Introduction
Breast cancer is the most commonly malignancy of
women worldwide (1), the primary
cause of the development and progression of breast cancer is
multiple genetic changes, whose accumulation results in the
metastatic invasion (2). Some
critical genes including HER-2/neu, p53, nm23, and cyclin D are
studied in clinical diagnosis for prognostic or predictive outcome
(3–6). Therefore, further understanding the
molecular etiology of breast cancer is helpful for estimating
disease prognosis and guiding treatment.
Eps8 extensively functions as an oncogene in various
types of human solid tumors, including squamous cell carcinoma,
pancreatic cancer and cervical cancer, as well as colon cancer,
pituitary tumors and glioma (7–13)
and even hematologic malignancies (14). High expression and concomitant
tyrosine phosphorylation of Eps8 were detected in a serial of human
tumor cell lines (15). Moreover,
elevated Eps8 in cancer patients might be a biomarker of poor
prognosis for decreased overall survival (8,14,16).
Eps8 could increase cell growth and motility, by modulating the
downstream pathways including the mTOR/STAT3/FAK, PI3K/AKT/
FOXM1/MMP9, p53/p21WAF1/CIP1-dependent pathway, EGFR signaling via
Rac, and Ras/Raf/MEK/ERK pathways in cancer cells, thus affecting
EGFR endocytosis, actin dynamics, RNA processing, cell cycle
progression, angiogenesis, cell migration and invasion (7–9,11,12).
Furthermore, the effects of downregulated expression of Eps8 on
chemotherapeutic agents are also studied. Mithramycin downregulates
Eps8 expression and inhibits human epithelial carcinoma cell
proliferation and migration (17).
Additionally, the cytotoxicity of cisplatin is increased in
Eps8-attenuated cervical cancer and lung cancer cells (8,18).
Moreover, the loss of Eps8 protein in colorectal adenoma and
carcinoma plays a role in the development of a subset of colorectal
cancers, suggesting the significance of personalized medicine in
patient treatment (19).
Although Eps8 plays a critical role in the
development of many malignancies (20), the expression and function of Eps8
in breast cancer are not clear. Combined cDNA array comparative
genomic hybridization (CGH) and serial analysis of gene expression
analysis (SAGE) identified candidate amplicon target Eps8 as novel
putative oncogene in breast cancer (21). In this study, we further determine
whether Eps8 is involved in breast cancer malignancy, including
cell proliferation and migration. Eps8 was found overexpressed in
breast cancer samples and highly invasive cell lines.
Overexpression of Eps8 promoted MCF7 cell growth, whereas
downregulation of Eps8 in MDA-MB-231 cells significantly inhibited
cell proliferation, migration and cell motility. These data suggest
that Eps8 serves as a novel growth regulator in breast cancer cells
and could be a target for the diagnosis and treatment of breast
cancer.
Materials and methods
Immunohistochemistry
Seventy-two breast cancer samples were examined and
3 adjacent normal mammary tissues were used as the control. The
study was approved by the Hunan Normal University Human Ethics
Committee, and informed consent was obtained from all patients. The
immunohistochemical analysis was performed on polyformalin-fixed
and paraffin-embedded tissues. Sections (5 μm) were deparaffinized
by two 10-min washes in xylene, then rehydrated through successive
graded ethanol solutions. Endogenous peroxidase was quenched with
3% H2O2 in methanol for 10 min and washed for
5 min in PBS. Antigen retrieval was achieved by microwaving
sections in 0.01 M citrate buffer (pH 6.0) for 10 min at 800 W. The
tissues were blocked in 5% bovine serum albumin (BSA) in PBS for 1
h before the addition of the mouse monoclonal anti-Eps8 antibody or
mouse IgG control (diluted 1:500, BD Biosciences, CA, USA) at 4°C
for overnight. The sections were incubated with HRP-conjugated goat
anti-mouse secondary antibody (diluted 1:100, Sigma-Aldrich, St.
Louis, MO, USA) for 45 min and then with 3,3-diaminobenzidine
(DAB)/H2O2 for 10 min. Sections were
counterstained with hematoxylin, mounted and photographed using an
optical microscope (Olympus CX41, Tokyo, Japan). The percentage of
tumor cells stained was scored as: 0 (no cell staining), + or 1
(≤30%), ++ or 2 (31–60%) and +++ or 3 (61–100%). The staining
between two score values was given 0.5.
Cell culture
Human breast cancer cells MDA-MB-231, MCF7 and T47D
were cultured in RPMI-1640 medium (Gibco BRL, Grand Island, NY,
USA) containing 100 U/ml penicillin, 100 μg/ml streptomycin and 10%
fetal bovine serum (FBS, Hyclone, Australia). MDA-MB-453,
MDA-MB-468 cells and normal mammary cells HBL100 were cultured in
Dulbecco’s modified Eagle’s medium (DMEM, Gibco) supplemented as
described above. All cells were kept at 37°C in a humidified
atmosphere with 5% CO2.
Establishment of MCF7 cell lines
overexpressing GFP and GFP-Eps8
MCF7 cells were transiently transfected with
constructed pEGFP-C3-Eps8 or pEGFP-C3 control plasmids using
Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) (13). Stably transfected cells were
screened by 400 μg/ml G418 (Sigma) (22,23).
The transfection efficiency was detected using an invert
fluorescence microscope (Olympus IX71, Tokyo, Japan).
Eps8 RNAI lentivirus generation
Efficient siRNA sequence targeting Eps8
(NM_004447.5) was from the position 93–111 relative to the start
codon, stem-loop DNA oligonucleotides were synthesized by Shanghai
GeneChem Co. Ltd., China and inserted into the lentivirus-based
RNAi vector pGCSIL-GFP (GeneChem) (13). A non-targeting stem-loop DNA was
also cloned into pGCSIL-GFP vector as a negative control (NC).
Lentiviral particles were prepared as described previously
(24). Briefly, the lentivirus
expression plasmid and packaging plasmids (pHelper 1.0 and pHelper
2.0) were cotransfected into 293T cells, supernatants were
harvested 48 h after transfection and filtered through a 0.45-μm
pore size filter (Millipore, Billerica, MA, USA) and concentrated
by ultracentrifugation. The infectious titer was determined using
hole-by-dilution titer assay. MDA-MB-231 cells at a density of
100,000 cells per well in 6-well plates were infected with
Eps8-RNAi-lentivirus or NC-RNAi-lentivirus and 5 μg/ml polybrene
(Sigma) at the multiplicity of infection (MOI) 10 and detected on
the 4th day by the invert fluorescence microscope.
Cell proliferation assays
For cell viability assay, 3,000 cells were seeded in
octuplicate in 96-well plates untreated or treated with 30 μM of
cisplatin (DDP, Sigma) for 24 h. On days 1, 3 and 4 or days 1, 3
and 6, cells were analyzed with 1 mg/ml
3-(4,5-dimethylthiazol-2-yl)-2, 5-diphenytetra zolium bromide (MTT,
Sigma) at 37°C for 4°h. Then 100 μl dimethylsulfoxide (DMSO)/well
was added to dissolve the formazan crystals. The absorbance at 490
nm was obtained using a spectrophotometer (UV-2102C, Changsha,
China).
