Introduction
Breast cancer is a genetically and clinically
heterogeneous disease, and the second most frequent cause of cancer
death among American women. It accounts for ~30% of overall cancer
deaths among women (1,2). Epidemiological data indicate that
women exposed to ionizing radiation such as radiation therapy,
X-ray diagnosis, A-bomb exposure had increased risk of breast
cancer incidence compared to normal population (3). The action of radiation on DNA is
damaging and consequently a mutagen, thus widely considered to be
the basis for its action as a carcinogen (4).
The progression of disease follows a complex
multi-step process that depends on various exogenous (socioeconomic
situation, diet, breast irradiation, oral contraception, geography
etc.) and endogenous (hormonal imbalances and family history of
breast cancer) factors that modulate the transformation of human
epithelial cells to a neoplastic stage (5). Recent experimental models also
suggest that the process of carcinogenesis can be driven by
abnormal interactions between cells and their microenvironment
(6). Thus, in addition to causing
DNA damage, radiation exposure also alters key regulators of
multi-cellular organization, which potentially disrupt the normal
intercellular communications and can promote neoplastic progression
in susceptible cells.
Recent progress in cancer research has shown the
importance of β-catenin, a molecule that plays pivotal roles in
cell-to-cell adhesion and as a transcriptional activator in Wnt/Wg
signal transduction pathway (7,8). The
Wnts are a family of highly conserved, secreted glycoproteins,
which have been implicated in stroma-epithelial interactions in the
breast and in neoplastic and benign breast disease (9,10).
The Wnt signaling pathway plays a critical role in organogenesis
and dysregulation of this pathway can contribute to tumorigenesis
(11,12). Wnt signaling via cytoplasmic
β-catenin plays a central role and critically control other Wnt
responsive genes during development and homeostasis (13).
The intracellular protein β-catenin remains as a
critical downstream factor in this pathway and plays an important
role in intercellular adhesion by linking the cytoplastic domain of
cadherins to α-catenin, which anchors the adhesion complex to the
cytoskeleton (14). It also
mediates the interactions between cadherins and other transmembrane
receptor protein, such as the epidermal growth factor receptor. Due
to this interaction, the cadherin-catenin complex becomes a target
for regulatory signals that govern cellular adhesiveness and
motility (15). These two
functions of β-catenin, E-cadherin binding and signal transduction
are interrelated (16).
The neoplastic transformation of human breast
epithelial cell lines in vitro represents a successful model
for obtaining step-by-step knowledge on the molecular and cellular
alterations that may contribute to the tumorigenic mechanisms.
There are few human cell culture models available to study the
mechanism of radiation carcinogenesis (17,18).
Therefore, to assess the effect of ionizing radiation, particularly
of high-LET (linear energy transfer) radiation, on the progression
of human breast carcinogenesis, a model system was used consisting
of irradiated, transformed and tumorigenic MCF-10F cell lines
treated with high-LET radiation (19). We have already reported the
involvement of c-Ha-ras oncogene and differential expression of a
series of other oncogene/tumor suppressor genes in this context
(20,21). Expression of various oncoproteins
related to different stages of neoplastic alterations was also
identified in this system (22,23).
The present study was undertaken to identify whether cell adhesion
molecules were involved in transformation induced by radiation and
estrogen in a breast cancer model.
Materials and methods
Cell lines
An established radiation-induced breast
carcinogenesis model was used (19). This spontaneously immortalized
breast epithelial cell line MCF-10F (24) retain all the characteristics of
normal epithelium in vitro, including anchorage-dependence,
non-invasiveness and non-tumorigenic in the nude mice (19). Cell lines were cultured on
Dulbecco’s modified Eagle’s media (DMEM) as described previously.
The breast cancer model consisted of the following cell lines: i)
control: MCF-10F; ii) MCF-10F treated with 17β-estradiol (E)
(10−8 M) (Sigma Chemical Co., St. Louis, MO, USA),
exposed to iii) single dose 60 cGy of α particle named Alpha1 (60
cGy); iii) single dose 60 cGy and treated with E, named Alpha2 (60
cGy plus E); iv) double doses of 60 cGy named Alpha3 (60/60 cGy)
that was anchorage-independent and non-tumorigenic in nude/SCID
mice; v) double dose of 60 cGy of α particle and treated with E,
named Alpha4 (60/60 cGy plus E); vi) double dose of 60 cGy of α
particle and treated with E before and after with radiation, named
Alpha5 (60 cGy plus E/60 cGy plus E), that was
anchorage-independent and formed tumors in nude/SCID mice, named
Tumor2 cell line. Phenotypic characteristics of these cell lines
and their genetic alterations including differentially expressed
genes were previously described (20–23).
Isolation and purification of total RNA
and mRNA
Total RNA was isolated from control MCF-10F and
experimental cell lines with TRIzol reagent (Invitrogen Corp., Long
Island, NY, USA). Each sample comprising 500 μg of total RNA was
treated with 5 μl of DNAse I (10 U/μl) (Roche Pharm., Indianapolis,
IN, USA) for 60 min at 37°C. Then 10X Termination Mix (0.1 M EDTA,
pH 8.0 and 1 mg/ml glycogen) (Clontech, CA, USA) was used to stop
the reaction. Each sample was then purified following established
procedure (20,25). The amount of purified RNA sample
was measured by a spectrophotometer (the ratio of absorbance
reading at 260/280 nm and then electrophoresed on denaturing
formaldehyde/agarose/ethidium bromide gel, to check its quality and
purity from proteins and free nucleotides. Each sample (500 μg) of
purified total RNA was subjected to polyA+ RNA analysis
with Oligotex mRNA Purification kit (Qiagen Inc., Valencia, CA,
USA). PolyA+ RNA was used following standard procedure
(25).
Northern blot analysis of β-catenin
mRNA
Total RNA (500 μg) was treated with 5 μl of DNAse I
(10 U/μl) (Roche Pharm.) for 60 min at 37°C. RNA was extracted and
precipitated using 7.5 M ammonium acetate, pH 5.2 (25). A sample of 0.5–1 μg of total RNA
was used for 1st-strand cDNA synthesis with the Advantage™ RT For-
PCR kit (Clontech) using oligo(dT)18 and random hexamer
primers. Then 100 ng of the 1st-strand cDNA synthesis product was
used for carrying out RT-PCR reactions using gene specific primers.
