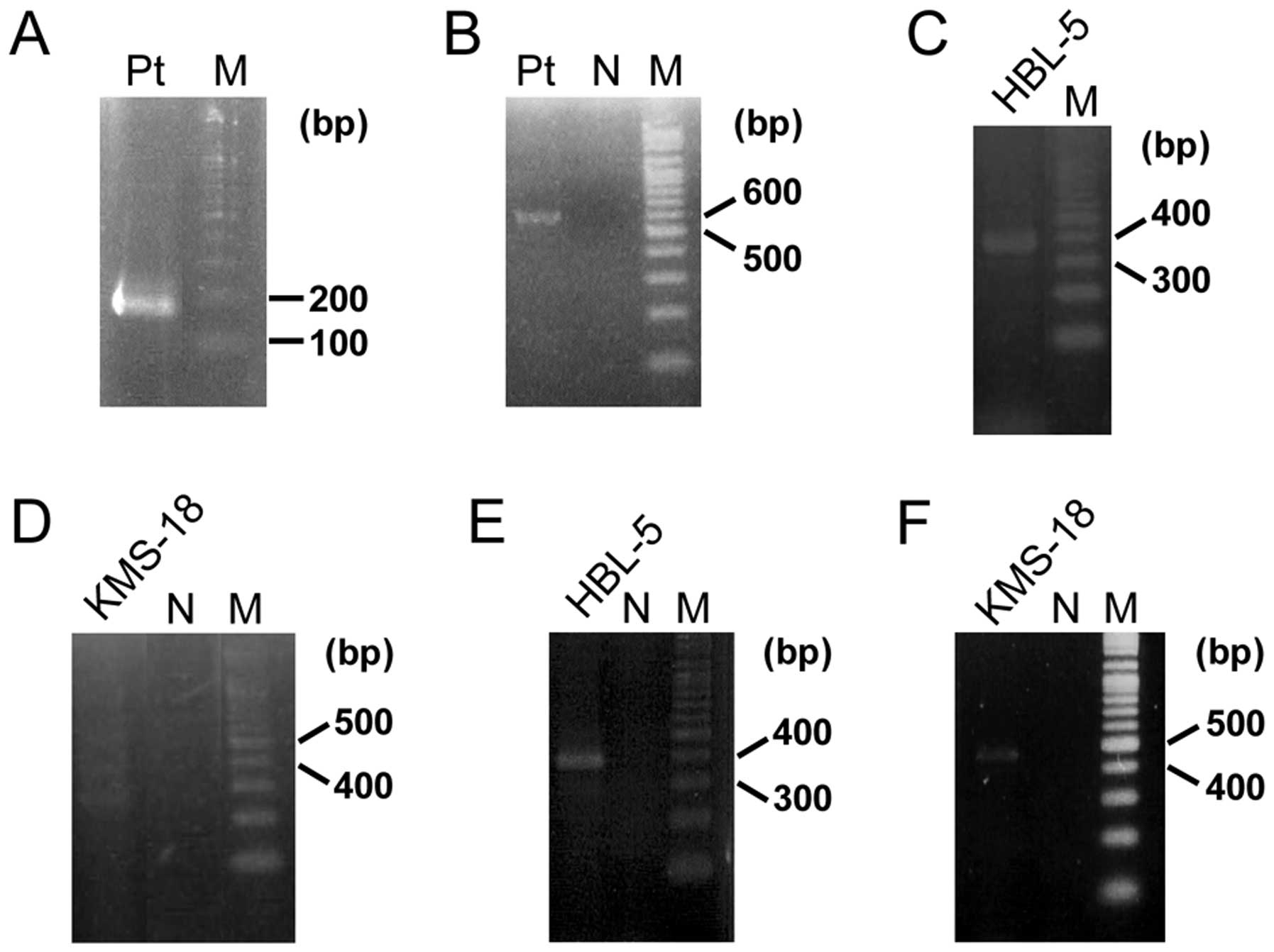
bubble PCR detected a product that contained a 34-bp sequence of
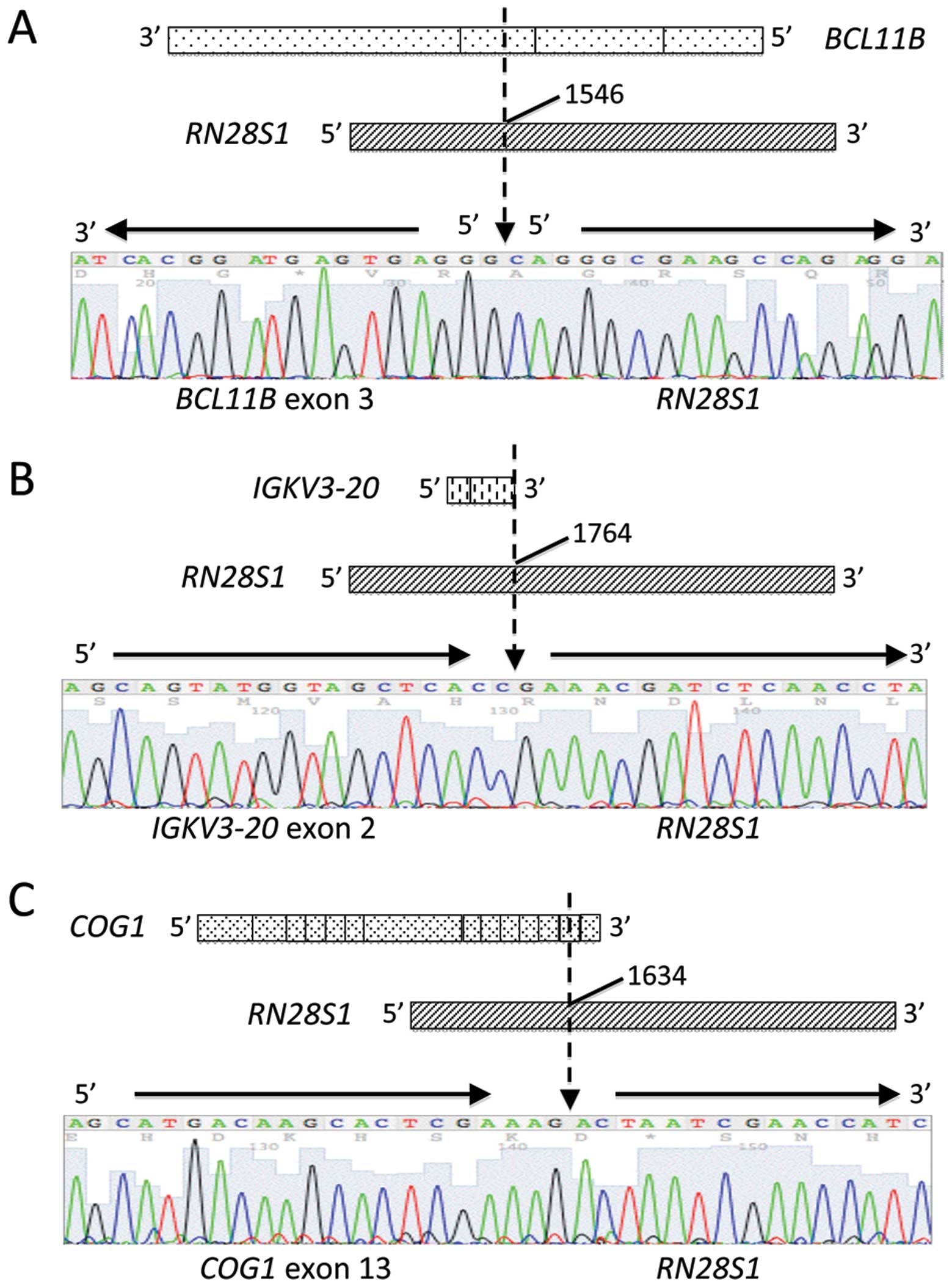
BCL11B exon 3 fused to an unknown 96-bp sequence (Fig. 3A). A BLAST search revealed that the
unknown sequence was 28S ribosomal DNA (RN28S1) (Fig. 4A). This fusion transcript was
confirmed by RT-PCR (Fig. 3B). The
transcriptional directions of the contributing genes in the fusion
transcript were opposed, and the fusion point of BCL11B was
within the exon (Fig. 4A). No
sequences from chromosome 6q25 were identified.
