Introduction
Cholangiocarcinoma (CCA) is a malignant neoplasm of
biliary tract epithelium, which shows progressively increased
incidence and mortality over the past decades (1). Northeast Thailand has the highest
incidence of CCA in the world (2),
where it is most often associated with Opisthorchis
viverrini infection (3). This
tumor is oftentimes fatal due to late stage diagnosis as well as
limited options and success with treatment. The most widely used
serum biomarker for CCA is carbohydrate antigen (CA) 19-9 and
carcinoembryonic antigen (CEA). However, CA19-9 is also increased
in non-malignant obstructive jaundice, severe hepatic injury, and
several cancers such as pancreatic and gastric cancer. In addition,
the sensitivity and specificity of CEA in serum is relatively low.
Novel potential serum and bile markers associated with CCA include
matrix metalloproteinase-7 (4),
interleukin-6 (5), and microRNA
(miR-9) (6). Unfortunately, none
of these biomarkers have yet to be validated in large clinical
studies. As diagnosis of CCA remains problematic, developing a more
effective biomarkers and therapeutic modalities could significantly
increase the chance of early detection for these patients.
Proteins secreted by cancer cells are ideal
specimens to investigate biomarkers since they have important roles
in cell signaling and pathological developments such as
differentiation, invasion and metastasis. Detection of these
secreted biomarkers is also non-invasive as they are detected in
bodily fluids such as blood and urine making secretomic study
beneficial for not only identifying biomarkers (7–9) but
also improve routine medical evaluations. However, secretome
analysis of monolayer cell culture has several limitations such as
low concentration of secreted proteins and contamination with
cellular proteins due to cell lysis. To improve success in
biomarker discovery, a more reliable and representative cell
culture model may help bridge the gap between in vitro
research and clinical studies (10).
Three-dimensional (3D) culture is being more
extensively used since it provides a more realistic
microenvironment in terms of natural physiology than conventional
monolayer culture (11). Several
techniques have been introduced to create in vitro 3D cancer
models such as scaffold-based, spheroid aggregation, liquid
overlay, and rotary cell culture systems. Scaffold-based 3D culture
offers advantages in providing a structural support for cellular
attachment and may modify the signaling responses of cells
(12). 3D culture systems have
been used in many applications including cancer biomarker discovery
studies (13). Thus, the present
study aimed to develop scaffold-based 3D culture of human
intrahepatic CCA and to use this model as a tool to study the
expression of secreted proteins by proteomic analysis. Comparative
secretome analysis of 3D culture to monolayer culture enabled us to
discover potential biomarkers for CCA.
Materials and methods
Cell cultures
Human cholangiocarcinoma cells (HuCCA-1), derived
from a Thai patient, were grown in Ham’s F-12 media
(Gibco®, Invitrogen, USA) (14). Hepatocellular carcinoma cells
HCC-S102 established from a Thai patient were grown in RPMI-1640
(Gibco) (15) while HepG2 and
SK-HEP-1 cell lines were purchased from American Type Culture
Collection (ATCC) and grown in DMEM (Gibco). All cell culture media
contained 10% fetal bovine serum (FBS, Hyclone Laboratories, USA),
100 U/ml penicillin, 100 mg/ml streptomycin, and 125 ng/ml
amphotericin B (Gibco) and were maintained at 37°C in a humidified
atmosphere, 95% air, 5% CO2. In serum-free culture, FBS
gradually decreased and was simultaneously replaced with chemically
defined medium for high density cell culture serum replacement
(CDM-HD; FiberCells® System, USA) (16).
Development of 3D culture
Scaffold-based 3D culture was established using
natural collagen type I fibrils named Lyostypt® (Braun,
Germany) (17). Briefly, cells
were seeded onto sterilized collagen scaffold, followed by
incubation for 2 h to allow cell attachment. Media was gently added
to completely submerge each scaffold piece then incubated overnight
and media was subsequently changed every alternate day throughout
the experiment.
Determination of viability of cells in
collagen scaffolds
Cell-seeded scaffolds were incubated with MTT in a
humidified incubator with 5% CO2 at 37°C for 2 h and the
number of cells was qualitatively determined under light
microscope. For Hoechst staining, cell-seeded scaffolds were
incubated in the dark with 1 μg/ml Hoechst dye for 2 min and
stained nuclei were detected by a fluorescence microscope.
Quantitative numbers of cells were determined by
PicoGreen® assay (Invitrogen) that binds to DNA and
measured by fluorescence microplate reader at wavelength of 485 nm
and emission at 520 nm. Metabolic activity was also determined to
confirm cell viability by assessing remaining glucose concentration
in the conditioned medium. The culture medium (2 μl) was assayed
daily using Medisafe-mini GR-102 blood glucose meter (Terumo,
Japan). The amount of depleted glucose in the medium each day was
calculated and reported as cumulative glucose consumption by viable
cells.
Histological and morphological
evaluation
Cells were fixed in 4% paraformaldehyde. Specimens
were stained with H&E and mucins for microscopic analysis. For
morphological evaluation, cells from both culture systems were
fixed with 2.5% glutaraldehyde (pH 7.4). Dried samples were
sputter-coated with platinum-palladium and observed under scanning
electron microscope (SEM) (Hitachi SEM S-2500, Japan).
2D-PAGE preparation and image
analysis
Conditioned medium was collected and cell debris was
removed by centrifugation. Supernatant was concentrated by
lyophilization and precipitated in 10% TCA at 4°C overnight.
Samples were centrifuged at 12,000 rpm, 4°C for 10 min, washed with
25% v/v acetone, and dried by speed vacuum. Specimens were
resuspended in lysis buffer containing 9 M urea, 2% CHAPS, 2% DTT,
2% ampholine pH 3.5–10.0, and 1:500 protease inhibitor
(Sigma-Aldrich, USA). Protein concentration was measured by
Bradford assay. IPG strips, 7 cm, non-linear, pH 3.0–10.0 gradient
(GE Healthcare, USA) were rehydrated with 150 mg protein overnight.
IEF was performed at 7,000 Vh, 55 mA per gel strip using an Ettan
IPGphor 3 (GE Healthcare). The IPG strips were equilibrated as
previously described (8) and
separated in 12.5% SDS-PAGE followed by CBB R-250 staining. Gels
were analyzed by ImageMaster 2D Platinum 7.0 software (GE
Healthcare).