The PCR amplified products were labeled by using respective primers
and Biotin-16-UTP as well as RT cocktail to generate the probes and
be used for Northern hybridization analysis. One μg of mRNA was
electrophoresed in a 1% (w/v) agarose-formaldehyde gel and
transferred to a nylon membrane (Hybond-N, Amersham-Pharmacia
Biotech, Piscataway, NJ, USA). RNA transfer was confirmed by
visualization of ethidium bromide-stained RNA under UV light. Blots
were UV cross-linked and stored at 4°C until hybridization. Human
β-actin control amplifier set probe was also used in
northern hybridization to confirm their similar expression in all
samples (25). The blot was then
exposed to Kodak X-OMAT AR film at −80°C for 24 h. The intensity
was assayed by densitometric scanning (Molecular Dynamics)
(20).
Isolation of DNA
All cell cultures were treated with 1 ml of lysis
buffer (100 mM NaCl, 20 mM Tris-HCl pH 8.0, 25 mM EDTA pH 8.0, 0.5%
sodium dodecyl sulfate) with 200 mg/ml of proteinase K and RNAse
(100 μg/ml), and incubated overnight at 37°C with constant gentle
agitation (26). Then they were
purified following two extractions with a phenol:chloroform (1:1)
mixture and the aqueous layer was adjusted to 0.75 M ammonium
acetate and DNA was spooled from two volumes of 100% ethanol, dried
and dissolved in TE buffer (10 mM Tris-HCl pH 8.0, 1 mM EDTA pH
8.0) as described (25).
β-catenin mutation assay
To screen for mutations in exon 3 of the β-catenin
gene, a portion of that exon was amplified from genomic DNA from
all cell lines using the following primer pairs: forward (F)
5′-ATTTGATGGAGTTGGACATGGC-3′ and reverse (R)
5′-GAGGAAGAGGATGTGGATACCTCC-3′ (27). Amplified DNA fragments were first
purified by electrophoresis on 1% agarose-TAE gel (Life
Technologies Inc., Grand Island, NY, USA) and eluted with 100 μl of
elution buffer from the QIAquick gel extraction kit (Qiagen Inc.)
(25). Sequencing was done using
the same β-catenin primers in an automated sequencer ABI PRISM 3100
Genetic Analyzer (Applied Biosystems/Hitachi, Foster City, CA,
USA). Each and every fragment was sequenced at least three times to
rule out contamination and PCR fidelity artifacts.
Immunoprecipitation and
immunoblotting
Whole cell extracts were prepared using a lysis
buffer containing 20 mM Tris-HCl (pH 7.8), 140 mM NaCl, 1 mM EDTA,
0.5% NP40, 1 mM phenylmthylsulfonyl fluoride, and complete protease
inhibitors mixture (Boehringer-Mannheim, Roche, Germany). Cells
were allowed to grow in the presence of 10−8 M DHT for
24 h before protein extraction. Cell lysates were passed several
times through a 30.5-gauge needle to disrupt the nuclei. Protein
extract (~400 μg) was washed four times with 0.5 ml of lysis
buffer, boiled for 5 min, and analysed in 8% Tris-glycine
acrylamide gel. After electrophoresis, proteins were transferred to
nitrocellulose membranes using a semidry blotter (Bio-Rad, USA).
Blots were probed and was incubated with the primary antibodies of
mouse β-catenin (E5; sc7963), rabbit GSK-3-β (H76; sc 9166), and
goat TCF-4 (C19; sc8632) (Santa Cruz, Biotechnology Inc., Santa
Cruz, CA, USA) at a 1:500 dilution overnight at 4°C followed by a
secondary antibody conjugated to horseradish peroxidase (1:1000).
Detection was by the ECL method (Amersham, Arlington Heights, IL,
USA) (28).
Protein expression by
immunocytochemistry
Exponentially growing cells were plated on a glass
chamber slide (Nunc Inc., Naperville, IL, USA) as previously
described at a density of 1×104 cells in 1 ml of medium
(29,30). The experiments were repeated three
times with similar passages. The primary mouse antibody β-catenin
(E5; sc7963), rabbit GSK-3β (H76; sc 9166) and goat TCF-4 (C19;
sc8632) (Santa Cruz Biotechnology Inc.) were used for protein
expression determination at a 1:500 dilution overnight at 4°C. The
secondary antibody used was Rhodamine conjugated secondary antibody
(Jackson ImmunoResearch Lab., West Grove, PA, USA) at a 1:1,000
dilution. Slides were mounted with Vectashield mounting media
(Vector Laboratories, Burlingame, CA, USA). Cells were quantified
as previously described (29,30)
and viewed on Zeiss Axiovert 100 TV microscope (Carl Zeiss,
Thornwood, NY, USA) using a 40× 11.3 NA objective lens equipped
with a laser scanning confocal attachment (LSM 410 Carl Zeiss). To
excite the fluorescent secondary antibody fluorescent images were
collected by argon/krypton mixed gas laser (488 nm). Composite
images were generated using Adobe Photoshop, 7.0 Program. Protein
expression was determined by the relative staining intensity of the
control and experimental cell lines. Such computer program gave the
area and the intensity of the staining of the cells present in the
cells. The number of immunoreactive cells per sample (30
cells/field) was counted in 5 randomly selected microscopy fields.
Statistical analysis was done with the F-test (randomized block)
and comparisons between groups with the Bonferroni-t-test with
significance at P-value of <0.05 (29,30).
cDNA expression array
GE Array Q Series Human Extracellular Matrix (ECM)
and Cell Adhesion array membranes were used in these studies (SA
Biosciences, Bethesda, MD, USA) and tested for control MCF-10F and
Tumor2 cell lines. These arrays were designed to profile gene
expression of a panel of 96 key genes important for cell-cell and
cell-matrix interactions compose of trans-membrane molecules, and
other adhesion molecules that included basement membrane,
extracellular matrix and collagens. Each of these genes was
amplified by polymerase chain reaction (PCR) with gene-specific
primers to generate 200- to 600-bp products. Approximately 100 ng
of each PCR product was spotted in quadruplicate onto a positively
charged membrane. Each GE Array Q series membrane was spotted with
a negative control of pUC18 DNA, blanks and housekeeping genes,
including β-actin, GAPDH, cyclophilin A and ribosomal protein L13A
(20).
