Introduction
The myocardin family members MRTF-A and MRTF-B (also
known as megakaryocytic acute leukemia and megakaryoblastic
leukemia-1/2) are well-established co-activators of serum response
factor (SRF) (1,2). Myocardin expression is restricted to
smooth and cardiac muscle lineages, whereas MRTF-A/B are expressed
ubiquitously. The activity of MRTF-A/B is regulated by their
translocation from the cytosol to the nucleus, which is triggered
by the activation of Rho family proteins (2). Like myocardin, MRTF-A/B are reported
to contribute to skeletal, cardiac and smooth muscle
differentiation (3). Their
importance in mammary myoepithelial differentiation (4,5) and
the epithelial-mesenchymal transition (6) has been established, but their
functions in cancer are absolutely different between myocardin and
MRTF-A/B. Myocardin functions as an effective inducer of growth
arrest and differentiation of some tumor and is frequently
repressed during human malignant transformation (3,7).
Interestingly, MRTF-A and MRTF-B have recently been implicated in
tumor cell invasion and metastasis (8). Suppression of MRTF-A and -B decreased
tumor cell motility in tumor xenografts in mice, while
proliferation of the cells was unaffected. In addition,
MRTF-depleted cancer cells failed to colonize the lungs after
injection in the tail vein of mice, probably due to a defect in
cell adhesion. Moreover, expression of an activated version of
MRTF-A was able to increase lung colonization of poorly metastatic
B16FO cells, leading to the conclusion that MRTF activity is a
critical step for tumor cell metastasis. However, roles of MRTF-A
and MRTF-B have not been reported in thyroid cancer up to now. In
this study, we found that MRTF-A expression was upregulated in
metastatic anaplastic thyroid cancer tissues, compared with primary
tumor tissues. MRTF-A overexpression promoted migration, invasion
and anoilds resistance in anaplastic thyroid cancer cells and
silencing inhibited its motility, invasion and anoilds
resistance.
MicroRNAs (miRNAs) are endogenous short non-coding
RNA molecules that regulate gene expression by repressing
translation or cleaving RNA transcripts in a sequence-specific
manner (9–11). Abnormal miRNA expression has been
linked to diseases, including cancer and it has been found
implicated in a multitude of cellular processes including
proliferation, differentiation, migration and apoptosis (11–17).
Many investigations have focused on the roles of
miR-206 in cancer. miR-206 is downregulated and inhibits cell
proliferation in breast cancer (18,19).
It blocks human rhabdomyosarcoma growth in xenotransplanted mice by
promoting myogenic differentiation (20). Downregulation of miR-206 promotes
proliferation and invasion of laryngeal cancer by regulating VEGF
expression (21) and it also
targets notch3, activates apoptosis, and inhibits tumor cell
migration and focus formation (22). Expression of the tumor suppressor
miR-206 is associated with cellular proliferative inhibition and
impairs invasion in ERα-positive endometrioid adenocarcinoma
(23). miR-206 is associated with
invasion and metastasis of lung cancer (24) and it can target c-Met
(25). Overexpression of miR-206
decreases the expression of metabolic genes and dramatically
impaired NADPH production, ribose synthesis, and in vivo
tumor growth in mice (26).
miR-206 inhibits gastric cancer proliferation in part by repressing
cyclin D2 (27). Herein, we found
that miR-206 was negatively associated with metastasis in
anaplastic thyroid cancer and it degraded MRTF-A by targeting its
3′-UTR in ARO anaplastic thyroid cancer cells. In addition, miR-206
overexpression inhibited invasion, migration and anoilds resistance
and silencing miR-206 promoted invasion, migration and anoilds
resistance in anaplastic thyroid cancer. More important,
restoration of MRTF-A abrogated pre-miR-206-mediated migration,
invasion and anoilds resistance regulation. Thus, we concluded that
miR-206 inhibited invasion, metastasis and anoilds-resistance by
degrading MRTF-A.
Materials and methods
Thyroid cancer tissues, ARO/FRO cell
lines
Nineteen thyroid cancer patients diagnosed with
anaplastic thyroid cancer were recruited from the Second Artillery
General Hospital of PLA and Tongji Hospital. The use of human
tissue samples followed internationally recognised guidelines as
well as local and national regulations. Research carried out on
humans follow international and national regulations. Medical
ethics committee approved the experiments undertaken. Informed
consent was obtained from each individual. Thyroid cancer cell
lines ARO and FRO was kindly donated by Dr Su Kai (Shanghai Cancer
Center, China). Briefly, cells were maintained in RPMI-1640 medium
supplemented with 5% fetal bovine serum (FBS) (Gibco, Grand Island,
NY, USA) and penicillin/streptomycin at 37°C in a humidified
atmosphere with 5% CO2.
MRTF-A expressing plasmids/empty vectors,
shMRTF-A/scramble, pre-miR-206/control miR, anti-miR-206/scramble
and transfection experiments
MRTF-A expressing plasmid/empty vector and shMRTF-A
plasmids/scramble were donated by Shao-Xin Huang (Boston
University, Boston, MA, USA) and was as described previously
(28). Pre-miR-206/control miR and
anti-miR-206/scramble were purchased from Ambion, Inc. (Ambion,
Austin, TX, USA). For transfection experiments, the cells were
cultured in serum-free medium without antibiotics at 60% confluence
for 24 h, and then transfected with transfection reagent
(Lipofectamine 2000, Invitrogen, Carlsbad, CA, USA) according to
the manufacturer’s instructions. After incubation for 6 h, the
medium was removed and replaced with normal culture medium for 48
h, unless otherwise specified.
Western blot analysis
Western blot analysis was performed as described
before (29). In brief, after
incubation with primary antibody anti-MRTF-A (1:500; Abcam,
Cambridge, MA, USA) and anti-β-actin (1:500; Abcam) overnight at
4°C, IRDye™-800 conjugated anti-rabbit secondary antibodies
(Li-COR, Biosciences, Lincoln, NE, USA) were used for 30 min at
room temperature. The specific proteins were visualized by Odyssey™
Infrared Imaging System (Gene Co., Lincoln, NE, USA).
Migration and invasion assay
For transwell migration assays,
2.5×104–5.3×104 cells were plated in the top
chamber with the non-coated membrane (24-well insert; pore size, 8
mm; BD Biosciences, San Jose, CA, USA). For invasion assays,
1.25×105 cells were plated in the top chamber with
Matrigel-coated membrane (24-well insert; pore size, 8 mm; BD
Biosciences). In both assays, cells were plated in medium without
serum or growth factors, and medium supplemented with serum was
used as a chemoattractant in the lower chamber. The cells were
incubated for 24 h and cells that did not migrate or invade through
the pores were removed by a cotton swab. Cells on the lower surface
of the membrane were stained with the Diff-Quick Staining Set
(Dade) and counted.
Wound healing assay
Cells (5×105) were seeded onto each 35-mm
glass bottom dish (MatTek Co., Ashland, MA, USA) and cultured at
37°C with 5% CO2 for 24 h. The confluent monolayer of
cells was wounded. Monolayers of cells were wounded with pipette
tips. After washing with warm PBS, the cells were incubated in
fresh culture medium. The wounded areas were photographed at the
beginning (0 h, top panels) and the end (10 h, bottom panels) of
the assay with Nikon inverted microscope (Eclipse TE-2000U, Nikon,
Japan) equipped with a video camera (DS-U1, Nikon, Japan).
