Introduction
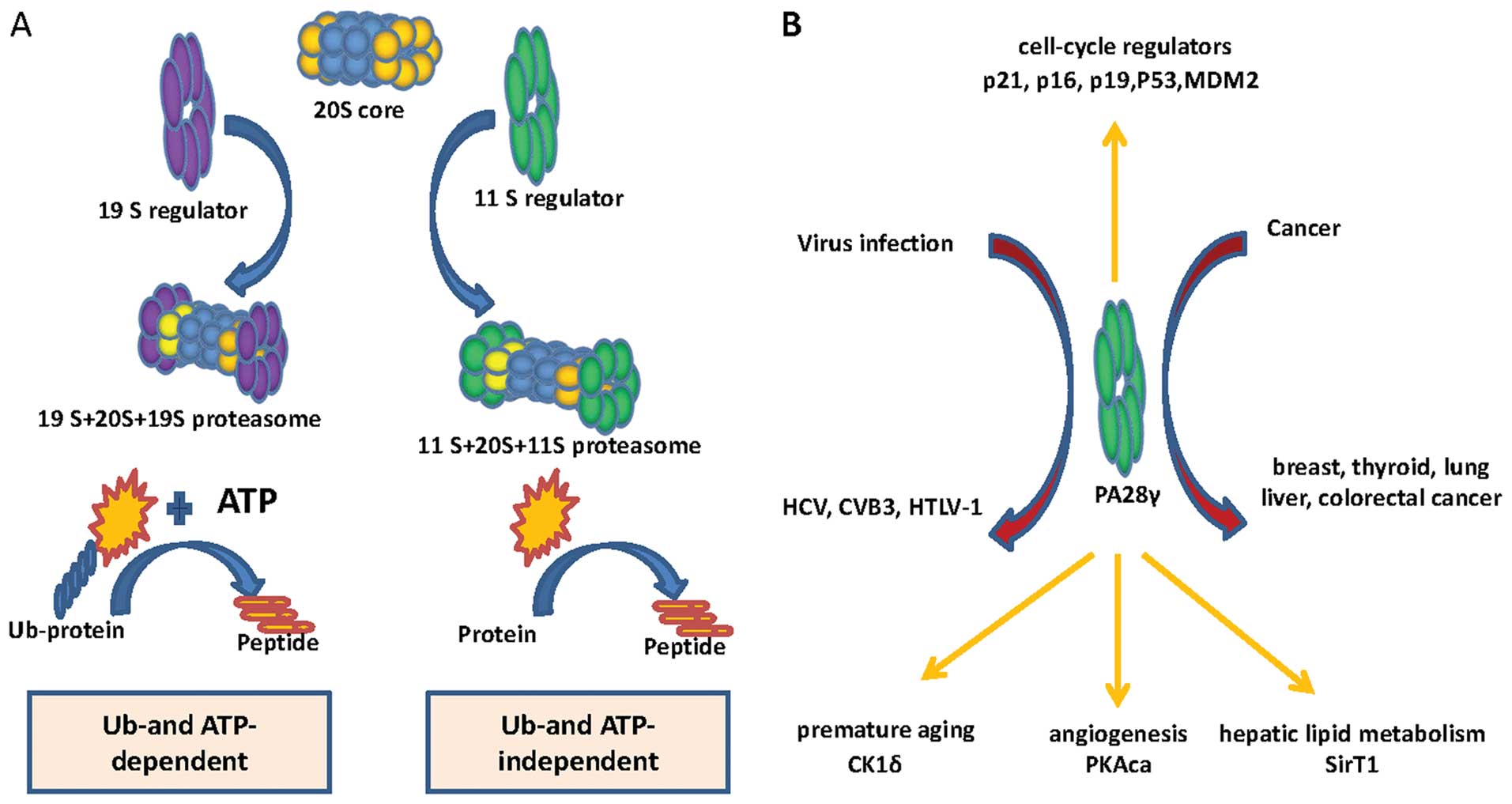
Proteasome activator 28γ (PA28γ) is a component of
the proteasome system, which is one of the most important
proteolytic systems in eukaryotes. It binds to the 20S core and
mainly promotes proteins degradation in a ubiquitin-and
ATP-independent manner (1–4). Strikingly, PA28γ not only plays a
role as a proteasome activator, but also has been shown to be
involved in cancer progression and virus infection (Fig. 1B). Loss of PA28γ expression in
PA28γ-deficient mice results in reduced body size and cell-specific
mitotic defects (5–7). It directs degradation of the steroid
receptor coactivator SRC-3, which is an oncogene frequently
amplified in breast cancer (8). It
is also a novel serum marker for human colorectal cancer (CRC)
because it can be detected in sera and is significantly elevated in
CRC patients compared with healthy donors and patients with benign
bowel disease (9). Moreover,
overexpression of PA28γ occurs in many different cancer types,
including thyroid, colon, liver, lung, ovary and gastric cancer
(10–14). PA28γ binds to and regulates the
stability and nuclear retention of hepatitis C core protein,
contributing to hepatitis C core protein-induced insulin resistance
and hepatocarcinoma (15–18). In addition, PA28γ promotes
coxsackievirus B3 (CVB3) replication (19–21)
and interacts with human T-lymphotropic virus type 1 (HTLV-1) p30
to increase viral spread (22,23).
A series of cell cycle, apoptosis, and cancer progression-related
PA28γ target proteins have been identified, including p21, p16,
p19, p53, Mdm2 (24–26). In recent years, more targets of
PA28γ have been identified using antibody array analysis. These
proteins include protein kinase A catalytic subuit-α (PKAca),
SirT1, and casein kinase (CK)1δ, which play important roles in
angiogenesis, hepatic lipid metabolism and premature aging,
respectively (27–29).