For cell survival assay, 100,000 MCF7 cells stably
expressing GFP-Eps8 or GFP and parental cells were plated in
triplicate in 6-well plates in complete medium containing 400 μg/ml
G418. MDA-MB-231 cells (50,000) infecting Eps8-RNAi-LV or
NC-RNAi-LV were plated in triplicate in 6-well plates. After 3–6
days, viable cell numbers were counted with a hemocytometer after
trypan blue staining of dead cells.
Liquid colony formation was performed, 2,000 cells
were seeded in triplicate in 6-well plates and grown in complete
culture medium for >10 days. Colonies were fixed with methanol,
stained with Giemsa (BBI International, Cardiff, UK) and
photographed with a digital camera (Canon IXUS 125 HS, Tokyo,
Japan). Only colonies containing >30 cells were counted under an
inverted microscope (Zeiss Axiovert 25, LLC, USA). All experiments
were carried out at least three times.
Cell cycle analysis
Lentivirus-infected and parental MDA-MB-231 cells
were plated onto 6-well plates for 24 h. Serum was withdrawn when
cells were 70% confluent. After 36 h, 10% FBS was added in the
medium for an additional 18 h. Cells were collected gently, fixed
in 70% ethanol and stained with propidium iodide (PI, BD
Pharmingen, San Diego, CA, USA), the DNA content was analyzed on a
BD FACSCalibur cytometer (Becton-Dickinson, San Jose, CA, USA)
using CellQuest Pro and ModFit software (BD).
Apoptosisassay
Lentivirus-infected and parental MDA-MB-231 cells
were harvested and washed with cold PBS. The pellet was resuspended
in 1X binding buffer and the sample solution was incubated with PI
and FITC-conjugated Annexin V (BD Pharmingen) for 15 min at room
temperature in the dark. The samples were analyzed by the BD
FACSCalibur cytometer using the CellQuest software.
Tumor formation in nude mice
The mouse experiments were carried out according to
the ethical guidelines for laboratory animal use and approved by
the Ethics Committee of Hunan Normal University. Approximately
107 of lentivirus-infected MDA-MB-231 cells in 0.2 ml of
sterile PBS were injected subcutaneously into the left and right
dorsal regions of 4- to 5-week-old female nude mice (BALb/c). Mice
were checked every 2 days and the formed tumors were measured with
a micrometer (25). After 26 days,
mice were sacrificed, and tumors were excised, weighed and
photographed.
Wound-healing migration assays
MDA-MB-231 cells were cultured in 24-well plates
until >90% confluence. A 100-μl pipette tip was used to generate
wounds. After wound creation, the medium was changed to remove
cellular debris. Three wounded areas in each well were marked on
the bottom of plates and photographed at 1 and 4 days with an
invert microscope.
Immunofluorescence assays
Stimulation of membrane ruffling with 60ng/ml of
human EGF (Sigma, Deisenhofen, Germany) in RPMI-1640 for 30 min was
performed after serum deprivation for 48 h. MDA-MB-231 cells were
fixed with 3.7% paraformaldehyde for 10 min, permeabilized with
0.1% Triton X-100 for 10 min, and blocked with 5% BSA in PBS. Alexa
fluor 595 conjugated to phalloidin (diluted 1:500, Molecular
Probes, Eugene, OR, USA) and 1 μg/ml Hoechst 33258 (Sigma) were
incubated for 20 min to stain F-actin and the nucleus in the dark.
All specimens were viewed with an upright fluorescence microscope
(Zeiss Axioskop 2, LLC, USA). At least 20 cells were measured in
three independent experiments.
Western blot analysis
Cells were lysed in RIPA buffer [50 mM Tris-HCl (pH
7.2), 150 mM NaCl, 1% Triton X-100, 1% sodium deoxycholate, 0.1%
SDS and cocktail protease inhibitors]. The lysates (50 μg) were
denatured in sample buffer and heated to 105°C for 5 min. Samples
were then separated on 10–15% SDS-PAGE gels and transferred to
polyvinylidene difluoride (PVDF) membranes (Millipore). The
membranes were blocked with 1% BSA in TBS-T [10 mM Tris-HCl (pH
7.5), 150 mM NaCl, 0.1% Tween-20] and incubated overnight with
rabbit polyclonal antibodies against GFP and MMP9, mouse monoclonal
antibodies against ERK and phosphorylated ERK, cyclin D1 (CCND1),
c-Myc, p53, N-cadherin, E-cadherin, vimentin, β-actin and GAPDH
(diluted 1:1,000, Santa Cruz Biotechnology, Santa Cruz, CA, USA) in
TBS-T containing 1% BSA with gentle shaking at 4°C. HRP-conjugated
goat anti-rabbit and goat anti-mouse secondary antibodies (diluted
1:5,000, Sigma) were used. The signals were detected with
SuperSignal West Pico chemiluminescent Substrate (Thermo Scientific
Pierce, Rockford, IL, USA) and visualized with tanon-5200 system
(Bio-tanon, Shanghai, China).
Statistical analysis
All statistical analyses were performed using the
SPSS 11.0 software (SPSS Inc., Chicago, IL, USA). Data are shown as
mean ± SD from at least 3 independent experiments. Statistical
significance was determined using Student’s t-test at P-values
<0.05.
