Introduction
Lung cancer is currently considered the leading
cause of malignancy-related death worldwide (1,2) with
a 5-year survival rate of ~15% (3). Metastasis is one of the most
devastating characteristics of cancer, which leads to almost 90% of
all lung cancer deaths (1,2). Although important advances have been
made in understanding the pathogenesis and treatment of primary
malignancy, little progress has been made in treating metastasis.
Exploring the still undiscovered mechanisms of it will improve
diagnosis and treatment methods. In this process, pivotal
importance is attached to developing metastatic models because they
can aid us in illustrating the underlying biological features of
metastasis in more detail.
An ideal lung cancer metastatic model should not
only mimic all aspects of tumor progression but should also be
generally practical, stable and optimal. Subcutaneous (s.c.)
injection, which has been widely used in the development of most
human tumor models (4,5), was also considered as the most
appropriate method for lung cancer in our study, because orthotopic
implantation is prone to complications, and its rate of
tumorigenicity and metastasis formation in vivo is always
unstable (6). Furthermore, unlike
breast cancer and melanoma, an orthotopic model for lung cancer is
relatively unavailable due to the inaccessibility of the anatomical
location. Compared with tail intravenous injection, s.c. injection
can adequately mimic the metastases of cancer, because it
recapitulates the early biological steps that a primary tumor must
undergo to form a distant metastatic site (7,8),
such as detachment of tumor cells from the primary site, invasion
into the surrounding tissue, lymphatic or blood vessels and
intravasation. Therefore, s.c. injection can credibly mimic the
selection and evolution of metastasis.
After having been artificially cultivated in
vitro for a long time, tumor cells often have altered
phenotypes and genotypes, no longer maintaining the initial
molecular characteristics and the heterogeneity of patient tumors
(9,10). Under such circumstances, s.c.
xenograft models may present poor predictive power because of the
inconsistencies between laboratory and clinical findings (11,12).
To circumvent these issues, new cell lines from patient-derived
tumor samples are urgently needed. To fill this need, we
established and characterized a new lung cancer cell line
(designated XL-2) that was isolated from the ascites fluid of a
Chinese patient with systemic lung cancer metastasis. Therefore,
this cell line can reliably reproduce some important clinical
features, such as metastatic behavior.
Through this in vivo metastatic model, we
successfully obtained a highly metastatic subpopulation of XL-2
cell line (termed XL-2sci). ITRAQ label technology together with
nano liquid chromatography-mass spectrometry (NanoLC-MS/MS) has
proven to be efficient in protein analysis (13–15).
Hence, the clinic-combined metastatic model and this technological
means will increase the likeliness of discovering new
metastasis-associated genes (16)
and proteins (17) for identifying
new antitumor targets.
Materials and methods
Cell lines and cell culture
The human lung cancer A549 cells were maintained in
our laboratory and cultured in Dulbecco's modified Eagle's medium
(DMEM, Invitrogen, Burlington, Ontario, Canada) supplemented with
10% fetal bovine serum (FBS) (Biowest, South America Origin), 100
U/ml penicillin (Sigma-Aldrich, St. Louis, MO, USA), and 100 μg/ml
streptomycin (Sigma-Aldrich) at 37°C in a humidified air atmosphere
containing 5% CO2.
Patient materials
A 45-year-old Chinese male patient from Ruijin
Hospital (Shanghai, China) was diagnosed with adenocarcinoma of the
right lung. Despite undergoing a right upper lobe lobectomy, he
still suffered from widespread metastases at one year following the
operation. The right adrenal gland, both kidneys, bilateral lungs,
retroperitoneal lymph nodes besides the liver were all involved in
the metastasis progression. The patient's clinical situation
continued to deteriorate, and predominant ascites was the
concomitant symptom. Written informed consent for the scientific
purpose of ascites specimen was obtained from him at the time of
surgery, according to an established protocol approved by the Human
Ethic Committee of Ruijin Hospital. All procedures adhered to the
generally accepted guidelines for utilization of human
materials.