Synthesis of cDNA probes from mRNA
The purified mRNAs were used for the synthesis of
cDNA probes with Biotin-16-dUTP (Roche Pharm.). Annealing mixture
was prepared by mixing ~1.0–5.0 μg of mRNA with 3 μl of Buffer A
(GE primer mix) (SA Biosciences) and the final volume was adjusted
to 10 μl. The mixture was then incubated in a preheated thermal
cycler at 70°C for 3 min, cooled to 42°C and kept at that
temperature for 2 min. Then 10 μl of RT cocktail was prepared by
mixing 4 μl of 5X buffer BN [for 50 μl 10X buffer, add 1 μl of 1 M
DTT and 50 μl of 10X dNTP mix (5 mM dATP, dCTP, dGTP and 500 μM
dTTP)], 2 μl of Biotin-16-UTP, 2 μl of RNase-free H2O, 1
μl of RNase inhibitor (Promega Corp., Madison, WI, USA) and 1 μl of
MMLV reverse transcriptase (Promega Corp.). RT cocktail was then
warmed at 42°C for 1 min and slowly mixed with 10 μl of pre-warmed
annealing mixture. The incubation continued for 90 min at 42°C and
then labeled cDNA probe was denatured by heating for 5 min at 94°C,
and quickly chilled on ice. mRNA was isolated and purified from the
cell lines, and cDNA probes were prepared and hybridized to the
respective membranes. Experiments using the same mRNA preparation
were repeated three times, and measurable, median-normalized
expression values of each gene were compared to avoid
false-positive signals (20).
Differential hybridization of cDNA
expression array
Each array membrane was pre-wetted with 5 ml of
de-ionized water and incubated at 60°C for 5 min. It was then
replaced with 2 ml of pre-warm (60°C) GEA pre-hybridized solution
(GEAprehyb) with a heat-denatured sheared salmon sperm DNA at a
final concentration of 100 μg/ml) (SA Biosciences) and mixed gently
for few seconds. Pre-hybridization was continued at 60°C for 1–2 h
with continuous gentle agitation. Approximately 0.75 ml solution of
GEAprehyb was prepared by adding the entire volume of denatured
cDNA probe onto GEAprehyb solution and kept at 60°C. Then GEAprehyb
solution was replaced by GEAhyb solution and hybridization
continued overnight at 60°C with continuous gentle agitation.
Subsequently, array membranes were washed twice in wash solution 1
(2X sodium chloride sodium citrate and 1% sodium dodecyl sulfate)
at 60°C for 15 min each with gentle agitation and then twice with
solution 2 (0.1X sodium chloride sodium citrate and 0.5% sodium
dodecyl sulfate) at 60°C for 15 min each with gentle agitation. To
assess the reproducibility of the hybridization array assays,
pair-wise comparisons between array data sets for each cell line
was tested by repeated hybridization and the mRNAs prepared in
different lots were analyzed in scatter plots with multiple
regression (20,31). In each case, expression levels of
95% of the genes had repeated values that were within 2-fold
(20).
Chemiluminescent detection of cDNA
probes
After discarding the last wash, 2 ml of GEA blocking
solution was added to each membrane and incubated for 40 min at
room temperature with continuous agitation. Then binding buffer was
prepared by diluting alkaline phosphatase-conjugated streptavidin
(AP) with 1X buffer F (SuperArray, Bethesda, MD, USA) in a 1:7,500
dilution. GEA blocking solution was replaced by 2 ml of binding
buffer and incubated for 10 min with continuous but gentle
agitation. Then membrane was washed for 4 times with 4 ml of 1X
binding buffer F for 5 min in each washing and rinse twice with 3
ml of rinsing buffer G (SuperArray). The membrane was covered with
1.0 ml of CDP-Star chemiluminescent substrate and incubated at room
temperature for 2–5 min. It was then exposed to X-ray film (Kodak
Bio Max MS Film; Kodak Corp., Rochester, NY, USA) with
corresponding intensifying screen at room temperature for multiple
exposures of 1–5 min.
Quantification of array
hybridization
Quantification of hybridization signals on the
expression array membranes was carried out by exposing the
autoradiographic film in a densitometric scanner (model 300A;
Molecular Dynamics, Sunnyvale, CA, USA). It was then estimated both
with the Image Quant (Molecular Dynamics) and Scan Analyze program
(Eisen Lab., California University, CA, USA). Volume quantification
was performed by calculating the volume under the surface created
by a three-dimensional plot of pixel locations and pixel values as
described (20,31). All raw signal intensities were
corrected for background by subtracting the signal intensity of a
negative control or blank. Results were also normalized to that of
a housekeeping gene. These corrected, normalized signals can then
be used to estimate the relative abundance of particular
transcripts. To delineate the potential signal interference between
adjacent strong hybridization signals, equal-sized ellipses were
drawn around each signal area (hybridization spots) using software
(Image Quant/Scan Alyze) and was then separately scanned and
compared with housekeeping genes so the chances of interference
between adjacent strong hybridization signals were minimized.
Normalization of the expression levels of different housekeeping
genes from multiple autoradiographic exposures between different
hybridization experiments were done by taking the average signals
of each of the housekeeping genes. Data from high intensity spots
were chosen for further use. Median background was subtracted, and
signals that were <2.0-fold above background level were
considered too low to accurately measure and were omitted from the
analysis. Signals for each individual gene were also normalized to
the geometric mean of the expression level of that gene across the
set of membranes being compared. Mean signals were calculated from
quadruplicate measurable spots, or if three of the four spots were
measurable. The change in the fold indicated whether a gene
exhibits increased, decreased, or unchanged expression based on
statistical criteria (31).
