Introduction
Lung cancer is the leading cause of cancer-related
death worldwide. Despite some advances in cancer diagnostics and
recent improvements in its treatment, the overall 5-year survival
rate is still only 15% (1).
Several oncogenic alterations, such as KRAS and EGFR
mutations, and EML4-ALK fusion genes as well as inactivation
of tumor suppressor gene of TP53 in lung cancer have been
reported, however the precise molecular mechanisms of pulmonary
carcinogenesis are still far from fully understood (2). Although several molecular
targeted-drugs such as gefitinib, bevacizumab and crizotinib have
been approved for lung cancer treatment, the portion of patients
who are able to have the benefit of these drugs is still limited
and several serious adverse reactions such as interstitial
pneumonia by gefitinib and hemorrhage by bevacizumab have been
reported (3,4). Hence, the development of molecular
targeted agents providing better clinical benefits with less
adverse events are eagerly required.
Systematic analysis of expression levels of
thousands of genes using a cDNA microarray is an effective
technique to identify molecules involved in carcinogenic pathways
(5); some of such genes or their
gene products may be good molecular targets for the development of
novel therapies and/or cancer biomarkers. To isolate potential
molecular targets for diagnosis, treatment, and/or prevention of
lung carcinomas, we performed a genome-wide analysis of gene
expression profiles of tumor tissues from 120 lung cancer cases by
means of cDNA microarray consisting of 27,648 genes or expressed
sequence tags (ESTs) (6–10). Among the transactivated genes, we
identified LRRC42 (Leucine-rich repeat containing 42) as a
potential therapeutic target for lung cancer. LRRC42 protein
contains two leucine-rich repeats (LRRs) which are widespread
structural motifs comprising 20–30 amino acids with a
characteristic repetitive sequence pattern rich in leucine
residues. Leucine-rich repeat domains are built from tandems of two
or more repeats and form curved solenoid structures that are
particularly suitable for protein-protein interactions.
LRR-containing proteins participate in many important biological
processes, including plant and animal immunity, hormone-receptor
interactions, cell adhesion, signal transduction, regulation of
gene expression and apoptosis (11–14).
However, the pathophysiological roles of LRRC42 in cancer cells
have not been reported. Herein we report identification of LRRC42
as a potential therapeutic target and also provide evidence that
LRRC42 could interact with GATAD2B (GATA zinc finger domain
containing 2B) and MBD3 (Methyl-CpG-binding domain protein 3)
proteins that are likely to play a significant role in human
pulmonary carcinogenesis.
Materials and methods
Lung cancer cell lines and tissue
samples
The human lung cancer cell lines used in this study
were as follows: A549, NCI-H1373, NCI-H1781, SKMES-1, NCI-H520,
NCI-H1703, NCI-H2170 and DMS114 were distributed from America Type
Culture Collection (ATCC, Manassas, VA, USA). LC319 was kindly
provided from Aichi Cancer Center (Aichi, Japan). PC14 was obtained
from RIKEN BioRsource Center (Ibaraki, Japan). LU61 and LX1 were
obtained from Central Institute for Experimental Animals (Kanagawa,
Japan). DMS273 was obtained from European Collection of Animal Cell
Cultures (ECACC, Salisbury, UK). SBC-3 and SBC-5 were obtained from
Japanese Collection of Research Bioresources (JCRB, Osaka, Japan).
All cells were grown in monolayers in appropriate medium
supplemented with 10% FCS and were maintained at 37°C in
atmospheres of humidified air with 5% CO2. Human small
airway epithelial cells (SAEC) were grown in optimized medium
purchased from Cambrex Bio Science, Inc. (Walkersville, MD, USA).
Primary lung cancer tissue samples were obtained with informed
consent as previously described (6,10).
This study was approved by individual institutional ethical
committees.
Quantitative real-time PCR
Total RNA was extracted from cultured cells using
the QIAshredder (Qiagen, Valencia, CA, USA) and RNeasy®
Plus mini kit (Qiagen) according to the manufacturer’s protocol.
Extracted RNAs were reverse transcribed using oligo (dT) primer
(Life Technologies, Carlsbad, CA, USA) and SuperScript III (Life
Technologies). Quantitative real-time PCR was conducted with the
SYBR Green I Master kit on a LightCycler 480 (Roche Diagnostics,
Mannheim, Germany) according to the manufacturer’s recommendations.
Each experiment was done in duplicate. GAPDH was used for
normalization of expression levels. For quantitative RT-PCR
reactions, specific primers for all human LRRC42, GATAD2B, MBD3,
p21Waf1/Cip1 and GAPDH were designed
as follows: LRRC42, 5′-TTGATCATAGTAACTGCAAGACAGAG-3′ and
5′-ACGCTCCCACTGCAGAAC-3′ GA R A D2B,
5′-CACCAACCGGCTGAAAAAT-3′ and 5′-GCTGCT GTAATCGCTGTTCA-3′,
MBD3, 5′-ACCATGGACC TCCCCAAG-3′ and
5′-CGACAGCAGCGTCTCATC-3′; p21Waf1/Cip1,
5′-GACCTGTCACTGTCTTGTACCC-3′ and 5′-AAGATCAGCCGGCGTTTG-3′
GAPDH, 5′-ACCATG GGGAAGGTGAAG-3′ and 5′-AATGAAGGGGTCATT
GATGG-3′.
Immunofluorescence analysis
Cells were plated onto glass coverslips (Becton
Dickinson, Mountain View, CA, USA), fixed with 4% paraformaldehyde,
and permeablilized with 0.1% Triton X-100 in PBS for 5 min at room
temperature. Non-specific binding was blocked by 5% skim milk for
10 min at room temperature. Cells were then incubated overnight at
4°C with primary antibodies for mouse monoclonal anti-Flag antibody
(Catalog no. F3165, Sigma, St. Louis, MO, USA) and anti-GATAD2B
antibody (Catalog no. HPA017015, ATLAS, Stockholm, Sweden) diluted
in PBS containing 1% BSA. After being washed with PBS, the cells
were stained with Alexa Fluor 488-conjugated secondary antibody
(Molecular Probes, Eugene, OR, USA) (Fig. 1C), or with Alexa Fluor
488-conjugated secondary antibody and Alexa Fluor 594-conjugated
secondary antibody (Molecular Probes) (Fig. 3C) for 60 min at room temperature.
After another wash with PBS, each specimen was mounted with
Vectashield (Vector Laboratories, Burlingame, CA, USA) containing
4′, 6′-diamidine-2′-phenylindolendihydrochrolide (DAPI) and
visualized with Spectral Confocal Scanning Systems (TSC SP2 AOBS:
Leica Microsystems, Wetzlar, Germany).
