Introduction
Breast cancer is the most common solid cancer in
women around the world and the leading cause of cancer-related
deaths. After initial surgery adjuvant treatment strategies include
cytotoxic chemotherapy, radiation therapy, and anti-hormonal
therapy (1). Advances in earlier
diagnosis and therapy have significantly improved outcomes.
However, recurrent metastatic breast cancer is still incurable and
only 3% of patients with metastatic disease achieve a complete
response for >5 years after combination chemotherapy (2,3); the
median survival time after therapy is ~2 years.
Endogenous estrogens are thought to play a major
role in the development of breast cancer, and estrogen receptors
(ER) are targets of hormonal therapy. These nuclear receptors are
ligand-dependent transcription factors that mediate the biological
effects of (anti-) estrogens. There are two types of specific
receptors: ERα and ERβ, which differ in their responses to agonists
and antagonist due to differences in their C-terminal ligand
binding domains (LBD) (4). The
receptors show different expression patterns in breast cancer
tissues and seem to have opposing roles in the proliferation of
breast cancer cells. ERα-positive tumours are related to a good
prognosis, and it is suggested that ERβ expression declines during
breast tumour genesis (5). Despite
high ERα levels in some primary tumours and in all patients with
metastatic disease resistance to endocrine therapies arise. The
potential mechanisms for either intrinsic or acquired endocrine
resistance are still poorly comprehended, but they include
cross-talk between the ER pathway and other growth factor and
kinase networks as well as ER-co-regulatory proteins (6). Increased expression of co-activator
proteins that mediate ER activity or downregulation of co-repressor
activity reducing the inhibitory potential of tamoxifen are
possible molecular mechanisms for resistance and the progression of
confined breast cancer to invasive disease (7–9).
The paxillin protein family, which comprises
paxillin, transforming growth factor β1 induced transcript 1
(TGFB1I1 or Hic-5) and leupaxin, is involved in the majority of the
steps during cell migration and invasion as part of the focal
adhesion complexes. It is also known, that all of them can interact
with different steroid hormone receptors and induce their
transcriptional activity in the nucleus (10,11),
thus serving as candidate proteins involved in the response to
hormonal therapy. All members of the paxillin protein family
contain two different protein-protein interaction domains, namely
LD motifs and LIM domains. LD motifs contain two invariant amino
acids, leucine and aspartate (LD). LIM domains are composed of two
zincfinger domains and were identified in the transcription factors
LIN -11,
ISL-1 and
MEC-3 (LIM). Due to
this protein structure paxillin proteins represent adaptor
platforms in transmitting signals from outside the cell to affect
transcriptional regulation in the nucleus. Originally identified in
hematopoietic cells, leupaxin was found to be expressed in a series
of other tissues, e.g., smooth muscle cells and prostate cancer
cells (12,13). Recently, it was shown that leupaxin
expression in human prostate cancer correlates with tumour stage
and that leupaxin downregulation in prostate cancer cells results
in decreased migratory ability and invasiveness. Furthermore, we
showed that leupaxin functioned as a co-activator of the androgen
receptor (13).
In the present study, we further elucidated the role
of leupaxin in different human cancers. We showed that leupaxin is
expressed in breast and endometrial carcinomas. Downregulation of
leupaxin expression in breast cancer cells decreases migration and
invasion. In addition, leupaxin shuttles between focal adhesion
sites and the nucleus and interacts with both ERs via their
N-terminal parts. This interaction results in the transcriptional
activation of ERα in the presence and absence of ER ligands. Taken
together, our results underline the role of leupaxin as an
important factor in the progression of breast cancers and give a
further basis to investigate the potential of leupaxin as a target
for the development of novel therapeutic strategies.
Materials and methods
Cancer profiling array
The Cancer Profiling Array I (BD Biosciences
Clontech, Heidelberg, Germany) was hybridized according to the
manufacturer’s instructions with a [32P]-labelled
leupaxin cDNA (nucleotide position 437-1748; NM_004811) probe,
using the Rediprime II labelling kit (GE Healthcare GmbH, Freiburg,
Germany). After overnight hybridization and a high-stringency wash,
the array was scanned and analysed with a Molecular Imager FX and
Quantity One Software (Bio-Rad, Hercules, CA, USA).
Patient material and
immunohistochemistry
Ethical approval was obtained from the ethics
committee of the University of Göttingen for the use of human
material in the present study. Immunohistochemistry was performed
as described previously (13). The
mouse monoclonal anti-leupaxin (clone 283 G) antibody was kindly
provided by Eli Lilly & Co. (Indianapolis, IN, USA). For
negative controls, blocking solution was used in place of the
primary antibody.
Semiquantitative analysis of leupaxin
immunoreactivity and statistical analysis
For quantification of the leupaxin immune signals in
tissue sections an additive immunoreactive score (IRS = SI + CN)
was applied comprising the average signal intensity (SI) and the
number of positive tumour cells (CN) (13). For comparative analyses of leupaxin
immunoreactivity with clinicopathologic features the χ2
test and Fisher’s exact test were applied.
Cell culture and transient
transfection
MDA-MB-231, MDA-MB-453, HCC70, ZR-77-1, MCF-7, T-47D
and NIH/3T3 cells were purchased from ATCC and grown in DMEM medium
(PAN-Systems, Nuremberg, Germany) containing 10% FCS and 1.2%
antibiotics. For MCF-7 and T-47D 1% non-essential amino acids was
added. Transient transfection experiments were performed using
FuGENE (Roche Diagnostics GmbH, Mannheim, Germany) according to the
manufacturer’s instructions.
Northern blot analysis
Northern blot analysis was performed as described
previously (14). Total RNA from
breast cancer cell lines was isolated using RNeasy MINI (Qiagen,
Hilden, Germany). Total RNA (5 μg) was separated and hybridized
with the leupaxin probe mentioned above.
Western blot analysis and
immunoprecipitation
Whole-cell lysates from parental and transfected
breast cancer cells were prepared using lysis buffer and subjected
to western blot analysis as described previously (13). The following primary antibodies
were used: mouse monoclonal anti-leupaxin 283 C (kindly provided by
Eli Lilly & Co.), mouse monoclonal anti-α-tubulin (Sigma,
Taufkirchen, Germany), rabbit polyclonal anti-ERα (MC-20, Santa
Cruz Biotechnology, Dallas, TX, USA) and rabbit polyclonal anti-ERβ
(Cell Signaling Technology, Danvers, MA, USA). For
coimmunoprecipitation assay cells were lysed with IP buffer (0.05 M
Tris pH 7.5, 0.15 M NaCl, 0.5% deoxycholic acid, 1% IGPAL) in the
presence of proteinase inhibitor cocktail (Roche) and 3 mg protein
was used for immunoprecipitation with 1 μg ERα and ERβ antibodies,
respectively. After incubation overnight at 4°C, 50 μl protein A/G
sepharose (Santa Cruz Biotechnology) was added and incubated for 2
h. Protein complexes were isolated by centrifugation and three
washes with IP buffer and final elution with 2X SDS sample buffer
(Cell Signaling Technology). Subsequently, western blotting was
performed. Immunoprecipitation was performed in three independent
experiments.