Results
Eps8 was overexpressed in breast cancer
tissues and high-invasive cell lines
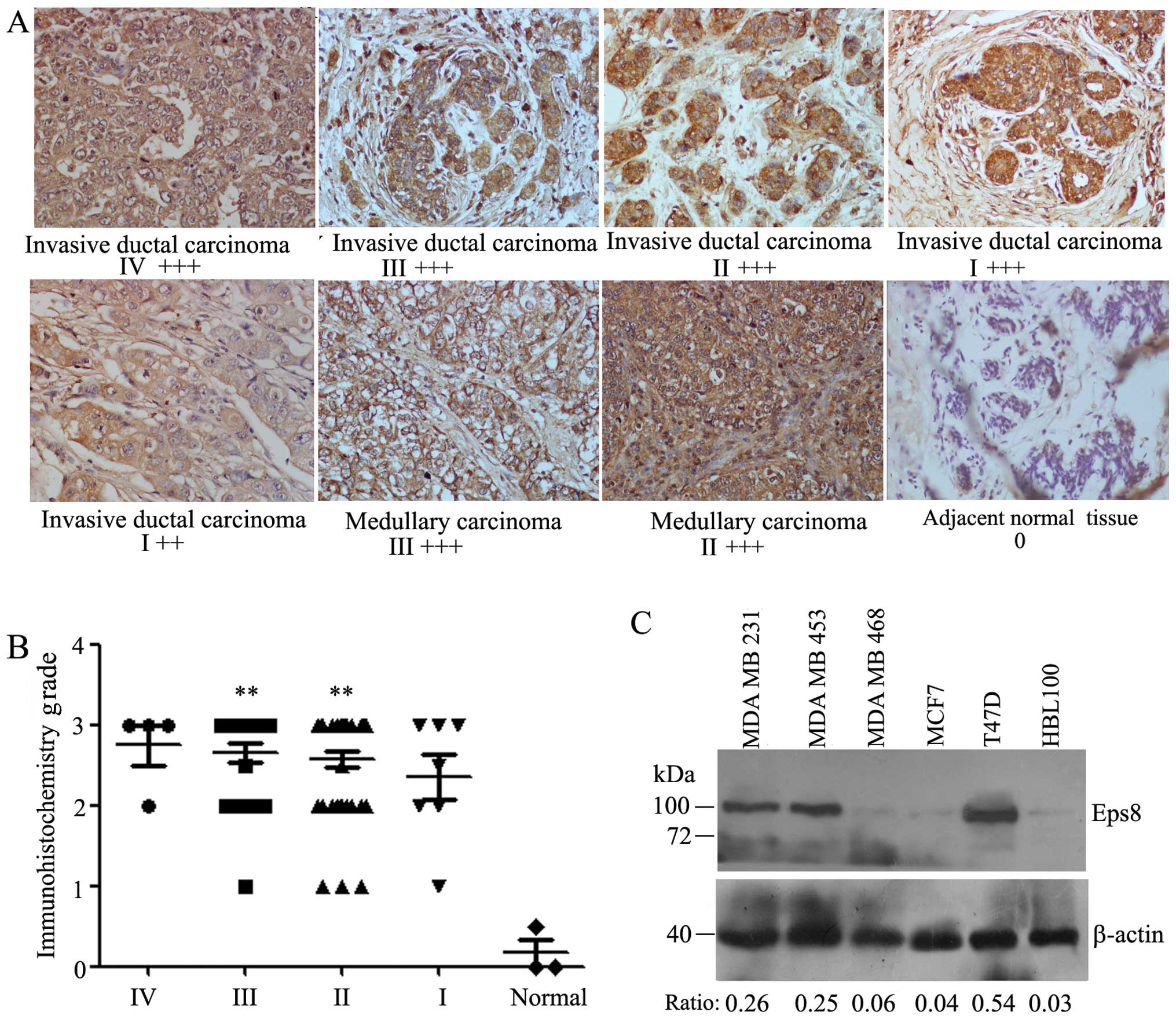
The expression level of Eps8 was examined in 4 stage
IV, 22 stage III, 39 stage II, 7 stage I human breast cancers and 3
adjacent normal mammary tissues by the immunohistochemistry
analysis using mouse monoclonal anti-Eps8 antibody. We found that
Eps8 was completely localized in the cytoplasm (Fig. 1A). Eps8 expression was detected in
46 (63.9%) of the 72 breast cancers with strong staining (3+), 21
(29%) of the 72 breast cancers were moderately stained (2+), and 5
(6.9%) were weakly stained or negative for Eps8 expression (+/0),
which indicated Eps8 was highly expressed in breast cancers
(P<0.001) according to Student’s t-test. A complete loss of Eps8
expression was observed in normal mammary tissues (Fig. 1B). Therefore, Eps8 expression was
significantly increased in human breast cancer samples. Patient
characteristics are summarized in Table I.
 | Table IPatient characteristics of
immunohistochemistry. |
Table I
Patient characteristics of
immunohistochemistry.
| Variable | No. of patients
(%) |
|---|
| Total number | 72 (100) |
| Age (median, 48
years) | |
| <48 years | 33 (45.8) |
| ≥48 years | 39 (54.2) |
| Histological
diagnosis (invasive carcinoma) | |
| Ductal | 61 (84.7) |
| Medullary | 11 (15.3) |
| Histological grade
(invasive carcinoma) | |
| Grade 1 | 14 (19.4) |
| Grade 2 | 40 (55.6) |
| Grade 3 | 18 (25) |
| TNM staging | |
| Stage IV | 4 (5.6) |
| Stage III | 22 (30.6) |
| Stage II | 39 (54.2) |
| Stage I | 7 (9.6) |
| Normal tissue | 3 |
We next analyzed the expression of Eps8 proteins in
five human breast cancer cell lines. Higher expression of Eps8
proteins was evident in MDA-MB-231, MDA-MB-453 and T47D cells than
in MDA-MB-468, MCF7 cells and normal breast epithelial cells HBL100
(Fig. 1C). Thus, the level of Eps8
expression was high in the highly invasive breast cancer cell line
MDA-MB-231 and low in the weakly invasive breast cancer cell line
MCF7. Both cell lines were further selected to knockdown and
overexpress Eps8 proteins, respectively.
Eps8 overexpression enhances the
proliferation of MCF7 cells
To further investigate the role of Eps8 in human
breast cancer cells, we overexpressed Eps8 and asked whether Eps8
upregulation enhances the proliferation of breast cancer cells. The
pEGFP-C3/Eps8 and pEGFP-C3 plasmids were transfected to established
MCF7 cell lines stably overexpressing GFP/Eps8 and GFP by G418
screening (Fig. 2A). As shown in
Fig. 2B, GFP and GFP/Eps8 target
proteins were expressed at high levels in MCF7 cells.