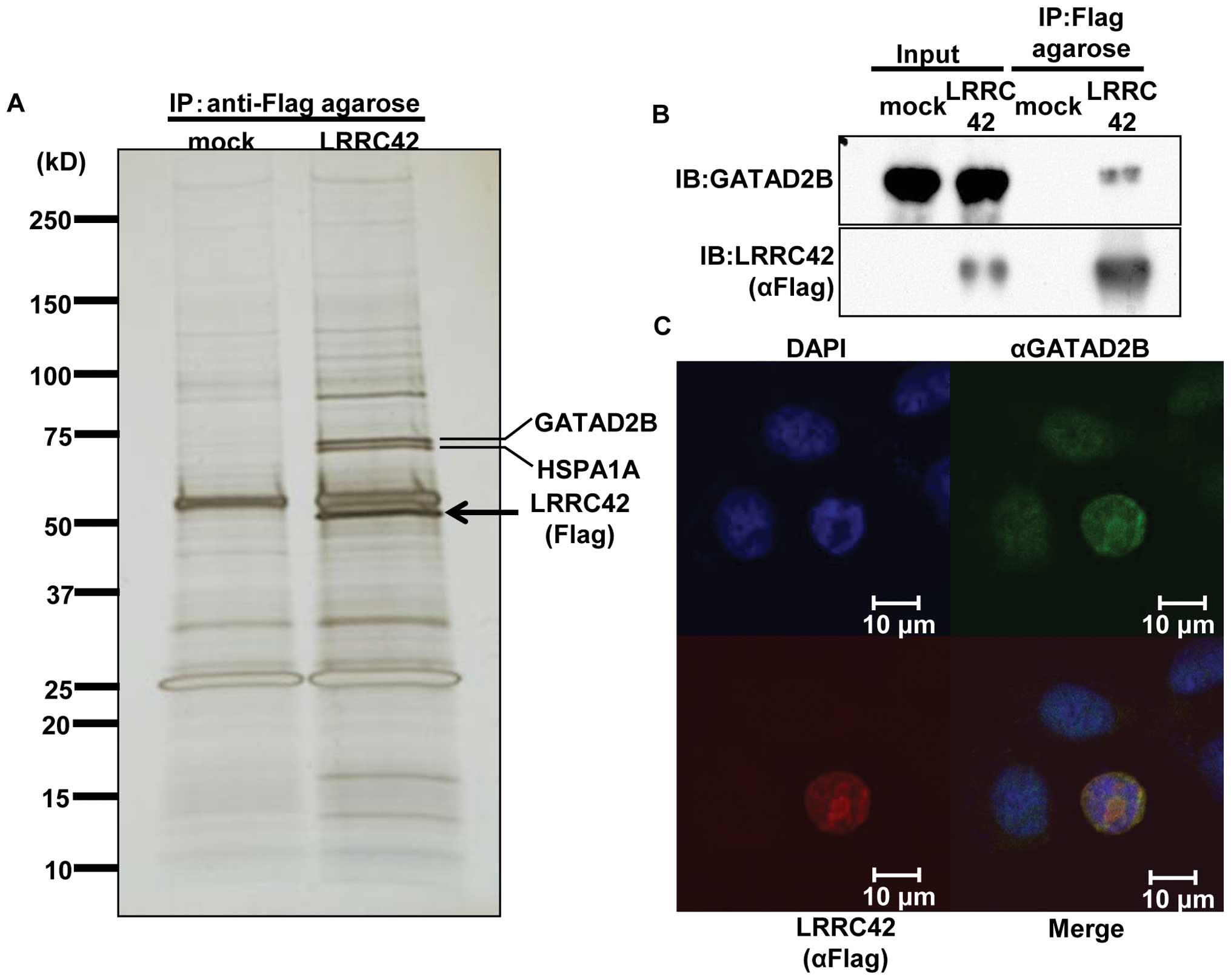 | Figure 3.Interaction of LRRC42 with GATAD2B.
(A) Silver staining of SDS-PAGE gels that contained
immunoprecipitated lysates of lung cancer SBC-3 cells, which were
transfected with Flag-tagged LRRC42 expression vector or mock
vector, using anti-Flag M2 agarose. (B) Interaction between
exogenous LRRC42 and endogenous GATAD2B in SBC-3 cells transfected
with LRRC42 expression vector by immunoprecipitation and western
blot analysis. (C) Colocalization of LRRC42 and GATAD2B in the
nucleus of SBC-3 cells transfected with LRRC42 expression vector,
detected by immunocytochemical staining. Interaction of LRRC42 with GATAD2B. (D and E) The
level of GATAD2B proteins detected by western blot analysis in
LC319 and SBC-3 cells transfected with si-EGFP or si-LRRC42. The
columns and bars represent the mean and SE, respectively. (F and G)
The level of GATAD2B mRNAs detected by quantitative real-time PCR
analysis in LC319 and SBC-3 cells transfected with si-EGFP or
si-LRRC42. The columns and bars represent the mean and SE,
respectively. (H and I) The level of LRRC42 mRNAs detected by
quantitative real-time PCR analysis in LC319 and SBC-3 cells
transfected with si-EGFP or si-LRRC42. The columns and bars
represent the mean and SE, respectively. (J) Interaction between
exogenous LRRC42 and endogenous GATAD2B or MBD3 in SBC-3 cells
transfected with LRRC42 expression vector by immunoprecipitation
and western blot analysis. (K and L) The level of MBD3 proteins
detected by western blot analysis in LC319 and SBC-3 cells
transfected with si-EGFP, si-LRRC42, GATAD2B or MBD3. (M and N) The
level of MBD3 mRNAs detected by quantitative real-time PCR analysis
in LC319 and SBC-3 cells transfected with si-EGFP, si-LRRC42,
si-GATAD2B or si-MBD3. The columns and bars represent the mean and
SE, respectively. |
Northern blot analysis
Human multiple-tissue blots (Clontech, Carlsbad, CA,
USA) were hybridized with 32P-labeled PCR products of
LRRC42. The cDNA probe of LRRC42 was prepared by
RT-PCR using following primers: LRRC42,
5′-GACCAGATCGTTCTGCAGTG-3′ and 5′-CCTCCCA CACCACAAAAGTA-3′.
Prehybridization, hybridization, and washing were performed
according to the supplier’s recommendations. The blots were
autoradiographed at −80°C for 14 days.