Immunocytochemistry
Cells were plated on culture slides coated with 10
μg/ml fibronectin (Sigma) under normal culture conditions. The
cells were then fixed with 4% formaldehyde in PBS, permeabilised
with 0.1% Triton X-100 in PBS and blocked with 3% BSA in PBS for 30
min at room temperature. After incubation with primary antibodies
(10 μg/ml mouse monoclonal anti-leupaxin) overnight at 4°C, cells
were washed with PBS and incubated for 2 h at RT with secondary
antibodies (1:500 sheep anti-mouse-IgG-Cy3, Sigma). Subsequently,
cells were washed in PBS, stained with FITC-phalloidin (Sigma) for
30 min and mounted using Vectashield/DAPI. Images were acquired
using the Olympus FluoView1000 confocal scanning microscope and
FluoView software (Olympus Deutschland GmbH, Hamburg, Germany).
Direct yeast two-hybrid experiments
The yeast two-hybrid experiments were carried out by
using the Matchmaker GAL4 two-hybrid system (Clontech Laboratories,
Inc., Saint-Germain-en-Laye, France). All procedures were performed
according to the manufacturer’s protocols. The plasmids
pGADT7-LPXN, pGADT7-LPXN-LD and pGADT7-LPXN-LIM were described
previously (13). The open reading
frames of ERα (EcoRI) and ERβ (NcoI/SalI) were
cloned into the pGBKT7 vector. Plasmids were co-transformed into
the yeast host strain AH109. Co-transformants were selected in the
presence or absence of 100 nM estradiol (Sigma) on minimal
synthetic dropout (SD) medium lacking the amino acids leucine,
tryptophan, histidine and adenine (SD-LTHA) containing 80 mg/l
X-Gal (ICN).
ERα transactivation assay
pcDNA-ERα was cloned by amplification of the ERα
open reading frame (361-2148, NM_000125) and cloning into the
EcoRI restriction site of pcDNAmyc/HisA vector (Life
Technologies, Darmstadt, Germany). Reporter gene assays were
performed as described previously (13) using charcoal-stripped FCS and
phenol-red free medium. Cells were transfected with the following
expression vector cocktail after 24 h: 0.05 μg pCMV-β-Gal, 0.05–0.4
μg GFP-LPXN (as indicated) and with 0.2 μg Vit-ERE-Luc and with 0.2
μg pcDNA-ERα vector. Thirty-six hours after transfection cell
lysates were prepared and luciferase activity was measured in a
microplate luminometer (LB953, Berthold) by injecting 100 μl of a
luciferin solution (P.J.K. GmbH, Kleinblittersdorf, Germany) per
well. The luciferase activity was normalized against the
β-galactosidase activity, which was measured by using the
Galacto-Light™ kit (BD Bioscience) according to the manufacturer’s
protocol.
RNA interference
Transfection of cells was accomplished using
Oligofectamine reagent (Life Technologies) according to the
manufacturer’s instructions with leupaxin gene-specific siRNA
duplexes as described previously (13). Control cells were transfected with
siRNA duplex oligonucleotides against the firefly luciferase gene
(15). At different time-points
after transfection (24, 48 and 72 h) cells were collected and used
in the following experiments.
Real-time RT-PCR analysis
Real-time RT-PCR analysis was performed as described
previously (13). Primers used for
quantitative RT-PCR were: PBGD-For-QGCAATGCGGCTGCAACGGCGGAAG;
PBGD-Rev-QCCTGTGGTGGACATAGCAATGATT;
TBP-For-QAGCCTGCCACCTTACGCTCAGTBP-Rev-QTGCTGCCTTTGTTGCTCTTCCA;
leupaxin-Q4-Fw AGTTCCTTTGCGGTCCTCTTCTTC; leupaxin-Q4b-Rev
GTCTCCTTTCTGGAATGCTGATCC.
Invasion and migration assay
Cell invasion was determined in BioCoat Matrigel
Invasion Chambers (BD Biosciences) as described previously
(13,15). siRNA transfected cells
(2.5×104 cells, respectively) were incubated in invasion
chambers for 22 h at 37°C. To estimate directional migration the
transfected MDA-MB-231 cells were plated on Millicell®
Cell Culture Inserts (Merck Millipore, Darmstadt, Germany) with
6×104 cells/well and incubated for 24 h. Invaded and
migrated cells were stained with haematoxylin and eosin and counted
from five randomly chosen fields under a BX60 microscope using the
analySIS software (Olympus). Data are expressed as the percentage
of cells with reduced leupaxin expression in comparison to control
transfected cells.
Statistical analyses
If not otherwise stated, experiments were performed
at least three independent times (biological replicates). For
statistical analysis Student’s t-test was applied.
*p≤0.05, significant; **p≤0.01, very
significant; ***p≤0.001, extremely significant; NS, not
significant.
Results
Leupaxin is expressed in different types
of cancer
Recent studies indicated that the expression of
leupaxin is not limited to cells of hematopoietic origin.