3-(4,5-Dimethylthiazol-2-yl),
3,5-diphenyl-2H-tetrazolium bromide (MTT) assay
Cells seeded on 96-well plates, were stained at
indicated time point with 100 ml sterile MTT dye (0.5 mg/ml, Sigma,
St. Louis, MO, USA) for 4 h at 37°C, followed by removal of the
culture medium and addition of 150 ml of dimethyl sulphoxide (DMSO)
(Sigma). The absorbance was measured at 570 nm, with 655 nm as the
reference wavelength.
miRNA microarray
Total RNA from cultured cells, with efficient
recovery of small RNAs, was isolated using the mirVana miRNA
Isolation kit (Ambion). cRNA for each sample was synthesized by
using 3′ IVT Express kit (Affymetrix, Santa Clara, CA, USA)
according to the manufacturer’s protocols. The purified cRNA was
fragmented by incubation in fragmentation buffer (provided in the
3′IVT express kit) at 95°C for 35 min and chilled on ice. The
fragmented labeled cRNA was applied to MicroRNA2.0 array
(Affymetrix) and hybridized in Genechip hybridization oven 640
(Affymetrix) at 45°C for 18 h. After washing and staining in
Genechip fluidics station 450 (Affymetrix), the arrays were scanned
by using Genechip scanner 3000 (Affymetrix). The gene expression
levels of samples were normalized and compared by using Partek GS
6.5 (Partek, Inc., St. Louis, MO, USA). Average-linkage
hierarchical clustering of the data was applied by using the
Cluster (Eisen et al, Stanford, Stanford University, CA,
USA; http://rana.lbl.gov) and the results were
displayed by using TreeView (Eisen et al, Stanford, Stanford
University, CA, USA; http://rana.lbl.gov).
Methods of bioinformatics
The analysis of potential microRNA target sites were
performed using common prediction algorithms - TargetScan
(http://www.targetscan.org) and PicTar
(http://pictar.mdc-berlin.de/).
Immunofluorescence analyses
For immunofluorescence analyses of cells were plated
on glass coverslips in 6-well plates and transfected with 30 nM
pre-miR-206 or control miR. At 36 h after transfection, coverslips
were stained with the mentioned anti-MRTF-A antibodies. Alexa Fluor
488 goat anti-rabbit IgG antibody was used as secondary antibody
(Invitrogen). Coverslips were counterstained with DAPI
(Invitrogen-Molecular Probes, Eugene, OR, USA) for visualization of
nuclei. Microscopic analysis was performed with a confocal
laser-scanning microscope (Leica Microsystems, Bensheim, Germany).
Fluorescence intensities were measured in a few viewing areas for
200–300 cells per coverslip and analyzed using ImageJ 1.37v
software (http://rsb.info.nih.gov/ij/index.html).
Reverse-transcription polymerase chain
reaction (RT-PCR) and quantitative real-time RT-PCR (qRT-PCR) for
MRTF-A
Total RNA was isolated from cells using TRIzol
reagent (Invitrogen). First-strand cDNA was synthesized from the
total RNA using M-MLV reverse transcriptase (Promega, Madison, WI,
USA) and random hexamer primers (Sangon, Shanghai, China). The
thermal cycle profile was as follows: denaturation for 30 sec at
95°C, annealing for 45 sec at 52–58°C depending on the primers
used, and extension for 45 sec at 73°C. PCR products were
visualized on 2% agarose gels stained with ethidium bromide under
UV transillumination. qRT-PCR was performed with a Power SYBR Green
PCR Master Mix (Applied Biosystems, Carlsbad, CA, USA) according to
the manufacturer’s protocol. The primer sequences for MRTF-A:
forward primer 5′-ATGGAGCTGGTGGAGAAGAATATC-3′ and reverse
5′-GAAGGAGGAACTGTCTGCTACC-3′.
Real-time PCR for miRNA
Total RNA from cultured cells, with efficient
recovery of small RNAs, was isolated using the mirVana miRNA
Isolation kit (Ambion). Detection of the mature form of miRNAs was
performed using the mirVana qRT-PCR miRNA Detection kit, according
to the manufacturer’s instructions (Ambion). The U6 small nuclear
RNA was used as an internal control.
Luciferase reporter assay
The 3′-untranslated region (3′-UTR) of human MRTF-A
mRNA was cloned in pRL-TK (Promega) using PCR-generated fragment.
Site-directed mutagenesis of the predicted target-site of miR-206
in the MRTF-A-3′-UTR was carried out using Quik change-mutagenesis
kit (Stratagene, Heidelberg, Germany), with MRTF-A-WT-luc as a
template. For reporter assays, cells was transiently transfected
with WT or mutant reporter plasmid and microRNA or anti-microRNA
using Lipofectamine 2000 (Invitrogen). Reporter assays were
performed 36 h post-transfection using the
Dual-luciferaseassay-system (Promega), and normalized for
transfection efficiency by cotransfected Renilla-luciferase.
Anoikis assays
Anoikis resistance was evaluated by seeding
7.5×104 cells in ultralow attachment plates (Corning).
After 24 h of anchorage-independent culture, cells were transfected
as indicated and resuspended in 0.4% trypan blue (Sigma) and cell
viability was assessed.
Statistical analysis
Data are presented as mean ± SEM Student’s t-test
(two-tailed) was used to compare two groups (P<0.05 was
considered significant), unless otherwise indicated (χ2
test).
Results
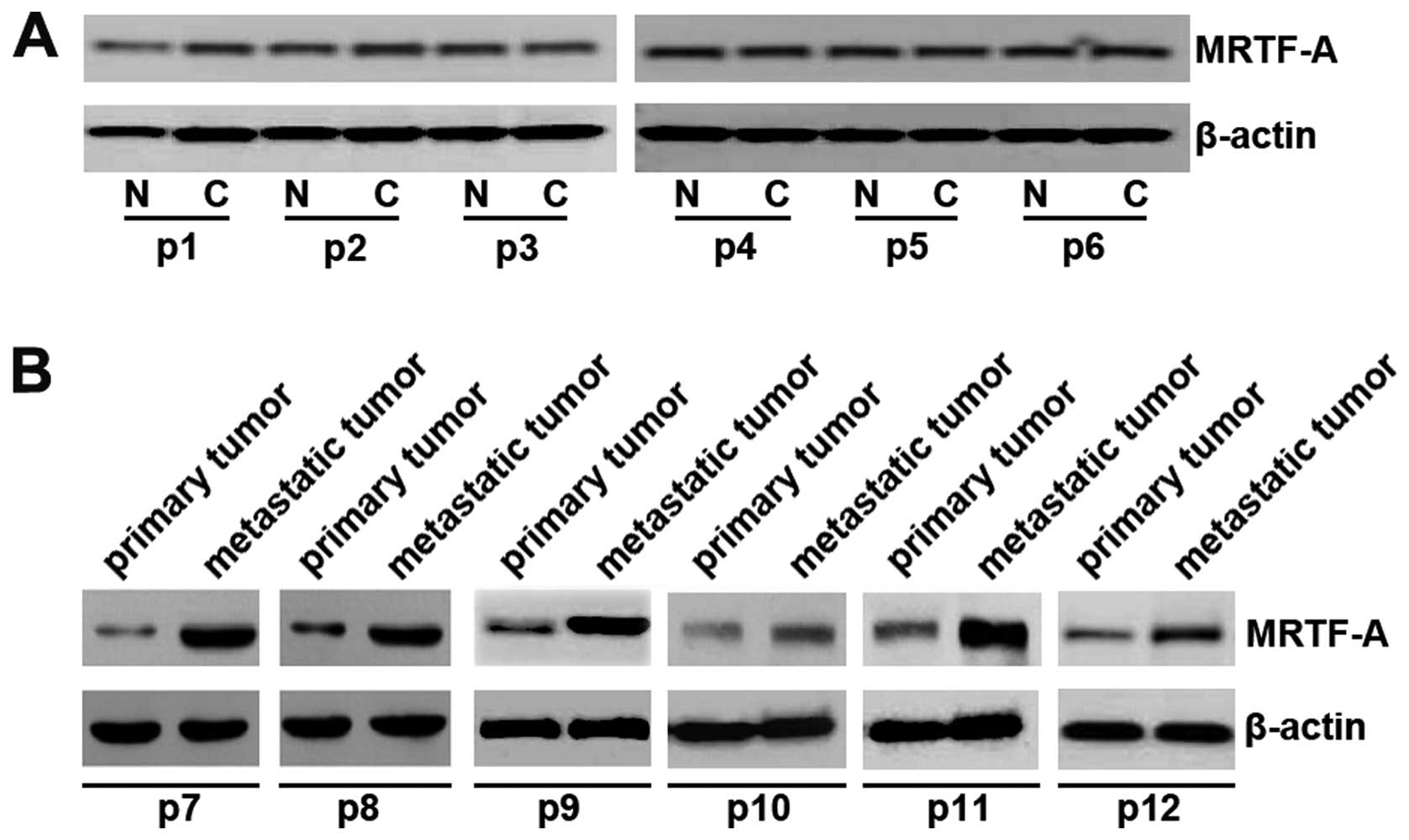
MRTF-A expression was upregulated in
metastatic anaplastic thyroid cancer
In an attempt to identify MRTF-A expression between
thyroid cancer tissues and adjacent normal tissues, we performed
western blotting in cancer tissues versus normal tissues. Protein
was isolated from 6 pairs of thyroid cancer tissues and normal
tissues (patients no. 1–6). However, we did not detect any
difference between cancer tissues and adjacent normal tissues
(Fig. 1A). Because MRTF-A was
recently implicated in tumor invasion and metastasis, we
hypothesized that MRTF-A was also associated with invasion and
metastasis in thyroid cancer (8).