RNA interference assay
To evaluate the biological functions of LRRC42 in
lung cancer cells, we used small interfering RNA (siRNA) duplexes
against the target genes (Sigma). The target sequences of the
synthetic oligonucleotides for RNA interference were as follows:
control-1: [EGFP, enhanced green fluorescence protein (GFP) gene, a
mutant of Aequorea gictoria GFP], 5′-GAAGCAGCACGACUUCUUC-3′
control-2 (LUC, luciferase gene from Photinus pyralis),
5′-CGUACG CGGAAUACUUCGA-3′ si-LRRC42-#1, 5′-CUUACUA CCUCAGCUCAGA-3′
si-LRRC42-#2, 5′-GACUUGUUA AAUUCCUAUU-3′ si-GATAD2B, 5′-GCCAAUAGCG
AGUUCAUCU-3′ si-MBD3, 5′-CACAGUCGAGGCACG UCAU-3′. Lung cancer cell
lines, LC319 and SBC-3, were plated onto 10-cm dishes
(5.0×105 per dish), and transfected with either of the
siRNA oligonucleotides (50 μM) using 30 μl of
Lipofectamine® RNAiMAX (Life Technologies) according to
the manufacturers’ instructions. After seven days of incubation,
the cells were stained by Giemsa solution to assess colony
formation, and cell numbers were assessed by Cell Counting Kit-8
(Dojindo Co., Kumamoto, Japan); briefly, Cell Counting Kit-8
solution was added to each dish at concentration of 1/10 volume,
and the plates were incubated at 37°C for additional 1 h.
Absorbance was then measured at 490 nm, and at 630 nm as a
reference, with a 2030 ARVO™ X3 (PerkinElmer, Courtaboeuf,
France).
Anti-LRRC42 antibody
Plasmids expressing partial LRRC42 that contained
His-tagged epitopes at their NH2 termini were prepared
using pET28 vector (Novagen, Darmstadt, Germany) and primers
LRRC42-F (5′-GGAATTCTGGGCTGA CCAGATCGTTCTGC-3′) and LRRC42-R
(5′-ATAGTTT AGCGGCCGCTTAGTTATTCTGTTCTGTCTCTGACT-3′. Recombinant
proteins were expressed in Escherichia coliBL21 codon-plus
strain (Stratagene, San Diego, CA, USA) and purified using Ni-NTA
(Qiagen) according to the supplier’s protocol. The protein was
inoculated into rabbits; immune sera were purified on affinity
columns according to standard methods. Affinity-purified
anti-LRRC42 antibodies were used for western blotting. We confirmed
that the antibody was specific to LRRC42 on western blots using
lysates from cell lines that had been transfected with LRRC42
expression vector.
Western blotting
Cells were lysed with immunoprecipitation assay
buffer [50 mmol/l Tris-HCl (pH 8.0), 150 mmol/l NaCl, 1% NP40, 0.5%
deoxychorate-Na, 0.1% SDS] containing Phosphatase Inhibitor
Cocktail Set II and Protease Inhibitor Cocktail Set III EDTA-Free
(Calbiochem, San Diego, CA, USA). Protein samples were separated by
SDS-polyacrylamide gels and electroblotted onto Hybond-P PVDF
membranes (GE Healthcare Bio-Sciences, Piscataway, NJ, USA). Blots
were incubated with a mouse monoclonal anti-Flag antibody, a rabbit
anti-Flag antibody (Catalog no. F7425, Sigma), an anti-GATAD2B
antibody, an anti-MBD3 antibody (Catalog no. sc-9402, Santa Cruz,
Santa Cruz, CA, USA), an anti-p21Waf1/Cip1 antibody
(Catalog no. 2947S, Cell Signaling, Danvers, MA, USA) and an
anti-p53 antibody (Catalog no. sc-126, Santa Cruz, Santa Cruz, CA,
USA). Antigen-antibody complexes were detected using secondary
antibodies conjugated to horse-radish peroxidase (GE Healthcare
Bio-Sciences). Protein bands were visualized by enhanced
chemiluminescence western blot detection reagents (GE Healthcare
Bio-Sciences).
Establishment of HEK293 cells expressing
exogenous LRRC42
T-REx HEK293 cells that could induce the expression
of exogenous LRRC42 (LRRC42-HEK293) cells were generated by Flp-In
expression system, where LRRC42 expression is under control of the
tetracycline-regulated cytomegalovirus/tetO2 hybrid
promoter, and the commercially available Flp-In T-REx 293 host cell
line, according to the manufacturer’s instructions (Invitrogen,
Carlsbad, CA, USA).
Cell growth assay
LRRC42-HEK293 cells were grown for four days in DMEM
containing 10% FBS, Blasticidin S HCl (Life Techonologies; 15
μg/ml) and Hygromycin B (Invitrogen; 100 μg/ml)
supplemented with or without doxycycline (Sigma; 100 ng/ml).
Viability of cells was evaluated by Cell Counting Kit-8.
Coimmunnoprecipitation and
matrix-assisted laser desorption/ionizing-time of flight mass
spectrometry mapping of LRRC42-associated proteins
Cell extracts from lung cancer cell line SBC-3 which
was transfected with LRRC42 expression (carboxyl-terminal
Flag-tagged pCAGGS plasmid vector) or mock vector were precleared
by incubation at 4°C for 1 h with 80 μl of protein G-agarose
beads in a final volume of 200 μl of immunoprecipitation
buffer (0.5% NP40, 50 mmol/l Tris-HCl, 150 mmol/l NaCl) in the
presence of phosphatase and proteinase inhibitors. After
centrifugation at 1,000 rpm for 5 min at 4°C, the supernatants were
incubated at 4°C with anti-Flag M2 agarose for 3 h. The beads were
then collected by centrifugation at 1,000 rpm for 1 min and washed
six times with 1 ml of each immunoprecipitation buffer. The washed
beads were resuspended in 30 μl of Laemmli sample buffer and
boiled for 5 min, and the proteins were separated using 5% to 20%
SDS PAGE gels (Bio-Rad Laboratories, Marnes-la-Coquette, France).
After electrophoresis, the gels were stained with SilverQuest
(Invitrogen). Protein bands specifically found in extracts which
was transfected with LRRC42 vector were excised and served for
matrix-assisted laser desorption/ionization-time of flight mass
spectrometry (MALDI-TOF-MS) analysis (AXIMA-CFR, Shimadzu-Biotech,
Kyoto, Japan).
Flow cytometry
After transfection of each siRNAs, cells were
treated with Aphidicolin (Sigma) at 1 μg/ml for 24 h. Then,
the cells were washed with PBS four times and growth medium was
added into the dish. The cells were collected in PBS every 3 h and
fixed in 70% cold ethanol for 30 min. After treatment with 100
μg/ml of RNase (Sigma), the cells were stained with 50
μg/ml propidium iodide (Sigma) in PBS. Flow cytometry was
analyzed by using FACScan (Becknman Coulter, Brea, CA, USA). The
cells selected from at least 20,000 ungated cells were analyzed for
DNA content.