Therefore, a cancer profiling array analysis with a human specific
leupaxin probe was performed to investigate the expression profile
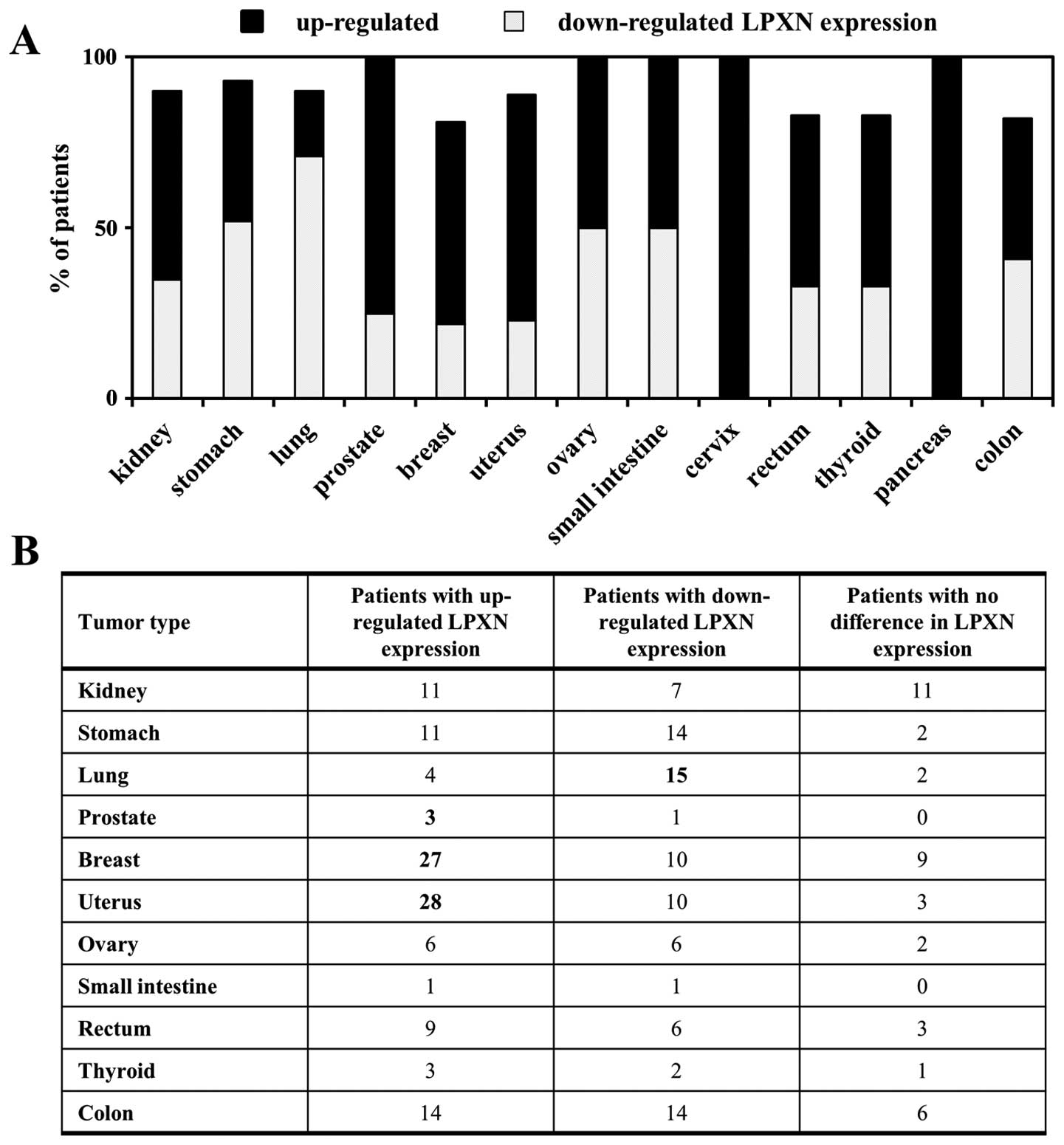
of leupaxin in normal and matched tumour tissue samples. As shown
in Fig. 1 most breast and
endometrial cancer patients displayed upregulation of leupaxin
expression in the tumour as compared to the normal tissue, whereas
in lung cancer patients downregulation of leupaxin expression was
clearly observed. It is noteworthy that upregulation of leupaxin
expression was detectable in three out of four prostate cancer
tissues confirming previous results of our group (13). Other cancer types showed an equal
distribution of patients with up or downregulated leupaxin
expression, e.g., ovarian and colon cancer, and were therefore
considered to be not relevant in this study.
Leupaxin is expressed in mammary cancer
tissue
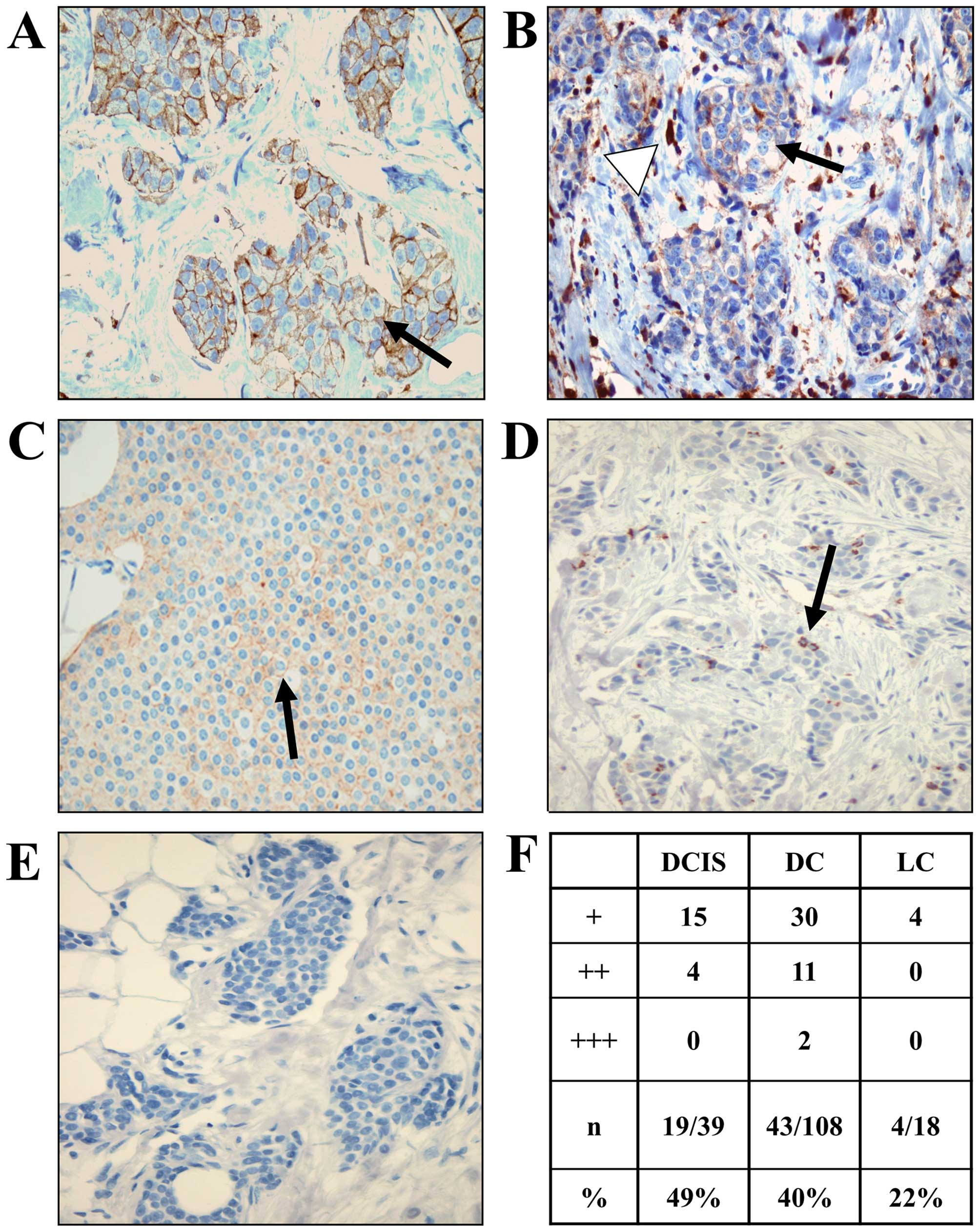
To evaluate the relevance of leupaxin expression in
breast cancer, 127 tissue sections from breast cancer patients were
stained with a leupaxin specific antibody and classified into low,
medium and high depending on the percentage of positive cancer
cells and the according leupaxin expression level. Mammary
carcinomas were classified in ductal carcinoma in situ
(DCIS), invasive ductal (DC) or invasive lobular (LC) carcinomas
(Fig. 2A–E). Different cancer
types in one sample were individually evaluated. As seen in
Fig. 2F, 49% of ductal carcinoma
in situ and 40% of invasive ductal carcinomas displayed
leupaxin expression. Only 22% of LC carcinomas showed staining for
leupaxin. There was no significant correlation between the
expression level of leupaxin and the tumour stage or hormone
receptor status of ERα and progesterone receptor (PR) as well as
HER2, respectively. However, we observed higher staining scores (++
and +++) only in more advanced breast cancers (DC).