Alternative splicing of mRNA allows many gene
products with different functions to be produced from a single
coding sequence according to the cell type, developmental stage, or
in response to acute stimuli. It explains how enormous mammalian
proteomic diversity can be achieved with the limited number of
genes found in higher eukaryotes. Based on deep sequencing of
alternative splicing complexity in the human transcriptome, it is
estimated that transcripts from ~95% of multiexon genes undergo
alternative splicing (30,31). A study involving probabilistic
analyses indicated that >60% of human disease-causing mutations
affect splicing rather than directly affecting coding sequences.
Another study concluded that one-third of all hereditary diseases
are likely to have a splicing component (32,33).
Abnormally spliced mRNAs are also found in a high proportion of
cancerous cells. Combined RNA-Seq and proteomics analyses
technology revealed striking divergent expression profile of splice
isoforms of key proteins in important cancer pathways. For example,
several abnormally spliced DNMT3B mRNAs are found in tumors and
cancer cell lines. In two separate studies, expression of two of
these abnormally spliced mRNAs in mammalian cells caused changes in
the DNA methylation patterns in those cells. Cells with one of the
abnormal mRNAs also grew twice as fast as control cells, indicating
a direct contribution to tumor development by this product
(34–37).
In a previous study, we identified 85 differentially
and constantly expressed proteins (>2-fold change, P<0.05) in
six pairs of oral leukoplakia tissues with dysplasia and oral
squamous cancer tissues via two dimensional electrophoresis (2-DE)
followed by ESI-Q-TOF-LC-MS/MS. Among them, three homologs of
proteasome activator PA28α, PA28β, and PA28γ were shown to have
upregulated mRNA levels in oral squamous cell carcinoma (OSCC)
cells relative to oral keratinocytes (38). In this study, we analyzed
alternative splicing of PA28γ using the alternative splicing
prediction data base ASPicDB (39,40),
and found that there are theoretically nearly 50 alternative
transcripts. We therefore tried cloning these predicted splice
variants. We successfully cloned a novel (the fifth) transcript
variant of PA28γ in oral cancer cells. This variant encodes a
truncated isoform that retains the most conserved residues of the
PA28 family. Furthermore, it is involved in regulation of p53 and
Mdm2.
Materials and methods
Bioinformatics prediction
The alternative splicing prediction data base
(ASPicDB) is a program designed to provide access to reliable
annotations of the alternative splicing pattern of human genes, and
to the functional annotation of predicted isoforms. Alternative
splicing prediction of PA28γ (PSME3) was performed in ASPicDB.
Cell type and cell culture
Six oral squamous cell carcinomaderived cell lines
(HSC-3, HOK, UM1, UM2, Cal27 and HN31) and human embryonic kidney
293 (HEK293) cells were cultured in DMEM with 10% fetal bovine
serum.
Gene cloning and DNA sequencing
Total RNA of each cell line was extracted with
TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the
manufacturer’s protocol. The RNA concentration was determined by
absorbance at 260 nm with a NanovueTM spectrophotometer
(GE Healthcare). Equal amounts of RNA (1.0 μg) were used as
template in each reverse transcription reaction (total volume, 30
μl) with the PrimeScript® RT Reagent kit with gDNA
Eraser (Takara, Shiga, Japan). The reaction conditions were 42°C
for 2 min to remove genomic DNA, 37°C for 15 min, followed by 85°C
for 5 sec to obtain total cDNA. The primers designed for PA28γ
amplification were: 5′-TTGTATTTCCAGGGCATGGCCTCGTTGCTG-3′ (forward
primer) and 5′-CAAGCTTCGTCATCATCAGTA CAGAGTCTC-3′ (reverse primer).
PCR was performed for 30 cycles with PrimeSTAR® HS DNA
Polymerase and 2 μl total cDNA was used as template in 50 μl
reaction volume. The reaction conditions were a denaturation step
at 98°C for 10 sec, an annealing step at 55°C for 15 sec, and an
elongation step at 72°C for 1 min. PCR product (10 μl) was
separated on a 1% agarose gel, and observed under ultraviolet light
(Bio-Rad, Hercules, CA, USA). DNA bands were eluted from the
agarose gel using a gel extraction kit (Doupson) and cloned into
the p15TV-L vector for sequencing.
Plasmid transfection
Plasmids for overexpressing human influenza
hemagglutinin A epitope (HA)-tagged PA28γ isoforms were constructed
by Chengdu Bio-atom Biotechnology. HEK293 cells were used as
experimental cells. The control and positive group cells were
transiently transfected with empty vector and HA-PA28γ isoform
5-expressing plasmids, respectively.
Western blotting
Seventy-two hours after treatment, cells were
harvested and protein was extracted with RIPA lysis buffer
(Beyotime, P0013B). Antibodies to GAPDH (XP® Rabbit mAb,
Cell Signaling Technology), p53 (mouse mAb, Cell Signaling
Technology), Mdm2 (Phospho-Mdm2 (Ser166) Antibody, Cell Signaling
Technology), PA28γ isoform1 (Purified mouse anti-PA28γ, BD
Transduction Laboratories), PA28γ isoform 5 (HA-tag antibody,
ZSGB-BIO) were used to test the amount of corresponding protein by
western blotting (Bio-Rad electrophoresis).
Quantitative PCR
The amount of mRNA of PA28γ transcript variant 1 was
measured by the ABI 7500 Real-time PCR (RT-PCR) System with One
Step SYBR® PrimeScript™ Plus RT-PCR kit (Takara,
RR096A). Gene-specific primers were designed as follows: PA28γ
transcript variant 1 forward primer, 5′-ATGGACTGGATGGTCCCACT-3′;
PA28γ transcript variant 1 reverse primer,
5′-ACAGCCGGATCTCAGGTTTC-3′; 18S rRNA forward primer,
5′-CTACCACATCCAAGGAA GGCA-3′; 18S rRNA reverse primer,
5′-TTTTTCGTCACTA CCTCCCCG-3′. Gene-specific primers for PA28γ
transcript variant 1 were designed crossing the lost sequences
which lack in PA28γ transcript variant 5. Mean Ct values for target
genes were normalized to mean Ct values for the endogenous control
18S [-ΔCt = Ct (18S)- Ct (target
gene)]. The ratio of mRNA expression of target gene versus 18S was
defined as 2−ΔCt. All experiments were repeated at least
three times.