Results
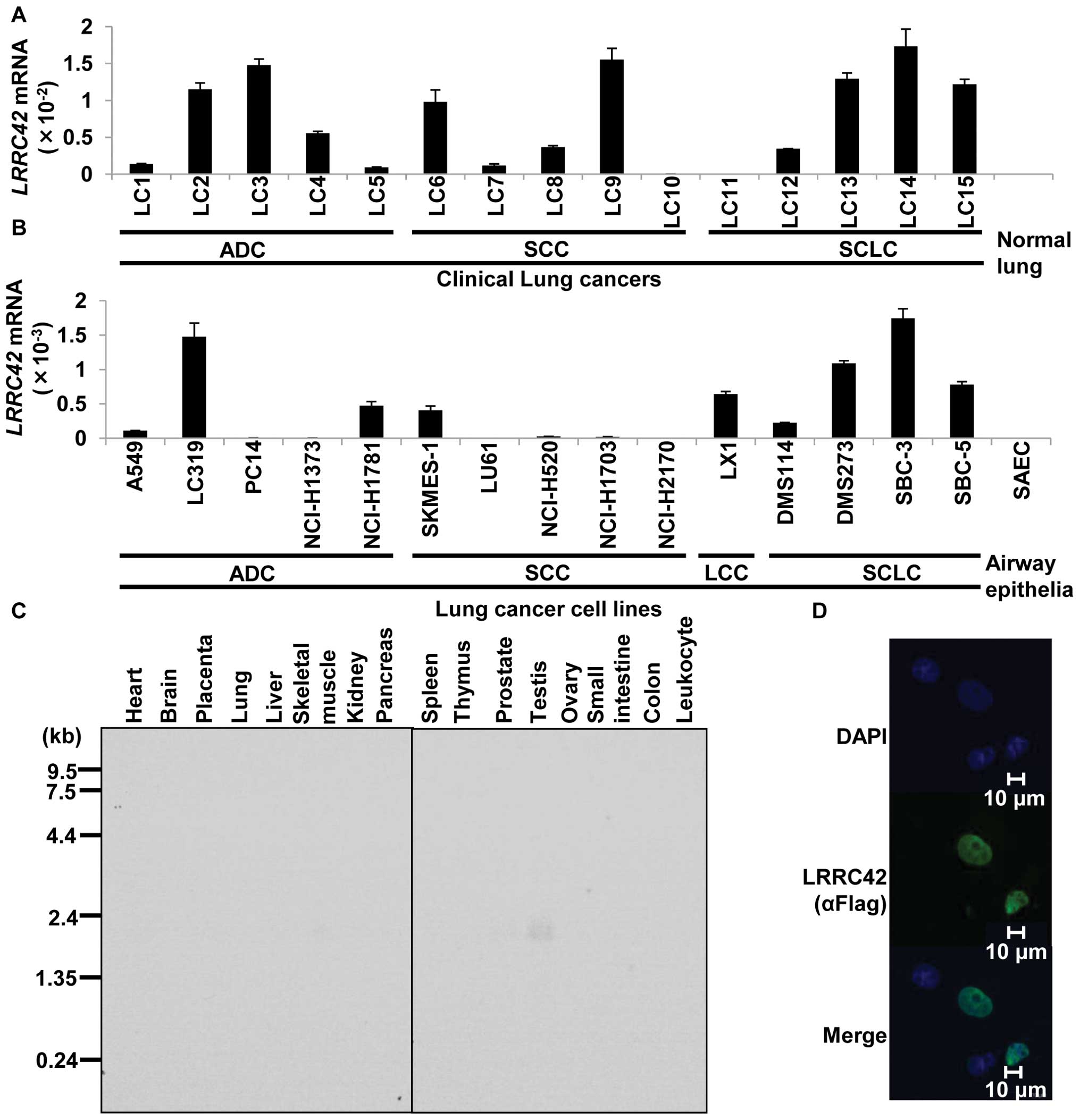
LRRC42 expression in lung cancers and
normal tissues
To identify novel target molecules for the
development of therapeutic agents and/or diagnostic biomarkers for
lung cancer, we previously performed gene expression profile
analysis of 120 lung carcinoma samples using cDNA microarray
containing 27,648 genes or expressed sequence tags (6–10),
and identified that LRRC42 was significantly transactivated (more
than 3 times higher than in their corresponding normal tissues) in
>50% of 120 lung cancer samples examined. We subsequently
confirmed its transactivation by quantitative real-time PCR
experiments in lung cancer tissues as well as lung cancer cell
lines (Fig. 1A and B). Northern
blot analysis with the LRRC42-specific probe identified a
1.7-kb transcript only in testis among 16 normal human tissues
examined (Fig. 1C). To determine
the subcellular localization of LRRC42 protein, we constructed a
plasmid expressing LRRC42 (carboxyl-terminal Flag-tagged pCAGGS
plasmid vector), transfected it into COS-7 cells and detected
exogenous LRRC42 protein in the nucleus of the cells using an
anti-flag antibody (Fig. 1D).
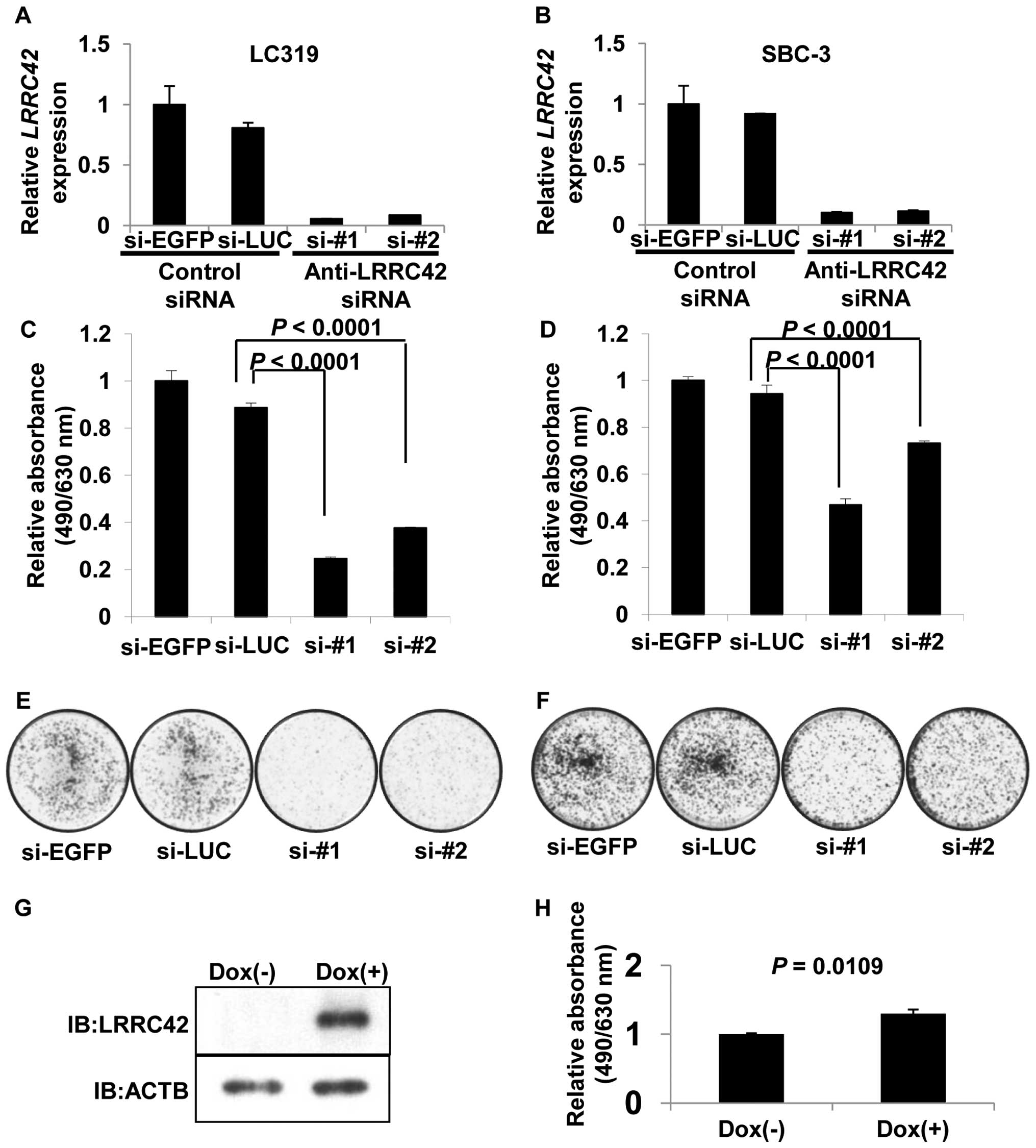
Effect of LRRC42 on cell growth
To assess whether LRRC42 is essential for growth or
survival of lung cancer cells, we transfected synthetic
oligonucleotide siRNAs against LRRC42 into lung adenocarcinoma
LC319 and small cell lung cancer SBC-3 cells in which LRRC42
was highly expressed. The mRNA levels of LRRC42 in the cells
transfected with si-LRRC42-#1 or -#2 were significantly decreased
in comparison with those transfected with either of the control
siRNAs (Fig. 2A and B). MTT and
colony formation assays revealed a significant reduction of cell
viability as well as the number of colonies in
si-LRRC42-transfected cells (Fig.
2C–F).
To further clarify a potential role of LRRC42 in
carcino-genesis, we established HEK293 cells using the Flp-In T-Rex
expression system, where LRRC42 expression was under the control of
the tetracycline-regulated cytomegalovirus/tetO2 hybrid
promoter. MTT assay demonstrated that the growth of the cells
treated with doxycycline was enhanced compared with that without
doxycycline, indicating the growth promoting activity of LRRC42
protein (Fig. 2G and H).
Interaction and colocalization of LRRC42
with GATAD2B
To elucidate the molecular mechanism of LRRC42 in