Results
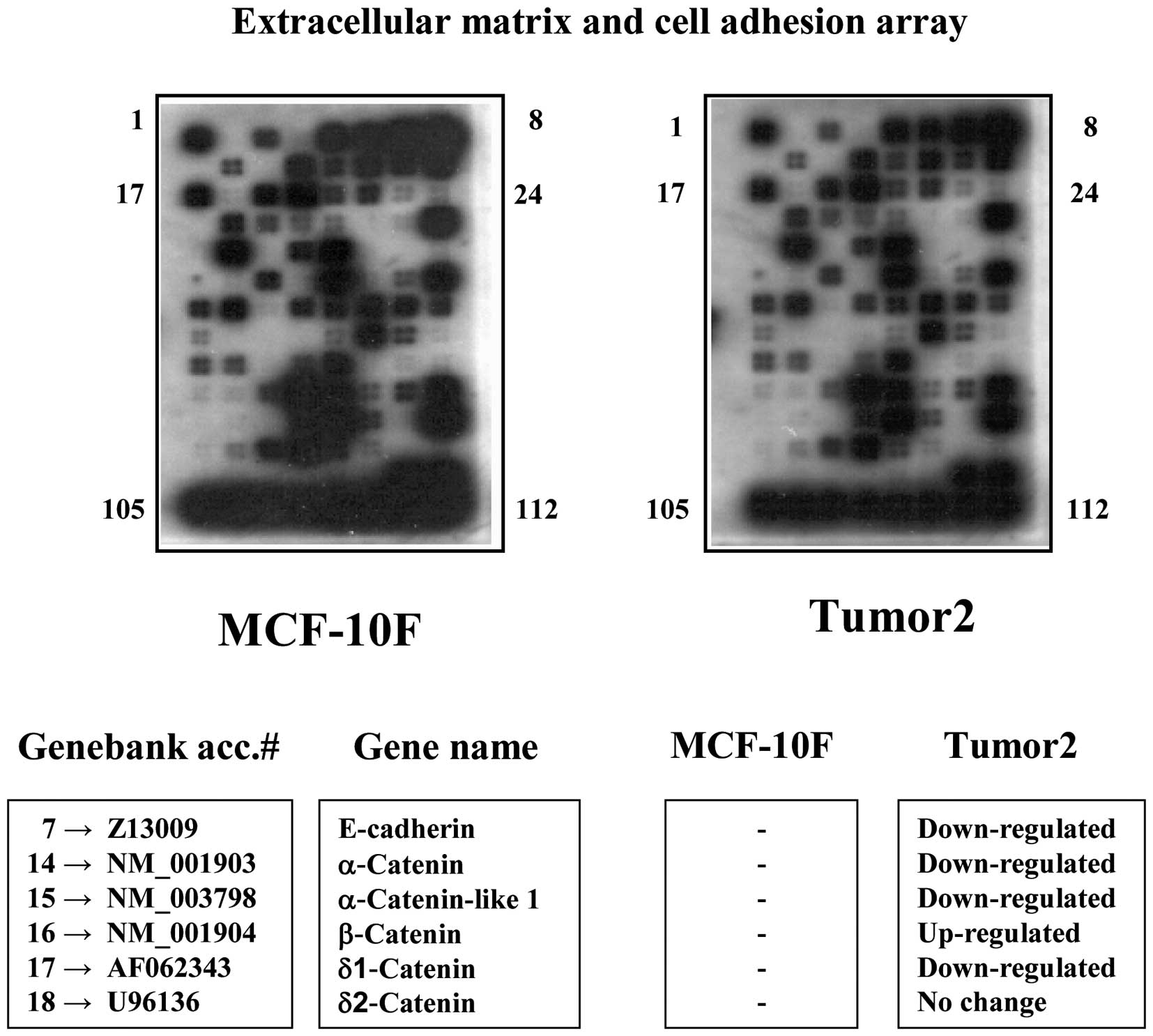
Differential expression of genes associated with
adhesion such as β-catenin, α-catenin, δ-catenins, E-cadherin were
identified through a cDNA expression array and comparing the
control MCF-10F and a tumor cell line as Tumor2 as seen in Fig. 1. Results showed downregulation of
genes as E-cadherin and catenins in Tumor2 cell line compared to
control. β-catenin gene was upregulated. Table I shows the detection of mutated
sequence of β-catenin gene of exon 3 from position 227 to 298.
These studies indicated mutation in Alpha5 and Tumor2 cell lines as
at positions 228 (G→C), 245 (G→T/C), 252 (T→C/A), 266(T→A), 276
(C→T), 283 (C→A), 285 (A→C), 288 (C→A), 290 (T→C), 293 (T→C/A).
 | Table IMutated sequences of β-catenin gene
at exon 3 in a breast cancer model. |
Table I
Mutated sequences of β-catenin gene
at exon 3 in a breast cancer model.
| MCF-10F |
CAGAATGCAGTTTTGAGAACTAAAAAGTTAGTGTCTAATAGTTTAAATAAAATGTTGCGGTGAACAAAACATACTCATAG |
| MCF10F+E |
CAGAATGCACTTTTGAGAACTAAAAATTTAGTGCCTAATAGTTTAAAAAAAATGTTGCGGTGAACAAAACATACCCATAG |
| Alpha1 |
CAGAATGCAGTTTTGAGAACTAAAAATTTAGTGACTAATAGTTTAAAAAAAATGTTGTGGTGAACAAAACATACCCATAG |
| Alpha2 |
CAGAATGCACTTTTGAGAACTAAAAATTTAGTGACTAATAGTTTAAAAAAAATGTTGTGGTGAACAAAACATACCCATAG |
| Alpha3 |
CAGAATGCAGTTTTGAGAACTAAAAATTTAGTGACTAATAGTTTAAAAAAAATGTTGTGGTGAAAAAAAAATACCCATAG |
| Alpha4 |
CAGAATGCACTTTTGAGAACTAAAAATTTAGTGACTAATAGTTTAAAAAAAATGTTGTGGTGAAAAAAAAATACCCATAG |
| Alpha5 |
CAGAATGCACTTTTGAGAACTAAAAATTTAGTGACTAATAGTTTAAATAAAATGTTGTGGTGAAAACAAAACACCCATAG |
| Tumor2 |
CAGAATGCAGTTTTGAGAACTAAAAACTTAGTGACTAATAGTTTAAATAAAATGTTGTGGTGAAAACAAAACACACATAG |
Fig. 2A corresponds
to β-catenin mRNA expression in all cell lines of this model when
compared to control MCF-10F. The β-catenin gene was amplified in
all irradiated- and estrogen-treated cell lines when compared to
MCF-10F. Fig. 2B shows a
β-catenin/GSK-3-β complex formed in non-malignant cell lines as
MCF-10F, Estrogen, Alpha1, Alpha3, and Alpha4 cell lines by
immunoprecipitation assays. However, Alpha5 and Tumor2 did not form
a complex in this assay. On the other hand, the β-catenin/TCF was
only found by co-precipitation in Alpha5 and Tumor2 (Fig. 2C). Fig. 2D shows a schematic flow chart of
the possible interaction of β-catenin with GSK-3-β and
β-catenin/TCF in cytoplasm and nucleus in different cell lines in
this particular breast cancer model.