Expression of leupaxin in mammary
carcinoma cell lines
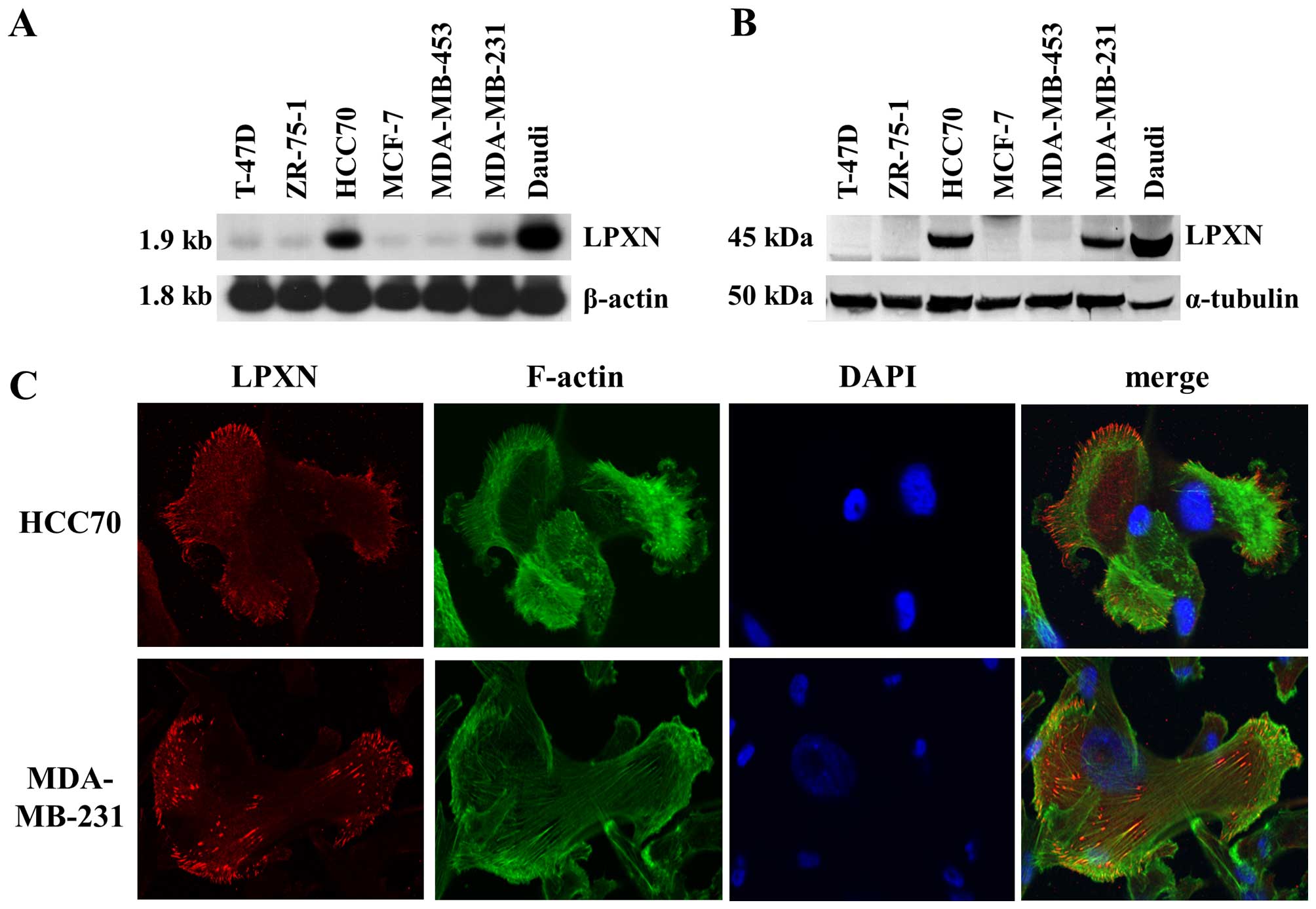
The expression of leupaxin was evaluated in seven
established breast cancer cell lines. Northern (Fig. 3A) and western blot (Fig. 3B) analyses demonstrated, that
leupaxin is highly expressed in the ER-negative MDA-MB-231 and in
the ER-positive HCC70 cell lines, whereas no expression was
detectable on the protein level independent of ER status or
invasive behaviour in the other analysed cell lines. Subcellularly,
leupaxin localized to the focal adhesion sites in MDA-MB-231 and
HCC70 cells (Fig. 3C).