Thus, using western blot assay, we detected MRTF-A expression
between primary thyroid cancer tissues and metastatic thyroid
cancer tissues obtained from further 6 patients (patients no.
7–12). Cancer metastasis has occurred in all the 6 patients.
Although we did not detect any difference between cancer tissues
and normal tissues, we found that MRTF-A protein was significantly
increased in metastatic thyroid cancer tissues, compared with
primary thyroid cancer (Fig. 1B).
All the results implied that MRTF-A was associated with metastasis
of anaplastic thyroid cancer.
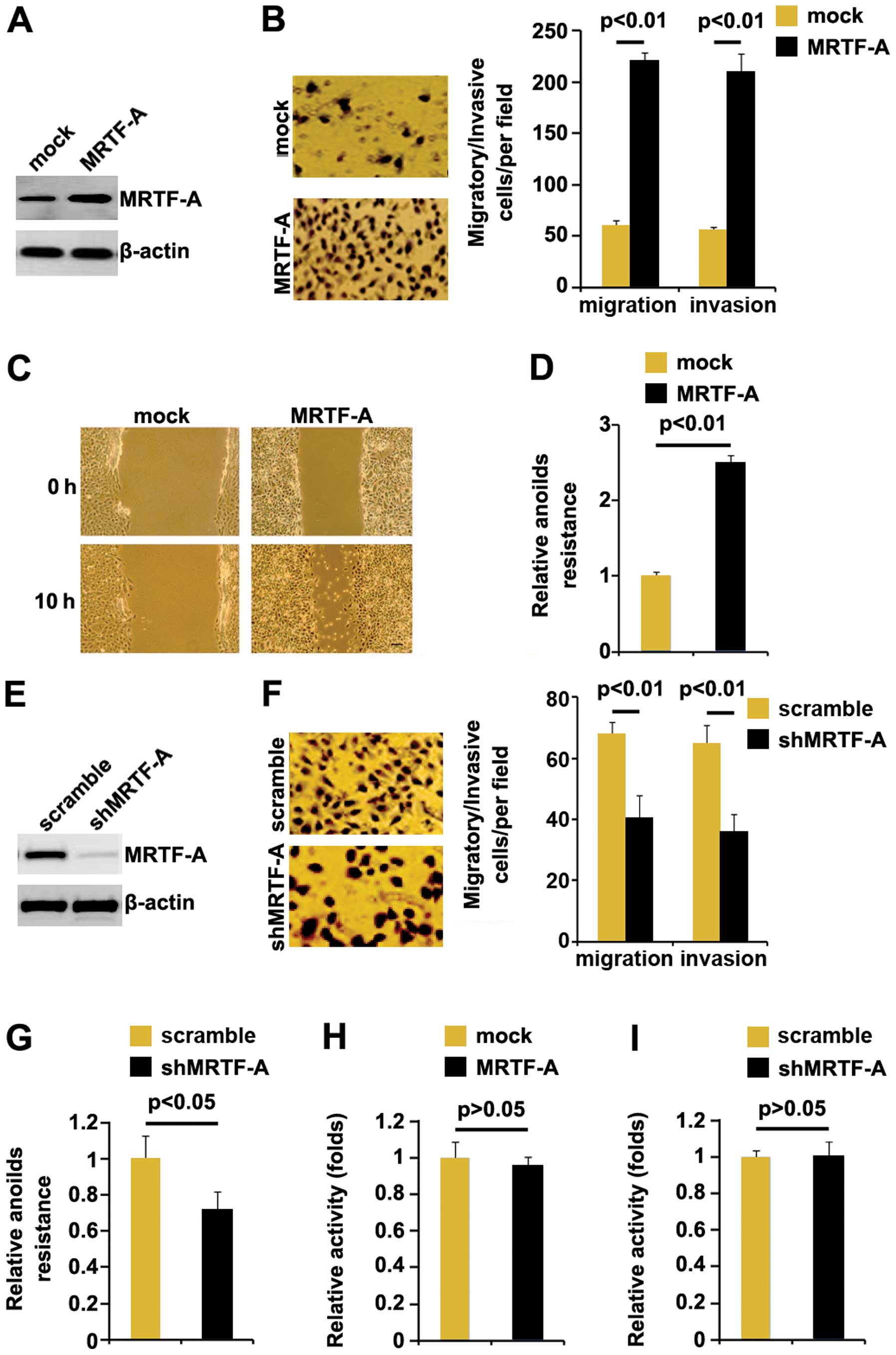
MRTF-A promotes metastasis-relevant
traits in thyroid cancer cells
In an attempt to identify the role of MRTF-A in
regulating migration and invasion of ARO cells, the cells were
transfected with MRTF-A expressing plasmids. After stable
transfection, MRTF-A protein expression was detected by western
blotting and the results showed that MRTF-A protein was increased
by MRTF-A expressing plasmids in the cells (Fig. 2A). Next, we performed migration and
invasion assay to detect migration and invasion of ARO cells
transfected with MRTF-A expressing plasmids and empty vectors.
Ectopic MRTF-A promoted motility and invasion by ~4-fold in the
cells (Fig. 2B). To confirm the
results, wound-healing assay was performed. Wound-healing assay
showed that MRTF-A significantly promoted motility in the cells
(Fig. 2C). To further show the
effects of MRTF-A on metastasis, we performed anoikis assays to
analyze its effects on anoikis resistance. Also, MRTF-A-transfected
cells exhibited ~150% increased resistance to anoikis-mediated cell
death (Fig. 2D). Having
demonstrated that MRTF-A overexpression promoted migration and
invasion in ARO cells, to provide further evidence that the roles
of MRTF-A were involved in migration and invasion, we studied the
effects of shMRTF-A, an inhibitor of MRTF-A. After stable
transfection, MRTF-A expression was detected by western blotting.