lung carcinogenesis, we screened a protein(s) that could interact
with LRRC42. Lysates of SBC-3 cells which was transfected with
LRRC42 expression vector (carboxyl-terminal Flag-tagged pCAGGS
plasmid vector) or mock vector were extracted, and
immunoprecipitated with anti-Flag M2 agarose. The protein complex
was separated by SDS-PAGE and visualized by silver staining
(Fig. 3A). A 65-kDa band, which
was detectable in lysates of cells transfected with LRRC42 vector,
but not in those with mock vector, was extracted. The peptide
sequence analysis determined by mass spectrometry indicated the
protein to be GATAD2B (GATA zinc finger domain containing 2B) that
is known to be a component of the MeCP1 complex that represses
transcription through preferential binding, remodeling and
deacetylation of methylated nucleosomes (15). We subsequently confirmed
interaction between exogenous LRRC42 and endogenous GATAD2B in
SBC-3 cells using anti-GATAD2B antibody by co-immunoprecipitation
experiment (Fig. 3B). We also
conducted immunofluorescence analysis and found colocalization of
exogenous LRRC42 with endogenous GATAD2B in the nucleus of SBC-3
cells which were transfected with the LRRC42 expression vector
(Fig. 3C).
To further investigate the biological significance
of the interaction between LRRC42 and GATAD2B in cancer cells, we
examined the protein level of GATAD2B after suppressing LRRC42
expression in LC319 and SBC-3 cells. Treatment of siRNA
oligonucleotides against LRRC42 (si-LRRC42) effectively knocked
down the expression of endogenous LRRC42, compared to the control
siRNA (si-EGFP). Interestingly, the protein level of GATAD2B was
also significantly decreased in cells transfected with si-LRRC42,
while the transcript level of GATAD2B was unchanged
(Fig. 3D-I). A previous study
indicated GATAD2B as a key component of the MeCP1 complex that
interacted with MBD3 (15).
Furthermore, GATAD2B was shown to possess an ability to target MBD3
protein to specific nuclear loci (15,46).
MBD3 was indicated to be recruited to a promoter region of the
21Waf1/Cip1 tumor suppressor gene and silence its
expression (47). Hence, we have
hypothesized that LRRC42 might have a very significant effect on
the function of the MeCP1 complex through the interaction with
GATAD2B as well as MBD3 (Fig. 3J).
We found that knockdown of LRRC42 or GATAD2B reduced the amount of
MBD3 protein while no change was observed in mRNA level of MBD3
(Fig. 3K–N).