Results
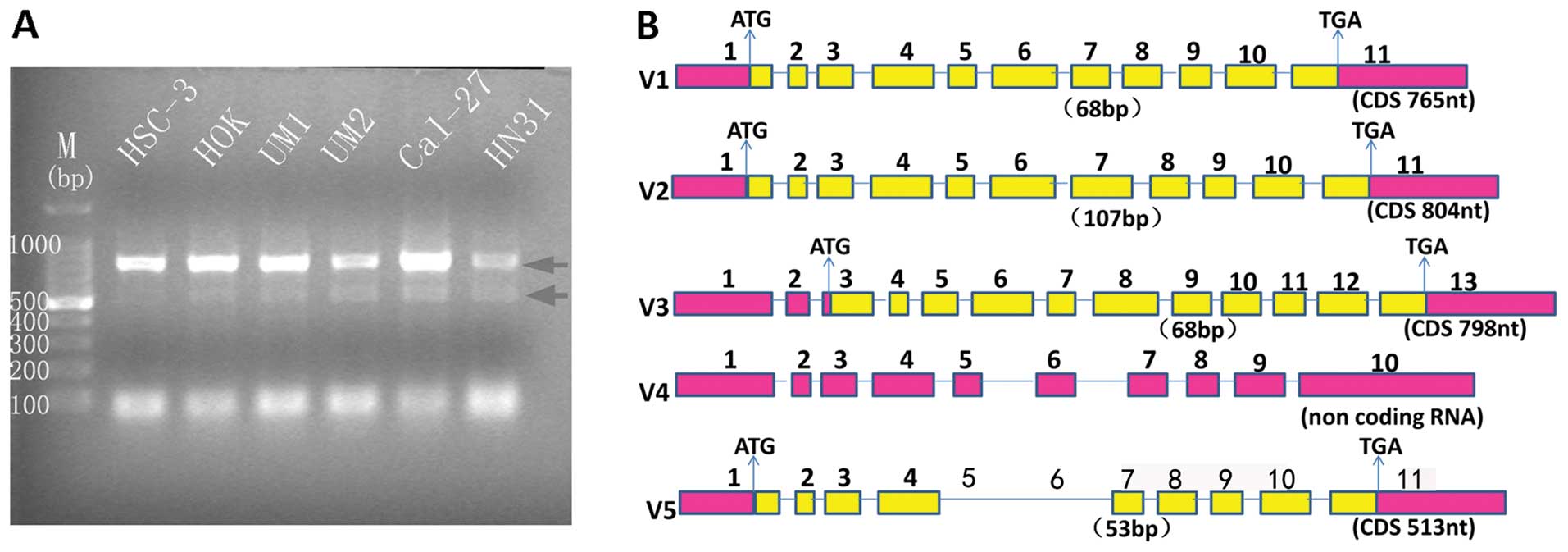
Alternative splicing prediction of PA28γ
via bioinformatics
The genomic location of human PA28γ is at chrosome
17: 40985423–40995777, with a size of 10,354 bp. Expression
sequence tags (EST) (1,180) exist in this cluster. Forty-nine
alternative transcripts are predicted and 45 of them are
protein-coding forms. To date, only four alternative transcripts
are reported in GenBank. PA28γ transcript variant 1 (Nucleotide
accession: NM_005789.3) encodes the predominant protein with 254
amino acids. Variant 2 (Nucleotide accession: NM_176863.2) uses an
alternative in-frame splice site; variant 2 is 13 amino acids
longer than variant 1. Variant 3 (Nucleotide accession:
NM_001267045.1) is distinguished in the 5′-untranslated region and
5′-coding region, and initiates translation at an alternative start
codon. Variant 3 has a distinct N-terminus and is 12 amino acids
longer than variant 1. Variant 4 (Nucleotide accession:
NR_049772.1) is a long non-coding RNA.
Cloning and identification of a novel
PA28γ transcript variant in oral cancer cells
As shown in Fig.