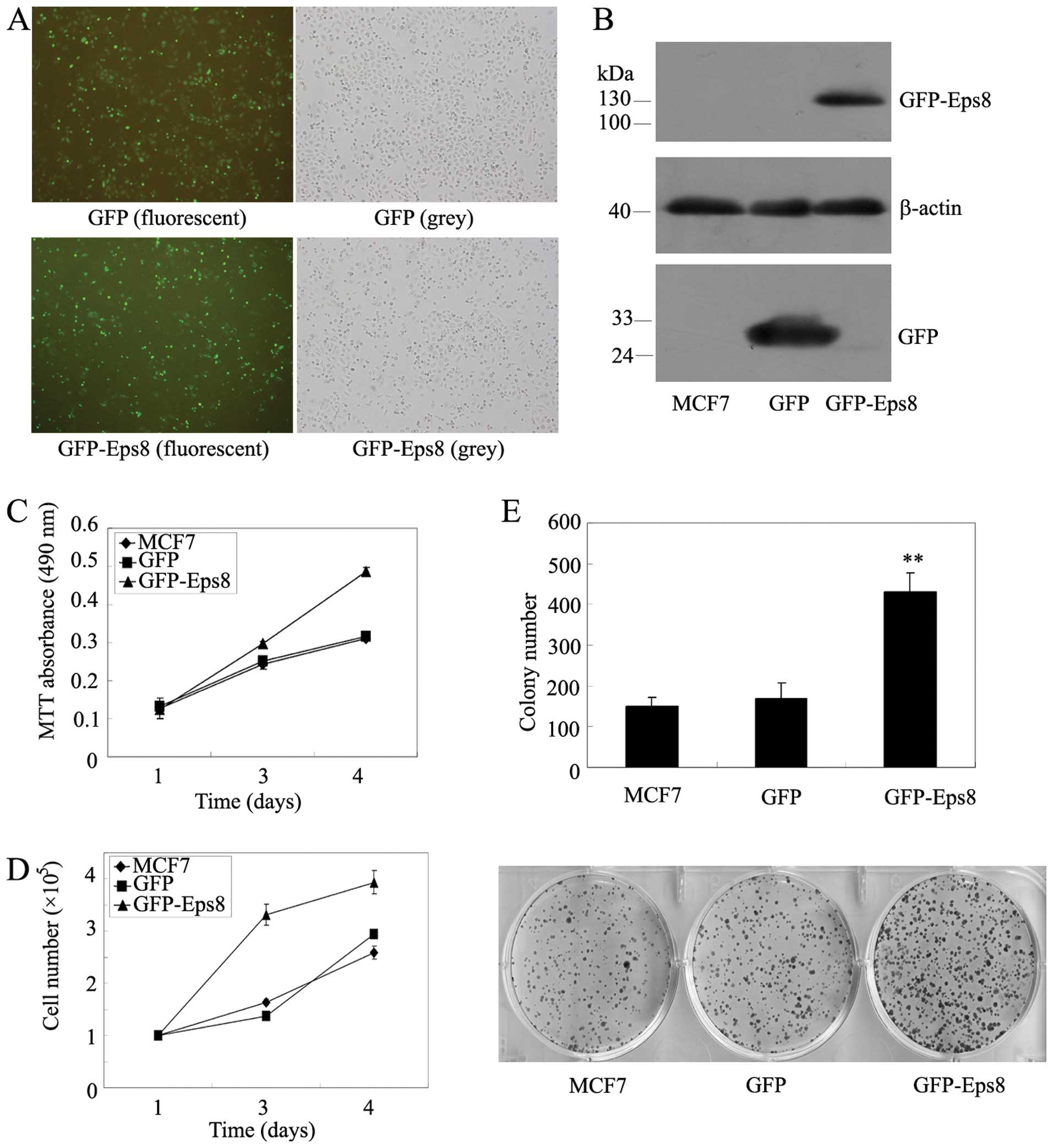 | Figure 2Effects of Eps8 overexpression on
MCF7 breast cancer cell proliferation. (A) Stable overexpression of
GFP-Eps8 and GFP in MCF7 cells by G418 screening was examined by
immunofluorescence assay. (B) GFP and GFP/Eps8 expression in MCF7
cells was confirmed by western blotting using anti-GFP antibodies.
β-actin served as a loading control. (C) MTT assays in mock or
transfected MCF7 cells. Cells (3,000) were plated in octuplicate in
96-well plates and grown in RPMI-1640 with 10% FBS. The absorbance
was analyzed for 1, 3 and 4 days. (D) Cell survival assay in
GFP-Eps8-transfected MCF7 cells as compared with GFP-transfected
and parental MCF7 cells. Cells (100,000) were plated into 6-well
plates in triplicate, grown in RPMI-1640 with 10% FBS for 1, 3 and
4 days and stained with trypan blue in PBS, viable cells were
counted. (E) Representative liquid colony formation analysis in
mock or transfected MCF7 cells. Cells (2,000) were seeded in
triplicate in 6-well plates, and grown for 13 days. Colonies were
fixed with methanol, stained with Giemsa, counted (upper panel) and
images were taken (lower panel). These data represent at least 3
independent experiments with similar results.
**p<0.01, compared with parental and control
cells. |
We then investigated whether Eps8 overexpression in
the breast cancer MCF7 cells leads to cell proliferation. The same
amount of cells was plated in 96-well plates, and we examined cell
viability using MTT assays, Eps8 overexpression resulted in a
remarkable increase in viable cells (Fig. 2C). The same amount of cells was
plated in triplicate in 6-well plates and cell number was counted
on days 1, 3 and 4. We found that the overexpression of Eps8 in
MCF7 cells shows an increased cellular growth compared with
controls (Fig. 2D). Further, the
liquid colony formation assays indicated a great increase in colony
number and size (Fig. 2E).
Therefore, these results suggested that Eps8 could contribute to
breast cancer cell survival and proliferation in vitro.
Eps8 knockdown inhibits the proliferation
and survival of MDA-MB-231 cells in vitro and in vivo
The above results indicated that Eps8 overexpression
significantly promotes MDA-MB-231 cellular proliferation. To gain
further supporting data, we attempted to knock down Eps8 expression
and asked whether shRNA-mediated Eps8 knockdown inhibits the
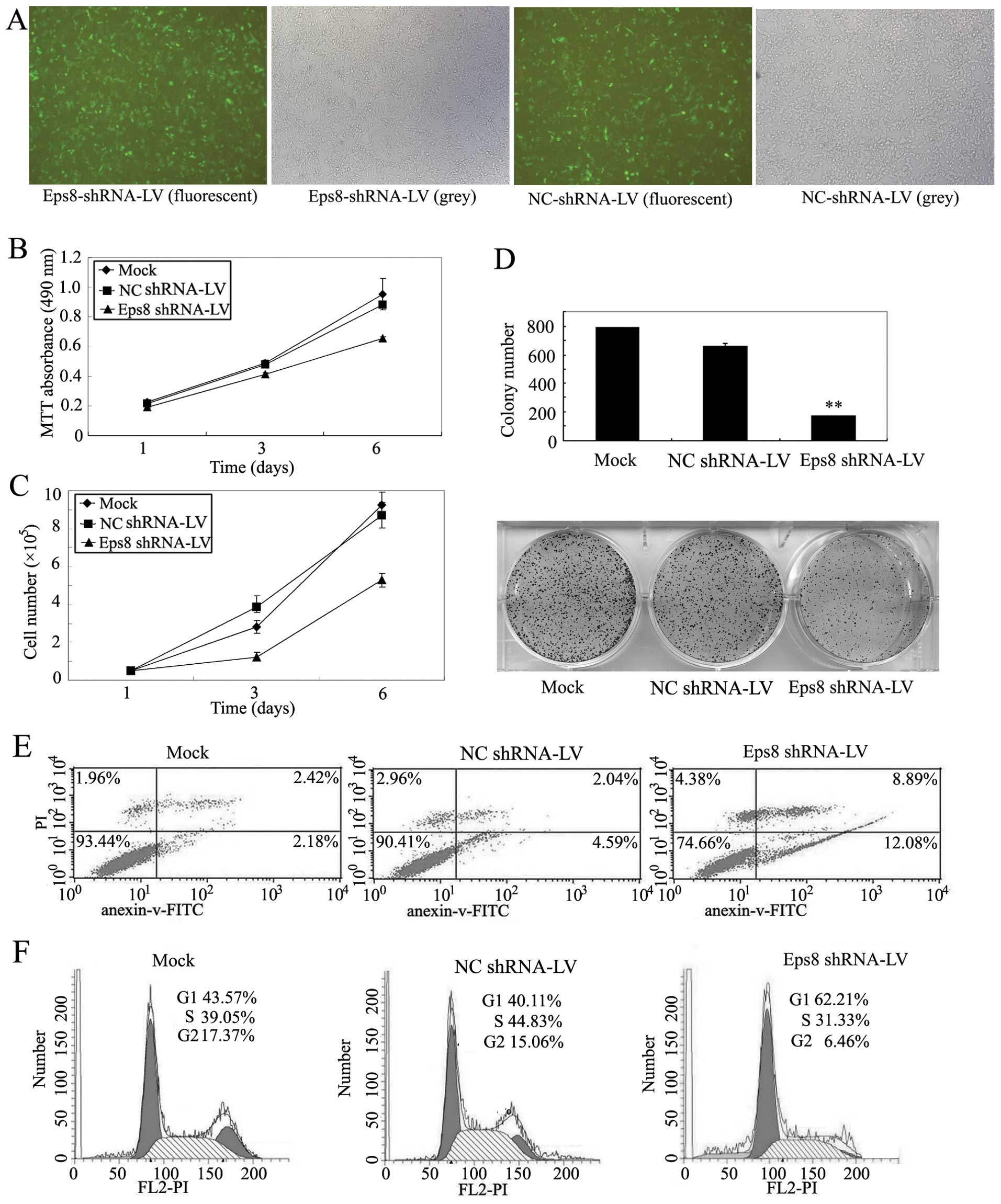
proliferation of breast cancer cells. Then, lentivirus-based RNAi
vector pGCSIL-Eps8 containing the efficient Eps8 shRNA1 and
packaging plasmids were cotransfected to 293T cells (13). Lentiviral particles were prepared
to infect MDA-MB-231 cells. The fluorescence intensity was markedly
increased 4 days after infection and the infection efficiency was
close to 90% in MDA-MB-231 cells (Fig.