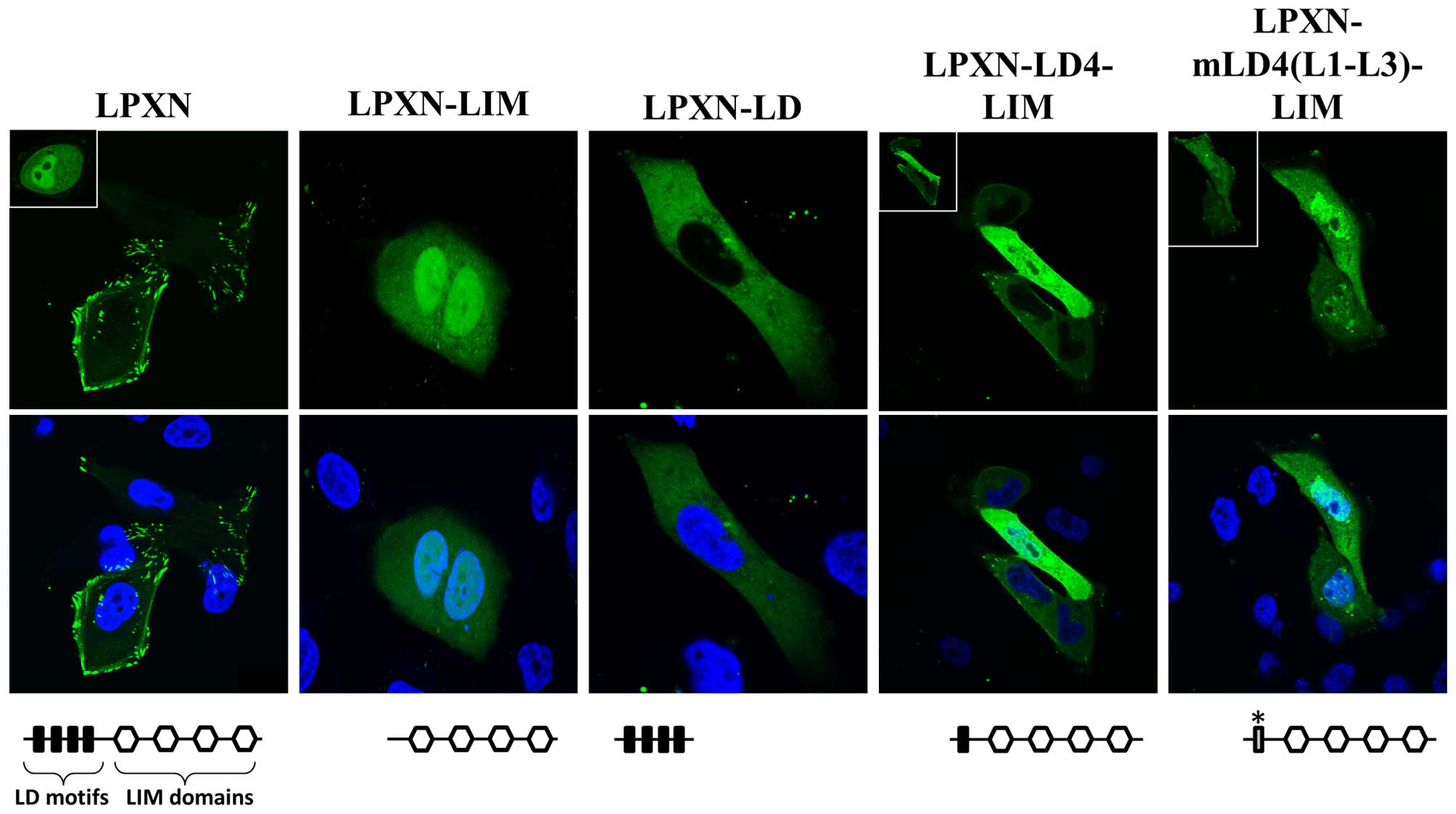
Furthermore, MDA-MB-231 cells were transfected with EGFP-constructs
coding for different EGFP-LPXN fusion proteins as indicated in
Fig. 4. The full-length EGFP-LPXN
fusion protein is located in the focal adhesion sites and in a
small proportion of the nucleus. EGFP-LPXN-LIM, which contains only
the LIM domains, localizes to the nucleus in 100% of the cells. If
the LD4 motif is present in the fusion protein (EGFP-LPXN-LD4-LIM)
only 53% of cells show a nuclear accumulation, but, if the LD4
motif is mutated, nuclear distribution is detectable in all
transfected cells (EGFP-LPXN-mLD4(L1-L3)-LIM) (Fig. 4). These studies clearly demonstrate
that leupaxin shuttles to the nucleus in breast cancer cells and
that mainly the LD4 is responsible for the nuclear export of
leupaxin.
Leupaxin interacts with the estrogen
receptors α and β
As it was shown that leupaxin interacts and
activates the androgen receptor in prostate cancer cells, a
putative interaction between leupaxin and the ERs α and β in breast
cancer cells was investigated using direct yeast-two-hybrid
experiments. Competent yeast cells of the strain AH109 were
transformed with plasmids coding for the full-length leupaxin
(LPXN), LPXN-LIM (containing only the LIM domains) or with LPXN-LD
(containing only the LD motifs) fused to the GAL4 activation domain
(AD) together with plasmids coding for the ERα and ERβ fused to the
DNA binding domain of the GAL4 transcription factor. Transformed
yeast cells were plated with or without estradiol on high
stringency drop-out plates containing α-Gal. An interaction of the
analysed proteins is reasoned upon growth of the yeasts and blue
colour development. As shown in Fig.
5 leupaxin interacts with the ERα only in the presence of
estradiol and via its LIM domains. In contrast, leupaxin binds to
the ERβ via its LIM domains in the absence and presence of
estradiol (Fig. 5). However, ERβ
showed interaction with the full-length leupaxin (LPXN) only in the
presence of estradiol.
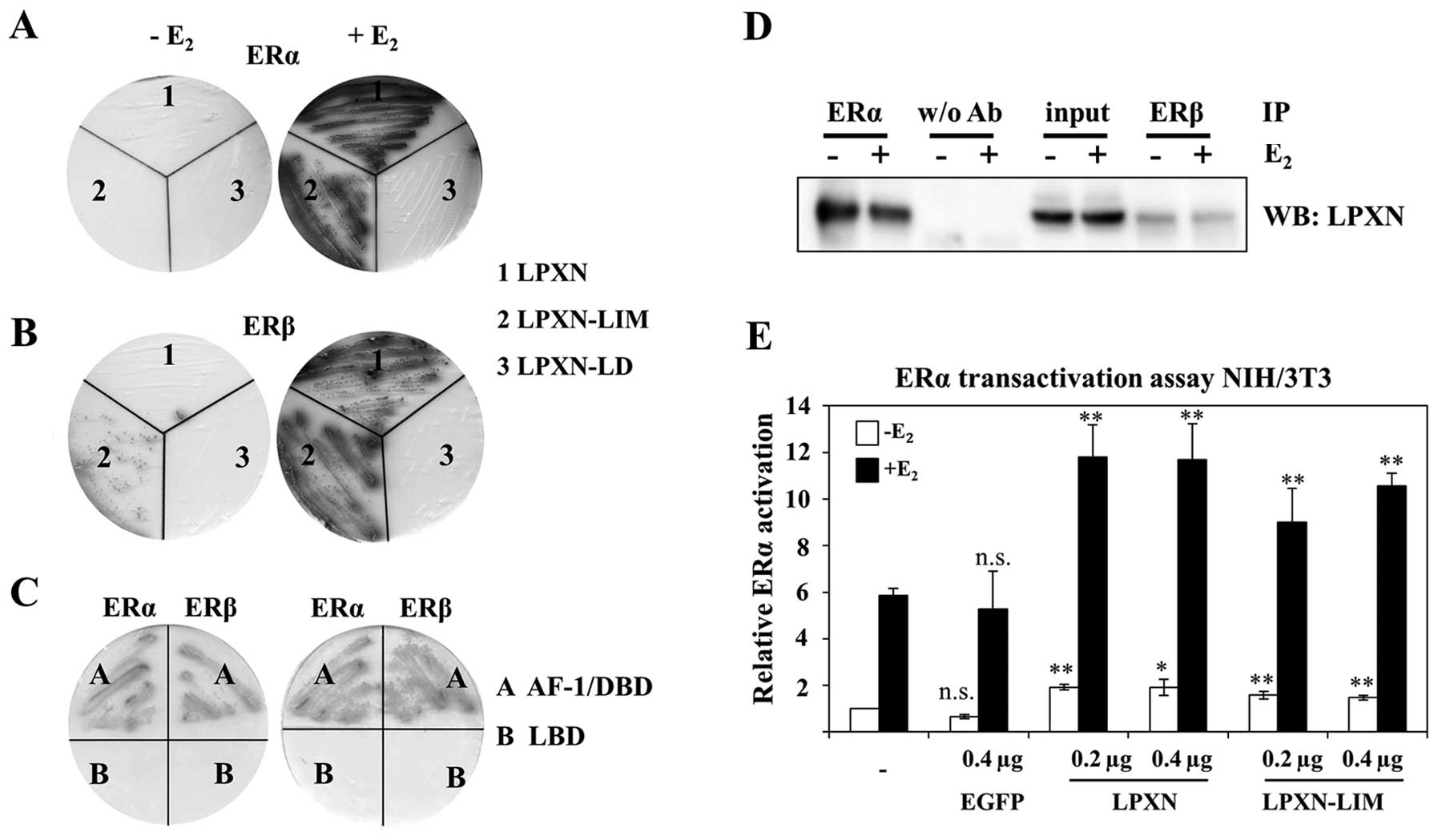 | Figure 5Leupaxin interacts with the estrogen
receptors α and β. (A and B) Direct yeast-two-hybrid experiments
were performed using full-length ERα (A) or full-length ERβ (B) and
leupaxin (1, full length), LPXN-LIM (2, containing all four LIM
domains) and LPXN-LD (3, containing all four LD motifs),
respectively. Transformed yeasts were plated on drop-out plates
(-LTHA) in the absence or presence of estrogen (E2).