2A, three DNA bands were observed on the agarose gel. According
to the DNA markers, from top to bottom, the first band likely
corresponds to PA28γ transcript variant 1, and the third band to
oligonucleotide primers. In addition, we observed a weak band
between the first and third bands. The sequencing data were
analyzed by the BLAST tool in NCBI. As expected, the first band was
confirmed as PA28γ transcript variant 1. We translated the
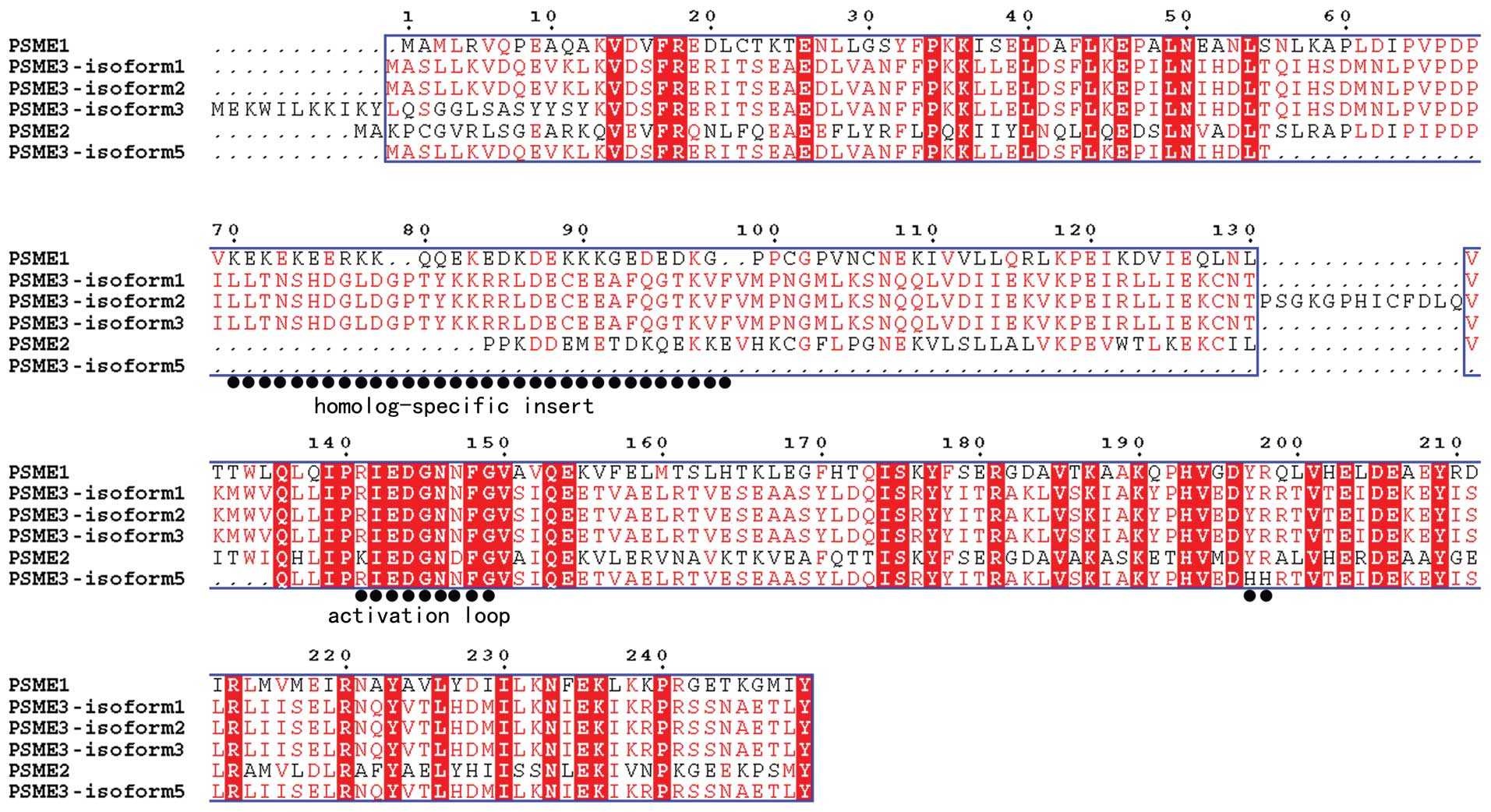
nucleotide acid sequence of the second band into amino acids and a
structure-based sequence alignment was performed. As shown in
Fig. 3, the new isoform lacks the
‘homolog-specific insert’ region (41), but it belongs to the PA28γ
subfamily according to the high identity in the C-terminal
sequence. Therefore, we termed the new alternative splicing as
transcript variant 5 and submitted it to GenBank (Nucleotide
accession: JX156303.1). Compared with PA28γ variant 1, PA28γ
variant 5 lacks the nucleotide acids regions in coding sequence
among 185–483 in PA28γ variant 1, which corresponds to the sequence
from exon 4 to exon 7 in variant 1 (Fig. 2B). Taken together, these findings
indicate that PA28γ transcript variant 5 is a novel transcript
variant in the PA28γ subfamily.
PA28γ isoform 5 involved in p53 and Mdm2
regulation
We predicted that PA28γ transcript variant 5 encodes
a truncated protein of 170 residues, which is 84 residues shorter
than PA28γ isoform1. However, structure-based sequence alignment
showed that isoform 5 retains most of the conserved residues and
the ‘activation loop’ of PA28 family (Fig. 3). As mentioned in the introduction,
PA28γ has a series of target proteins and plays an important role
in cell cycle regulation. It was reported that PA28γ regulates p53
by enhancing Mdm2-mediated degradation (25). This prompted us to explore whether
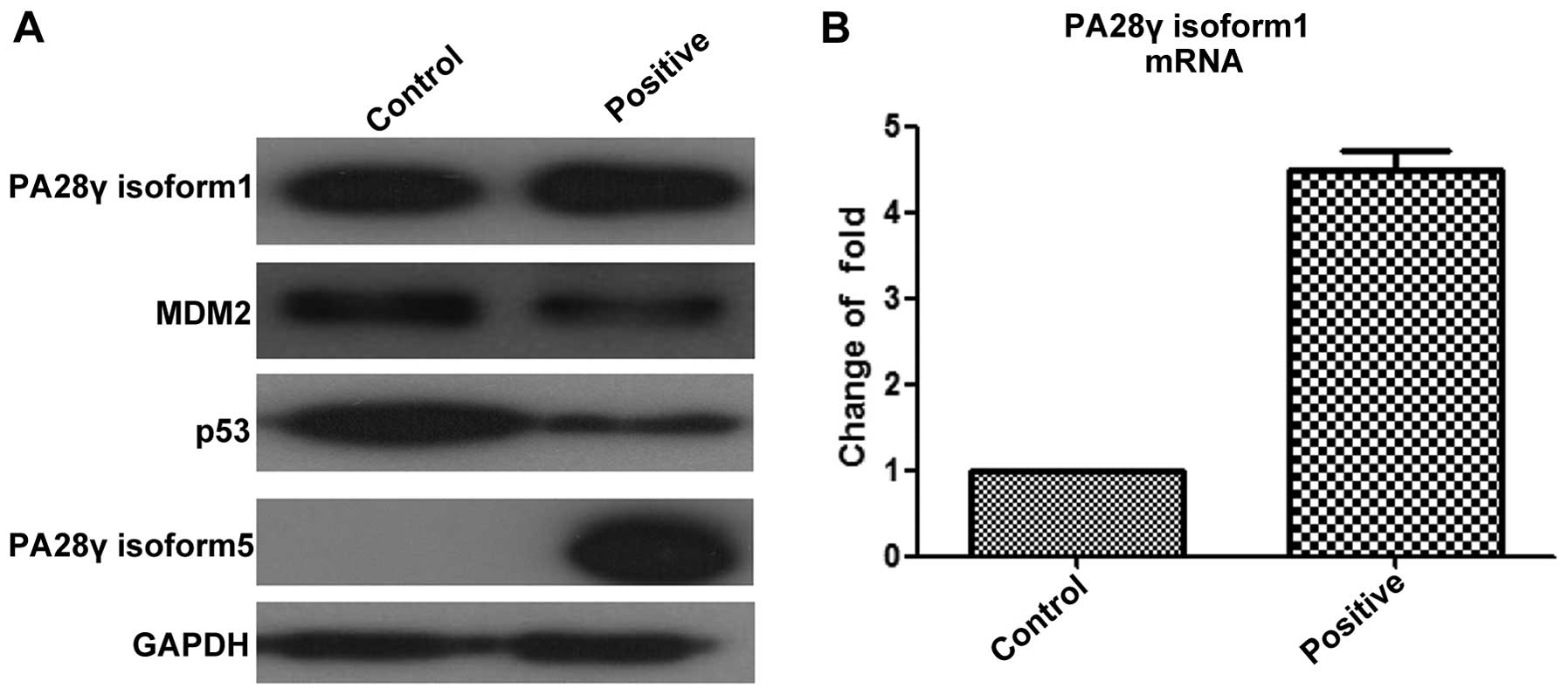
PA28γ isoform 5 could play a similar role in this process. As shown
in Fig. 4, the positive group
overexpressed PA28γ isoform 5, which resulted in significant
decrease of p53 and Mdm2 at protein level. Furthermore, we tested
the amount of PA28γ isoform1 at both mRNA and protein levels.
Strikingly, the mRNA level of PA28γ isoform1 in the positive group
was 4.5-fold higher than in the control group. The change in
protein levels of PA28γ isoform1 between positive and negative
groups was not as great as for mRNA levels, but still increased. To
explain these changes, we propose two possibilities: i) increased
PA28γ isoform 5 triggers the transcription of PA28γ alternative
transcript1, and the increased PA28γ isoform1 regulates the
degradation of p53 and Mdm2, as reported before; or ii)
overexpressed PA28γ isoform 5 maintains a pool of functional PA28γ,
and represses the translation of PA28γ alternative transcript1 as
feedback regulation. In either case, the downregulation of p53 and
Mdm2 at the protein level resulted from overexpression of PA28γ
isoform 5, which indicates that PA28γ isoform 5 is a functional