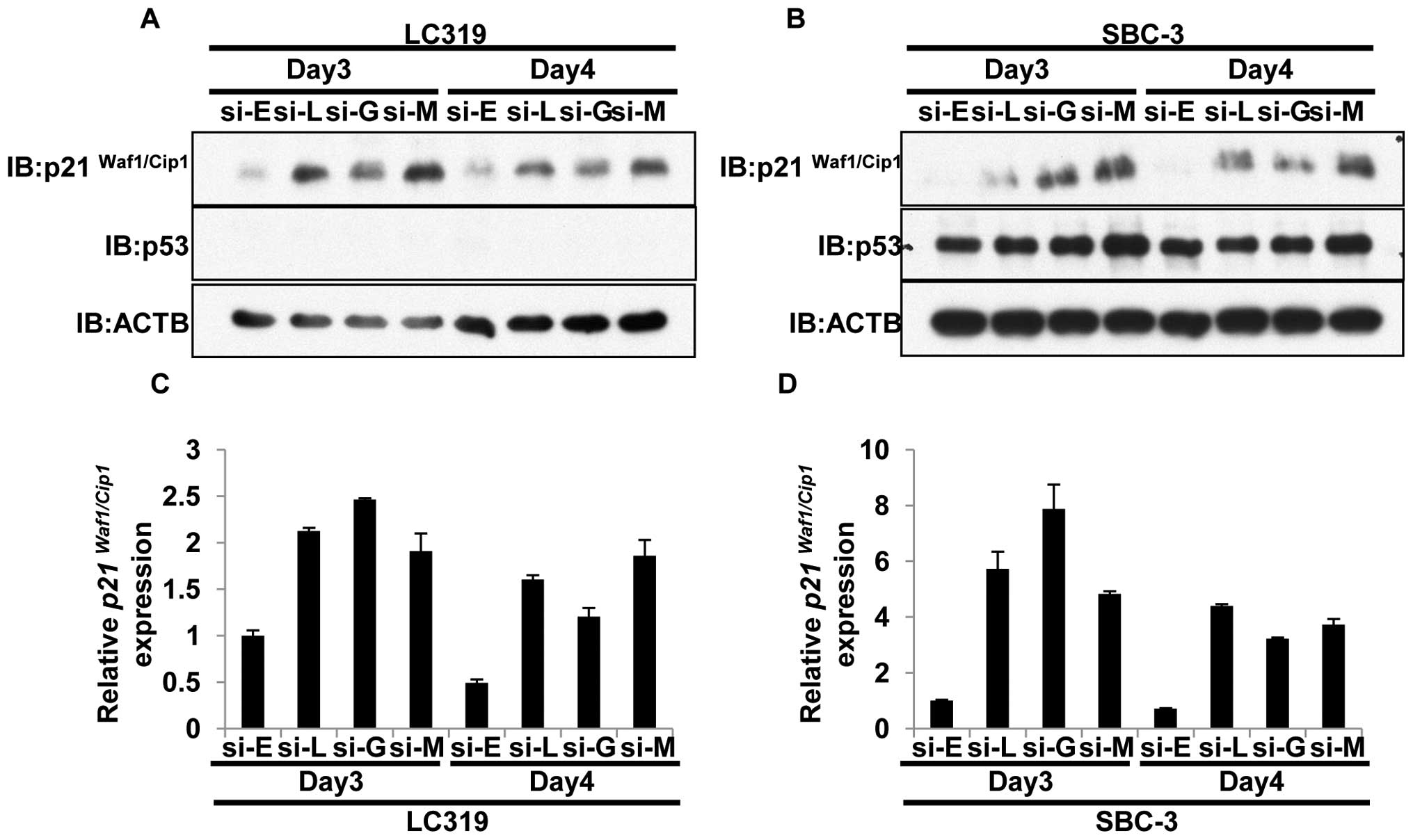
LRRC42-GATAD2B-MBD3 axis regulates
p21Waf1/Cip1 expression
We then examined the downstream target of the
LRRC42-GATAD2B-MBD3 complex in lung cancer cells. As described
above, MBD3 might be recruited at the p21Waf1/Cip1
promoter and silence its expression (47). Therefore, we firstly assessed the
knockdown effect of either of LRRC42, GATAD2B or MBD3 on
p21Waf1/Cip1 expression by quantitative real-time PCR
and western blot analysis. Suppression of either of LRRC42, GATAD2B
or MBD3 by siRNA appeared to increase p21Waf1/Cip1 at
transcriptional and protein levels in LC319 (p53 null) and SBC-3
(p53 wild-type) cells (Fig. 4A–D).
However, this effect on p21Waf1/Cip1 was not clear in
other lung cancer cell lines examined (NCI-H1781, NCI-H358,
NCI-H1299 and DMS273; data not shown).
Since p21Waf1/Cip1 expression was known
to cause the G0/G1 arrest, we performed FACS
(Fluorescence Activated Cell Sorter) analysis to evaluate the
knockdown effect on the cell cycle in these cell lines. After
synchronization of the cancer cells at G1 phase by
aphidicolin, we removed aphidicolin from the cell culture medium
and monitored the cell cycle progression process (Fig. 4E). The cells transfected with
si-EGFP progressed rapidly into the S and G2/M phases.
However, the cells treated with siRNA against LRRC42,
GATAD2B or MBD3 revealed significant delay in entering
into S phase although the delay from the G1 to S
transition in the cells treated with si-MBD3 was less significant
to those treated with siLRRC42 or si-GATAD2B. These data implied
that the LRRC42-GATAD2B interaction may regulate MBD3 protein as
well as the MeCP1 complex, and enhance the growth of cancer
cells.
Discussion
Recent advances in understanding the molecular
mechanisms underlying cancer development/progression have driven
the design of new therapeutic approaches, termed ‘molecular
targeted therapies’, that selectively interfere with molecules or
pathways involved in tumor growth and progression. inactivation of
growth factors and/or their receptors on tumor cells as well as the
inhibition of oncogenic tyrosine kinase pathways that play crucial
roles in cancer cells constitute the main rationale of new cancer
treatments and also lead to the way for the personalized treatment
for individual patients. Small-molecule inhibitors and monoclonal
antibodies are at present major components of these targeted
approaches for various types of human caner (16). Molecular targeted cancer therapies
hold the promise of being very selective to cancer cells, but not
affecting normal cells. Hence, they are expected to be less harmful
to normal cells, reduce severe side effects, and improve the
quality of life of cancer patients. Toward identification of
molecular targets for drug development, we had performed
whole-genome expression profiles of 120 clinical lung cancer
samples using cDNA microarray data and subsequent loss-of-function
phenotype analyses by means of RNA interference systems (17–38).
On the basis of this approach, we found LRRC42 to be highly
over-expressed in the majority of clinical lung cancer cases as
well as in most of the lung cancer cell lines examined, while its
expression was absent in normal tissues except testis.
Furthermore, we demonstrated that knockdown of
LRRC42 expression suppressed the growth of lung cancer cells. In
addition, induction of LRRC42 expression by a T-REx system resulted
in enhancement of cell growth, suggesting that LRRC42 is likely to
be an important growth promoting factor in lung cancer cells and
could serve as a valuable target for the development of an
anticancer agent for lung cancer.
LRR family members were reported to participate in
many biologically important processes such as hormone-receptor
interactions, enzyme inhibition, cell adhesion and cellular
trafficking. A number of studies clarified the involvement of LRR
proteins in early mammalian development (39), development of neuron (40), cell polarization (41), regulation of gene expression
(42) and apoptosis (43). It was also shown that LRR domains
may be critical for the cell morphology as well as the cytoskeleton
dynamics (44,45). In these processes, the LRR motifs
are probably essential in mediating protein-protein interactions.
However, there has been no report describing the involvement of
LRRC42 in human carcinogenesis.
Our data indicated that LRRC42 was able to interact
with GATAD2B. GATAD2B is an important component of the MeCP1
complex that represses transcription of genes through preferential
binding to, remodeling, and deacetylating methylated nucleosomes.