Isolation of primary cell line from
ascites
Approximately 60 ml of ascites fluid was collected
and immediately centrifuged at 1,500 × g for 5 min. The upper
supernatant was discarded and the remaining sediment washed with
phosphate-buffered saline (PBS, Sangon Biotech, Shanghai, China) at
1,500 × g for 5 min; the deposit was then resuspended in fresh PBS.
Subsequently, density gradient centrifugation was performed using
Percoll (Biodee Biotechnology, Beijing, China) to isolate the cells
as previously described (18,19)
with slight modifications. The cell suspension was processed using
a 60 and 40% Percoll gradient (1:1:1 volume ratio) at 3,000 × g for
30 min at room temperature. The cream-colored layer at the
interface of the two Percoll gradients was collected and washed
with PBS. After centrifugation at 1,500 × g for 5 min, the cell
pellets were resuspended with DMEM, containing 10% FBS, 100 U/ml
penicillin and 100 μg/ml streptomycin; then incubated them at 37°C
in a humidified atmosphere of 5% CO2. Several days
later, the bottom of the cell culture flask began to fill with
numerous cells. Once the mixed cells reached confluence of ~80%,
differential trypsinization was used to minimize the fibroblasts
and mesothelial cells. Finally, a pure cancer cell line named XL-2
by our laboratory was harvested. Aliquots of cells in low passages
were regularly stored in liquid nitrogen. Some others were further
passaged for future study.
Morphological characteristics
For light microscopy, the morphology of XL-2 cells
was observed using phase-contrast microscopy in bright fields as
80% confluent cultures (Olympus, Tokyo, Japan).
Determination of chromosome numbers
For chromosome preparation, XL-2 and XL-2sci cells
were seeded in a 75-cm2 culture flask (Corning, Lowell,
MA, USA). The cells at 70% confluence were treated with colchicine
(0.2 μg/ml) (Biodee Biotechnology) for 2 h before harvesting. After
centrifugation, 75 mM KCL hypotonic solution was added to the
incubated cell pellets at a concentration of 1×104
cells/ml for 25 min at 37°C, prior to fixation with
methanol/glacial acetic acid (3:1) following routine methods
(20). The cell suspension was
then dropped onto ice-cold slides and stained with Giemsa solution.
Approximately 50 mitotic figures per cell were randomly analyzed
and recorded.
Immunofluorescence
Immunofluorescence was performed to detect the
expression of pan-cytokeratin (21–23)
to identify the epithelial origin of XL-2. Because surfactant
protein C (SP-C) is considered a marker of mature type II alveolar
epithelial cells (AEC-II) (24–27),
immunofluorescence was also used to evaluate its expression level.
Cells were cultured on glass coverslips for 18–20 h, fixed with 4%
paraformaldehyde for 30 min, and permeabilized in 0.25% Triton
X-100 for 15 min. After blocking with immunostaining blocking
buffer (Beyotime, Jiangsu, China), cells were subsequently
incubated with the primary antibodies: rabbit polyclonal anti-SP-C
(1:150, sc-13979; Santa Cruz Biotechnology, Santa Cruz, CA, USA)
and anti-pan-cytokeratin (1:100, sc-15367; Santa Cruz
Biotechnology) overnight at 4°C. Slides were then incubated with a
mixture of FITC-labeled goat anti-rabbit IgG (1:100, Sigma) and
Alexa-Fluor-labeled chicken anti-rabbit IgG (1:100, Invitrogen) at
37°C for 1 h. Finally, they were incubated with DAPI (1:200,
Invitrogen) for 2 min. Slides were viewed with a Fluoview FV1000
microscope (Olympus).