In-gel digestion
Protein spots having a volume ratio change of
>1.5-fold were excised and subjected to in-gel digestion
according to the protocol by Srisomsap et al (9). Briefly, gels were destained with 50%
ACN in 0.1 M NH4HCO3, reduced with 10 mM DTT,
and alkylated with 100 mM iodoacetamide, respectively. Gel pieces
were dried then 0.3 μg of trypsin (Promega, USA) was added,
followed by incubation at 37°C overnight and digested peptides were
collected for protein identification.
Mass spectrometry
Nanoflow liquid chromatography coupled with the
amaZon speed ion trap mass spectrometry (Bruker, USA) was utilized
to identify protein spots. A 75 μm id × 100 mm C18 EASY-nLC™ column
(Thermo Scientific, USA) was used. Gradient separation was
performed using 0.1% formic acid in water (solution A) and 0.1%
formic acid in ACN (solution B) followed by MS/MS equipped with
CaptiveSpray™ source. Parent mass peaks with a range from 50 to
3,000 m/z were selected for MS/MS analysis in which collision
energy was fixed at 1,300 V. MS/MS data were processed by Bruker
Compass 1.4 software and proteins were then identified using MASCOT
with similar search parameters as in a previous study (18). Proteins with molecular weight and
pI consistent to gel spot with MASCOT score >25 using p-value
≤0.05 were considered positively identified.
Western blot analysis
Proteins were resolved in 10% SDS-PAGE and
electrophoretically transferred to PVDF membranes (Millipore, USA).
The membranes were probed with antibody against human TPI, SFN
(1:2,000, Abcam, USA), PGAM1, ENO1, LCP1 (1:1,000, Abcam),
β-tubulin (1:2,000, Cell Signaling Technology, USA), and actin
(1:5,000, Sigma, USA) at 4°C overnight. Membranes were washed and
incubated with corresponding secondary antibody conjugated with HRP
(DakoCytomation, Denmark) at room temperature for 1 h. Membranes
were probed with ECL (GE Healthcare) and detected by ImageQuant™
LAS 4000 (GE Healthcare). Then, membranes were stained with CBB
R-250 and band intensity was determined to show equal protein
loadings.
Statistical analysis
The differences between monolayer and 3D cell
culture were analyzed with STATA 10.1 using unpaired t-test.
p<0.05 was considered statistically significant.
Results
Development of HuCCA-1 in scaffold-based
3D culture
The serum-free media cell culture was completely
switched to CDM-HD and morphology observed under a light
microscope. There was no significant difference in percentage of
viable cells between serum-containing and serum-free cultures. Cell
viability was consistently maintained at >95%.
The serum-free cells were then grown in natural
collagen-based scaffold to mimic the dense 3D microenvironment of
the in vivo tumor. Growth and localization of cells were
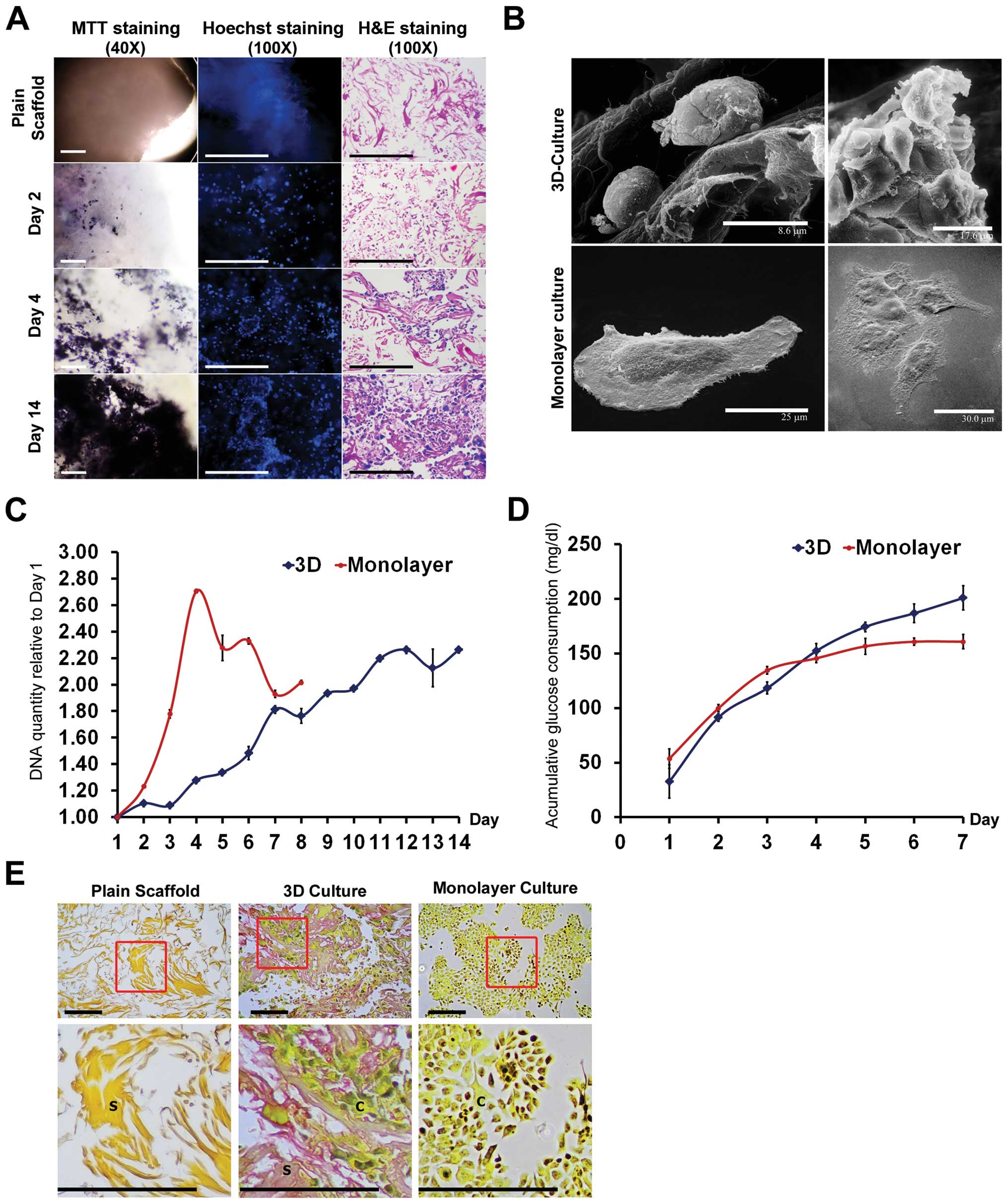
studied (Fig. 1A). At the initial
phase (day 2), single cells were dispersed throughout the scaffold.