isoform that may play a complementary role in regulating p53 and
Mdm2.
Discussion
Based on the results of bioinformatics analysis via
ASPicDB, 30 alternative transcripts of PA28α, one alternative
transcripts of PA28β, and 49 alternative transcripts of PA28γ were
predicted (39,40). We checked the alternative
transcripts of these PA28 members deposited in GenBank, and found
that four transcript variants of PA28α, one transcript variant of
PA28β and four transcript variants of PA28γ have been reported.
Therefore, the number of reported transcript variants of PA28β is
consistent with the bioinformatics prediction. However, there are
many more alternative transcripts of PA28α and PA28γ that are not
yet defined. Typically, alternatively spliced transcripts have been
found by comparing expression sequence tags (ESTs), but this
requires sequencing of very large numbers of ESTs. Most EST
libraries come from a very limited number of tissues, so
tissue-specific splice variants are likely to be missed in any
case. However, high-throughput approaches to examine splicing have
been developed, such as DNA microarray-based analyses, RNA-binding
assays, and deep sequencing. When combined with splicing assays,
including in vivo reporter gene assays, the functional
effects of polymorphisms or mutations on the splicing of pre-mRNA
transcripts can be analyzed (33,42–44).
Herein, utilizing bioinformatics, RT-PCR technology and gene
overexpression in model cells, we successfully predicted, cloned,
and confirmed a novel transcript variant of PA28γ. Moreover, we
found PA28γ isoform 5 lacks long regions compared with other
reported isoforms of PA28γ. Therefore, the siRNA target to this
region of PA28γ will affect other reported isoforms of PA28γ but
not on PA28γ transcript variant 5, which may result in a
significant off-target effect of siRNA interference.
Generally speaking, the diversity of transcript
variant corresponds to the functional significance of the gene
regardless of whether the transcript variant has the same function
or not. The variants with the same function will compensate the
loss-of-function mutation to maintain homeostasis, whereas patterns
of divergent functional variants are likely to be indicators for
some disease. In a retrospective analysis of 80 individuals with
gastrointestinal sarcoma, predominant expression of the
immunosuppressive NKp30c isoform (over the immunostimulatory NKp30a
and NKp30b isoforms) was associated with reduced survival of the
subjects (45). Previously, we
identified three alternative transcripts of oral cancer
overexpressed 1 gene (ORAOV1), and an inverse correlation was found
between the expression frequency of ORAOV1-A and the degree of
differentiation in OSCC (46). In
future studies, we will assess the clinical value of PA28γ isoform
5 in tissue samples from normal, precancerous to infiltrative OSCC.
Moreover, the relationships of immunostaning with survival rate and
recurrence will be analyzed.