The results showed that shMRTF-A significantly downregulated MRTF-A
expression in ARO cells (Fig. 2E).
Next, we performed migration and invasion assay to detect migration
and invasion of ARO cells transfected with shMRTF-A plasmids and
scramble. Silencing MRTF-A inhibited motility and invasion by
~0.6-fold (Fig. 2F). To further
confirm the roles of shMRTF-A on metastasis, we performed anoikis
assays to analyze its effects on anoikis resistance. Contrary to
MRTF-A overexpression, shMRTF-A-transfected cells exhibited ~30%
decreased resistance to anoikis-mediated cell death (Fig. 2G).
We also performed MTT assay to detect whether it
affected proliferation in ARO cells. Although MRTF-A was associated
with metastasis in thyroid cancer patients and promoted migration
and invasion in thyroid cancer ARO cells, neither overexpressing
MRTF-A nor silencing it affected the cell viability (Fig. 2H and I).
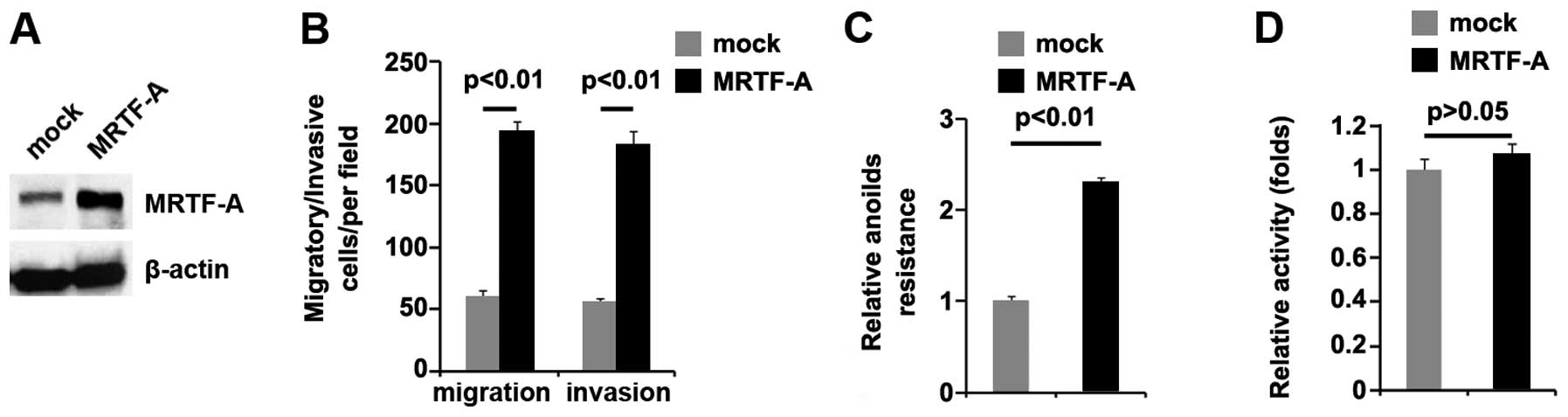
The consequences of MRTF-A expression were not
unique to ARO cells: MRTF-A protein was not only increased by
MRTF-A expressing plasmids (Fig.
3A), but also promoted invasion (Fig. 3B), motility (Fig. 3B), and anoikis resistance (Fig. 3C), yet did not affect proliferation
(Fig. 3D), in FRO human thyroid
cancer cells. Hence, MRTF-A promotes in vitro surrogates of
metastatic ability in thyroid cancer.
miR-206 degrades MRTF-A in thyroid cancer
cells
Having demonstrated that MRTF-A expression is
specifically upregulated in metastatic thyroid cancer and it
promotes metastasis-relevant traits in vitro, MRTF-A
expression in metastatic thyroid cancer was studied for clarifying
the mechanisms promoting MRTF-A expression in the disease.
MicroRNAs (miRNAs) are a new class of small (~22 nucleotide)
noncoding RNAs and negatively regulate protein-coding gene
expression by targeting mRNA degradation or translation inhibition
(9–11). Downregulation of specific miRNA can
contribute to oncogene overexpression (30). Thus we evaluated whether MRTF-A was
upregulated by defection of specific miRNA in metastatic thyroid
cancer.
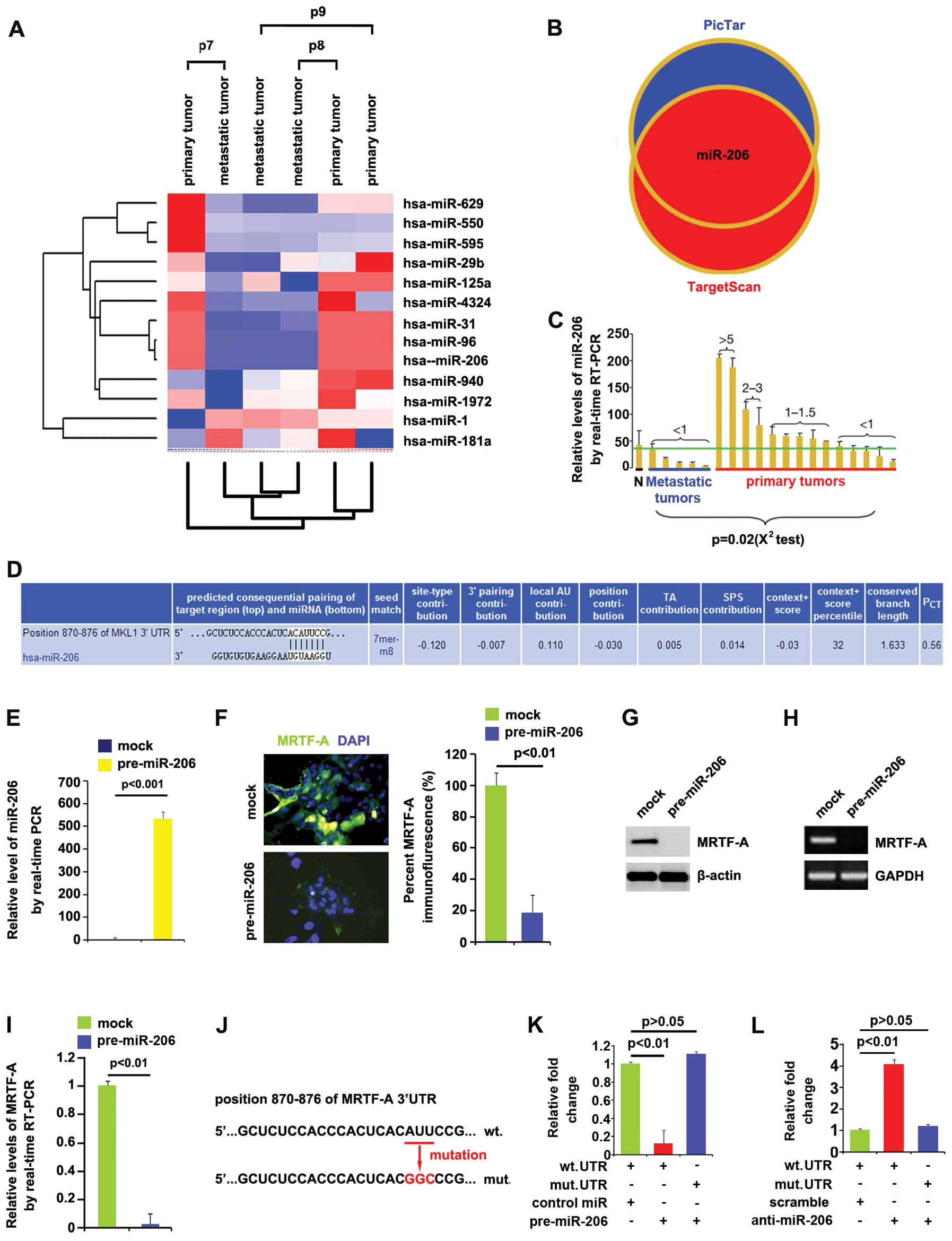
In an attempt to identify the level of miRNA
expression in primary thyroid cancer and metastatic thyroid
tissues, we performed miRNA profiling. RNAs isolated from 3 pairs
of primary tumors and metastatic tumors were hybridized to a custom
miRNA microarray platform. After hybridization, quantification, and
normalization, we found that miR-206, miR-31 and miR-96 were
significantly decreased in the metastatic tumors compared with
primary tumors >100-fold (Fig.