It has the ability to translocate MBD3 protein to specific nuclear
foci (15,46). We demonstrated that LRRC42 could
activate this transcription-repressive complex through interacting
with and stabilizing GATAD2B and MBD3 proteins. We also revealed
that the LRRC42-GATAD2B-MBD3 interaction is likely to play a
significant role in transcriptional regulation of the
cyclin-dependent kinase inhibitor p21Waf1/Cip1 which is
a well-known tumor suppressor and an inhibitor of cell cycle
progression from G1 to S phase. p21Waf1/Cip1
negatively regulates DNA replication through the interaction with
PCNA (Proliferative cell nuclear antigen), and also binds to the
CDK (Cyclin-dependent kinase) complex and inhibits the
G1-S transition (48).
Several transcriptional regulators, activators or
repressors of p21Waf1/Cip1 have been reported. p53 is an
activator of p21Waf1/Cip1 and the activation of the
p53-p21Waf1/Cip1 pathway is critically important when
cells need to arrest the cell cycle and repair the DNA damage. Myc,
one of p21Waf1/Cip1 repressors, was reported to induce
the expression of AP4 (Transcription Factor AP-4) that has the
ability to repress p21Waf1/Cip1 expression. Myc may also
repress p21Waf1/Cip1 expression through the interaction
with MIZ1 (ZBTB17) (49). MBD3 is
known to be one of the repressors which directly regulate
p21Waf1/Cip1 expression and play important roles in
oncogenic transformation and proliferation (50). FACS analysis clearly demonstrated
that suppression of MBD3 caused the delay of G1-S transition
although its effect was not as significant as the knockdown of
LRRC42 or GATAD2B.
In conclusion, human LRRC42 is essential for growth
and survival of lung cancer cells. Our data imply that targeting
LRRC42 and/or the LRRC42-GATAD2B interaction may be a good approach
for development of new treatment of lung cancer with specific
activity and minimum toxicity.
Acknowledgements
This study was supported in part by
Grant-in-Aid for Scientific Research (B) and Grant-in-Aid for
Scientific Research on Innovative Areas from The Japan Society for
the Promotion of Science to Y.D. Y.D. is a member of Shiga Cancer
Treatment Project supported by Shiga Prefecture (Japan).
References
|
1.
|
Jemal A, Siegel R, Xu J and Ward E: Cancer
statistics, 2010. CA Cancer J Clin. 60:277–300. 2010. View Article : Google Scholar
|
|
2.
|
Daigo Y, Takano A, Teramoto K, Chung S and
Nakamura Y: A systematic approach to the development of novel
therapeutics for lung cancer using genomic analyses. Clin Pharmacol
Ther. 94:218–223. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
3.
|
Inoue A, Saijo Y, Maemondo M, et al:
Severe acute interstitial pneumonia and gefitinib. Lancet.
361:137–139. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
4.
|
Sandomenico C, Costanzo R, Carillio G, et
al: Bevacizumab in non small cell lung cancer: development, current
status and issues. Curr Med Chem. 19:961–971. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5.
|
Daigo Y and Nakamura Y: From cancer
genomics to thoracic oncology: discovery of new biomarkers and
therapeutic targets for lung and esophageal carcinoma. Gen Thorac
Cardiovasc Surg. 56:43–53. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
6.
|
Kikuchi T, Daigo Y, Katagiri T, et al:
Expression profiles of non-small cell lung cancers on cDNA
microarrays: identification of genes for prediction of lymph-node
metastasis and sensitivity to anti-cancer drugs. Oncogene.
22:2192–2205. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7.
|
Kakiuchi S, Daigo Y, Tsunoda T, Yano S,
Sone S and Nakamura Y: Genome-wide analysis of organ-preferential
metastasis of human small cell lung cancer in mice. Mol Cancer Res.
1:485–499. 2003.PubMed/NCBI
|
|
8.
|
Kakiuchi S, Daigo Y, Ishikawa N, et al:
Prediction of sensitivity of advanced non-small cell lung cancers
to gefitinib (Iressa, ZD1839). Hum Mol Genet. 13:3029–3043. 2004.
View Article : Google Scholar
|
|
9.
|
Kikuchi T, Daigo Y, Ishikawa N, et al:
Expression profiles of metastatic brain tumor from lung
adenocarcinomas on cDNA microarray. Int J Oncol. 28:799–805.
2006.PubMed/NCBI
|
|
10.
|
Taniwaki M, Daigo Y, Ishikawa N, et al:
Gene expression profiles of small-cell lung cancers: molecular
signatures of lung cancer. Int J Oncol. 29:567–575. 2006.PubMed/NCBI
|
|
11.
|
Bella J, Hindle KL, McEwan PA and Lovell
SC: The leucine-rich repeat structure. Cell Mol Life Sci.
65:2307–2333. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12.
|
Chai L, Dai L, Che Y, et al: LRRC19, a
novel member of the leucine-rich repeat protein family, activates
NF-κB and induces expression of proinflammatory cytokines. Biochem
Biophys Res Commun. 388:543–548. 2009.PubMed/NCBI
|
|
13.
|
Kajava AV: Structural diversity of
leucine-rich repeat proteins. J Mol Biol. 277:519–527. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
14.
|
Pancer Z and Cooper MD: The evolution of
adaptive immunity. Annu Rev Immunol. 24:497–518. 2006. View Article : Google Scholar
|
|
15.
|
Feng Q, Cao R, Xia L, Erdjument-Bromage H,
Tempst P and Zhang Y: Identification and functional
characterization of the p66/p68 components of the MeCP1 complex.
Mol Cell Biol. 22:536–546. 2002. View Article : Google Scholar
|
|
16.
|
Ciavarella S, Milano A, Dammacco F and
Silverstris F: Targeted therapies in cancer. BioDrugs. 24:77–88.
2010. View Article : Google Scholar
|
|
17.
|
Suzuki C, Daigo Y, Ishikawa N, et al: ANLN
plays a critical role in human lung carcinogenesis through the
activation of RHOA and by involvement in the phosphoinositide
3-kinase/AKT pathway. Cancer Res. 65:11314–11325. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
18.
|
Ishikawa N, Daigo Y, Takano A, et al:
Characterization of SEZ6L2 cell-surface protein as a novel
prognostic marker for lung cancer. Cancer Sci. 97:737–745. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
19.
|
Kato T, Sato N, Takano A, et al:
Activation of placenta-specific transcription factor distal-less
homeobox 5 predicts clinical outcome in primary lung cancer
patients. Clin Cancer Res. 14:2363–2370. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
20.
|
Dunleavy EM, Roche D, Tagami H, et al:
HJURP is a cell-cycle-dependent maintenance and deposition factor
of CENP-A at centromeres. Cell. 137:485–497. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21.
|
Hirata D, Yamabuki T, Miki D, et al:
Involvement of epithelial cell transforming sequence-2 oncoantigen
in lung and esophageal cancer progression. Clin Cancer Res.
15:256–266. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22.
|
Sato N, Koinuma J, Fujita M, et al:
Activation of WD repeat and high-mobility group box DNA binding
protein 1 in pulmonary and esophageal carcinogenesis. Clin Cancer
Res. 16:226–239. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23.
|
Sato N, Koinuma J, Ito T, et al:
Activation of an oncogenic TBC1D7 (TBC1 domain family, member 7)
protein in pulmonary carcinogenesis. Genes Chromosomes Cancer.