Growth and blue staining of yeast show interaction of ERα and ERβ
with the indicated leupaxin protein, respectively. (C) Interaction
of leupaxin with ERs was further analysed with different ER
proteins. The N-terminal part of ERα and ERβ containing AF-1 and
the DNA binding domain (A) interacts with full-length leupaxin in
the absence and presence of E2 whereas no interaction
was observed using leupaxin and the ligand binding domain (LBD) (B)
of ERα and ERβ, respectively. (D) Co-immunoprecipitation
experiments verified interaction of leupaxin with ERα and ERβ in
HCC70 cells. In contrast to the yeast experiment an involvement of
estrogen was not observed. (E) Leupaxin enhances ERα
transcriptional activity. NIH3T3 cells were transfected with
plasmids pCMV-β-Gal, Vit-ERE-luc, pCDNA-ERα and the indicated
amount (0.2 or 0.4 μg DNA) of EGFP and EGFP-LPXN constructs in the
absence or presence of E2. Luciferase activity was
measured 36 h after transfection and normalized against
β-galactosidase activity. Three independent experiments were
performed. For statistical analysis Student’s t-test was applied to
compare with parental cells. *p≤0.05;
**p≤0.01; ***p≤0.001; NS, not
significant. |
To verify the interaction of leupaxin and ERα and
ERβ, respectively, coimmunoprecipitation experiments were
performed. HCC70 cells were incubated in the presence or absence of
estradiol and total protein was isolated. For immunoprecipitation
an ERα and an ERβ-specific antibody, respectively, and for western
blotting a leupaxin specific antibody were applied. An interaction
of leupaxin and the ERα and ERβ was observed in the presence and
absence of estradiol, contrary to the yeast-two-hybrid results. No
interaction was detectable in the control setting (without primary
antibody).
Leupaxin enhances the transcriptional
function of the ERα
To analyse the biological relevance of leupaxin-ERα
interactions a transactivation assay using pVit-ERE-Luc as reporter
construct was performed. NIH/3T3 cells were chosen for the assay to
rule out any influence of endogenous active estrogen receptors.
Cells were transfected with pVit-ERE-Luc along with the plasmids
EGFP-LPXN or EGFP-LPXN-LIM, respectively, and pcDNA-ERα.
Measurement of luciferase activity clearly demonstrates that
leupaxin increases the transcriptional activity of the ERα mainly
in the presence of estradiol. There is a slight but significant
increase of transcriptional activity visible without estradiol when
using the full-length leupaxin construct supporting the
co-immunoprecipitation studies.
Leupaxin influences migration and
invasion of breast cancer cells
To analyse the function of leupaxin in breast cancer
cells, MDA-MB-231 and HCC70 cells were transfected with two
leupaxin specific siRNAs (si-LPXN and si-LPXNst) and as a control
with siRNA against the firefly luciferase gene (si-Luc).
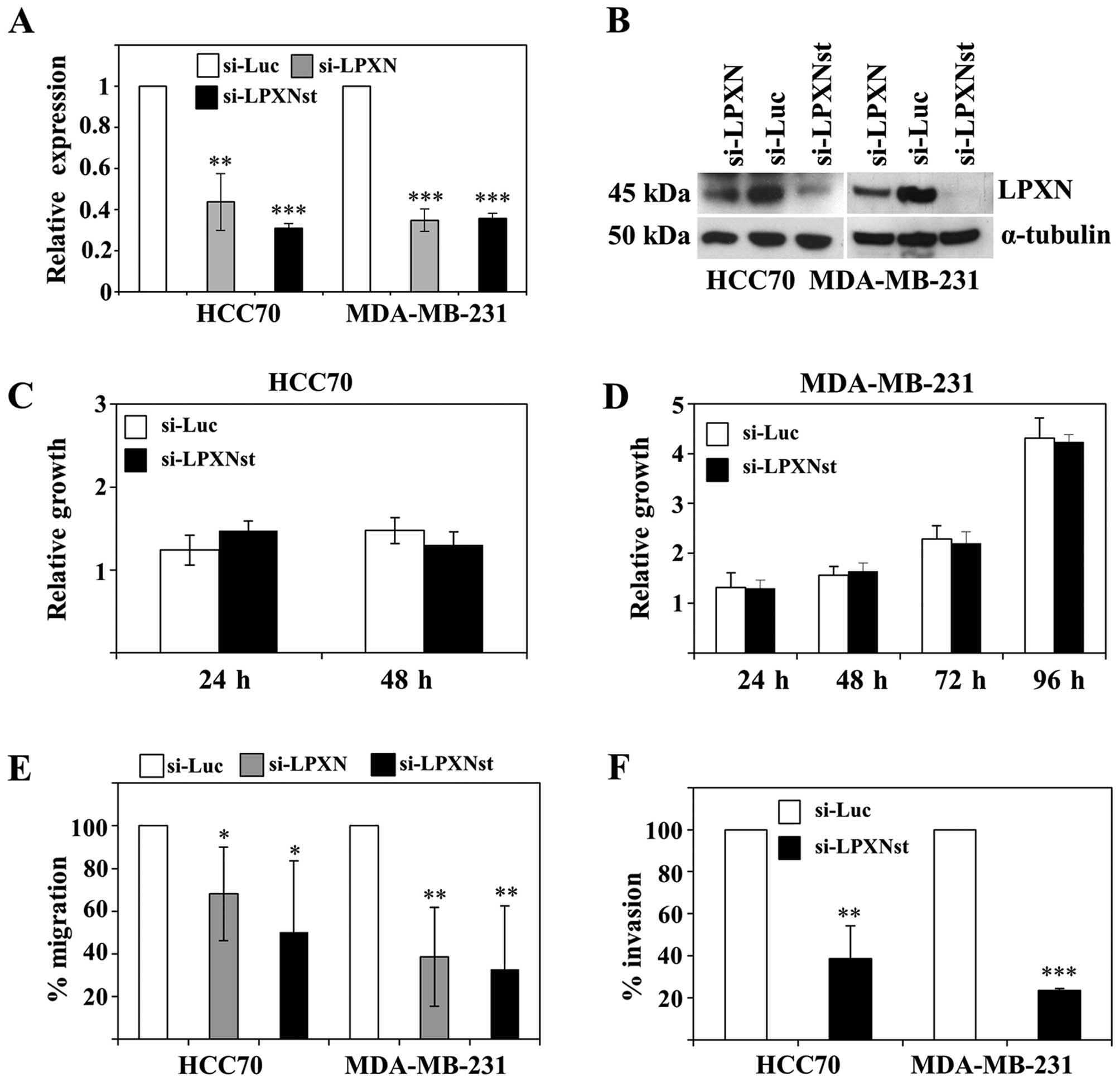
Downregulation of leupaxin expression was confirmed on the RNA and
on the protein level by using quantitative RT-PCR and western
blotting, respectively (Fig. 6A and
B), showing highest efficiency for si-LPXNst.