Then, the cells aggregated into colonies and expanded between the
spaces of the spun fibers (days 4 and 14). When cell-seeded
scaffolds were stained with MTT (left column), black spots were
visible representing cells or small colonies of cells (day 2).
Hoechst staining of dsDNA (middle column) showed increasing numbers
of blue spots under a fluorescence microscope, indicating increased
number of nuclei of cells at increasing time-points (days 2, 4, and
14). H&E staining (right column) indicated cells with deep blue
nuclei and faint pink cytoplasm throughout the pink collagen
fibers. These three different methods confirmed progressive growth
of HuCCA-1 cells inside collagen scaffolds.
Morphological study under SEM (Fig. 1B) showed alteration of cell shape
in 3D culture when compared to monolayer culture. The appearance of
cells in the scaffold was round, growing into a dense cluster,
whereas the attached cells were flat and expanded as a single thin
layer during the 14 days of culture. Cell-cell contacts as well as
cell-collagen scaffold contacts were observed in our 3D culture
system.
Differential growth patterns between the
3D and monolayer cultures
The growth pattern of cells in different culture
systems was determined by PicoGreen assay. Results showed
alterations in growth pattern between 3D and monolayer cultures
(Fig. 1C). Cells in monolayer
culture showed rapid increase in cell proliferation but decreased
after reaching the maximum growth in day 4, while in the 3D culture
system, cells showed slower proliferation but maintained a longer
growth phase covering a period of over 14 days.
The levels of glucose were monitored and cumulative
glucose consumption was calculated compared to starting glucose
level in the culture media (Fig.
1D). The pattern of cumulative glucose consumption of cells in
3D culture continuously increased until day 7, while cumulative
glucose consumption of cells in monolayer culture became constant
after day 4, reflecting the cessation of growth in monolayer
culture after 4 days (Fig. 1C).
Thus, cells at day 4 of both culture systems were selected and used
for further comparative analyses.
Extracellular matrix deposition in 3D
culture
Histological analysis revealed that 3D culture
produced increased level of secreted ECM when compared to monolayer
culture. Compared to plain collagen scaffold, a positive result
(pink) of mucicarmine staining represented accumulation of mucins
in 3D culture, which was not found in monolayer culture (Fig. 1E).
The quantity and quality of secreted
proteins
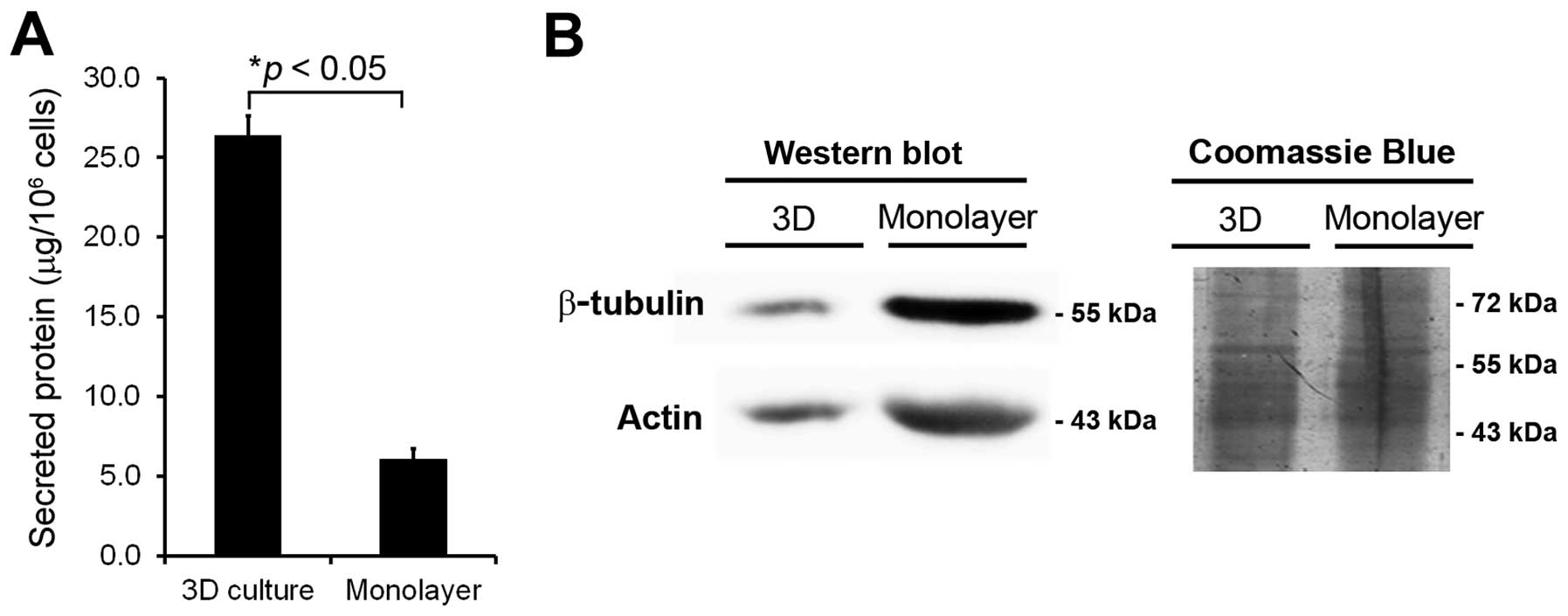
The protein concentrations extracted from 3D culture
were significantly increased (>5-fold, p<0.05) compared to
monolayer culture (Fig. 2A). Thus,
the secreted proteins from 3D culture were more extensively
enriched and sufficient for further studies.