Western blot analysis
An equal amount of protein samples extracted from
XL-2 and A549 (positive control) cells was separated using 15%
SDS-PAGE gels. Following electrophoresis, the proteins were
transferred onto PVDF filter membranes (Millipore, Billerica, MA,
USA). After blocking with 5% (w/v) non-fat dry milk for 1.5 h at
room temperature, the membrane was incubated with rabbit polyclonal
anti-SP-C (1:400) antibody overnight at 4°C. HRP-conjugated
anti-rabbit IgG (1:4,000, Sigma-Aldrich) was used as a secondary
antibody for a 2-h incubation. A 10-min PBST wash was conducted
three times after each antibody incubation step. Subsequent
visualization was performed using SuperSignal West Femto Maximun
Sensitivity Substrate (Thermo Fisher Scientific, Waltham, MA, USA)
and the loading control was β-actin (1:30,000, Sigma-Aldrich).
Animal experiments
Athymic male BALB/c-nu/nu mice, 6–8 weeks of age at
experiments were provided by Shanghai Cancer Institute and
maintained under specific pathogen-free (SPF) conditions. Housing
and all procedures involving animals were performed in strict
compliance with protocols approved by the Shanghai Medical
Experimental Animal Care Commission at Shanghai Jiaotong University
[approval ID SYXK (S) 2012-0001].
The tumorigenicity assay for XL-2 cell line was
conducted as previously described (28). Ten nude mice were used for the s.c.
route of inoculation. A total of 2×106 XL-2 cells mixed
with 0.2 ml saline were injected into the right upper flank of each
mouse, and tumor growth was monitored regularly. When the s.c.
tumors developed and reached 2 cm in largest dimension, they were
removed, preserved in 10% buffered formalin and embedded in
paraffin. Five non-sequential serial sections per animal were
sampled for hematoxylin and eosin (H&E) staining to evaluate
tumor morphology and for subsequent immunohistochemistry (IHC)
examination. Because A549 cell line was chosen as the positive
control in IHC analysis, thus, slices derived from the A549 s.c.
tumors were prepared in the same way as depicted above.
The model for testing drug-sensitivity of XL-2 was
generated as previously reported (29). Briefly, thirty-six mice was
randomly divided into two groups. The XL-2 group and A549 positive
control group were respectively subcutaneously injected with
2×106 of XL-2 or A549 cells per mouse. When tumor volume
reached ~100 mm3, the corresponding group was equally
divided into three subgroups (n=6), and this day was designated as
day 0. The three subgroups were treated with three different
formulations via intravenous injection: PBS (negative control),
doxorubicin (15 mg/kg) (twice a week) and docetaxel (4 mg/kg) (once
a week). Tumor size were regularly measured twice per week, and
tumor volume (mm3) was calculated using the formula:
tumor volume = 0.5 × length × width2 (mm3).
When body weight of most of the mice in one subgroup declined by
20%, the test of all the three subgroups were stopped, the mice
were sacrificed, the tumors excised, and photographed.
To establish the metastatic model, a consecutive
in vivo metastasis selection was carried out using XL-2 cell
line via s.c. injection as previously described (30,31).
Briefly, XL-2 cells were injected subcutaneously into the mice, and
primary tumors were excised 4 weeks later to improve the life-span
of each mouse, and for checking the appearance of spontaneous lung
metastasis. When the mice became moribund, macroscopic metastatic
lesions were removed and implanted into the s.c. region of new
recipient mice. Then the new flank-grown tumor was dissected to
initiate the daughter cells. When a certain number of cells were
present, they were re-injected subcutaneously into another
recipient for the next round. This procedure was repeated three
times. At the fourth and fifth rounds of selection, cells were
generated directly from the isolated pulmonary nodules. Nodules
were finely minced and then seeded in tissue culture dishes to
generate cells. Routinely, primary tumors were no longer needed to
be excised during the last two rounds for the shortened metastasis
time in this stage.