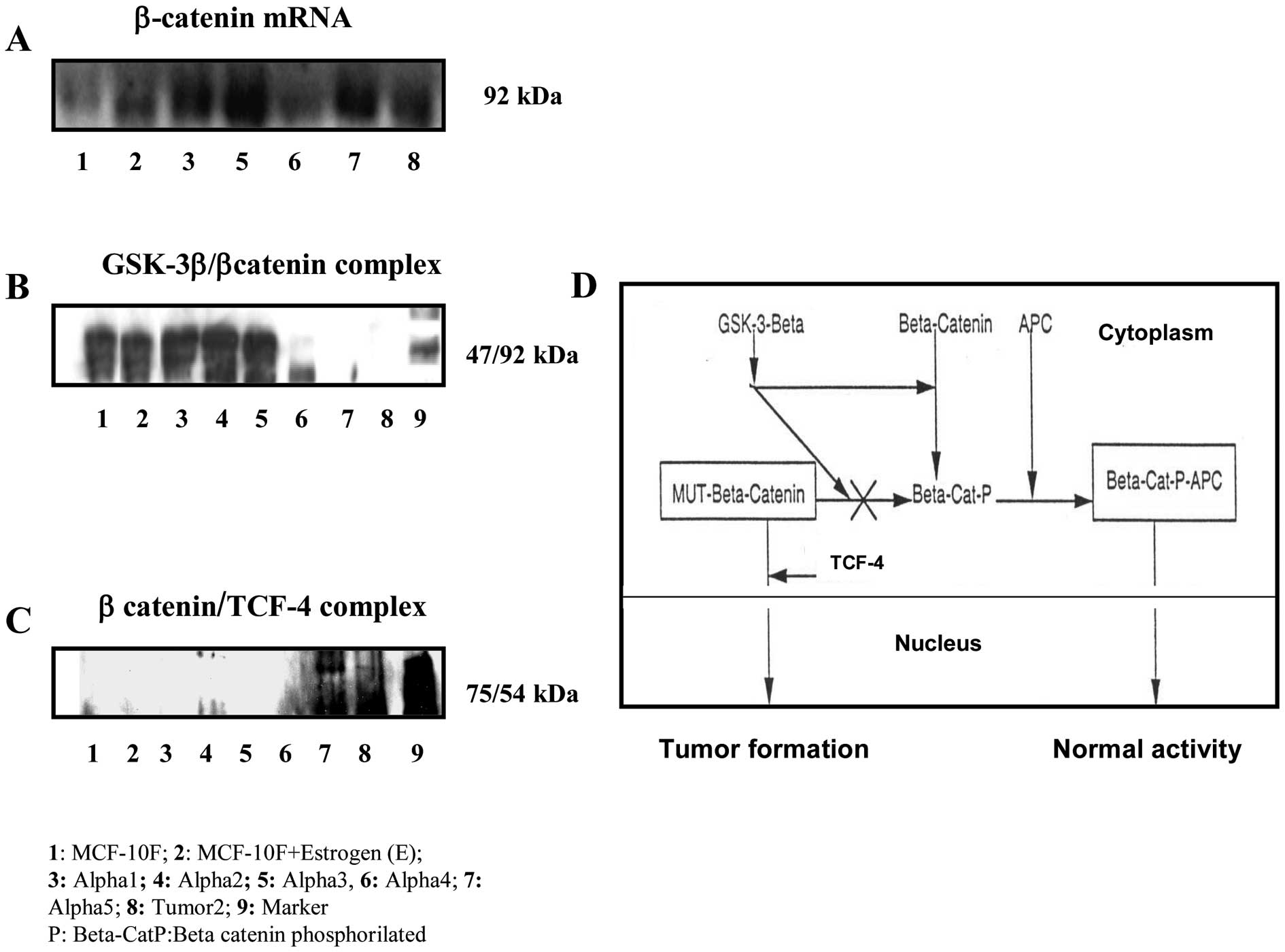 | Figure 2Immunoprecipitation assay with the
following cell lines. 1, MCF-10F; 2, MCF-10F+Estrogen (E); 3,
Alpha1; 4, Alpha2; 5, Alpha3; 6, Alpha4; 7, Alpha5; 8, Tumor2; 9,
Marker. (A) β-catenin mRNA of MCF-10F, Estrogen, Alpha1, Alpha2,
Alpha3, Alpha4, Alpha5 and Tumor2 cell lines. (B) β-catenin and
GSK-3-β complex formed by immunoprecipitation in MCF-10F, Estrogen,
Alpha1, Alpha2, Alpha3, Alpha4 cell lines. (C) β-catenin/TCF-4
complex formed by immunoprecipation in Alpha5 and Tumor2 cell cell
lines. (D) Proposed scheme for β-catenin, GSK-3-β and
β-catenin/TCF-4 interaction in a breast cancer model. |
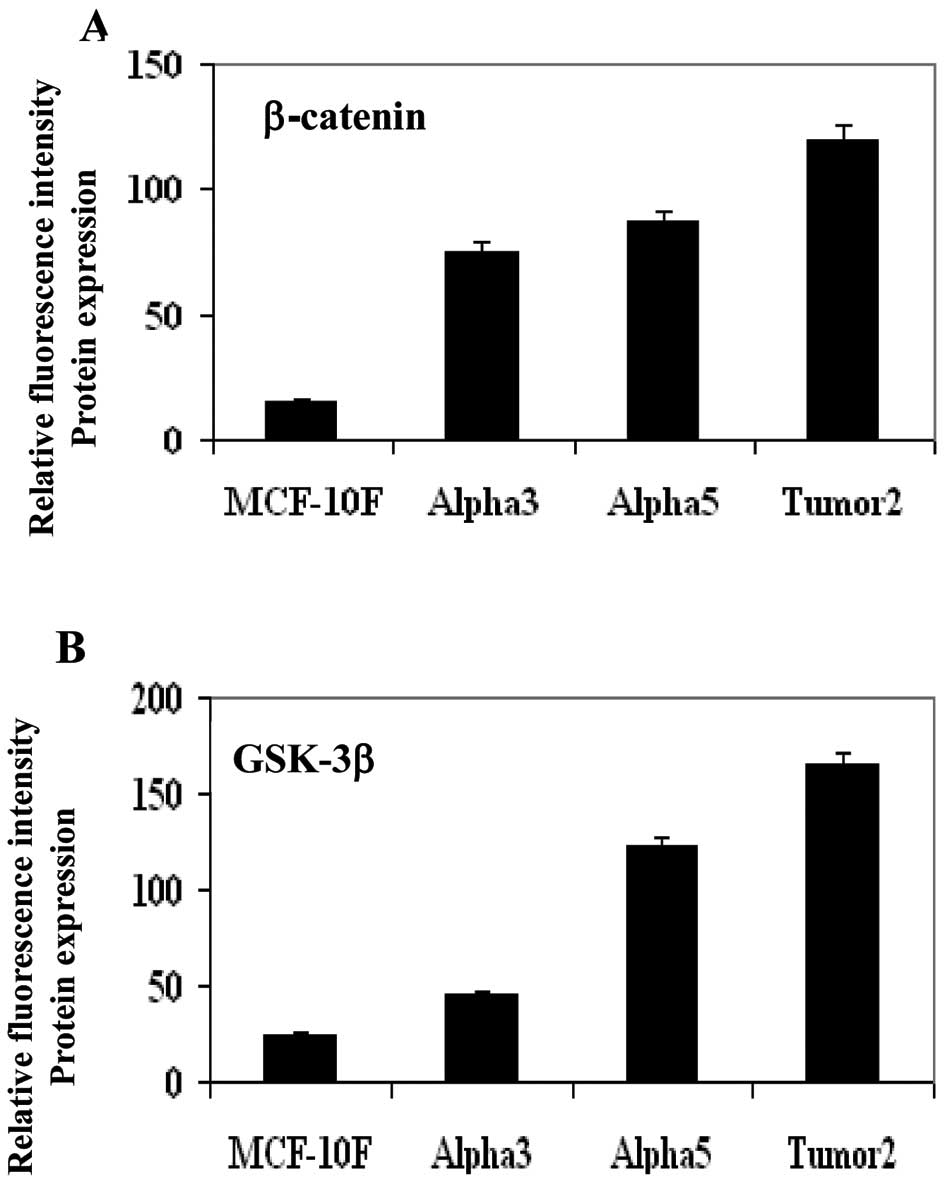