Accidental detection of IGK-RN28S1 and
COG1-RN28S1 fusion transcripts in B-cell malignancy cell lines
RN28S1 is one of three ribosomal DNAs
encoding the 18S, 5.8S and 28S rRNAs, which exist in nucleolar
organizer regions on the five acrocentric chromosomes (13p, 14p,
15p, 21p and 22p). Therefore, in this case, the
RN28S1-BCL11B fusion transcript was considered not to be
generated by the chromosome translocation of t(6;14)(q25;q32).
Thus, we could infer that a mechanism other than chromosome
abnormalities was involved in the creation of the
RN28S1-BCL11B fusion transcript. To analyze whether the
RN28S1-BCL11B fusion transcripts were expressed in other
hematologic malignancy cell lines, RT-PCR using RN28S1 and
BCL11B primers was performed. PCR products were detected in
several cell lines. In the Burkitt lymphoma cell line HBL-5, a
367-bp PCR product (Fig. 3C),
contained 119-bp of immunoglobulin κ variable 3-20
(IGKV3-20) exon 2 fused to an RN28S1 sequence
(Fig. 4B). The multiple myeloma
cell line KMS-18, amplified multiple PCR products (Fig. 3D) including a 441-bp product that
resulted from a 62-bp sequence of the component of oligomeric Golgi
complex 1 (COG1) gene fused to an RN28S1 sequence
(Fig. 4C). These fusions both
occurred within the exons of IGKV3-20 and COG1, as in
the RN28S1-BCL11B fusion, and they were confirmed by RT-PCR
using each specific primer (Fig. 3E
and F).
Comparison of the sequence of BCL11B primer with
those of IGKV3-20 and COG1 found similarities between
them, with six mismatches in 20 nucleotides, suggesting that
annealing of the primer to the similar sequences of IGK and
COG1 made it possible to amplify these fusion genes by
chance (Fig. 5). Other products
amplified by RT-PCR for the detection of RN28S1-BCL11B were
either the normal sequence of RN28S1 or non-specific
sequences.
Discussion
In the present study, we identified three novel
fusion transcripts involving RN28S1. Only one
RN28S1-related fusion gene, RN28S1-BCL6, has been
previously reported, and this was in a case of gastric lymphoma
(13). RN28S1 is a gene
that would not translate to protein, and the breakpoints in partner
genes of RN28S1 were within the coding exons. Notably,
BCL11B and RN28S1 were fused with opposite
transcription directions. These findings suggest that fusion genes
involving RN28S1 do not produce fusion proteins, but that
disruption of the normal functions of fusion partners contribute to
tumorigenesis in these cases.
Several studies have noted an association of
ribosomal DNA (RNA) with tumorigenesis. rRNA transcription and
ribosome biogenesis are controlled by several cancer-related genes
through the PI3 kinase/mTOR, MYC or RAS/ERK pathways (14). Proto-oncoprotein MYC controls
ribosome biogenesis through the regulation of transcription by all
three RNA polymerases (15).
Another cancer-related gene, nucleophosmin (NPM1), which is
frequently mutated in acute myeloid leukemia with a normal
karyotype and creates fusion genes with ALK in anaplastic
large cell lymphoma with t(2;5) (p23;q35), is necessary for
MYC-mediated rRNA synthesis (16).
RN28S1 has three MYC-binding sites (17), and the breakpoints within
RN28S1 in our cases were to the 5’ side of the MYC-binding
sites (Fig. 6), suggesting that
the RN28S1-fusion transcripts we detected are associated
with tumorigenesis through the dysregulation of MYC-mediated rRNA
synthesis. In other correlations of rDNA with tumorigenesis, a high
frequency of rDNA rearrangements was noted in lung and colon
cancers (18) and overexpression
of rDNA was seen in prostatic cancer (19). The breakpoints of RN28S1 in
our three cases were within an ∼600 bp region (Fig. 6); these may be related to
recombinational hot spots in repetitive sequences.