Several studies have evaluated the interaction of
PA28γ variant 1, p53 and Mdm2. P53 encodes a tumor suppressor
protein and responds to diverse cellular stresses to regulate
expression of target genes, thereby inducing cell cycle arrest,
apoptosis, senescence, DNA repair, or changes in metabolism.
Mutations in this gene are associated with a variety of human
cancers. Thus, it is a focus of numerous investigations for
reversing tumor progression. MDM2 encodes a nuclear-localized E3
ubiquitin ligase. The protein can promote tumor formation by
targeting tumor suppressor proteins, such as p53, for proteasomal
degradation. Amplification of this locus is detected in a variety
of different cancers. Noteworthy, the polymer form of PA28γ variant
1 interacts with both p53 and Mdm2, which facilitates
ubiquitination and Mdm2-dependent proteasomal degradation of p53.
The decreased p53 attenuated apoptosis stimulation after DNA damage
(47,48). In our study, we confirmed that
PA28γ isoform 5 also mediates the downregulation of p53 and Mdm2,
which may serve as a complementary mechanism for PA28γ isoform1 in
regulating p53 and Mdm2. Given that PA28γ is involved in cancer
progression, it suggests to examine the PA28γ isoform 5 before we
determine PA28γ isoform1 as a therapy target via regulating p53 and
Mdm2 pathway.
The proteasome is a primary proteolytic system in
eukaryotes. This system is a multi-subunit protease complex
composed of 20S catalytic core and proteasome activators (Fig. 1A). The 20S core is a cylindrical
stack of four heptameric rings with two outer α rings and two inner
β rings. The PA700 (19S) activator binds to the 20S core and
primarily mediates degradation of ubiquitinated proteins in
ATP-dependent manner. In contrast, the PA28 (11S) activator binds
to the 20S core and mainly promotes protein degradation in Ub- and
ATP-independent manner (1–4). To date, three classes of PA28 have
been identified: PA28α, PA28β, and PA28γ. PA28α and β form a
heteroheptamer, which is mainly localized in the cytosol. PA28γ
exists as a homoheptamer and is primarily found in the nucleus.
PA28α and β mediate proteolytic cleavage after basic, acidic, and
hydrophobic residues. PA28γ stimulates proteasomal hydrolysis of
peptides with basic residues (41,49–56).
Thus, the subcellular location of PA28γ isoform 5 and its effect on
proteolytic activity in the proteasome system need to be further
studied.
In this study, we made an attempt to transiently
transfect HA-tagged PA28γ isoform 5 overexpressing plasmids into
oral cancer cells. However, we failed because oral cancer cells are
highly keratinized. Although we determined the primary function of
PA28γ isoform 5 in HEK293 cells, further investigation of its
effects on cell cycle, apoptosis and its correlation with
progression of oral squamous cell carcinoma will be performed in
oral cancer cells using lentivirus transfection.
Acknowledgements
We acknowledge Min Zhou, at State Key Laboratory of
Oral Diseases, West China Hospital of Stomatology, Sichuan
University, for preparing some experimental materials. This project
was supported by grants from National Natural Science Foundations
of China (nos. 81321002, 81472533, 81302371, and 81072218) and 111
Project of MOE, ISTCPC (2012DFA31370).
References
|
1
|
Ciechanover A: The ubiquitin-proteasome
proteolytic pathway. Cell. 79:13–21. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ciechanover A: The ubiquitin-proteasome
pathway: On protein death and cell life. EMBO J. 17:7151–7160.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Baumeister W, Walz J, Zühl F and Seemüller
E: The proteasome: Paradigm of a self-compartmentalizing protease.
Cell. 92:367–380. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Groll M, Ditzel L, Löwe J, Stock D,
Bochtler M, Bartunik HD and Huber R: Structure of 20S proteasome
from yeast at 2.4 A resolution. Nature. 386:463–471. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Barton LF, Runnels HA, Schell TD, Cho Y,
Gibbons R, Tevethia SS, Deepe GS Jr and Monaco JJ: Immune defects
in 28-kDa proteasome activator gamma-deficient mice. J Immunol.
172:3948–3954. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Preckel T, Fung-Leung WP, Cai Z, Vitiello
A, Salter-Cid L, Winqvist O, Wolfe TG, Von Herrath M, Angulo A,
Ghazal P, et al: Impaired immunoproteasome assembly and immune
responses in PA28-/- mice. Science. 286:2162–2165. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Murata S, Kawahara H, Tohma S, Yamamoto K,
Kasahara M, Nabeshima Y, Tanaka K and Chiba T: Growth retardation
in mice lacking the proteasome activator PA28gamma. J Biol Chem.
274:38211–38215. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li X, Lonard DM, Jung SY, Malovannaya A,
Feng Q, Qin J, Tsai SY, Tsai MJ and O’Malley BW: The SRC-3/AIB1
coactivator is degraded in a ubiquitin- and ATP-independent manner
by the REGgamma proteasome. Cell. 124:381–392. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Roessler M, Rollinger W, Mantovani-Endl L,
Hagmann ML, Palme S, Berndt P, Engel AM, Pfeffer M, Karl J,
Bodenmüller H, et al: Identification of PSME3 as a novel serum
tumor marker for colorectal cancer by combining two-dimensional
polyacrylamide gel electrophoresis with a strictly mass
spectrometry-based approach for data analysis. Mol Cell Proteomics.
5:2092–2101. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
He J, Cui L, Zeng Y, Wang G, Zhou P, Yang
Y, Ji L, Zhao Y, Chen J, Wang Z, et al: REGγ is associated with
multiple oncogenic pathways in human cancers. BMC Cancer.