As already shown for prostate cancer cells leupaxin
has no influence on the proliferation of HCC70 and MDA-MB-231 cells
(Fig. 6C and D). Leupaxin
knockdown cells were subsequently analysed for their migratory
capability. HCC70 and MDA-MB-231 cells with downregulated leupaxin
expression show an ≤70% diminished migratory ability than control
transfected cells (Fig. 6E).
Furthermore, a Matrigel invasion assay revealed a 63 and 77%
reduced invasiveness of HCC70 and MDA-MB-231 cells, respectively,
with reduced leupaxin expression (Fig.
6F).
Discussion
Breast cancer development and progression is
critically influenced by ERs. Especially ERα, which is expressed in
~75% of breast cancers, is the most important target in endocrine
treatment strategies. Serial treatment at tumour progression with
different endocrine agents is the standard therapy strategy, often
resulting in a long period of disease control (6). However, most patients with advanced
breast cancer will develop resistance to endocrine therapy and
different mechanisms of resistance have been described (6,16).
In addition to modifications of the ERα itself, crosstalk with
growth factor receptor signalling pathways and the deregulation of
co-factors involved in the proper ERα signalling have been
described (7–9). In the present study, we provide
evidence that the focal adhesion protein leupaxin is involved in
the regulation of ER action as a co-factor and that it is expressed
in breast cancers, but not in normal breast epithelial cells. We
obtained the first hints of an involvement of leupaxin in breast
cancer from an array study containing patient matched tumour and
normal tissues of a variety of cancer types. Leupaxin was found to
be overexpressed in >50% of tumours in the breast and uterus,
whereas in lung cancer 71% of tumours showed downregulation of
leupaxin expression. Further analysis of leupaxin expression in 127
breast cancer specimens showed expression of leupaxin in 49, 40 and
22% of DCIS, DC and LC, respectively. However, there was no
significant correlation of leupaxin expression with the nodal, HER2
or ER/PR status. This result was also reflected in the analysis of
leupaxin expression in breast cancer cell lines with different
receptor status (Fig. 3). However,
more pronounced staining of leupaxin was observed mainly in DCs,
which represents the more advanced breast cancer stage. Paxillin
was also previously studied in breast cancers but with different
outcomes. Whereas no correlation of paxillin expression with ER, PR
and HER2 status in imprint smears of aggressive breast cancers was
found (17), Short et al
provided evidence that paxillin is overexpressed in 28% of breast
cancers, and that this correlates with HER2 status (18). In vitro studies concerning
paxillin function during breast cancer lung metastasis identified
paxillin as a key regulator of 3D adhesion assembly, stabilization
and disassembly (19). This
afore-mentioned study also showed a contribution of Hic-5 (TGFB1I1)
in the metastatic process, but up to date, to our knowledge, there
are no data of Hic-5 expression in human breast cancer
available.
To further evaluate if leupaxin plays an important
role also during breast cancer progression we used HCC70 and
MDA-MB-231 showing highest leupaxin expression for further
analyses. Neither paxillin nor Hic-5 interaction with ERs was
described (10,11). In contrast, leupaxin interacts via
its LIM domains with the N-terminal part of the ERs [comprising
activation function-1 (AF-1) and DNA binding domain]. However,
whereas the interaction of leupaxin with ERα in our
yeast-two-hybrid experiments was highly estrogen-dependent,
co-immunoprecipitation studies could not verify this observation.
Of note, the observed estrogen dependence of ERα/leupaxin
interaction was abrogated when we performed the yeast-two-hybrid
experiments with only the N-terminal part of the ERα which does not
contain the estrogen binding site. In addition, reporter gene
studies in NIH3T3 cells, which do not express ERα, showed that in
the presence of estrogens the activation of ERα through leupaxin is
enhanced, but the overexpression of leupaxin in the absence of
estrogens is also sufficient to increase ERα transcriptional
activity (Fig. 5E). These results
lead to the idea that the interaction of leupaxin with the ERα in
the cellular context might be regulated also through other factors
thereby determining the estrogen dependence of the interaction.
To influence transcriptional activity of steroid
hormone receptors paxillin proteins have to shuttle to the nucleus.
The precise import mechanisms are not fully understood, but it is
known that paxillin, Hic-5 and leupaxin contain a nuclear export
signal (NES) within the N-terminal LD motifs (13,20,21).
Staining of HCC70 and MDA-MB-231 cells with a leupaxin specific
antibody revealed mainly localization of leupaxin at focal adhesion
sites (Fig. 3C). A few cells with
overexpression of leupaxin as a EGFP fusion protein showed strong
accumulation of leupaxin in the nucleus as well (Fig. 4). From prostate cancer cells it is
known that the leupaxin-LD4 motif is most important for nuclear
export of leupaxin. Mutation of important amino acids within this
motif led to the accumulation of leupaxin in the nucleus in
MDA-MB-231 cells demonstrating that leupaxin also shuttles between
cytoplasm and nucleus in breast cancer cells.
To further explore the involvement of leupaxin in
breast cancer we knocked down leupaxin expression using previously
established siRNAs (13). We
showed that reduction of leupaxin expression did not result in
diminished cell proliferation. Instead, a clear reduction of
migration and invasion was observed. Of note, primary
cancer-derived cell lines MDA-MB-231 and HCC70 showed highest
expression of leupaxin. All other cell lines derived from breast
cancer metastases showed low or even no expression of leupaxin.