Protein expression of GSK-3-β and
β-catenin by immunofluorescent studies
Measurement of relative fluorescence intensity of
protein expression indicated that β-catenin and GSK-3β had greater
gene expression in the tumorigenic cell line Alpha5 and Tumor2
tumor cell line than control MCF-10F and non-malignant Alpha3 cell
lines. Fig. 3 shows a graph of
relative fluorescent intensity of β-catenin (Fig. 3A) and GSK-3-β (Fig. 3B), respectively. Representative
images of the corresponding graphs of β-catenin and GSK-3-β protein
expression in MCF-10F, Alpha3, Alpha5 and Tumor2 are shown in
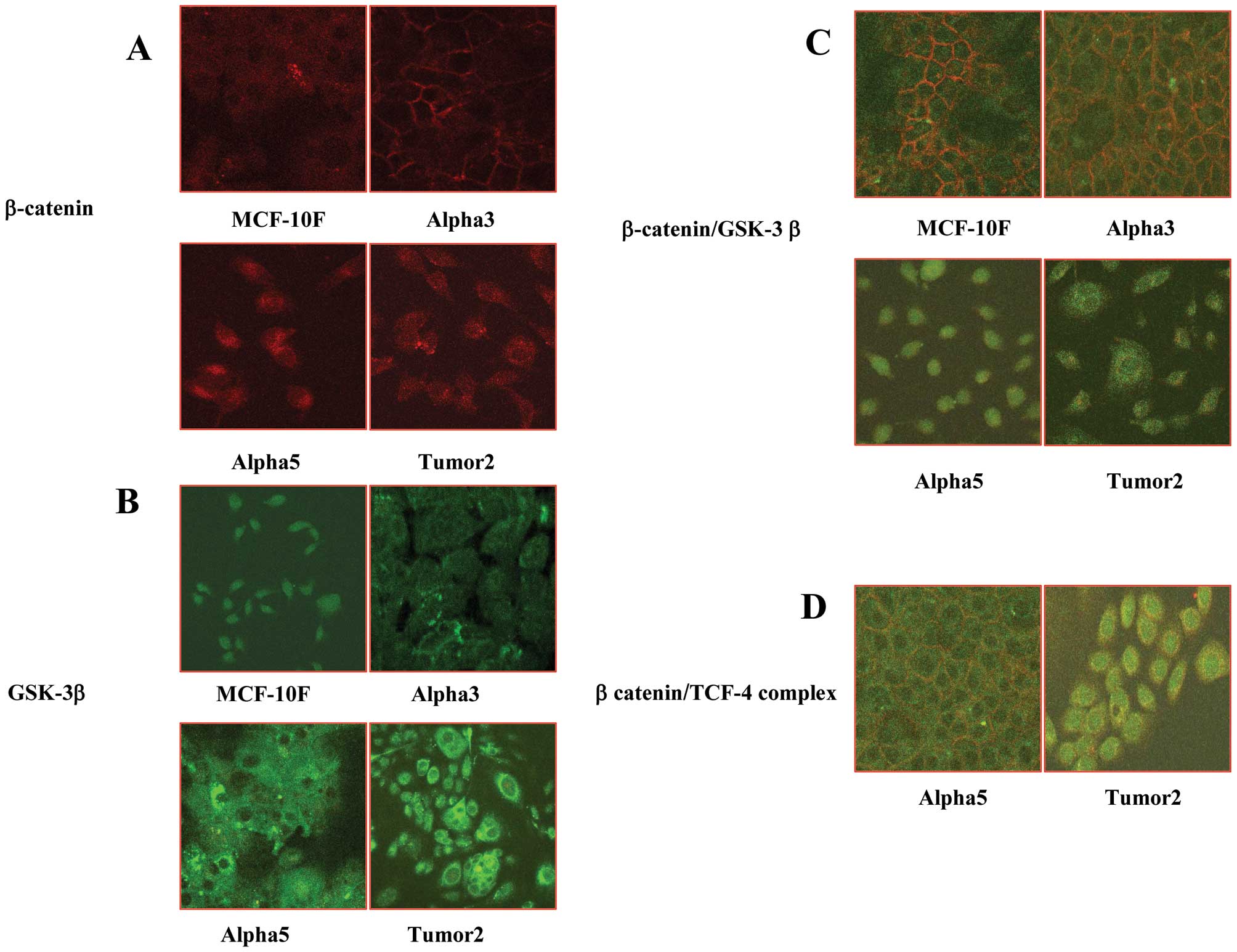
Fig. 4A and B, respectively.