Contamination of intracellular protein leakage from
damaged or dead cells is also a major concern in secretome
analysis. To evaluate interference by cell lysis, western blotting
of two major cytosolic proteins (actin and β-tubulin) was performed
in conditioned media from both culture systems at day 4. The data
indicated significant levels of actin and β-tubulin in conditioned
media obtained from monolayer culture, but decreased presence of
these proteins in conditioned media prepared from 3D culture
(Fig. 2B). Thus, the lack of these
cytoplasmic protein contaminations confirmed the high quality of
secreted proteins obtained from 3D culture, making samples from 3D
culture ideal for secretome analysis.
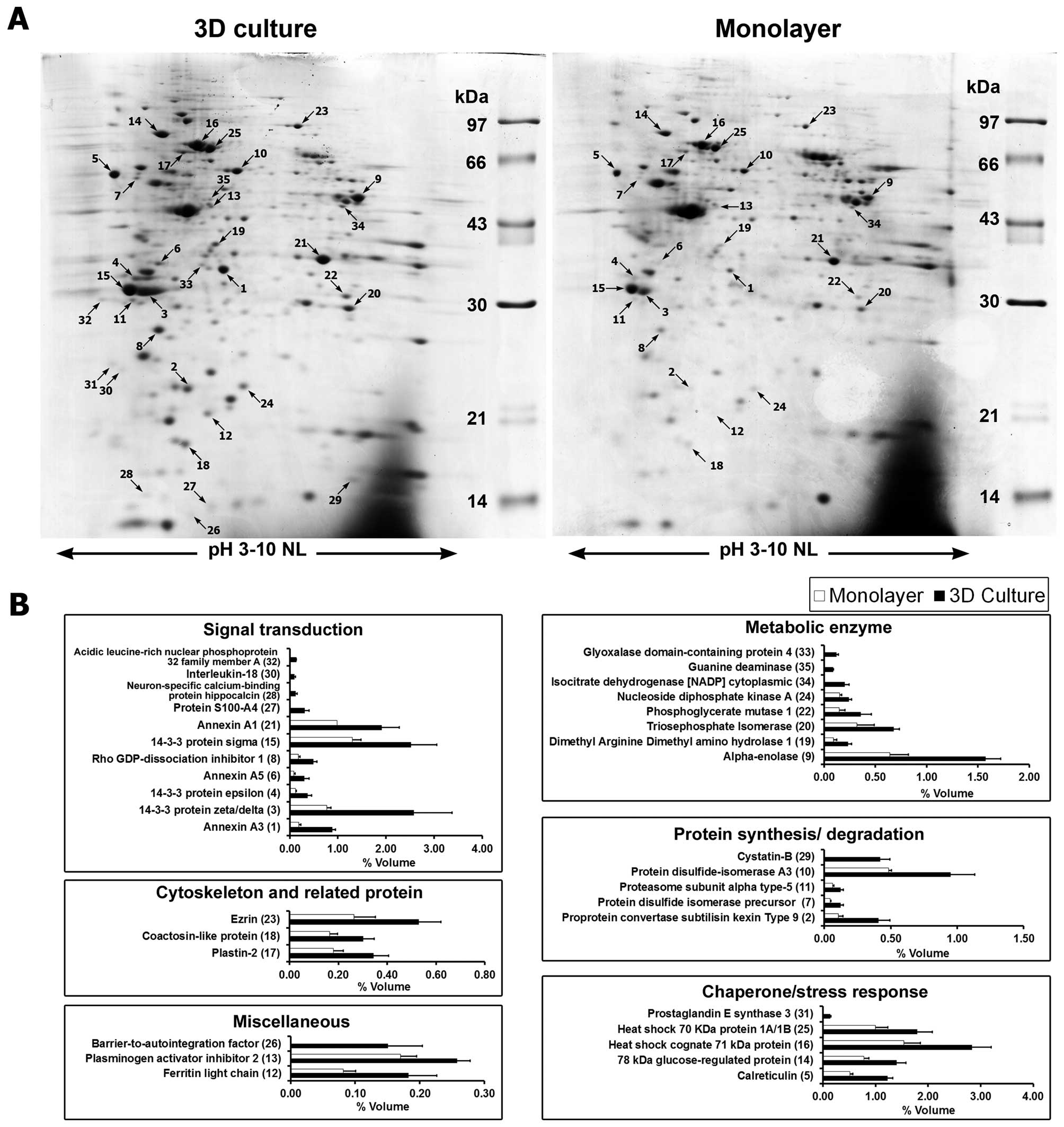
Secretome analysis
The secreted proteins from cells grown under the two
culture conditions were compared by 2DE (Fig. 3A). Overall, 431±14 protein spots
were found in 3D culture while 410±20 spots were found in monolayer
culture. Image analysis showed that 35 secreted protein spots were
differentially expressed in 3D culture compared to monolayer
culture. Of these, 25 spots were significantly increased in 3D
culture and 10 spots were found to be expressed only in 3D culture
system (Table I). These proteins
were categorized according to their functions, namely: signal
transduction, metabolic enzymes, chaperone and stress response,
protein synthesis and degradation, cytoskeleton and related, and
miscellaneous function proteins (Fig.
3B).
 | Table ISecreted proteins identified in the
conditioned media by LC-MS/MS analysis. |
Table I
Secreted proteins identified in the
conditioned media by LC-MS/MS analysis.