12:752012. View Article : Google Scholar
|
|
11
|
Yu G, Zhao Y, He J, Lonard DM, Mao CA,
Wang G, Li M and Li X: Comparative analysis of REG{gamma}
expression in mouse and human tissues. J Mol Cell Biol. 2:192–198.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Tian M, Xiaoyi W, Xiaotao L and Guosheng
R: Proteasomes reactivator REG gamma enchances oncogenicity of
MDA-MB-231 cell line via promoting cell proliferation and
inhibiting apoptosis. Cell Mol Biol (Noisy-le-grand). 55(Suppl):
OL1121–OL1131. 2009.
|
|
13
|
Chen D, Yang X, Huang L and Chi P: The
expression and clinical significance of PA28γ in colorectal cancer.
J Investig Med. 61:1192–1196. 2013.PubMed/NCBI
|
|
14
|
Wang X, Tu S, Tan J, Tian T, Ran L, Rodier
JF and Ren G: REG gamma: A potential marker in breast cancer and
effect on cell cycle and proliferation of breast cancer cell. Med
Oncol. 28:31–41. 2011. View Article : Google Scholar
|
|
15
|
Miyamoto H, Moriishi K, Moriya K, Murata
S, Tanaka K, Suzuki T, Miyamura T, Koike K and Matsuura Y:
Involvement of the PA28gamma-dependent pathway in insulin
resistance induced by hepatitis C virus core protein. J Virol.
81:1727–1735. 2007. View Article : Google Scholar :
|
|
16
|
Moriishi K, Mochizuki R, Moriya K,
Miyamoto H, Mori Y, Abe T, Murata S, Tanaka K, Miyamura T, Suzuki
T, et al: Critical role of PA28gamma in hepatitis C
virus-associated steatogenesis and hepatocarcinogenesis. Proc Natl
Acad Sci USA. 104:1661–1666. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Moriishi K, Shoji I, Mori Y, Suzuki R,
Suzuki T, Kataoka C and Matsuura Y: Involvement of PA28gamma in the
propagation of hepatitis C virus. Hepatology. 52:411–420. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Suzuki R, Moriishi K, Fukuda K, Shirakura
M, Ishii K, Shoji I, Wakita T, Miyamura T, Matsuura Y and Suzuki T:
Proteasomal turnover of hepatitis C virus core protein is regulated
by two distinct mechanisms: A ubiquitin-dependent mechanism and a
ubiquitin-independent but PA28gamma-dependent mechanism. J Virol.
83:2389–2392. 2009. View Article : Google Scholar :
|
|
19
|
Gao G, Wong J, Zhang J, Mao I, Shravah J,
Wu Y, Xiao A, Li X and Luo H: Proteasome activator REGgamma
enhances coxsackieviral infection by facilitating p53 degradation.
J Virol. 84:11056–11066. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Luo H, Zhang J, Cheung C, Suarez A,
McManus BM and Yang D: Proteasome inhibition reduces coxsackievirus
B3 replication in murine cardiomyocytes. Am J Pathol. 163:381–385.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Si X, McManus BM, Zhang J, Yuan J, Cheung
C, Esfandiarei M, Suarez A, Morgan A and Luo H: Pyrrolidine
dithiocarbamate reduces coxsackievirus B3 replication through
inhibition of the ubiquitin-proteasome pathway. J Virol.
79:8014–8023. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ko NL, Taylor JM, Bellon M, Bai XT,
Shevtsov SP, Dundr M and Nicot C: PA28γ is a novel corepressor of
HTLV-1 replication and controls viral latency. Blood. 121:791–800.
2013. View Article : Google Scholar :
|
|
23
|
Anupam R, Datta A, Kesic M, Green-Church
K, Shkriabai N, Kvaratskhelia M and Lairmore MD: Human
T-lymphotropic virus type 1 p30 interacts with REGgamma and
modulates ATM (ataxia telangiectasia mutated) to promote cell
survival. J Biol Chem. 286:7661–7668. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen X, Barton LF, Chi Y, Clurman BE and
Roberts JM: Ubiquitin-independent degradation of cell-cycle
inhibitors by the REGgamma proteasome. Mol Cell. 26:843–852. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang Z and Zhang R: Proteasome activator
PA28 gamma regulates p53 by enhancing its MDM2-mediated
degradation. EMBO J. 27:852–864. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li X, Amazit L, Long W, Lonard DM, Monaco
JJ and O’Malley BW: Ubiquitin- and ATP-independent proteolytic
turnover of p21 by the REGgamma-proteasome pathway. Mol Cell.
26:831–842. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Liu S, Lai L, Zuo Q, Dai F, Wu L, Wang Y,
Zhou Q, Liu J, Liu J, Li L, et al: PKA turnover by the
REGγ-proteasome modulates FoxO1 cellular activity and VEGF-induced
angiogenesis. J Mol Cell Cardiol. 72:28–38. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Dong S, Jia C, Zhang S, Fan G, Li Y, Shan
P, Sun L, Xiao W, Li L, Zheng Y, et al: The REGγ proteasome
regulates hepatic lipid metabolism through inhibition of autophagy.
Cell Metab. 18:380–391. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li L, Zhao D, Wei H, Yao L, Dang Y, Amjad
A, Xu J, Liu J, Guo L, Li D, et al: REGγ deficiency promotes
premature aging via the casein kinase 1 pathway. Proc Natl Acad Sci
USA. 110:11005–11010. 2013. View Article : Google Scholar
|
|
30
|
Black DL: Protein diversity from
alternative splicing: A challenge for bioinformatics and
post-genome biology. Cell. 103:367–370. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Luco RF, Allo M, Schor IE, Kornblihtt AR
and Misteli T: Epigenetics in alternative pre-mRNA splicing. Cell.