4A).
As further confirmation, we used two common
prediction algorithms - TargetScan (http://www.targetscan.org) and PicTar (http://pictar.mdc-berlin.de/) to analyze 3′-UTR of
MRTF-A. All 2 algorithms predicted that miR-206 could target 3′-UTR
of MRTF-A (Fig. 4B). In order to
further study whether that miR-206 expression was associated with
metastasis of thyroid cancer, we performed real-time PCR to detect
miR-206 expression in 5 metastatic tumors and 14 primary tumors.
Consistent with the results of miRNA microarray, real-time PCR
demonstrated that miR-206 expression was significantly
downregulated in metastatic tumors (Fig. 4C). Target sites on 3′-UTR of MRTF-A
are shown in Fig. 4D. We reasoned
that miR-206 could downregulate MRTF-A expression by targeting its
3′-UTR in thyroid cancer and that MRTF-A was overexpressed in
thyroid cancer, due to lack of miR-206.
In an attempt to identify the role of miR-206 in
regulating MRTF-A expression in ARO cells, cells were transfected
with pre-miR-206 and control miR. After transfection, miR-206
expression was detected by real-time PCR and the results showed
that miR-206 was increased by the pre-miR-206 in the cells
(Fig. 4E). To confirm the reason,
we performed immunofluorescence analyses in ARO cells transfected
with pre-miR-206 or control miR. The results showed that MRTF-A
protein was evidently suppressed in the cells transfected with
pre-miR-206 (Fig. 4F). We next
performed RT-PCR and western blotting to detect MRTF-A expression
in ARO cells transfected with pre-miR-206 or control miR. The
results showed that MRTF-A protein (Fig. 4G) and mRNA (Fig. 4H) were significantly downregulated
in the cells transfected with pre-miR-206. Consistent with the
results of RT-PCR, real-time PCR demonstrated that MRTF-A mRNA was
reduced in ARO cells transfected with pre-miR-206, compared with
control miR-transfected groups (Fig.
4I).
To further demonstrate the direct regulation of
MRTF-A by miR-206, we constructed luciferase reporters with the
targeting sequences of wild-type (MRTF-A-WT-luc) and mutated MRTF-A
3′-UTRs (MRTF-A-MUT-luc) (Fig.
4J). Both the wild-type and mutant reporters were introduced
into ARO cells.
Luciferase reporter assay showed that the luciferase
activities of MRTF-A-WT-luc plasmids were significantly suppressed
in the cells transfected with pre-miR-206, implying that miR-206
targeted 3′-UTR of MRTF-A mRNA (Fig.
4K). In order to further identify that miR-206 targeted 3′-UTR
of MRTF-A by the predicted sites, we mutated 3 bases in the
predicted sites (Fig. 4J). In
addition, mutant reporters were introduced into ARO cells, as
expected the luciferase activities of MRTF-A-MUT-luc were not
suppressed by miR-206 in ARO cells (Fig. 4K).
Having demonstrated that miR-206 overexpression
inhibited MRTF-A-WT-luc plasmids by the predicted sites, we next
studied whether silencing miR-206 could affect activity of the
MRTF-A-WT-luc plasmids. Thus luciferase reporters assay was
performed again and the results showed that contrary to
pre-miR-206, anti-miR-206 significantly promoted luciferase
activity of MRTF-A-WT-luc in ARO cells (Fig. 4L). Moreover, mutant reporters were
also introduced into ARO cells, but the luciferase activities of
MRTF-A-MUT-luc were not affected by anti-miR-206 in ARO cells
(Fig. 4L). All the data
illustrated that miR-206 degraded MRTF-A by targeting the specific
sites predicted in silico in anaplastic thyroid cancer
cells.
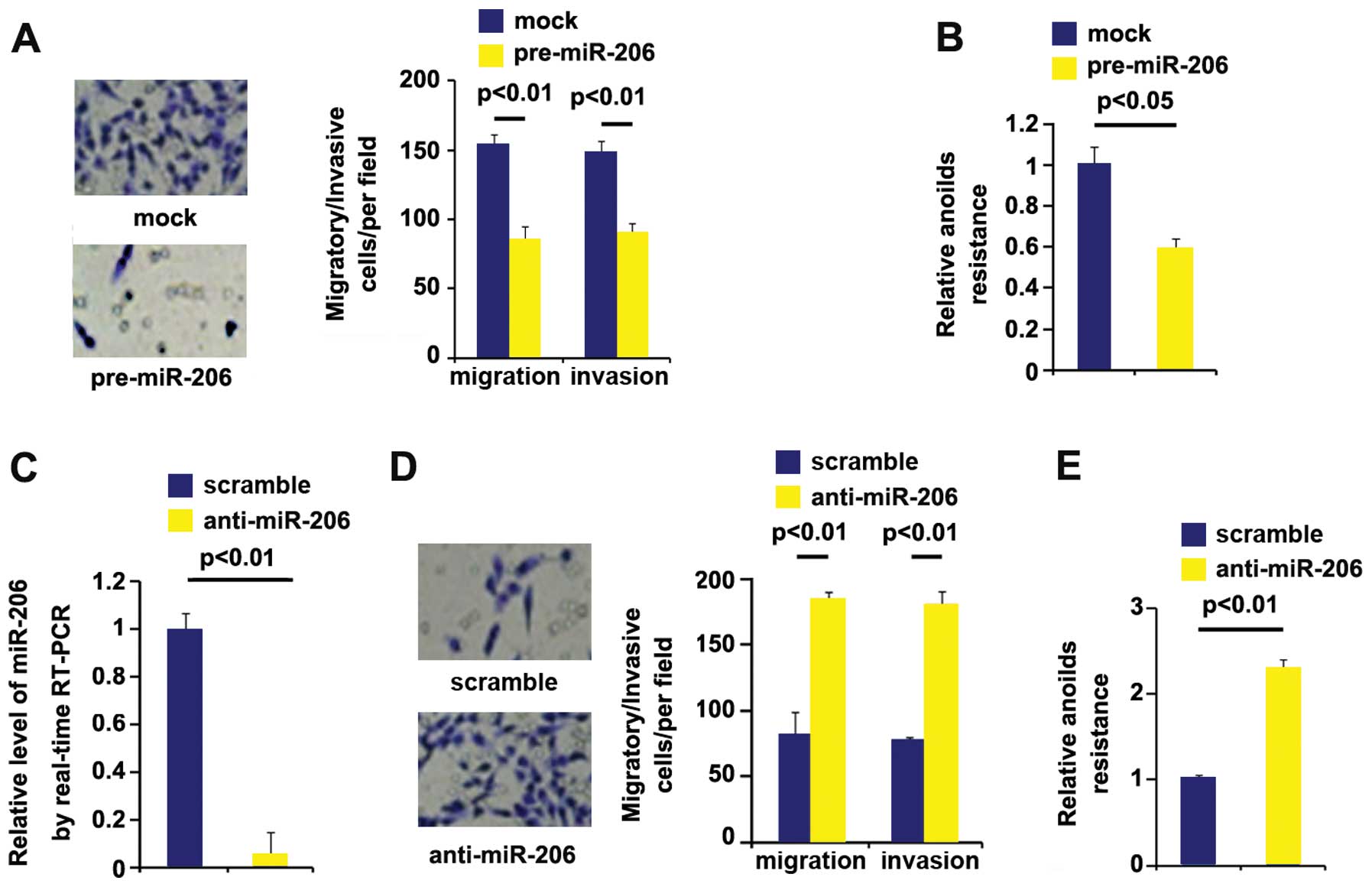
miR-206 overexpression inhibits invasion
and migration and silencing miR-206 promotes migration and invasion
in thyroid cancer
Having demonstrated that miR-206 was decreased in
the metastatic tumors compared with primary tumors and it could
degrade metastasis-relevant MRTF-A expression, we reasoned that it
was also associated with metastasis-relevant traits in thyroid
cancer cells. It was confirmed that miR-206 could be increased by
pre-miR-206 (Fig. 4E). Next, we
performed migration and invasion assay to detect migration and
invasion of ARO cells transfected with pre-miR-206 and control miR.