| Spot no.a | Accession no. | Protein
description | Gene name | Theoritical
(MW/pI)b | Scorec | No. of
peptided | Coverage (%) | Fold changee |
|---|
| 1 | P12429 | Annexin A3 | ANXA3 | 36353/5.63 | 38 | 5 | 13 | ↑4.71±0.25 |
| 2 | Q8NBP7 | Proprotein
convertase subtilisin type 9f | PCSK9 | 14080/5.04 | | | | ↑3.79±0.38 |
| 3 | P63104 | 14-3-3 protein
ζ/δ | YWHAZ | 27728/4.73 | 47 | 5 | 20 | ↑3.35±0.33 |
| 4 | P62258 | 14-3-3 protein
ɛ | YWHAE | 29155/4.63 | 21 | 2 | 5 | ↑3.19±0.31 |
| 5 | P27797 | Calreticulin | CALR | 48112/4.29 | 65 | 3 | 11 | ↑2.39±0.13 |
| 6 | P08758 | Annexin A5 | ANXA5 | 35914/4.94 | 27 | 3 | 7 | ↑3.36±0.41 |
| 7 | P07237 | Protein disulfide
isomerase precursor (PDI)f | P4HB | 57100/4.78 | | | | ↑2.63±0.23 |
| 8 | P52565 | Rho
GDP-dissociation inhibitor 1 | ARHGDIA | 23193/5.02 | 40 | 3 | 14 | ↑2.71±0.27 |
| 9 | P06733 | α-enolase | ENO1 | 47139/7.01 | 70 | 13 | 24 | ↑2.45±0.29 |
| 10 | P30101 | Protein
disulfide-isomerase A3 | PDIA3 | 56747/5.98 | 25 | 4 | 7 | ↑1.95±0.20 |
| 11 | P28066 | Proteasome subunit
α type-5 | PSMA5 | 26394/4.74 | 76 | 2 | 7 | ↑1.94±0.22 |
| 12 | P02792 | Ferritin light
chain | FTL | 20007/5.51 | 46 | 1 | 4 | ↑2.24±0.34 |
| 13 | P05120 | Plasminogen
activator inhibitor 2 | SERPINB2 | 46566/5.46 | 92 | 5 | 12 | ↑1.51±0.17 |
| 14 | P11021 | 78-kDa
glucose-regulated protein | HSPA5 | 72288/5.07 | 301 | 13 | 20 | ↑1.79±0.17 |
| 15 | P31947 | 14-3-3 protein
σ | SFN | 27757/4.68 | 172 | 5 | 16 | ↑1.93±0.26 |
| 16 | P11142 | Heat shock cognate
71-kDa protein | HSPA8 | 70854/5.37 | 382 | 17 | 22 | ↑1.84±0.24 |
| 17 | P13796 | Plastin-2 | LCP1 | 70244/5.29 | 363 | 10 | 18 | ↑1.91±0.28 |
| 18 | Q14019 | Coactosin-like
protein | COTL1 | 15935/5.54 | 107 | 7 | 33 | ↑1.82±0.25 |
| 19 | O94760 | Dimethyl arginine
dimethyl amino-hydrolase 1 | DDAH1 | 31102/5.53 | 240 | 7 | 26 | ↑2.54±0.36 |
| 20 | P60174 | Triosephosphate
isomerase | TPI1 | 30772/5.65 | 386 | 9 | 37 | ↑2.12±0.53 |
| 21 | P04083 | Annexin A1 | ANXA1 | 38690/6.57 | 253 | 12 | 29 | ↑1.94±0.31 |
| 22 | P18669 | Phosphoglycerate
mutase 1 | PGAM1 | 28786/6.67 | 76 | 3 | 10 | ↑2.43±0.51 |
| 23 | P15311 | Ezrin | EZR | 69370/5.94 | 335 | 21 | 26 | ↑2.00±0.37 |
| P26038 | Moesin | MSN | 67778/6.08 | 128 | 9 | 10 | |
| 24 | P15531 | Nucleoside
diphosphate kinase A | NME1 | 17138/5.38 | 95 | 5 | 30 | ↑1.58±0.16 |
| 25 | P08107 | Heat shock 70-kDa
protein 1A/1B | HSPA1A | 70009/5.48 | 556 | 15 | 24 | ↑1.79±0.29 |
| 26 | O75531 |
Barrier-to-autointegration factor | BANF1 | 10052/5.81 | 125 | 2 | 21 | N/A |
| 27 | P26447 | Protein
S100-A4 | S100A4 | 11721/5.85 | 57 | 1 | 7 | N/A |
| 28 | P84074 | Neuron-specific
calcium-binding protein hippocalcin | HPCA | 22413/4.87 | 27 | 1 | 5 | N/A |
| 29 | P04080 | Cystatin-B | CSTB | 11133/6.96 | 29 | 2 | 12 | N/A |
| 30 | Q14116 | Interleukin-18 | IL18 | 22312/4.54 | 78 | 3 | 12 | N/A |
| 31 | Q15185 | Prostaglandin E
synthase 3 | PTGES3 | 18685/4.35 | 82 | 2 | 12 | N/A |
| 32 | P39687 | Acidic leucine-rich
nuclear phospho-protein 32 family member A | ANP32A | 28568/3.99 | 92 | 5 | 16 | N/A |
| 33 | Q9HC38 | Glyoxalase
domain-containing protein 4 | GLOD4 | 34771/5.40 | 124 | 5 | 17 | N/A |
| 34 | O75874 | Isocitrate
dehydrogenase (NADP) cytoplasmic | IDH1 | 46630/6.53 | 151 | 7 | 16 | N/A |
| 35 | Q9Y2T3 | Guanine
deaminase | GDA | 50971/5.44 | 104 | 6 | 11 | N/A |
Validation of differential protein
expression
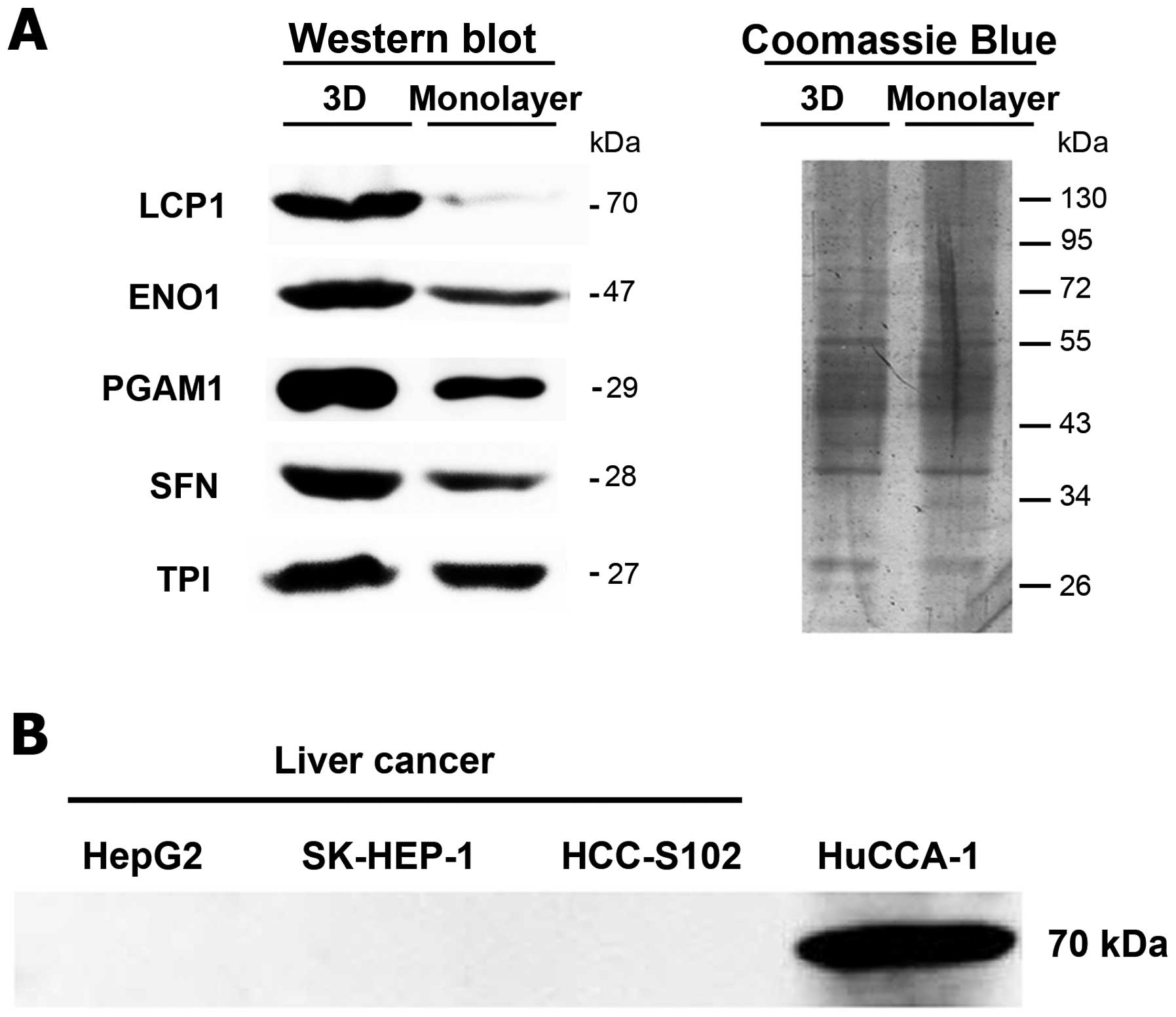
Western blot analyses were used to verify the
expression of selected secreted proteins which differed