Fig. 4C shows the fluorescence
images that indicated that there was co-localization of β-catenin
and GSK-3-β in the non-malignant cell lines MCF-10F and Alpha3, but
not in Alpha5 and Tumor2 cell lines. On the other hand, there was
only β-catenin/TCF-4 co-localization in Alpha5 and Tumor2 cell
lines (Fig. 4D).
Discussion
Gene regulation of adhesion molecules is very
important as are all catenins and E-cadherin in cancer progression.
Differential expression of genes associated with adhesion such as
β-catenin, α-catenin, δ-catenins and E-cadherin, were identified
and compared in Tumor2, a tumor and MCF-10F, a control cell line.
Mutation and amplification of β-catenin gene in this process has a
vital role. β-catenin and GSK-3-β were highly expressed in the
tumorigenic cell line Alpha5 and Tumor2 but not in the control
MCF-10F and the non-malignant Alpha3 cell lines. These two genes
are important in the cancer progression since β-catenin/GSK-3-β
complex was identified in the non-malignant cell lines as MCF-10F,
estrogen, Alpha1, Alpha3, and Alpha4 cell lines. Lack of
co-localization in the tumorigenic cell lines Alpha5 and Tumor2
corroborated the crucial role of genes in the signaling
pathway.
The β-catenin/TCF complex observed only in Alpha5
and Tumor2 confirmed the significance of genes in this cancer
progression. A schematic flow chart is proposed to suggest a
possible action of mutated β-catenin with GSK-3-β or TCF-4 applied
in this particular breast cancer model. Authors have shown that
regulation of β-catenin turnover requires the
NH2-terminal region of the protein containing the
potential GSK-3-β phosphorylation site, since phosphorylated
β-catenin is targeted for degradation. Loss of control of
intracellular β-catenin levels through mutation or to any other
associated factors has been proposed as an important oncogenic step
in tumorigenic progression (32).
It has been proposed that mutated β-catenin gene at
exon 3 probably prevents phosphorylation of GSK-3-β (33). Inhibition of this activity is
probably due to unbound β-catenin to accumulate in the cytoplasm
and translocate into the nucleus by activating TCF-dependent
transcription that occurs during tumorigenic progression. The TCF
protein is a weak activator of transcription, therefore the binding
of β-catenin induces a significant increase in transcriptional
activity.
It can be concluded that it is very interesting to
find a complex formed between β-catenin and GSK-3-β only in the
non-tumorigenic cell lines and GSK-3-β complex only in the tumor
ones compared to MCF-10F. These findings corroborated the role of
β-catenin/TCF in mammary cancer which acts as a switch to determine
cell fate and promote cell survival and proliferation at several
stages of mammary gland development and cancer progression
(34,35). It is known that in the absence of a
Wnt signal the β-catenin levels are low in the cells due to the
association with a cytoplasmic protein complex comprised of
GSK-3-β, APC and axin (27). It
has been shown that phosphorylation of β-catenin by GSK-3-β leads
to proteosomal degradation of β-catenin. Wnt stimulation inhibits
GSK-3-β, resulting in stabilization of β-catenin translocation to
the nucleus, and activation of target genes and enhancement of its
association with the TCF family of transcription factors, can
result in transcriptional activation of multiple target genes
(11).
It has been also been reported that deregulation of
β-catenin leads to constitutive formation of the β-catenin-Tcf/LEF
complex and altered expression of Tcf target genes such as, Wnt/Tcf
target genes in cancer cells (36–38).
Altered expression was also noted in various adenocarcinomas in
breast, gastric and prostate cancer (39–41).
Recently, translocation and altered expression of β-catenin were
also reported in irradiated-colonic mucosa as well as in colon
cancer of mouse mammary progenitor cells (42,43).
The present report also provides evidence that of β-catenin
signaling pathway may play a role during progression in radiation
and estrogen induced human breast cancer. It can be concluded that
mutation of β-catenin and its interaction with other associated
proteins may be an early event during radiation and estrogen
induced human breast cancer progression.
Acknowledgements
The support given by FONDECYT no. 1120006 (GMC) and
MINEDUC-Universidad de Tarapacá, Chile (GMC) and assistance given
by Richard Ponce-Cusi is greatly appreciated.
References
|
1
|
Greenlee RT, Murray T, Bolden S and Wingo
PA: Cancer Statistics. CA Cancer J Clin. 50:7–33. 2000. View Article : Google Scholar
|
|
2
|
American Cancer Society (ACS). Breast
cancer facts and figures, 2013–2014. https://www.cancer.org/research/cancer-facts-statistics/breast-cancer-facts-figures.html.
|
|
3
|
Park CC, Henshall-Powell RL, Erickson AC,
Talhouk R, Parvin B, Bissell MJ and Barcellos-Hoff MH: Ionizing
radiation induces heritable disruption of epithelial cell
interactions. Proc Natl Acad Sci USA. 100:10728–10733. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Grosovsky AJ: Radiation-induced mutations
in unirradiated DNA. Proc Natl Acad Sci USA. 96:5346–5347. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Loeb LA: Mutator phenotype may be required
for multistage carcinogenesis. Cancer Res. 51:3075–3079.
1991.PubMed/NCBI
|
|
6
|
Bissell MJ and Radisky D: Putting tumours
in context. Nat Rev Cancer. 1:46–54. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Bullions LC and Levine AJ: The role of
beta-catenin in cell adhesion, signal transduction and cancer. Curr
Opin Oncol. 10:81–87. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Willert K and Nusse R: Beta-catenin: a key
mediator of Wnt signaling. Curr Opin Genet Dev. 8:95–102. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huguet EL, McMahon JA, McMahon AP,
Bicknell R and Harris AL: Differential expression of human Wnt
genes 2, 3, 4, and 7B in human breast cell lines and normal and
disease states of human breast tissue. Cancer Res. 54:2615–2621.
1994.PubMed/NCBI
|
|
10
|
Lejeune S: Wnt5a cloning, expression, and
up-regulation in human primary breast cancers. Clin Cancer Res.
1:215–222. 1995.PubMed/NCBI
|
|
11
|
Wodarz A and Nusse R: Mechanisms of Wnt
signaling in development. Annu Rev Cell Dev Biol. 14:59–88. 1998.