3A). Additionally, western blot analysis showed that Eps8
shRNA1 could significantly suppress the expression of endogenous
Eps8 proteins when compared with negative control shRNA (Fig. 6A).
Next, we examined whether Eps8 is a critical
regulator of breast cancer cell proliferation and investigated the
effect of Eps8 knockdown on MDA-MB-231 breast cancer cell growth.
From Fig. 3B and C, we observed
that Eps8 deletion significantly inhibits the proliferation of
MDA-MB-231 cells, whereas control shRNA has no effect. The liquid
colony formation assays showed that Eps8 knockdown displays much
fewer and smaller colonies in MDA-MB-231 cells, while control shRNA
has no effect compared with uninfected parental cells (Fig. 3D). We further explored the
molecular mechanism involved in Eps8-mediated cancer cell survival,
the effects of Eps8 knockdown on cell cycle and apoptosis were
investigated. The flow cytometry analysis showed that Eps8
knockdown results in a significant decrease from mean 91.9 to
74.66% in living MDA-MB-231 cells and exhibits an increase in early
and late apoptotic cells compared to control shRNA-infected and
parental cells (Fig. 3E).
Moreover, we compared the DNA content between Eps8 repressed and
control cells and found that the proportion of S-phase cells was
significantly decreased in Eps8 knockdown cells (31.33%) compared
with MDA-MB-231 cells (39.05%), and the proportion of S-phase cells
were even lower in Eps8 repressed cells compared with NC control
cells (44.83%) (Fig. 3F). These
observations suggested that Eps8 knockdown arrested cells at the
G1/S checkpoint. Therefore, the above data showed that Eps8
knockdown suppresses proliferation, and cell cycle progression
enhancing cell apoptosis of MDA-MB-231 cells.
The inhibitory effect of Eps8 knockdown on breast
cancer cell proliferation in vitro suggested that Eps8
deletion might suppress tumor growth in vivo. Tumorigenicity
assays were performed by subcutaneous injection of Eps8 shRNA cells
into nude mice, and NC shRNA cells were used as controls. Within 4
weeks, solid tumors were readily visible in left and right dorsal
regions of all mice (Fig. 4A), but
the average tumor volume and weight of the Eps8 shRNA group were
markedly reduced by >60% compared with negative controls
(Fig. 4B and C). Thus, the above
data showed Eps8 knockdown could decrease breast cancer cell
proliferation in vivo.
Eps8 knockdown suppresses breast cancer
cell migration, decreases the number and size of EGF-induced
filopodia and increases the sensitivity of breast cancer cells to
cisplatin
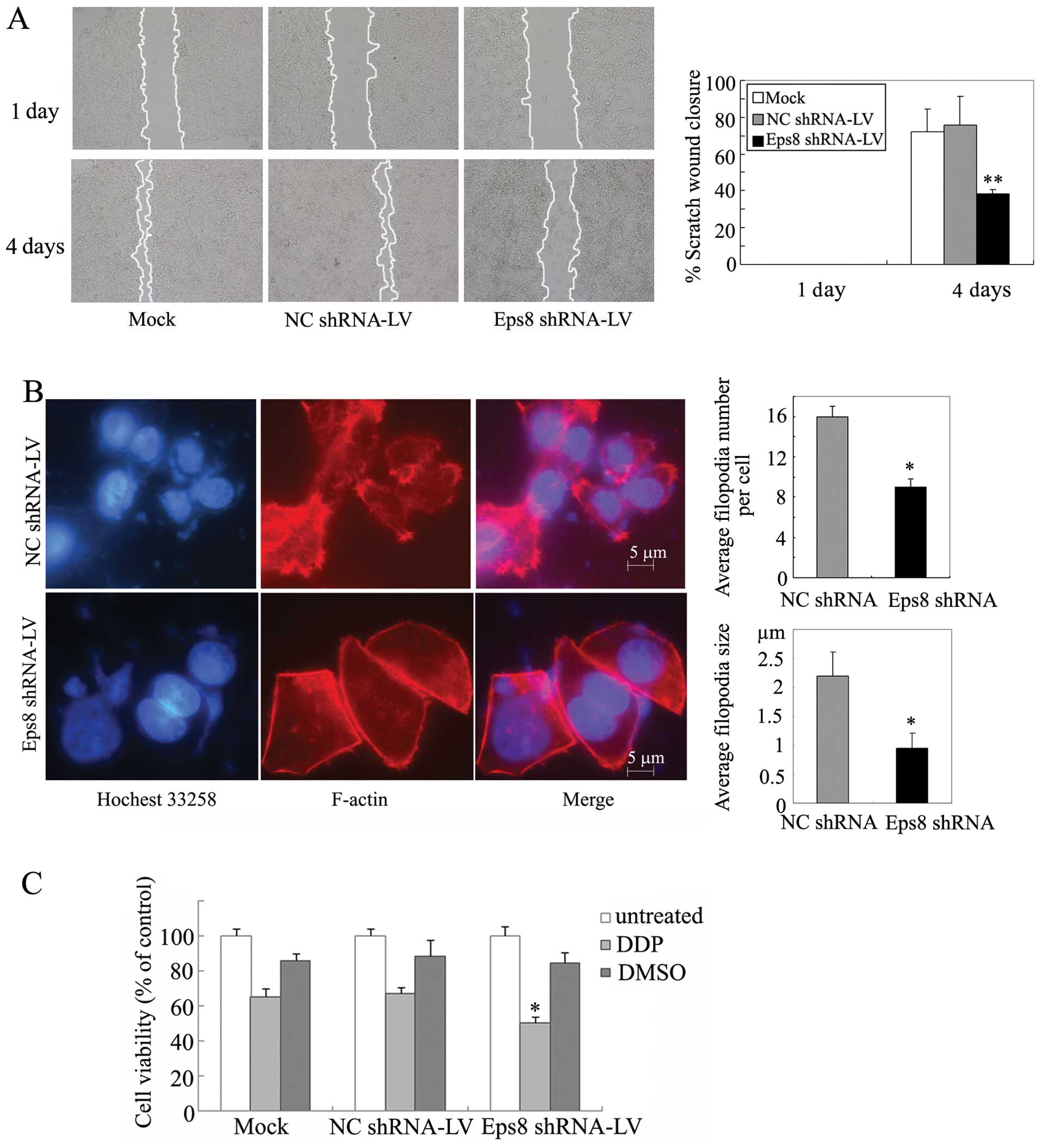
We next assessed the effect of Eps8 on breast cancer
cell migration by wound-healing assays. As shown in Fig. 5A, Eps8 depletion produced 62%
inhibition of cell migration in MDA-MB-231 cells. In contrast,
control groups dramatically promoted cell migration. Thus, Eps8
knockdown was able to significantly inhibit cell motility, which
confirmed the role of Eps8 in breast cancer cell migration.