Ectopic miR-206 inhibited motility and invasion by about 2-fold
(Fig. 5A). To further show the
effects of miR-206 on metastasis, we performed anoikis assays to
analyze its effects on anoikis resistance. Also, miR-206
overexpressing cells exhibited 40% decreased resistance to
anoikis-mediated cell death (Fig.
5B).
Having demonstrated that miR-206 overexpression
inhibits metastasis-relevant traits in ARO cells, to provide
further evidence that miR-206 was involved in ARO cell migration
and invasion, we studied the effects of anti-miR-206, an inhibitor
of miR-206. After stable transfection, miR-206 expression was
detected by real-time PCR. The results showed that anti-miR-206
significantly downregulated miR-206 expression in ARO cells
(Fig. 5C). Next, we performed
migration and invasion assay to detect migration and invasion of
ARO cells transfected with anti-miR-206 and scramble. Silencing
miR-206 promotes motility and invasion by more than 2-fold
(Fig. 5D). To further confirm that
the roles of anti-miR-206 on migration and invasion, we performed
anoikis assays to analyze its effects on anoikis resistance.
Contrary to miR-206 overexpression, anti-miR-206-transfected cells
exhibited more than 2-fold increased resistance to anoikis-mediated
cell death (Fig. 5E).
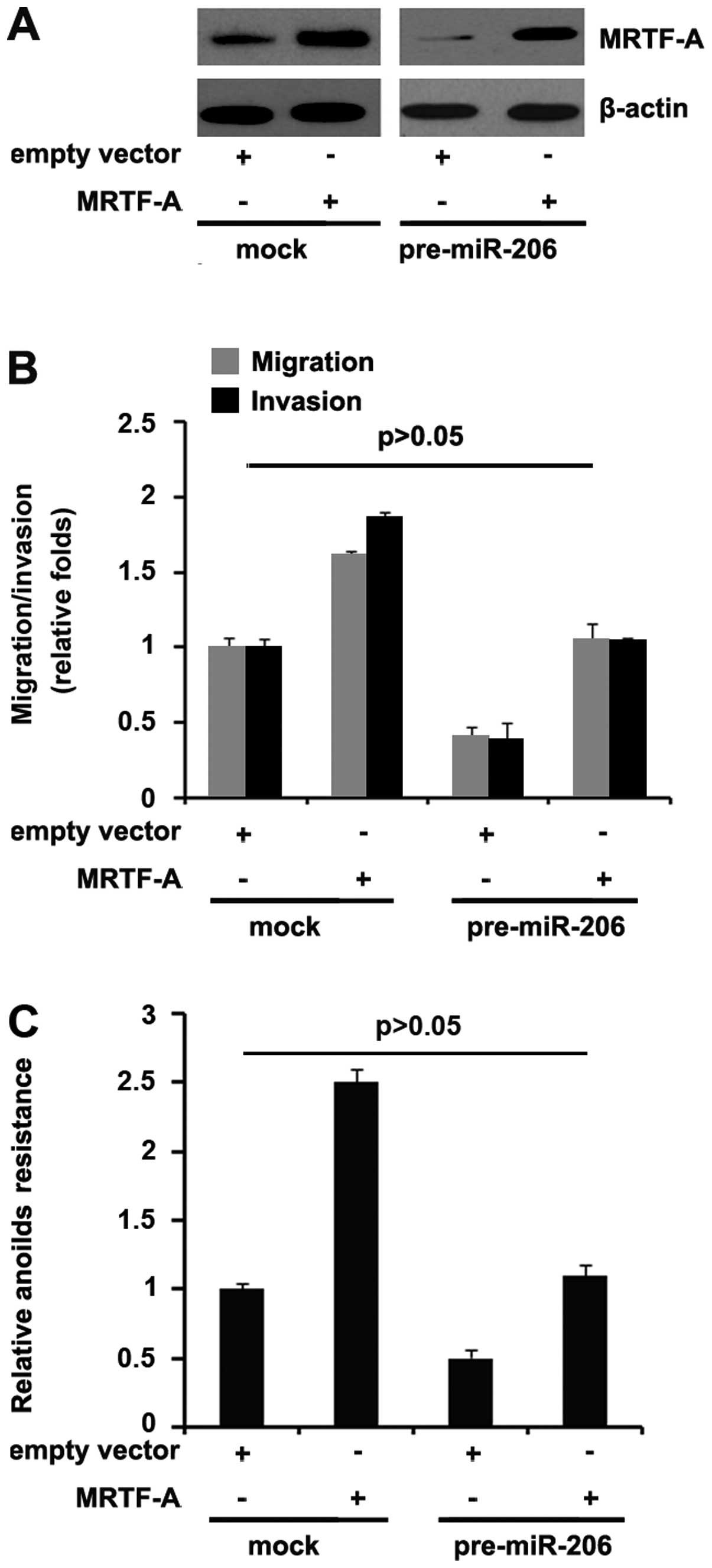
MRTF-A abrogates the regulation of
pre-miR-206-mediated migration and invasion
Because miR-206 directly degraded MRTF-A through its
3′-UTR, we reasoned that ectopic expression of MRTF-A by
transfection of the cDNA that did not contain the predicted target
of 3′-UTR should escape the regulation of miR-206 and thus
attenuate or eliminate miR-206 function. To this end, we
transfected MRTF-A expressing plasmids or empty vector (mock) into
control miR or pre-miR-206-treated ARO cells. Immunoblot analysis
revealed that transfection of MRTF-A expressing plasmids eliminated
the effect of miR-206 on MRTF-A protein (Fig. 6A).
Because overexpression of miR-206 in ARO cells
inhibited motility and invasion, and in order to identify whether
MRTF-A could abrogate or attenuate the roles of miR-206 on motility
and invasion, control miR or pre-miR-206-treated ARO cells were
transfected with either MRTF-A expressing plasmids or empty vectors
(mock), we performed migration and invasion assay and then found
that pre-miR-206-treated ARO cells displayed >50% decrease in
migration and invasion compared to control miR-treated cells
(Fig. 6B). Restoration of MRTF-A
sufficed to reverse the loss of migration and invasion (Fig. 6B) as well as resistance to
anoikis-mediated cell death (Fig.