This fact points to the hypothesis, that leupaxin-mediated
invasiveness is more important for the extravasation, but not for
the intravasation of cells at metastatic sites. Expression analyses
for leupaxin in metastases of breast cancers may strengthen this
hypothesis. This second function of leupaxin is independent of its
ERα co-activation function as we observed the effects in
ERα-negative MDA-MB-231 cells as well.
In conclusion, we showed in the present study that
leupaxin is overexpressed in human breast cancers. Supported by the
results of the in vitro studies we postulate leupaxin to be
an important player during breast cancer progression. Therefore,
leupaxin and its involved genes and pathways could serve as
potential targets in the development of new therapeutic strategies
for breast cancer.
Acknowledgements
We thank N. Putzer and L.-M. Hartmund for excellent
technical assistance. This study was supported in part by the
Deutsche Forschungsgemeinschaft (KA 2942/1-1, 2942/1-2, BU 992/2-1,
BU 992/2-2).
References
|
1
|
Carlson RW, Anderson BO, Burstein HJ,
Carter WB, Edge SB, Farrar WB, Goldstein LJ, Gradishar WJ, Hayes
DF, Hudis CA, et al: Invasive breast cancer. J Natl Compr Canc
Netw. 5:246–312. 2007.PubMed/NCBI
|
|
2
|
Cufer T: Reducing the risk of late
recurrence in hormone-responsive breast cancer. Ann Oncol. 18(Suppl
8): viii18–viii25. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Greenberg PA, Hortobagyi GN, Smith TL,
Ziegler LD, Frye DK and Buzdar AU: Long-term follow-up of patients
with complete remission following combination chemotherapy for
metastatic breast cancer. J Clin Oncol. 14:2197–2205.
1996.PubMed/NCBI
|
|
4
|
Heldring N, Pike A, Andersson S, Matthews
J, Cheng G, Hartman J, Tujague M, Ström A, Treuter E, Warner M, et
al: Estrogen receptors: How do they signal and what are their
targets. Physiol Rev. 87:905–931. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Roger P, Sahla ME, Mäkelä S, Gustafsson
JA, Baldet P and Rochefort H: Decreased expression of estrogen
receptor beta protein in proliferative preinvasive mammary tumors.
Cancer Res. 61:2537–2541. 2001.PubMed/NCBI
|
|
6
|
Zhao M and Ramaswamy B: Mechanisms and
therapeutic advances in the management of endocrine-resistant
breast cancer. World J Clin Oncol. 5:248–262. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ali S and Coombes RC: Endocrine-responsive
breast cancer and strategies for combating resistance. Nat Rev
Cancer. 2:101–112. 2002. View
Article : Google Scholar
|
|
8
|
Musgrove EA and Sutherland RL: Biological
determinants of endocrine resistance in breast cancer. Nat Rev
Cancer. 9:631–643. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
9
|
Osborne CK, Bardou V, Hopp TA, Chamness
GC, Hilsenbeck SG, Fuqua SA, Wong J, Allred DC, Clark GM and Schiff
R: Role of the estrogen receptor coactivator AIB1 (SRC-3) and
HER-2/neu in tamoxifen resistance in breast cancer. J Natl Cancer
Inst. 95:353–361. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kasai M, Guerrero-Santoro J, Friedman R,
Leman ES, Getzenberg RH and DeFranco DB: The Group 3 LIM domain
protein paxillin potentiates androgen receptor transactivation in
prostate cancer cell lines. Cancer Res. 63:4927–4935.
2003.PubMed/NCBI
|
|
11
|
Fujimoto N, Yeh S, Kang HY, Inui S, Chang
HC, Mizokami A and Chang C: Cloning and characterization of
androgen receptor coactivator, ARA55, in human prostate. J Biol
Chem. 274:8316–8321. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Sundberg-Smith LJ, DiMichele LA, Sayers
RL, Mack CP and Taylor JM: The LIM protein leupaxin is enriched in
smooth muscle and functions as an serum response factor cofactor to
induce smooth muscle cell gene transcription. Circ Res.
102:1502–1511. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kaulfuss S, Grzmil M, Hemmerlein B, Thelen
P, Schweyer S, Neesen J, Bubendorf L, Glass AG, Jarry H, Auber B,
et al: Leupaxin, a novel coactivator of the androgen receptor, is
expressed in prostate cancer and plays a role in adhesion and
invasion of prostate carcinoma cells. Mol Endocrinol. 22:1606–1621.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Grzmil M, Kaulfuss S, Thelen P, Hemmerlein
B, Schweyer S, Obenauer S, Kang TW and Burfeind P: Expression and
functional analysis of Bax inhibitor-1 in human breast cancer
cells. J Pathol. 208:340–349. 2006. View Article : Google Scholar
|
|
15
|
Grzmil M, Thelen P, Hemmerlein B, Schweyer
S, Voigt S, Mury D and Burfeind P: Bax inhibitor-1 is overexpressed
in prostate cancer and its specific down-regulation by RNA
interference leads to cell death in human prostate carcinoma cells.
Am J Pathol. 163:543–552. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Renoir JM, Marsaud V and Lazennec G:
Estrogen receptor signaling as a target for novel breast cancer
therapeutics. Biochem Pharmacol. 85:449–465. 2013. View Article : Google Scholar
|
|
17
|
Panousis D, Patsouris E, Lagoudianakis E,
Pappas A, Kyriakidou V, Voulgaris Z, Xepapadakis G, Manouras A,
Athanassiadou AM and Athanassiadou P: The value of TOP2A, EZH2 and
paxillin expression as markers of aggressive breast cancer:
Relationship with other prognostic factors. Eur J Gynaecol Oncol.
32:156–159. 2011.PubMed/NCBI
|
|
18
|
Short SM, Yoder BJ, Tarr SM, Prescott NL,
Laniauskas S, Coleman KA, Downs-Kelly E, Pettay JD, Choueiri TK,
Crowe JP, et al: The expression of the cytoskeletal focal adhesion
protein paxillin in breast cancer correlates with HER2
overexpression and may help predict response to chemotherapy: A
retrospective immunohistochemical study. Breast J. 13:130–139.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Deakin NO and Turner CE: Distinct roles
for paxillin and Hic-5 in regulating breast cancer cell morphology,
invasion, and metastasis. Mol Biol Cell. 22:327–341. 2011.
View Article : Google Scholar :
|
|
20
|
Shibanuma M, Kim-Kaneyama JR, Ishino K,
Sakamoto N, Hishiki T, Yamaguchi K, Mori K, Mashimo J and Nose K:
Hic-5 communicates between focal adhesions and the nucleus through
oxidant-sensitive nuclear export signal. Mol Biol Cell.
14:1158–1171. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Dong JM, Lau LS, Ng YW, Lim L and Manser
E: Paxillin nuclear-cytoplasmic localization is regulated by
phosphorylation of the LD4 motif: Evidence that nuclear paxillin
promotes cell proliferation. Biochem J. 418:173–184. 2009.
View Article : Google Scholar
|