144:16–26. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
López-Bigas N, Audit B, Ouzounis C, Parra
G and Guigó R: Are splicing mutations the most frequent cause of
hereditary disease? FEBS Lett. 579:1900–1903. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lim KH, Ferraris L, Filloux ME, Raphael BJ
and Fairbrother WG: Using positional distribution to identify
splicing elements and predict pre-mRNA processing defects in human
genes. Proc Natl Acad Sci USA. 108:11093–11098. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Skotheim RI and Nees M: Alternative
splicing in cancer: Noise, functional, or systematic? Int J Biochem
Cell Biol. 39:1432–1449. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
He C, Zhou F, Zuo Z, Cheng H and Zhou R: A
global view of cancer-specific transcript variants by subtractive
transcriptome-wide analysis. PLoS One. 4:e47322009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Omenn GS, Guan Y and Menon R: A new class
of protein cancer biomarker candidates: Differentially expressed
splice variants of ERBB2 (HER2/neu) and ERBB1 (EGFR) in breast
cancer cell lines. J Proteomics. 107:103–112. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Fackenthal JD and Godley LA: Aberrant RNA
splicing and its functional consequences in cancer cells. Dis Model
Mech. 1:37–42. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang Z, Feng X, Liu X, Jiang L, Zeng X, Ji
N, Li J, Li L and Chen Q: Involvement of potential pathways in
malignant transformation from oral leukoplakia to oral squamous
cell carcinoma revealed by proteomic analysis. BMC Genomics.
10:3832009. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Martelli PL, D’Antonio M, Bonizzoni P,
Castrignanò T, D’Erchia AM, D’Onorio De Meo P, Fariselli P, Finelli
M, Licciulli F, Mangiulli M, et al: ASPicDB: A database of
annotated transcript and protein variants generated by alternative
splicing. Nucleic Acids Res. 39:Database. D80–D85. 2011. View Article : Google Scholar :
|
|
40
|
Castrignanò T, D’Antonio M, Anselmo A,
Carrabino D, D’Onorio De Meo A, D’Erchia AM, Licciulli F, Mangiulli
M, Mignone F, Pavesi G, et al: ASPicDB: A database resource for
alternative splicing analysis. Bioinformatics. 24:1300–1304. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li J and Rechsteiner M: Molecular
dissection of the 11S REG (PA28) proteasome activators. Biochimie.
83:373–383. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Wang Z and Burge CB: Splicing regulation:
From a parts list of regulatory elements to an integrated splicing
code. RNA. 14:802–813. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fairbrother WG, Yeh RF, Sharp PA and Burge
CB: Predictive identification of exonic splicing enhancers in human
genes. Science. 297:1007–1013. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Jiang WP, Sima ZH, Wang HC, Zhang JY, Sun
LS, Chen F and Li TJ: Identification of the involvement of LOXL4 in
generation of keratocystic odontogenic tumors by RNA-Seq analysis.
Int J Oral Sci. 6:31–38. 2014. View Article : Google Scholar :
|
|
45
|
Delahaye NF, Rusakiewicz S, Martins I,
Ménard C, Roux S, Lyonnet L, Paul P, Sarabi M, Chaput N, Semeraro
M, et al: Alternatively spliced NKp30 isoforms affect the prognosis
of gastrointestinal stromal tumors. Nat Med. 17:700–707. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Jiang L, Yang HS, Wang Z, Zhou Y, Zhou M,
Zeng X and Chen QM: ORAOV1-A correlates with poor differentiation
in oral cancer. J Dent Res. 88:433–438. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Chène P: Inhibiting the p53-MDM2
interaction: An important target for cancer therapy. Nat Rev
Cancer. 3:102–109. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
48
|
Shangary S and Wang S: Small-molecule
inhibitors of the MDM2-p53 protein-protein interaction to
reactivate p53 function: A novel approach for cancer therapy. Annu
Rev Pharmacol Toxicol. 49:223–241. 2009. View Article : Google Scholar :
|
|
49
|
Ahn JY, Tanahashi N, Akiyama K, Hisamatsu
H, Noda C, Tanaka K, Chung CH, Shibmara N, Willy PJ, Mott JD, et
al: Primary structures of two homologous subunits of PA28, a
gamma-interferon-inducible protein activator of the 20S proteasome.
FEBS Lett. 366:37–42. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Kohda K, Ishibashi T, Shimbara N, Tanaka
K, Matsuda Y and Kasahara M: Characterization of the mouse PA28
activator complex gene family: Complete organizations of the three
member genes and a physical map of the approximately 150-kb region
containing the alpha- and beta-subunit genes. J Immunol.
160:4923–4935. 1998.PubMed/NCBI
|
|
51
|
Mao I, Liu J, Li X and Luo H: REGgamma, a
proteasome activator and beyond? Cell Mol Life Sci. 65:3971–3980.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Realini C, Jensen CC, Zhang Z, Johnston
SC, Knowlton JR, Hill CP and Rechsteiner M: Characterization of
recombinant REGalpha, REGbeta, and REGgamma proteasome activators.
J Biol Chem. 272:25483–25492. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Schwarz K, Eggers M, Soza A, Koszinowski
UH, Kloetzel PM and Groettrup M: The proteasome regulator
PA28alpha/beta can enhance antigen presentation without affecting
20S proteasome subunit composition. Eur J Immunol. 30:3672–3679.
2000. View Article : Google Scholar
|
|
54
|
Stohwasser R, Salzmann U, Giesebrecht J,
Kloetzel PM and Holzhütter HG: Kinetic evidences for facilitation
of peptide channelling by the proteasome activator PA28. Eur J
Biochem. 267:6221–6230. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Wilk S, Chen WE and Magnusson RP:
Properties of the nuclear proteasome activator PA28gamma
(REGgamma). Arch Biochem Biophys. 383:265–271. 2000. View Article : Google Scholar
|
|
56
|
Yamano T, Murata S, Shimbara N, Tanaka N,
Chiba T, Tanaka K, Yui K and Udono H: Two distinct pathways
mediated by PA28 and hsp90 in major histocompatibility complex
class I antigen processing. J Exp Med. 196:185–196. 2002.
View Article : Google Scholar : PubMed/NCBI
|