For comparison of the primary tumor growth and
spontaneous metastatic ability of XL-2 and XL-2sci cells, twenty
mice were randomly divided equally into two groups. Both cell lines
were implanted into the mice as described above. Tumor size was
measured, and tumor volume was calculated. Nine weeks
post-implantation, all individuals in these two parallel groups
were euthanized. Liver, spleen and lung were all harvested at
necropsy and processed according to the standard protocol for
H&E staining to analyze the presence of metastases.
IHC
Sections from primary s.c. tumors of XL-2 were
stained with pan-cytokeratin antibody (1:150) to confirm its
epithelial origin. The following antibodies were used to identify
the lung adenocarcinoma characteristics: thyroid transcription
factor-1 (TTF-1) (1:150, sc-13040; Santa Cruz Biotechnology) and
carcinoembryonic antigen (CEA) (1:100, 60053-1-Ig; Santa Cruz
Biotechnology). All procedures were performed according to the
manufacturer's protocols (Polymer HRP Detection System, Polink-2
plus, ZSGB-BIO, Beijing, China). Results were photographed and
observed with a full-automatic slide scanning system (Leica, Solms,
Germany).
Cells transduction
The GFP-Luc lentiviral vector was constructed by
recombining the plasmid eGFP-2A-CBGr99 that encodes the GFP-Luc
fusion gene (donated by Professor G.J. Hammerling) and a pWPXL
vector (Addgene). Following the instructions described by Addgene
(http://www.addgene.org), the XL-2 and XL-2sci
cells were transfected with the recombinant vector expressing the
fusion protein of GFP-Luc. Cell sorting (Epics Altra, Beckman
Coulter) was further introduced to purify the stable transfectants
expressing high levels of EGFP, and then the isolated cells were
amplified by conventional culture methods.
Luciferase imaging and GFP imaging
A Berthold LB983 NC320 NightOwl System (Berthold,
Bad Wildbad, Germany) was used in the s.c. xenografts model to
monitor the primary and metastatic tumor growth of XL-2 and XL-2sci
cells. All these procedures were conducted as previously described
(32,33). For in vivo bioluminescence
imaging (in vivo BLI), the animals were given D-luciferin
(150 mg/kg body weight in PBS) (Promega, San Luis Obispo, CA, USA)
by intra-peritoneal injection and anesthetized with 1%
pentobarbital sodium (10 ml/kg body weight) (Sigma-Aldrich).
Imaging was completed 15 min after the injection. In monitoring
pulmonary lesions, mice were euthanized, and their lungs were
excised because signals from the s.c. tumor would disturb the
observation. Ex vivo biofluorescence imaging (ex vivo
BFI) was adopted in this stage. The exposure time was 20 and 5 sec,
respectively, for in vivo BLI and ex vivo BFI.
Micro-computed tomography (micro-CT)
scanning
To provide three-dimensional non-destructive
detection of the pulmonary lesions, micro-CT (Skyscan1076, Kontich,
Belgium) was performed with the following parameters: 50 kV, 200
μA, 130-ms exposure, 0.7 degree at angle of increment and 360
views. A normal mouse was also scanned as a negative control. The
mice were anesthetized with 1% pentobarbital sodium (10 ml/kg body
weight) prior to the scanning. After being placed on its back on
the animal bed and banded across the chest area, each mouse was
imaged. Images of the chest were reconstructed.
Migration and Matrigel invasion
assays
Analyses assessing the migration and invasion of
cells were performed in 24-well Transwell plates (8-μm pore size,
Corning). For the invasion assay, we first prepared Matrigel-coated
Transwell chambers. Subsequently, 1×105 cells were added
into the upper chamber of each well. For the migration assay, the
number of cells seeded into upper chamber lined with a non-coated
membrane was 5×104. Cells for both assays were
resuspended with serum-free culture medium. To induce cell
invasion, 800 μl of 10% FBS medium was added to the lower chambers.
The incubation lasted 16 h for migration assays and 24 h for
invasion assays. Cells that had moved to the basal surface of the
chamber were fixed, stained with 0.1% crystal violet and imaged
with a CKX41 microscope (Olympus). Images of ten random fields from
three replicate wells were obtained. The number of cells were
counted, and the differences were analyzed. Assays were conducted
three independent times.