6C) observed in pre-miR-206-treated cells. Thus, we concluded
that miR-206 inhibited migration and invasion via degrading MRTF-A
expression.
Discussion
Gene expression is fine-tuned and tightly regulated
through complex transcriptional signaling networks involving
interactions of miRNA and target genes (31–36).
Megakaryoblastic leukemia 1 (MKL1) is a member of the
myocardin-related transcription factor (MRTF) family and functions
as a co-activator for serum response factor (SRF), which regulates
essential biological processes ranging from gastrulation and
development to cell survival and apoptosis (37–39).
Myocardin-related transcription factor-A (MRTF-A),
also termed megakaryocytic acute leukemia (MAL), basic, SAP and
coiled-coil (BSAC), and MKL1, was originally identified as a
chromosome 22 encoded fusion partner of the t(1;22)(p13;q13)
translocation causing acute megakaryoblastic leukemia (AMKL) in
infants and children (40,41). As a result of the translocation,
the MRTF-A gene is fused to the RNA-binding motif protein 15
(RBM15), also known as OTT gene, on chromosome 1 (40,41).
This fusion gene encodes the deregulated protein RBM15-MKL1 with
potential oncogenic properties (42,43).
MRTF-A was also identified in a screen for genes that protect
against tumor necrosis factor-induced cell death, and named BSAC
(44). However, its roles still
keep emerging in thyroid cancer. Consistent with previous reports
that MRTF-A promoted tumor cell invasion and metastasis (8,45,46),
we found that MRTF-A expression was not only upregulated in
metastatic thyroid cancer tissues, but its overexpression promoted
metastasis-relevant traits in thyroid cancer cells. To further
indentify that MRTF-A also promotes the metastasis-relevant traits
in vivo will strengthen the conclution. First steps in the
development of novel pharmacological tools targeting
transcriptional responses of MRTF-A signaling in cancer were made
with the recent development of compounds that selectively target
the Rho/MKL1 signaling and inhibit the invasion of prostate cancer
cells in a Matrigel model of metastasis (47). Given the incontestable involvement
of MRTF-A in tumorigenesis and the effects it exerts on tumor cell
invasion and metastasis, it will be important to assess how MRTF-A
is integrated within the major signaling pathways that communicate
extracellular signals and mechanical cues to the transcriptional
machinery to alter the motility of cancer cells.
Elucidating the mechanism by which miR-206 inhibits
metastasis-relevant traits in anaplastic thyroid cancer by
downregulating MRTF-A will help us to better understand the
mechanism of invasion and metastasis. We found miR-206 was
significantly downregulated in metastatic thyroid cancer and had
potential to target 3′-UTR of MRTF-A mRNA. Following studies
confirmed that miR-206 degraded MRTF-A by targeting its 3′-UTR.
Thus, restoration of miR-206 may represent a promising therapeutic
way to suppress MRTF-A-mediated metastasis. However, the roles of
miR-206 also need to be further confirmed in vivo.
Recognition of the differential regulation and
function of MRTF-A in tumors will ultimately provide a better
understanding of the signaling pathways that can be therapeutically
modulated.
References
|
1
|
Wang DZ, Li S, Hockemeyer D, Sutherland L,
Wang Z, Schratt G, Richardson JA, Nordheim A and Olson EN:
Potentiation of serum response factor activity by a family of
myocardin-related transcription factors. Proc Natl Acad Sci USA.
99:14855–14860. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Miralles F, Posern G, Zaromytidou AI and
Treisman R: Actin dynamics control SRF activity by regulation of
its coactivator MAL. Cell. 113:329–342. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Pipes GC, Creemers EE and Olson EN: The
myocardin family of transcriptional coactivators: Versatile
regulators of cell growth, migration, and myogenesis. Genes Dev.
20:1545–1556. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Li S, Chang S, Qi X, Richardson JA and
Olson EN: Requirement of a myocardin-related transcription factor
for development of mammary myoepithelial cells. Mol Cell Biol.
26:5797–5808. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sun Y, Boyd K, Xu W, Ma J, Jackson CW, Fu
A, Shillingford JM, Robinson GW, Hennighausen L, Hitzler JK, et al:
Acute myeloid leukemia-associated Mkl1 (Mrtf-a) is a key regulator
of mammary gland function. Mol Cell Biol. 26:5809–5826. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Morita T, Mayanagi T and Sobue K: Dual
roles of myocardin-related transcription factors in epithelial
mesenchymal transition via slug induction and actin remodeling. J
Cell Biol. 179:1027–1042. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xu WG, Shang YL, Cong XR, Bian X and Yuan
Z: MicroRNA-135b promotes proliferation, invasion and migration of
osteosarcoma cells by degrading myocardin. Int J Oncol.
45:2024–2032. 2014.PubMed/NCBI
|
|
8
|
Medjkane S, Perez-Sanchez C, Gaggioli C,
Sahai E and Treisman R: Myocardin-related transcription factors and
SRF are required for cytoskeletal dynamics and experimental
metastasis. Nat Cell Biol. 11:257–268. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lee RC, Feinbaum RL and Ambros V: The C.
elegans heterochronic gene lin-4 encodes small RNAs with antisense
complementarity to lin-14. Cell. 75:843–854. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Pasquinelli AE, Reinhart BJ, Slack F,
Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B,
Müller P, et al: Conservation of the sequence and temporal
expression of let-7 heterochronic regulatory RNA. Nature.
408:86–89. 2000. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Reinhart BJ, Slack FJ, Basson M,
Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR and Ruvkun G:
The 21-nucleotide let-7 RNA regulates developmental timing in
Caenorhabditis elegans. Nature. 403:901–906. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kloosterman WP and Plasterk RH: The
diverse functions of microRNAs in animal development and disease.
Dev Cell. 11:441–450. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Meng F, Henson R, Wehbe-Janek H, Ghoshal
K, Jacob ST and Patel T: MicroRNA-21 regulates expression of the
PTEN tumor suppressor gene in human hepatocellular cancer.
Gastroenterology. 133:647–658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jovanovic M and Hengartner MO: miRNAs and
apoptosis: RNAs to die for. Oncogene. 25:6176–6187. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Esquela-Kerscher A and Slack FJ: Oncomirs
- microRNAs with a role in cancer. Nat Rev Cancer. 6:259–269. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Calin GA and Croce CM: MicroRNA signatures
in human cancers. Nat Rev Cancer. 6:857–866. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kondo N, Toyama T, Sugiura H, Fujii Y and
Yamashita H: miR-206 expression is down-regulated in estrogen
receptor alpha-positive human breast cancer. Cancer Res.
68:5004–5008. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhou J, Tian Y, Li J, Lu B, Sun M, Zou Y,
Kong R, Luo Y, Shi Y, Wang K, et al: miR-206 is down-regulated in
breast cancer and inhibits cell proliferation through the
up-regulation of cyclin D2. Biochem Biophys Res Commun.
433:207–212. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Taulli R, Bersani F, Foglizzo V, Linari A,
Vigna E, Ladanyi M, Tuschl T and Ponzetto C: The muscle-specific
microRNA miR-206 blocks human rhabdomyosarcoma growth in
xenotransplanted mice by promoting myogenic differentiation. J Clin
Invest. 119:2366–2378. 2009.PubMed/NCBI
|
|
21
|
Zhang T, Liu M, Wang C, Lin C, Sun Y and
Jin D: Down-regulation of MiR-206 promotes proliferation and
invasion of laryngeal cancer by regulating VEGF expression.