considerably between two culture conditions including 14-3-3 σ
(SFN), triosephosphate isomerase (TPI), α-enolase (ENO1),
phosphoglycerate mutase 1 (PGAM1), and L-plastin (LCP1). These
results were consistent with the 2DE data in which the expressions
of these secreted proteins were higher in 3D than monolayer culture
(Fig. 4A). Moreover, the
expression of LCP1 in secretomes of CCA cells was used to compare
with three liver cancer cell lines (HepG2, SK-HEP-1, and HCC-S102).
The result showed that LCP1 was found only in the conditioned media
of CCA, but not in the conditioned media of the other liver cell
lines (Fig. 4B).
Discussion
We suggest that the developed 3D culture model of
CCA is suitable for secretome analysis and may be a useful method
for discovery of novel biomarkers. Here, scaffold-based 3D culture
of HuCCA-1 cells in serum-free condition was analyzed. The result
suggests the advantage of 3D culture model over monolayer culture
in providing long-term culture. Since CCA is an extremely
heterogeneous cancer (19),
long-term culture might facilitate differentiation and
proliferation of different cell types providing a more realistic
tumor population. SEM technique also revealed that the morphology
of HuCCA-1 cells inside the scaffold were ellipsoid in shape
similar to simple cuboidal epithelium, the morphology of
cholangiocytes in vivo. Moreover, cells in 3D culture mimic
a more natural cell-cell and cell-ECM behavior. Interestingly,
Chiarini et al reported the induction of ductal-like
structure of cholangiocytes in vitro using the collagen gel
scaffold (20). Taken together,
this evidence indicates the induction of in vivo-like cell
morphology, which shows promising advantage over the flat cell
morphology found in monolayer culture. The cell-ECM interaction in
3D culture also allows the collagen fiber to absorb extracellular
matrix mucin secreted by HuCCA-1 cells. The capability of cancer
cells grown in 3D culture to enhance ECM deposition was reported by
Pruksakorn et al using this scaffold for 3D culture of HepG2
cells and they found increased levels of
sulfated-glycosaminoglycans (17).
Mucin expression appears to be associated with intrahepatic bile
duct development and relate to progression of CCA (21). Thus, culture of HuCCA-1 cells in a
3D scaffold makes it feasible to accumulate crucial cell-ECM
interaction, which may confer a better in vivo-like
environment than conventional cell culture.
Our findings provide that scaffold-based 3D cell
culture yields superior quantity and quality of secreted proteins.
Our group previously reported the analysis of secreted protein from
HuCCA-1 cell culture in hollow fiber bioreactor (16). Consistent with this study, secreted
proteins from hollow fiber system, which also aims to mimic in
vivo microenvironment, were enhanced compared to monolayer
culture. In agreement with this notion, rabbit mammary cells will
synthesize, store, and secrete fat and milk proteins in 3D culture
but cannot do so in monolayer culture (22). In this case, researchers propose
that cell polarity and differentiation, which are important for
milk secretion, were influenced by cell-matrix interaction in 3D
culture. Since cholangiocytes are polarized cell-like mammary
cells, it is possible that cell shape and cell-collagen
interactions might affect cell polarity and secretion ability of
the cells. Thus, further supporting our study that scaffold-based
3D cell cultures are better suited for secretome analysis and the
differentially expressed proteins would be a more accurate
reflection of proteins secreted in vivo.