The three genes fused to RN28S1 that we found
in our study are related to tumorigenesis in certain types of
malignancies. IGK is one of the immunoglobulin light chain
genes that is frequently rearranged by chromosome translocations,
such as t(2;8)(p11;q24) in B-cell malignancies (20). Rearrangement of the BCL11B
gene locus is observed in T-cell malignancies, and three fusion
transcripts involving BCL11B have been identified:
TLX3-BCL11B fusion gene by cryptic t(5;14)(q35;q32.2) in
T-ALL (21), BCL11B-TRDC by
inv(14) (q11.2q32.31) in T-ALL (8)
and HELIOS-BCL11B by t(2;14) (q34;q32) in adult T-cell
leukemia (22). COG1 is a
component of the conserved oligomeric Golgi (COG) complex, Golgi
transport complex, that is involved in glycosylation reactions and
vesicular transport (23).
Although the COG1 gene has not been firmly associated with
tumorigenesis to date, one COG family gene, COG5, was
found fused to HMGA2 in uterine leiomyoma (24), suggesting a possible link with
tumorigenesis. Although the fusion genes we identified in this
study were not related to chromosome translocations, each gene is
involved in tumorigenesis in some way, suggesting that
RN28S1-related fusion genes play some roles in
tumorigenesis.
Molecular analysis of ribosomal DNA is challenging
due to its repetitive nature. In this study, we attempted to
confirm the three fusions at the genomic level; however, genomic
PCR was unsuccessful. A possible explanation for the failure of
amplification of genomic junctions is the quantitative imbalance of
genomic DNAs between partner genes and RN28S1 due to the
∼400 copies of ribosomal DNA in a diploid human genome.
Whole-transcriptome analysis by next-generation sequencing is
usually a powerful tool for the detection of fusion transcripts.
However, detection of RN28S1 fusion transcripts using this
method is difficult, because ribosomal RNAs, which comprises
>95% of total RNA, is usually removed from total RNA prior to
sequencing. While further analysis is needed to clarify the role of
rDNA in tumorigenesis, our findings provide an important insight
into the role of rDNA function in human genomic architecture and
tumorigenesis.
Acknowledgements
We express appreciation to Akari
Kazami for the outstanding technical assistance. This study was
supported by a research program of the Project for Development of
Innovative Research on Cancer Therapeutics (P-Direct), a
Grant-in-Aid for Cancer Research from the Ministry of Health, Labor
and Welfare of Japan, and a Grant-in-Aid for Scientific Research
(C) from the Ministry of Education, Culture, Sports, Science and
Technology of Japan.
References
|
1.
|
Taki T and Taniwaki M: Chromosomal
translocations in cancer and their relevance for therapy. Curr Opin
Oncol. 18:62–68. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2.
|
Tomlins SA, Rhodes DR, Perner S, et al:
Recurrent fusion of TMPRSS2 and ETS transcription factor genes in
prostate cancer. Science. 310:644–648. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Euhus DM, Timmons CF and Tomlinson GE:
ETV6-NTRK3 - Trk-ing the primary event in human secretory breast
cancer. Cancer Cell. 2:347–348. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Soda M, Choi YL, Enomoto M, et al:
Identification of the transforming EML4-ALK fusion gene in
non-small-cell lung cancer. Nature. 448:561–566. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Kobayashi S, Taki T, Chinen Y, et al:
Identification of IGHC5-BACH2 fusion transcripts resulting from
cryptic chromosomal rearrangements of 14q32 with 6q15 in aggressive
B-cell lymphoma/leukemia. Genes Chromosomes Cancer. 50:207–216.
2011.
|
|
6.
|
Chesi M, Nardini E, Lim RS, Smith KD,
Kuehl WM and Bergsagel PL: The t(4;14) translocation in myeloma
dysregulates both FGFR3 and a novel gene, MMSET, resulting in
IgH/MMSET hybrid transcripts. Blood. 92:3025–3034. 1998.PubMed/NCBI
|
|
7.
|
Hatzivassiliou G, Miller I, Takizawa J, et
al: IRTA1 and IRTA2, novel immunoglobulin superfamily receptors
expressed in B cells and involved in chromosome 1q21 abnormalities
in B cell malignancy. Immunity. 14:277–289. 2001. View Article : Google Scholar
|
|
8.