As Eps8 was previously described to be essential for
actin dynamics and cytoskeletal organization in pancreatic cancer
(12), we examined the effects of
Eps8 knockdown on filopodia in MDA-MB-231 cells. Cells were
cultivated for 48 h in serum-free medium and then EGF was added for
30 min. Eps8-deletion cells exhibited reduction of filopodial
density and length and inhibited the filopodial growth (Fig. 5B), indicating that Eps8 is involved
in a rearrangement of F-actin in breast cancer cells.
We further investigated the implication of Eps8 in
chemosensitivity of breast cancer cells. MDA-MB-231 cells were
treated with cisplatin for 24 h. Sixty-two percent reduction of MTT
absorbance was detected in Eps8 shRNA cells compared with 48%
inhibition in their control cells (Fig. 5C). Thus, Eps8 knockdown was able to
sensitize MDA-MB-231 cells to the cytotoxicity of cisplatin as
reported in cervical cancer cells (8).
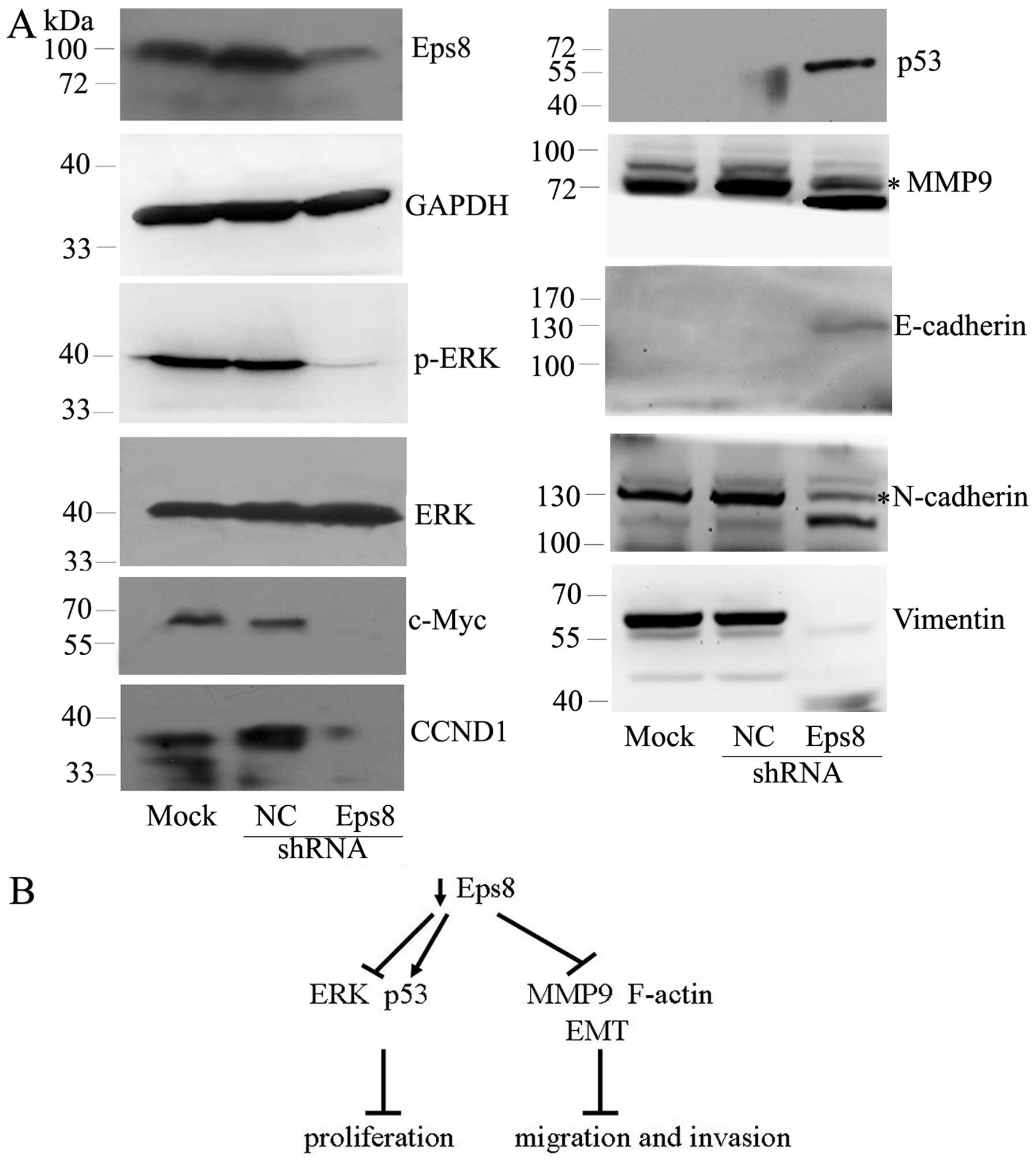
Eps8 affects the expression of ERK
signaling, MMP9, p53 and EMT markers
Because Eps8 influences many genes involved in the
development and progression of human cancers, we decided to examine
whether Eps8 regulates these target genes in breast cancer cells.