Anticancer Res. 31:3859–3863. 2011.PubMed/NCBI
|
|
22
|
Song G, Zhang Y and Wang L: MicroRNA-206
targets notch3, activates apoptosis, and inhibits tumor cell
migration and focus formation. J Biol Chem. 284:31921–31927. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Chen X, Yan Q, Li S, Zhou L, Yang H, Yang
Y, Liu X and Wan X: Expression of the tumor suppressor miR-206 is
associated with cellular proliferative inhibition and impairs
invasion in ERα-positive endometrioid adenocarcinoma. Cancer Lett.
314:41–53. 2012. View Article : Google Scholar
|
|
24
|
Wang X, Ling C, Bai Y and Zhao J:
MicroRNA-206 is associated with invasion and metastasis of lung
cancer. Anat Rec (Hoboken). 294:88–92. 2011. View Article : Google Scholar
|
|
25
|
Yan D, Dong XE, Chen X, Wang L, Lu C, Wang
J, Qu J and Tu L: MicroRNA-1/206 targets c-Met and inhibits
rhabdomyosarcoma development. J Biol Chem. 284:29596–29604. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Singh A, Happel C, Manna SK,
Acquaah-Mensah G, Carrerero J, Kumar S, Nasipuri P, Krausz KW,
Wakabayashi N, Dewi R, et al: Transcription factor NRF2 regulates
miR-1 and miR-206 to drive tumorigenesis. J Clin Invest.
123:2921–2934. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang L, Liu X, Jin H, Guo X, Xia L, Chen
Z, Bai M, Liu J, Shang X, Wu K, et al: miR-206 inhibits gastric
cancer proliferation in part by repressing cyclin D2. Cancer Lett.
332:94–101. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Chen CH, Wu ML, Lee YC, Layne MD and Yet
SF: Intronic CArG box regulates cysteine-rich protein 2 expression
in the adult but not in developing vasculature. Arterioscler Thromb
Vasc Biol. 30:835–842. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Liao XH, Lu DL, Wang N, Liu LY, Wang Y, Li
YQ, Yan TB, Sun XG, Hu P and Zhang TC: Estrogen receptor α mediates
proliferation of breast cancer MCF-7 cells via a
p21/PCNA/E2F1-dependent pathway. FEBS J. 281:927–942. 2014.
View Article : Google Scholar
|
|
30
|
Kong WQ, Bai R, Liu T, Cai CL, Liu M, Li X
and Tang H: MicroRNA-182 targets cAMP-responsive element-binding
protein 1 and suppresses cell growth in human gastric
adenocarcinoma. FEBS J. 279:1252–1260. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ma L, Teruya-Feldstein J and Weinberg RA:
Tumour invasion and metastasis initiated by microRNA-10b in breast
cancer. Nature. 449:682–688. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Frankel LB, Christoffersen NR, Jacobsen A,
Lindow M, Krogh A and Lund AH: Programmed cell death 4 (PDCD4) is
an important functional target of the microRNA miR-21 in breast
cancer cells. J Biol Chem. 283:1026–1033. 2008. View Article : Google Scholar
|
|
33
|
Yan LX, Huang XF, Shao Q, Huang MY, Deng
L, Wu QL, Zeng YX and Shao JY: MicroRNA miR-21 overexpression in
human breast cancer is associated with advanced clinical stage,
lymph node metastasis and patient poor prognosis. RNA.
14:2348–2360. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Valastyan S, Reinhardt F, Benaich N,
Calogrias D, Szász AM, Wang ZC, Brock JE, Richardson AL and
Weinberg RA: A pleiotropically acting microRNA, miR-31, inhibits
breast cancer metastasis. Cell. 137:1032–1046. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Shimono Y, Zabala M, Cho RW, Lobo N,
Dalerba P, Qian D, Diehn M, Liu H, Panula SP, Chiao E, et al:
Downregulation of miRNA-200c links breast cancer stem cells with
normal stem cells. Cell. 138:592–603. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Miller TE, Ghoshal K, Ramaswamy B, Roy S,
Datta J, Shapiro CL, Jacob S and Majumder S: MicroRNA-221/222
confers tamoxifen resistance in breast cancer by targeting
p27Kip1. J Biol Chem. 283:29897–29903. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yun CH, Choi SC, Park E, Kim SJ, Chung AS,
Lee HK, Lee HJ and Han JK: Negative regulation of Activin/Nodal
signaling by SRF during Xenopus gastrulation. Development.
134:769–777. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Schratt G, Philippar U, Hockemeyer D,
Schwarz H, Alberti S and Nordheim A: SRF regulates Bcl-2 expression
and promotes cell survival during murine embryonic development.
EMBO J. 23:1834–1844. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Cao XL, Hu XM, Hu JQ and Zheng WX:
Myocardin-related transcription factor-A promoting neuronal
survival against apoptosis induced by hypoxia/ischemia. Brain Res.
1385:263–274. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ma Z, Morris SW, Valentine V, Li M,
Herbrick JA, Cui X, Bouman D, Li Y, Mehta PK, Nizetic D, et al:
Fusion of two novel genes, RBM15 and MKL1, in the t(1;22)(p13;q13)
of acute mega-karyoblastic leukemia. Nat Genet. 28:220–221. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Mercher T, Coniat MB-L, Monni R,
Mauchauffe M, Nguyen Khac F, Gressin L, Mugneret F, Leblanc T,
Dastugue N, Berger R, et al: Involvement of a human gene related to
the Drosophila spen gene in the recurrent t(1;22) translocation of
acute megakaryocytic leukemia. Proc Natl Acad Sci USA.
98:5776–5779. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Descot A, Rex-Haffner M, Courtois G,
Bluteau D, Menssen A, Mercher T, Bernard OA, Treisman R and Posern
G: OTT-MAL is a deregulated activator of serum response
factor-dependent gene expression. Mol Cell Biol. 28:6171–6181.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Mercher T, Raffel GD, Moore SA, Cornejo
MG, Baudry-Bluteau D, Cagnard N, Jesneck JL, Pikman Y, Cullen D,
Williams IR, et al: The OTT-MAL fusion oncogene activates
RBPJ-mediated transcription and induces acute megakaryoblastic
leukemia in a knock-in mouse model. J Clin Invest. 119:852–864.
2009.PubMed/NCBI
|
|
44
|
Sasazuki T, Sawada T, Sakon S, Kitamura T,
Kishi T, Okazaki T, Katano M, Tanaka M, Watanabe M, Yagita H, et
al: Identification of a novel transcriptional activator, BSAC, by a
functional cloning to inhibit tumor necrosis factor-induced cell
death. J Biol Chem. 277:28853–28860. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Brandt DT, Baarlink C, Kitzing TM, Kremmer
E, Ivaska J, Nollau P and Grosse R: SCAI acts as a suppressor of
cancer cell invasion through the transcriptional control of
beta1-integrin. Nat Cell Biol. 11:557–568. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Huet G, Mérot Y, Percevault F, Tiffoche C,
Arnal JF, Boujrad N, Pakdel F, Métivier R and Flouriot G:
Repression of the estrogen receptor-alpha transcriptional activity
by the Rho/mega-karyoblastic leukemia 1 signaling pathway. J Biol
Chem. 284:33729–33739. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Evelyn CR, Wade SM, Wang Q, Wu M,
Iñiguez-Lluhí JA, Merajver SD and Neubig RR: CCG-1423: A
small-molecule inhibitor of RhoA transcriptional signaling. Mol
Cancer Ther. 6:2249–2260. 2007. View Article : Google Scholar : PubMed/NCBI
|