49:353–367. 2010.PubMed/NCBI
|
|
24.
|
Nguyen MH, Koinuma J, Ueda K, et al:
Phosphorylation and activation of cell division cycle associated 5
by mitogen-activated protein kinase play a crucial role in human
lung carcinogenesis. Cancer Res. 70:5337–5347. 2010. View Article : Google Scholar
|
|
25.
|
Takano A, Ishikawa N, Nishino R, et al:
Identification of nectin-4 oncoprotein as a diagnostic and
therapeutic target for lung cancer. Cancer Res. 69:6694–6703. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
26.
|
Sato N, Yamabuki T, Takano A, et al: Wnt
inhibitor Dickkopf-1 as a target for passive cancer immunotherapy.
Cancer Res. 70:5326–5336. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
27.
|
Mizukami Y, Kono K, Daigo Y, et al:
Detection of novel cancer-testis antigen-specific T-cell responses
in TIL, regional lymph nodes, and PBL in patients with esophageal
squamous cell carcinoma. Cancer Sci. 99:1448–1454. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28.
|
Harao M, Hirata S, Irie A, et al:
HLA-A2-restricted CTL epitopes of a novel lung cancer-associated
cancer testis antigen, cell division cycle associated 1, can induce
tumor-reactive CTL. Int J Cancer. 123:2616–2625. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29.
|
Kono K, Mizukami Y, Daigo Y, et al:
Vaccination with multiple peptides derived from novel cancer-testis
antigens can induce specific T-cell responses and clinical
responses in advanced esophageal cancer. Cancer Sci. 100:1502–1509.
2009. View Article : Google Scholar
|
|
30.
|
Yokomine K, Senju S, Nakatsura T, et al:
The forkhead Box M1 transcription factor as a candidate of target
for anti-cancer immunotherapy. Int J Cancer. 126:2153–2163.
2010.PubMed/NCBI
|
|
31.
|
Tomita Y, Imai K, Senju S, et al: A novel
tumor-associated antigen, cell division cycle 45-like can induce
cytotoxic T-lymphocytes reactive to tumor cells. Cancer Sci.
102:697–705. 2011. View Article : Google Scholar
|
|
32.
|
Aragaki M, Takahashi K, Akiyama H, et al:
Characterization of a cleavage stimulation factor, 3′ pre-RNA,
subunit 2, 64 kDa (CSTF2) as a therapeutic target for lung cancer.
Clin Cancer Res. 17:5889–5900. 2011.
|
|
33.
|
Nishino R, Takano A, Oshita H, et al:
Identification of Epstein-Barr virus-induced gene 3 as a novel
serum and tissue biomarker and a therapeutic target for lung
cancer. Clin Cancer Res. 17:6272–6286. 2011. View Article : Google Scholar
|
|
34.
|
Masuda K, Takano A, Oshita H, et al:
Chondrolectin is a novel diagnostic biomarker and a therapeutic
target for lung cancer. Clin Cancer Res. 17:7712–7722. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
35.
|
Fujitomo T, Daigo Y, Matsuda K, Ueda K and
Nakamura Y: Critical function for nuclear envelope protein TMEM209
in human pulmonary carcinogenesis. Cancer Res. 72:4110–4118. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
36.
|
Koinuma J, Akiyama H, Fujita M, et al:
Characterization of an Opa interacting protein 5 involved in lung
and esophageal carcinogenesis. Cancer Sci. 103:577–586. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
37.
|
Nguyen MH, Ueda K, Nakamura Y and Daigo Y:
Identification of a novel oncogene, MMS22L, involved in lung
and esophageal carcinogenesis. Int J Oncol. 41:1285–1296. 2012.
|
|
38.
|
Oshita H, Nishino R, Takano A, et al:
RASEF is a novel diagnostic biomarker and a therapeutic target for
lung cancer. Mol Cancer Res. 11:937–951. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39.
|
Tong ZB, Nelson LM and Dean J: Mater
encodes a maternal protein in mice with a leucine-rich repeat
domain homologous to porcine ribonuclease inhibitor. Mamm Genome.
11:281–287. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
40.
|
Mutai H, Toyoshima Y, Sun W, Hattori N,
Tanaka S and Shiota K: PAL31, a novel nuclear protein, expressed in
the developing brain. Biochem Biophys Res Commun. 274:427–433.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
41.
|
Bilder D and Perrimon N: Localization of
apical epithelial determinants by the basolateral PDZ protein
Scribble. Nature. 403:676–680. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
42.
|
Linhoff MW, Harton JA, Cressman DE, Martin
BK and Ting JP: Two distinct domains within CIITA mediate
self-association: involvement of the GTP-binding and leucine-rich
repeat domains. Mol Cell Biol. 21:3001–3011. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
43.
|
Inohara N, Koseki T, del Peso L, et al:
Nod1, an Apaf-1-like activator of caspase-9 and nuclear
factor-kappaB. J Biol Chem. 274:14560–14567. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
44.
|
Wu H, Maciejewski MW, Marintchev A,
Benashski SE, Mullen GP and King SM: Solution structure of a dynein
motor domain associated light chain. Nat Struct Biol. 7:575–579.
2000. View Article : Google Scholar
|
|
45.
|
Xu P, Mitchelhill KI, Kobe B, Kemp BE and
Zot HG: The myosin-I-binding protein Acan125 binds the SH3 domain
and belongs to the superfamily of leucine-rich repeat proteins.
Proc Natl Acad Sci USA. 94:3685–3690. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
46.
|
Brackertz M, Boeke J, Zhang R and
Renkawitz R: Two highly related p66 proteins comprise a new family
of potent transcriptional repressors interacting with MBD2 and
MBD3. J Biol Chem. 277:40958–40966. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
47.
|
Noh EJ, Lim DS and Lee JS: A novel role
for methyl CpG-binding domain protein 3, a component of the histone
deacetylase complex, in regulation of cell cycle progression and
cell death. Biochem Biophys Res Commun. 378:332–337. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
48.
|
Abbas T and Dutta A: p21 in cancer:
intricate networks and multiple activities. Nat Rev Cancer.
9:400–414. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
49.
|
Wu S, Cetinkaya C, Munoz-Alonso MJ, et al:
Myc represses differentiation-induced p21CIP1 expression via
Miz-1-dependent interaction with the p21 core promoter. Oncogene.
22:351–360. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
50.
|
Reese KJ, Lin S, Verona RI, Schultz RM and
Bartolomei MS: Maintenance of paternal methylation and repression
of the imprinted H19 gene requires MBD3. PLoS Genet. 3:e1372007.
View Article : Google Scholar : PubMed/NCBI
|