As shown in Fig. 6A, Eps8
knockdown had no effect on ERK protein, but it decreased the
phosphorylated level of ERK and downregulated the expression of
c-Myc and cyclin D1 (CCND1) as downstream mitogenic targets of ERK
signaling (26). Moreover, Eps8
knockdown exhibited inhibited MMP9 and enhanced the tumor
suppressor p53. The level of EMT epithelial marker E-cadherin was
concomitantly increased in Eps8-depleted cells. In contrast, Eps8
knockdown decreased the levels of mesenchymal markers N-cadherin
and vimentin. Taken together, these data showed that Eps8 regulates
breast cancer cell proliferation and migration, at least in part,
by affecting ERK signaling, MMP9, p53 and EMT markers.
Discussion
Eps8 is important in regulating the development and
progression of many human cancers. However, the role of Eps8 in
human breast cancer has not been reported. In the present study, we
examined the expression level of Eps8 in histological samples from
a large number of patients with previously untreated breast cancer.
As detected in the immunohistochemistry analysis, Eps8 was
overexpressed in most of breast cancer tissues. Moreover, Eps8
protein was significantly higher in 55.6% of stages II to III
breast cancer samples compared to normal mammary tissues.
Furthermore, 5 breast cancer cell lines were detected to confirm
that the protein level of Eps8 is high in highly invasive breast
cancer cells, and low or lost in weakly invasive breast cancer
cells. Thus, Eps8 expression may predict cancer recurrence and
outcome in stages II–III breast cancer patients.
Evidence presented here demonstrated the effect of
Eps8 on breast cancer cell proliferation, we overexpressed Eps8/GFP
in MCF7 cell line with low expression of Eps8 to investigate the
role of Eps8 in breast cancer cellular growth in vitro. Eps8
expression significantly promoted cell proliferation. In contrast,
we used lentivirus-based RNAi system to knock down Eps8 expression
in MDA-MB-231 cell line with high expression of Eps8. Eps8
attenuation reversed the growth phenotype of Eps8-overexpressing
breast cancer cells and increased the sensitivity of breast cancer
cells to cisplatin. Our data indicated that Eps8 expression is
critical for cell growth and proliferation of breast cancer cells
in vitro and in vivo and possibly involved in the
development and progression in human breast cancer.
Actin polymerization and formation of lamellipodia
are believed to play important roles in cell migration during
metastasis (27). Eps8, Abi-1 and
Sos1 form a tricomplex, induce Rac-specific guanine nucleotide
exchange factor (GEF) activity and transduce signals from Ras to
Rac leading to actin remodeling (28–30).
Moreover, Eps8 binds with F-actin and colocalizes with actin-based
membrane protrusions such as lamellipodia, filopodia and membrane
ruffles (28,29). Likely, Eps8 knockdown reduces
filopodial density and length and inhibits cell migration in breast
cancer cells, suggesting the important role of Eps8 in cellular
movement and migration of tumor cells.
Eps8 is an important signal molecule and integrates
multiple pathways. For example, Eps8 could control the
Ras-Raf-MEK-ERK signaling cascade (31), which plays a crucial role in
regulating cellular processes including differentiation,
proliferation, survival and apoptosis (32). We demonstrated that Eps8 affects
the expression of phosphorylated ERK and downstream genes c-Myc and
CCND1 in cellular growth of breast cancer. Apart from regulating
cell survival, the ERK pathway promoted invasiveness in tumor cells
by upregulation of MMPs such as MMP9 for extracellular matrix
remodeling (33). We provided
evidence that the levels of MMP9 are decreased in Eps8 shRNA
infected cells. In addition, the tumor suppressor gene p53 inhibits
the development and growth of the majority of human tumors
(34). Eps8 knockdown also
enhanced p53 upregulation, resulting in growth arrest or apoptosis
of breast cancer cells. The results supported the conclusion that
Eps8 regulates the expression of c-Myc, CCND1 and MMP9 through the
ERK pathway as well as p53.
Cancer progression toward malignancy is mostly
associated with the loss of epithelial differentiation and a switch
toward a mesenchymal phenotype. Epithelial-mesenchymal transition
(EMT) is considered a critical process for tumor invasion and
metastasis (35). Many epithelial
proteins including E-cadherin, α-catenin and β-catenin were
down-regulated, whereas mesenchymal proteins were upregulated, such
as fibonectin, N-cadherin, α-SMA and vimentin (36). Loss of E-cadherin promotes
metastasis by disrupting inter-cellular contacts in metastatic
dissemination (37). N-cadherin
promotes proliferation, adhesion and invasion of prostate cancer
cells (38). Moreover, vimentin is
associated with a highly invasive cellular phenotype (39). In the present study we found that
Eps8 knockdown has a significant impact on EMT by increasing
E-cadherin expression and decreasing N-cadherin and vimentin, and
might achieve lower motility and invasiveness (Fig. 6B). Therefore, these data suggested
that Eps8 might partly mediate EMT-like transition and invasion in
breast cancer.
In conclusion, our results revealed the pivotal role
of Eps8 in breast cancer progression. Eps8 was highly expressed in
breast cancer tissues and highly-invasive cell lines. Eps8
regulated the proliferation, migration and invasion of breast
cancer cells in vitro and in vivo. Eps8 knockdown
inhibited cell migration, decreased the number and size of
EGF-induced filopodin and increased the cytotoxicity of cisplatin
in breast cancer cells, at least in part, by affecting the
expression of ERK signaling, MMP9, p53 and EMT markers. The
oncoprotein Eps8 might have an important role as a molecular
therapeutic target and even prognostic marker in human breast
cancer.
Acknowledgements
This study was supported by the National Natural
Science Foundation of China (nos. 81272318 and 81272190), and the
Scientific Research Fund of Hunan Provincial Education Department
(no. 13B068).
References
|
1
|
Bray F, McCarron P and Parkin DM: The
changing global patterns of female breast cancer incidence and
mortality. Breast Cancer Res. 6:229–239. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Gupta GP and Massague J: Cancer
metastasis: building a framework. Cell. 127:679–695. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Dahiya R and Deng G: Molecular prognostic
markers in breast cancer. Breast Cancer Res Treat. 52:185–200.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ross JS and Fletcher JA: The HER-2/neu
oncogene in breast cancer: prognostic factor, predictive factor,
and target for therapy. Oncologist. 3:237–252. 1998.
|
|
5
|
Sgouros J, Galani E, Gonos E, et al:
Correlation of nm23-H1 gene expression with clinical outcome in
patients with advanced breast cancer. In Vivo. 21:519–522.
2007.PubMed/NCBI
|
|
6
|
Barnes DM and Gillett CE: Cyclin D1 in
breast cancer. Breast Cancer Res Treat. 52:1–15. 1998. View Article : Google Scholar
|
|
7
|
Wang H, Teh MT, Ji Y, et al: EPS8
upregulates FOXM1 expression, enhancing cell growth and motility.