In this investigation, upregulation of 14-3-3 σ or
stratifin (SFN) in 3D culture compared to monolayer culture was
confirmed by Western blot analysis. This signaling protein is an
intracellular, phosphoserine binding protein that is thought to be
involved in cancer development of several organs. We recently
reported the upregulation of this protein and its important role in
anoikis resistance of CCA cells (18). Interestingly, overexpression of SFN
appears to be correlated with increased tumor progression and poor
prognosis in colorectal (23) and
gastric cancer (24). Since SFN
was intensely secreted in 3D culture of HuCCA-1, this protein might
also play an important role in the tumor microenvironment of the
cells and may be a potential biomarker in CCA. Additionally,
upregulation of selected glycolytic proteins such as TPI, PGAM1,
and ENO1 in 3D culture were confirmed. We hypothesize that this is
due to the enhanced Warburg phenomenon as reported in other studies
such as osteosarcoma (25) and
hepatocellular carcinoma (17).
Multiple proteins associated with aggressive cancer
phenotypes were found to be upregulated. TPI is a glycolytic enzyme
that catalyzes the reversible interconversion of dihydroxyacetone
phosphate (DHAP) and D-glyceraldehyde 3-phosphate (G3P).
Significant upregulation of TPI has been found in various cancers
including breast cancer (26) and
brain metastatic tissues of endometrial and ovarian cancers
(27). Another glycolytic enzyme
considerably upregulated in 3D culture is PGAM1 which reversibly
catalyzes the conversion of 3-phosphoglycerate (3PG) into
2-phosphoglycerate (2PG). Marked upregulation of PGAM1 is strongly
correlated with poor differentiation of hepatocellular carcinoma
patients (28). We also found high
expression of ENO1. This enzyme catalyzes 2-phosphoglycerate (2-PG)
to phosphoenolpyruvate (PEP) in the glycolysis pathway. Upregulated
ENO1 in CCA tissue is significantly associated with poor prognosis
and tumor invasion (29).
Promising biomarkers need to be significantly
expressed and ideally specific or capable of differentiating
between types of cancer. In our study, one such protein was
identified to be LCP1. This protein is generally expressed in
rapidly movable cells such as leukocytes and cancer cells may gain
the ability to metastasize to other parts of the body by expressing
LCP1 (30). This protein is
specifically expressed in non-hematopoietic cancers including
ovarian (31) and colorectal
cancer (32). In addition, Lin
et al reported that 68% of cancers derived from epithelia
express LCP1 (33). Accordingly,
LCP1 was a main focus among validated secreted proteins because of
its functional involvement in cancer metastasis and lack of
research on the role of LCP1 in CCA. Though LCP1 is expressed in
several types of cancers, the level of LCP1 mRNA expression in
HepG2 could not be detected by northern blot analysis (33) and RT-PCR (34) while SK-HEP-1 showed only trace
levels of LCP1 mRNA detectable by RT-PCR (34). Thus, LCP1 has the potential to be
used as biomarker for the discrimination of CCA from HCC and needs
to be evaluated further in clinical specimens such as serum and
tissues from CCA patients.
Additionally, our data revealed that some proteins
can be detected only in the secreted fraction from 3D culture,
suggesting that they may require the 3D microenvironment for
expression and/or secretion. Interleukin-18 has been shown to be
associated with various cancers including renal cancer (35), and hepatocellular carcinoma
(36). By using 3D culture based
secretome analysis, this is the first evidence of IL-18 secretion
in the HuCCA-1 cell line. Therefore, this protein might be another
promising candidate as a biomarker of CCA and should be studied
further.
In conclusion, we are the first group to report the
successful development of serum-free intrahepatic CCA cell line
cultured in a collagen scaffold-based 3D system. Our study shows
that characteristics of HuCCA-1 cells were modified in response to
the 3D environment allowing cells to behave in a more in
vivo-like manner where cell contact (cell-cell or cell-ECM) is
drastically different from conventional cell culture. Consequently,
the cells in this environment should behave and secrete proteins
that mimic in vivo CCA more accurately allowing for
discovery of promising biomarkers for CCA.
Acknowledgements
We would like to thank Dr Titipatima Sakulterdkiat
for constructive feedback in writing this manuscript. This study
was supported by the Chulabhorn Research Institute, Chulabhorn
Graduate Institute, and the Center of Excellence on Environmental
Health, Toxicology and Management of Chemicals, Bangkok,
Thailand.
Abbreviations:
|
CCA
|
cholangiocarcinoma
|
|
CDM-HD
|
chemically defined medium for high
density cell culture
|
|
ECM
|
extracellular matrix
|
|
ENO1
|
α-enolase
|
|
HuCCA-1
|
human cholangiocarcinoma cells
|
|
LCP1
|
L-plastin
|
|
PGAM1
|
phosphoglycerate mutase 1
|
|
SFN
|
14-3-3 σ
|
|
TPI
|
triosephosphate isomerase
|
References
|
1
|
Khan SA, Thomas HC, Davidson BR and
Taylor-Robinson SD: Cholangiocarcinoma. Lancet. 366:1303–1314.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Khan SA, Toledano MB and Taylor-Robinson
SD: Epidemiology, risk factors, and pathogenesis of
cholangiocarcinoma. HPB. 10:77–82. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sripa B, Kaewkes S, Sithithaworn P, et al:
Liver fluke induces cholangiocarcinoma. PLoS Med. 4:e2012007.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Leelawat K, Narong S, Wannaprasert J and
Ratanashu-ek T: Prospective study of MMP7 serum levels in the
diagnosis of cholangiocarcinoma. World J Gastroenterol.
16:4697–4703. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Mott JL and Gores GJ: Targeting IL-6 in
cholangiocarcinoma therapy. Am J Gastroenterol. 102:2171–2172.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Shigehara K, Yokomuro S, Ishibashi O, et
al: Real-time PCR-based analysis of the human bile microRNAome
identifies miR-9 as a potential diagnostic biomarker for biliary
tract cancer. PLoS One. 6:e235842011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Makridakis M and Vlahou A: Secretome
proteomics for discovery of cancer biomarkers. J Proteomics.
73:2291–2305. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Srisomsap C, Sawangareetrakul P,
Subhasitanont P, et al: Proteomic analysis of cholangiocarcinoma
cell line. Proteomics. 4:1135–1144. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Srisomsap C, Sawangareetrakul P,
Subhasitanont P, et al: Proteomic studies of cholangiocarcinoma and
hepatocellular carcinoma cell secretomes. J Biomed Biotechnol.