View Article : Google Scholar
|
|
12
|
Taipale J and Beachy PA: The Hedgehog and
Wnt signalling pathways in cancer. Nature. 411:349–354. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Polakis P: Wnt signaling and cancer. Genes
Dev. 14:1837–1851. 2000.
|
|
14
|
Gumbiner BH: Signal transduction of
beta-catenin. Curr Opin Cell Biol. 7:634–640. 1995. View Article : Google Scholar
|
|
15
|
Kinch MS: Tyrosine phosphorylation
regulates the adhesions of ras-transformed breast epithelia. J Cell
Biol. 130:461–471. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Fagotto F, Funayama N, Gluck U and
Gumbiner BM: Binding to cadherins antagonizes the signaling
activity of beta-catenin during axis formation in Xenopus. J
Cell Biol. 132:1105–1114. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Thraves PJ, Salehi Z, Dritschilo A and
Rhim JS: Neoplastic transformation of immortalized human epidermal
keratinocytes by ionizing radiation. Proc Natl Acad Sci USA.
87:1174–1177. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hei TK, Piao CQ, Willey JC, Thomas S and
Hall EJ: Malignant transformation of human bronchial epithelial
cells by radonsimulated alpha-particles. Carcinogenesis.
15:431–437. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Calaf GM and Hei TK: Establishment of a
radiation- and estrogen-induced breast cancer model.
Carcinogenesis. 21:769–776. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Roy D, Calaf GM and Hei TK: Profiling of
differentially expressed genes induced by high linear energy
transfer radiation in breast epithelial cells. Mol Carcinog.
31:192–203. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Roy D, Calaf GM and Hei TK: Allelic
imbalance at 11p15.5–15.4 correlated with c-Ha-ras mutation during
radiation-induced neoplastic transformation of human breast
epithelial cells. Int J Cancer. 103:730–737. 2003. View Article : Google Scholar
|
|
22
|
Calaf GM, Roy D and Hei TK: Immunochemical
analysis of protein expression in breast epithelial cells
transformed by estrogens and high linear energy transfer (LET)
radiation. Histochem Cell Biol. 124:261–274. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Calaf G and Hei TK: Oncoprotein expression
in human breast epithelial cells transformed by high-LET radiation.
Int J Rad Biol. 77:31–40. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Soule HD, Vazguez J, Long A, Albert S and
Brennan M: A human cell line from a pleural effusion derived from a
breast carcinoma. J Natl Cancer Inst. 51:1409–1413. 1973.PubMed/NCBI
|
|
25
|
Sambrook J and Green MR: Molecular Cloning
- A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold
Spring Harbor; NY: 1989
|
|
26
|
Gross-Bellard M, Oudet P and Chambon P:
Isolation of high-molecular-weight DNA from mammalian cells. Eur J
Biochem. 36:32–38. 1973. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Morin PJ: Beta-catenin signaling and
cancer. Bioessays. 21:1021–1030. 1999. View Article : Google Scholar
|
|
28
|
Truica CI, Byers S and Gelmann EP:
β-catenin affects androgen receptor transcriptional activity and
ligand specificity. Cancer Res. 60:4709–4713. 2000.
|
|
29
|
Calaf GM and Roy D: Gene and protein
expressions induced by 17β-estradiol and parathion in cultured
breast epithelial cells. Mol Med. 13:255–265. 2007. View Article : Google Scholar
|
|
30
|
Calaf GM, Alvarado ME and Hei TK:
β-catenin is associated with breast cancer progression in
vitro. Int J Oncol. 26:913–921. 2000.
|
|
31
|
Lui WM, Mei R, Di X, et al: Analysis of
high density expression microarrays with signed-rank call
algorithms. Bioinformatics. 18:1596–1599. 2002.PubMed/NCBI
|
|
32
|
Cowin P, Rowlands TM and Hatsell SJ:
Cadherins and catenins in breast cancer. Curr Opin Cell Biol.
17:499–508. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Miller JR and Moon RT: Signal transduction
through β-catenin and specification of cell fate during
embryogenesis. Genes Dev. 10:2527–2539. 1996. View Article : Google Scholar
|
|
34
|
Hatsell S, Rowlands T, Hiremath M and
Cowin P: Beta-catenin and Tcfs in mammary development and cancer. J
Mammary Gland Biol Neoplasia. 8:145–158. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gebeshuber CA, Sladecek S and Grunert S:
Beta-catenin/LEF-1 signalling in breast cancer-central players
activated by a plethora of inputs. Cell Tissues Organs. 185:51–60.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
He TC, Sparks AB, Rago C, et al:
Identification of c-MYC as a target of the APC pathway. Science.
281:1509–1512. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tetsu O and McCormick F: Beta-catenin
regulates expression of cyclin D1 in colon carcinoma cells. Nature.
398:422–426. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
38
|
Crawford HC, Fingleton B, Gustavson MD, et
al: The PEA3 subfamily of Ets transcription factors synergizes with
beta-catenin-LEF-1 to activate matrilysin transcription in
intestinal tumors. Mol Cell Biol. 21:1370–1383. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Pierceall WE, Woodard AS, Morrow JS, Rimm
D and Fearon ER: Frequent alterations in E-cadherin and alpha- and
beta-catenin expression in human breast cancer cell lines.
Oncogene. 11:1319–1326. 1995.PubMed/NCBI
|
|
40
|
Jawhari A, Jordan S, Poole S, Browne P,
Pignatelli M and Farthing MJ: Abnormal immunoreactivity of the
E-cadherin-catenin complex in gastric carcinoma: relationship with
patient survival. Gastroenterology. 112:46–54. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Paul R, Ewing CM, Jarrard DF and Isaacs
WB: The cadherin cell-cell adhesion pathway in prostate cancer
progression. Br J Urol. 79:37–43. 1997. View Article : Google Scholar
|
|
42
|
Nakashima M, Meirmanov S, Matsufuji R,
Hayashida M, Fukuda E, Naito S, Matsuu M, Shichijo K, Kondo H, Ito
M, Yamashita S and Sekine I: Altered expression of beta-catenin
during radiation-induced colonic carcinogenesis. Pathol Res Pract.
198:717–724. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Woodward WA, Chen MS, Behbod F, Alfaro MP,
Buchholz TA and Rosen JM: WNT/beta-catenin mediates radiation
resistance of mouse mammary progenitor cells. Proc Natl Acad Sci
USA. 104:618–623. 2007. View Article : Google Scholar : PubMed/NCBI
|