Carcinogenesis. 31:1132–1141. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen YJ, Shen MR, Chen YJ, Maa MC and Leu
TH: Eps8 decreases chemosensitivity and affects survival of
cervical cancer patients. Mol Cancer Ther. 7:1376–1385. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Maa MC, Lee JC, Chen YJ, et al: Eps8
facilitates cellular growth and motility of colon cancer cells by
increasing the expression and activity of focal adhesion kinase. J
Biol Chem. 282:19399–19409. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang H, Patel V, Miyazaki H, Gutkind JS
and Yeudall WA: Role for EPS8 in squamous carcinogenesis.
Carcinogenesis. 30:165–174. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang W, Wang L, Liu Y, et al: Structure
of human lanthionine synthetase C-like protein 1 and its
interaction with Eps8 and glutathione. Genes Dev. 23:1387–1392.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Welsch T, Endlich K, Giese T, Buchler MW
and Schmidt J: Eps8 is increased in pancreatic cancer and required
for dynamic actin-based cell protrusions and intercellular
cytoskeletal organization. Cancer Lett. 255:205–218. 2007.
View Article : Google Scholar
|
|
13
|
Ding X, Zhou F, Wang F, et al: Eps8
promotes cellular growth of human malignant gliomas. Oncol Rep.
29:697–703. 2013.PubMed/NCBI
|
|
14
|
Kang H, Wilson CS, Harvey RC, et al: Gene
expression profiles predictive of outcome and age in infant acute
lymphoblastic leukemia: a Children’s Oncology Group study. Blood.
119:1872–1881. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Matoskova B, Wong WT, Salcini AE, Pelicci
PG and Di Fiore PP: Constitutive phosphorylation of eps8 in tumor
cell lines: relevance to malignant transformation. Mol Cell Biol.
15:3805–3812. 1995.PubMed/NCBI
|
|
16
|
Chu PY, Liou JH, Lin YM, et al: Expression
of Eps8 correlates with poor survival in oral squamous cell
carcinoma. Asia Pac J Clin Oncol. 8:e77–e81. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Yang TP, Chiou HL, Maa MC and Wang CJ:
Mithramycin inhibits human epithelial carcinoma cell proliferation
and migration involving downregulation of Eps8 expression. Chem
Biol Interact. 183:181–186. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gorsic LK, Stark AL, Wheeler HE, et al:
EPS8 inhibition increases cisplatin sensitivity in lung cancer
cells. PLoS One. 8:e822202013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Abdel-Rahman WM, Ruosaari S, Knuutila S
and Peltomaki P: Differential roles of EPS8 in carcinogenesis: loss
of protein expression in a subset of colorectal carcinoma and
adenoma. World J Gastroenterol. 18:3896–3903. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Li YH, Xue TY, He YZ and Du JW: Novel
oncoprotein EPS8: a new target for anticancer therapy. Future
Oncol. 9:1587–1594. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yao J, Weremowicz S, Feng B, et al:
Combined cDNA array comparative genomic hybridization and serial
analysis of gene expression analysis of breast tumor progression.
Cancer Res. 66:4065–4078. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mangravite LM, Lipschutz JH, Mostov KE and
Giacomini KM: Localization of GFP-tagged concentrative nucleoside
transporters in a renal polarized epithelial cell line. Am J
Physiol Renal Physiol. 280:F879–F885. 2001.PubMed/NCBI
|
|
23
|
Schmid JA, Birbach A, Hofer-Warbinek R, et
al: Dynamics of NF kappa B and Ikappa Balpha studied with green
fluorescent protein (GFP) fusion proteins. Investigation of GFP-p65
binding to DNa by fluorescence resonance energy transfer. J Biol
Chem. 275:17035–17042. 2000. View Article : Google Scholar
|
|
24
|
Lois C, Hong EJ, Pease S, Brown EJ and
Baltimore D: Germline transmission and tissue-specific expression
of transgenes delivered by lentiviral vectors. Science.
295:868–872. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ding X, Yang Z, Zhou F, et al:
Transcription factor AP-2alpha regulates acute myeloid leukemia
cell proliferation by influencing Hoxa gene expression. Int J
Biochem Cell Biol. 45:1647–1656. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Pan CC, Bloodworth JC, Mythreye K and Lee
NY: Endoglin inhibits ERK-induced c-Myc and cyclin D1 expression to
impede endothelial cell proliferation. Biochem Biophys Res Commun.
424:620–623. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ridley AJ: Rho GTPases and cell migration.
J Cell Sci. 114:2713–2722. 2001.PubMed/NCBI
|
|
28
|
Scita G, Tenca P, Areces LB, et al: An
effector region in Eps8 is responsible for the activation of the
Rac-specific GEF activity of Sos-1 and for the proper localization
of the Rac-based actin-polymerizing machine. J Cell Biol.
154:1031–1044. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Scita G, Nordstrom J, Carbone R, et al:
EPS8 and E3B1 transduce signals from Ras to Rac. Nature.
401:290–293. 1999. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen H, Wu X, Pan ZK and Huang S:
Integrity of SOS1/EPS8/ ABI1 tri-complex determines ovarian cancer
metastasis. Cancer Res. 70:9979–9990. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hecquet C, Lefevre G, Valtink M, Engelmann
K and Mascarelli F: Activation and role of MAP kinase-dependent
pathways in retinal pigment epithelial cells: ERK and RPE cell
proliferation. Invest Ophthalmol Vis Sci. 43:3091–3098.
2002.PubMed/NCBI
|
|
32
|
Chang F, Steelman LS, Lee JT, et al:
Signal transduction mediated by the Ras/Raf/MEK/ERK pathway from
cytokine receptors to transcription factors: potential targeting
for therapeutic intervention. Leukemia. 17:1263–1293. 2003.
View Article : Google Scholar
|
|
33
|
McCawley LJ, Li S, Wattenberg EV and
Hudson LG: Sustained activation of the mitogen-activated protein
kinase pathway. A mechanism underlying receptor tyrosine kinase
specificity for matrix metalloproteinase-9 induction and cell
migration. J Biol Chem. 274:4347–4353. 1999. View Article : Google Scholar
|
|
34
|
Oren M: Decision making by p53: life,
death and cancer. Cell Death Differ. 10:431–442. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Thiery JP: Epithelial-mesenchymal
transitions in development and pathologies. Curr Opin Cell Biol.
15:740–746. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Grunert S, Jechlinger M and Beug H:
Diverse cellular and molecular mechanisms contribute to epithelial
plasticity and metastasis. Nat Rev Mol Cell Biol. 4:657–665. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Onder TT, Gupta PB, Mani SA, et al: Loss
of E-cadherin promotes metastasis via multiple downstream
transcriptional pathways. Cancer Res. 68:3645–3654. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tanaka H, Kono E, Tran CP, et al:
Monoclonal antibody targeting of N-cadherin inhibits prostate
cancer growth, metastasis and castration resistance. Nat Med.
16:1414–1420. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lang SH, Hyde C, Reid IN, et al: Enhanced
expression of vimentin in motile prostate cell lines and in poorly
differentiated and metastatic prostate carcinoma. Prostate.
52:253–263. 2002. View Article : Google Scholar : PubMed/NCBI
|