2010:4371432010. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hutmacher DW, Loessner D, Rizzi S, Kaplan
DL, Mooney DJ and Clements JA: Can tissue engineering concepts
advance tumor biology research? Trends Biotechnol. 28:125–133.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Pampaloni F, Reynaud EG and Stelzer EH:
The third dimension bridges the gap between cell culture and live
tissue. Nat Rev Mol Cell Biol. 8:839–845. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yamada KM and Cukierman E: Modeling tissue
morphogenesis and cancer in 3D. Cell. 130:601–610. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Weigelt B and Bissell MJ: The need for
complex 3D culture models to unravel novel pathways and identify
accurate biomarkers in breast cancer. Adv Drug Deliv Rev.
69–70:42–51. 2014.PubMed/NCBI
|
|
14
|
Sirisinha S, Tengchaisri T, Boonpucknavig
S, Prempracha N, Ratanarapee S and Pausawasdi A: Establishment and
characterization of a cholangiocarcinoma cell line from a Thai
patient with intrahepatic bile duct cancer. Asian Pac J Allergy
Immunol. 9:153–157. 1991.PubMed/NCBI
|
|
15
|
Laohathai K and Bhamarapravati N:
Culturing of human hepatocellular carcinoma. A simple and
reproducible method. Am J Pathol. 118:203–208. 1985.PubMed/NCBI
|
|
16
|
Weeraphan C, Diskul-Na-Ayudthaya P,
Chiablaem K, et al: Effective enrichment of cholangiocarcinoma
secretomes using the hollow fiber bioreactor culture system.
Talanta. 99:294–301. 2012. View Article : Google Scholar
|
|
17
|
Pruksakorn D, Lirdprapamongkol K,
Chokchaichamnankit D, et al: Metabolic alteration of HepG2 in
scaffold-based 3-D culture: proteomic approach. Proteomics.
10:3896–3904. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Khongmanee A, Lirdprapamongkol K, Tit-oon
P, Chokchaichamnankit D, Svasti J and Srisomsap C: Proteomic
analysis reveals important role of 14-3-3sigma in anoikis
resistance of cholangiocarcinoma cells. Proteomics. 13:3157–3166.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Cardinale V, Carpino G, Reid L, Gaudio E
and Alvaro D: Multiple cells of origin in cholangiocarcinoma
underlie biological, epidemiological and clinical heterogeneity.
World J Gastrointest Oncol. 4:94–102. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chiarini LB, Takiya CM, Borojevic R and
Monteiro AN: Long-term culture of cholangiocytes from liver
fibro-granulomatous lesions. BMC Gastroenterol. 6:132006.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mall AS, Tyler MG, Ho SB, et al: The
expression of MUC mucin in cholangiocarcinoma. Pathol Res Pract.
206:805–809. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Haeuptle MT, Suard YL, Bogenmann E, Reggio
H, Racine L and Kraehenbuhl JP: Effect of cell shape change on the
function and differentiation of rabbit mammary cells in culture. J
Cell Biol. 96:1425–1434. 1983. View Article : Google Scholar
|
|
23
|
Perathoner A, Pirkebner D, Brandacher G,
et al: 14-3-3sigma expression is an independent prognostic
parameter for poor survival in colorectal carcinoma patients. Clin
Cancer Res. 11:3274–3279. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhou WH, Tang F, Xu J, et al: Aberrant
upregulation of 14-3-3σ expression serves as an inferior prognostic
biomarker for gastric cancer. BMC Cancer. 11:3972011.
|
|
25
|
Santini MT, Rainaldi G, Romano R, et al:
MG-63 human osteosarcoma cells grown in monolayer and as
three-dimensional tumor spheroids present a different metabolic
profile: a (1)H NMR study. FEBS Lett. 557:148–154. 2004. View Article : Google Scholar
|
|
26
|
Thongwatchara P, Promwikorn W, Srisomsap
C, Chokchaichamnankit D, Boonyaphiphat P and Thongsuksai P:
Differential protein expression in primary breast cancer and
matched axillary node metastasis. Oncol Rep. 26:185–191.
2011.PubMed/NCBI
|
|
27
|
Yoshida A, Okamoto N, Tozawa-Ono A, et al:
Proteomic analysis of differential protein expression by brain
metastases of gynecological malignancies. Hum Cell. 26:56–66. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ren F, Wu H, Lei Y, et al: Quantitative
proteomics identification of phosphoglycerate mutase 1 as a novel
therapeutic target in hepatocellular carcinoma. Mol Cancer.
9:812010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Yonglitthipagon P, Pairojkul C,
Bhudhisawasdi V, Mulvenna J, Loukas A and Sripa B: Proteomics-based
identification of alphaenolase as a potential prognostic marker in
cholangiocarcinoma. Clin Biochem. 45:827–834. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Shinomiya H: Plastin family of
actin-bundling proteins: its functions in leukocytes, neurons,
intestines, and cancer. Int J Cell Biol. 2012:2134922012.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kang S, Shim HS, Lee JS, et al: Molecular
proteomics imaging of tumor interfaces by mass spectrometry. J
Proteome Res. 9:1157–1164. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ang CS and Nice EC: Targeted in-gel MRM: a
hypothesis driven approach for colorectal cancer biomarker
discovery in human feces. J Proteome Res. 9:4346–4355. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lin CS, Park T, Chen ZP and Leavitt J:
Human plastin genes. Comparative gene structure, chromosome
location, and differential expression in normal and neoplastic
cells. J Biol Chem. 268:2781–2792. 1993.
|
|
34
|
Park T, Chen ZP and Leavitt J: Activation
of the leukocyte plastin gene occurs in most human cancer cells.
Cancer Res. 54:1775–1781. 1994.PubMed/NCBI
|
|
35
|
Saenz-Lopez P, Carretero R, Vazquez F, et
al: Impact of interleukin-18 polymorphisms-607 and -137 on clinical
characteristics of renal cell carcinoma patients. Hum Immunol.
71:309–313. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Tangkijvanich P, Thong-Ngam D, Mahachai V,
Theamboonlers A and Poovorawan Y: Role of serum interleukin-18 as a
prognostic factor in patients with hepatocellular carcinoma. World
J Gastroenterol. 13:4345–4349. 2007.PubMed/NCBI
|