|
Przybylski GK, Dik WA, Wanzeck J, et al:
Disruption of the BCL11B gene through inv(14)(q11.2q32.31) results
in the expression of BCL11B-TRDC fusion transcripts and is
associated with the absence of wild-type BCL11B transcripts in
T-ALL. Leukemia. 19:201–208. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9.
|
Nagoshi H, Taki T, Hanamura I, et al:
Frequent PVT1 rearrangement and novel chimeric genes PVT1-NBEA and
PVT1-WWOX occur in multiple myeloma with 8q24 abnormality. Cancer
Res. 72:4954–4962. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10.
|
Pleasance ED, Stephens PJ, O’Meara S, et
al: A small-cell lung cancer genome with complex signatures of
tobacco exposure. Nature. 463:184–190. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11.
|
Northcott PA, Shih DJ, Peacock J, et al:
Subgroup-specific structural variation across 1,000 medulloblastoma
genomes. Nature. 488:49–56. 2012. View Article : Google Scholar
|
|
12.
|
Chinen Y, Taki T, Nishida K, et al:
Identification of the novel AML1 fusion partner gene, LAF4, a
fusion partner of MLL, in childhood T-cell acute lymphoblastic
leukemia with t(2;21) (q11;q22) by bubble PCR method for cDNA.
Oncogene. 27:2249–2256. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13.
|
Chen YW, Hu XT, Liang AC, et al: High BCL6
expression predicts better prognosis, independent of BCL6
translocation status, translocation partner, or BCL6-deregulating
mutations, in gastric lymphoma. Blood. 108:2373–2383. 2006.
View Article : Google Scholar
|
|
14.
|
Montanaro L, Treré D and Derenzini M:
Changes in ribosome biogenesis may induce cancer by down-regulating
the cell tumor suppressor potential. Biochim Biophys Acta.
1825:101–110. 2012.PubMed/NCBI
|
|
15.
|
Dai MS and Lu H: Crosstalk between c-Myc
and ribosome in ribosomal biogenesis and cancer. J Cell Biochem.
105:670–677. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
16.
|
Li Z and Hann SR: Nucleophosmin is
essential for c-Myc nucleolar localization and c-Myc-mediated rDNA
transcription. Oncogene. 32:1988–1994. 2013. View Article : Google Scholar
|
|
17.
|
Grandori C, Gomez-Roman N, Felton-Edkins
ZA, et al: c-Myc binds to human ribosomal DNA and stimulates
transcription of rRNA genes by RNA polymerase I. Nat Cell Biol.
7:311–318. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Stults DM, Killen MW, Williamson EP, et
al: Human rRNA gene clusters are recombinational hotspots in
cancer. Cancer Res. 69:9096–10104. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Uemura M, Zheng Q, Koh CM, Nelson WG,
Yegnasubramanian S and De Marzo AM: Overexpression of ribosomal RNA
in prostate cancer is common but not linked to rDNA promoter
hypomethylation. Oncogene. 31:1254–1263. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Croce CM and Nowell PC: Molecular basis of
human B cell neoplasia. Blood. 65:1–7. 1985.
|
|
21.
|
MacLeod RA, Nagel S, Kaufmann M, Janssen
JW and Drexler HG: Activation of HOX11L2 by juxtaposition with
3’-BCL11B in an acute lymphoblastic leukemia cell line (HPB-ALL)
with t(5;14) (q35;q32.2). Genes Chromosomes Cancer. 37:84–91.
2003.PubMed/NCBI
|
|
22.
|
Fujimoto R, Ozawa T, Itoyama T, Sadamori
N, Kurosawa N and Isobe M: HELIOS-BCL11B fusion gene involvement in
a t(2;14) (q34;q32) in an adult T-cell leukemia patient. Cancer
Genet. 205:356–364. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Ungar D, Oka T, Brittle EE, et al:
Characterization of a mammalian Golgi-localized protein complex,
COG, that is required for normal Golgi morphology and function. J
Cell Biol. 157:405–415. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
24.
|
Velagaleti GV, Tonk VS, Hakim NM, et al:
Fusion of HMGA2 to COG5 in uterine leiomyoma. Cancer Genet
Cytogenet. 202:11–16. 2010. View Article : Google Scholar : PubMed/NCBI
|