In vitro growth kinetics
For growth kinetics, subconfluent cells were seeded
at 2,000 cells/well in 96-well plates (Corning). After being
incubated for 24 h, the cells were counted at a regular time every
day for one week. A mixture of 10 μl of CCK8 (Dojindo, Kumamoto,
Japan) and 100 μl of DMEM was added to each well, which were then
incubated for 2 h. Three corresponding wells were measured
together, and the average number of cells were calculated based on
the absorbance at 450 nm of the triplicate wells. The cell doubling
time (Td) was calculated using the formula:
Td = T × lg2/(lgNt - lgN0), where
N0 represents the number of cells at the beginning,
Nt means the number of total living cells at the time of
counting and T represents the culturing time at the counting. The
experiments were independently repeated three times.
Cell cycle analysis
Both XL-2 and XL-2sci cells were fixed with 70%
ethanol at −20°C overnight. After fixation, cells were rinsed twice
with PBS, and then stained according to the manufacturer's protocol
(cell cycle detection kit, KeyGena FACSCalibur flow cytometer (BD
Biosciences, San Jose, CA, USA). Results were processed using
ModFit software (BD Biosciences). Every cell line was independently
analyzed three times.
Soft agar colony formation assays
For the soft agar colony formation assay, we used a
two-layer technique as described elsewhere (34) with some modifications, which
included a base layer consisting of 1% agar (Sigma-Aldrich) (0.25
ml/well) and a second layer containing XL-2 or XL-2sci cells (1,000
cells/well) with 0.6% agar in a 24-well plate. The plate was
subsequently cultured for 14 days at 37°C under 5% CO2.
Finally, colonies formed were observed using an Axioskop 2
microscope (Carl Zeiss, Oberkochen, Germany) and a DP70 Imaging
system (Olympus). This experiment was independently performed three
times.
ITRAQ labeled quantitative proteomics
analysis of XL-2 and XL-2sci cells
To comprehensively compare the proteome profile of
highly metastatic XL-2sci cells and its parental XL-2 cells, iTRAQ
quantitative proteomics approach was introduced (17). The first step was to prepare
proteins as previously described (17). Second, the iTRAQ 4-Plex reagents
(Applied Biosystems, Foster City, CA, USA) were applied to label
the samples. Cell lysates of XL-2 and XL-2sci cells were,
respectively, labeled with iTRAQ labeling reagents 114 and 115.
Briefly, comparable amount of each lysate (100 μg) go through
reducing, alkylating, digestion, plus desalination. Then, peptides
were separated by means of Agilent HPLC with a high PH RP column
(Durashell, C18, 250 mm × 4.6 mm, 5 μm). All these procedures were
conducted as previously reported by our lab (35). Third, a NanoLC system (NanoLC-2D
Ultra, Eksigent) was introduced to separate fractions and Triple
TOF 5600 mass spectrometer (AB SCIEX, USA) was used for analysis.
ProteinPilot4.1 software (AB SCIEX), was used for identification
and quantification of protein was performed via database searching.
The difference of the protein expression levels was considered
significant if iTRAQ ratio (115:114) was >1.5 or <0.67.
Finally, CapitalBio MAS 3.0 Molecule Annotation System (CapitalBio
Corp., Beijing, http://bioinfo.capitalbio.com/mas3/) was used in Gene
Ontology (GO) analysis of the upregulated and down-regulated
proteins. The analysis was performed mainly in three aspects:
cellular component (CC), biological processes (BP) and molecular
functions (MF).
Statistical analysis
Quantitative variables were represented as the mean
± SD and analyzed using Student's t-test. Qualitative variables
were compared by χ2 test. The significance level was set
at <0.05. All the analyses were performed with SPSS 19.0
software (IBM Corp., New York, NY, USA).
Results
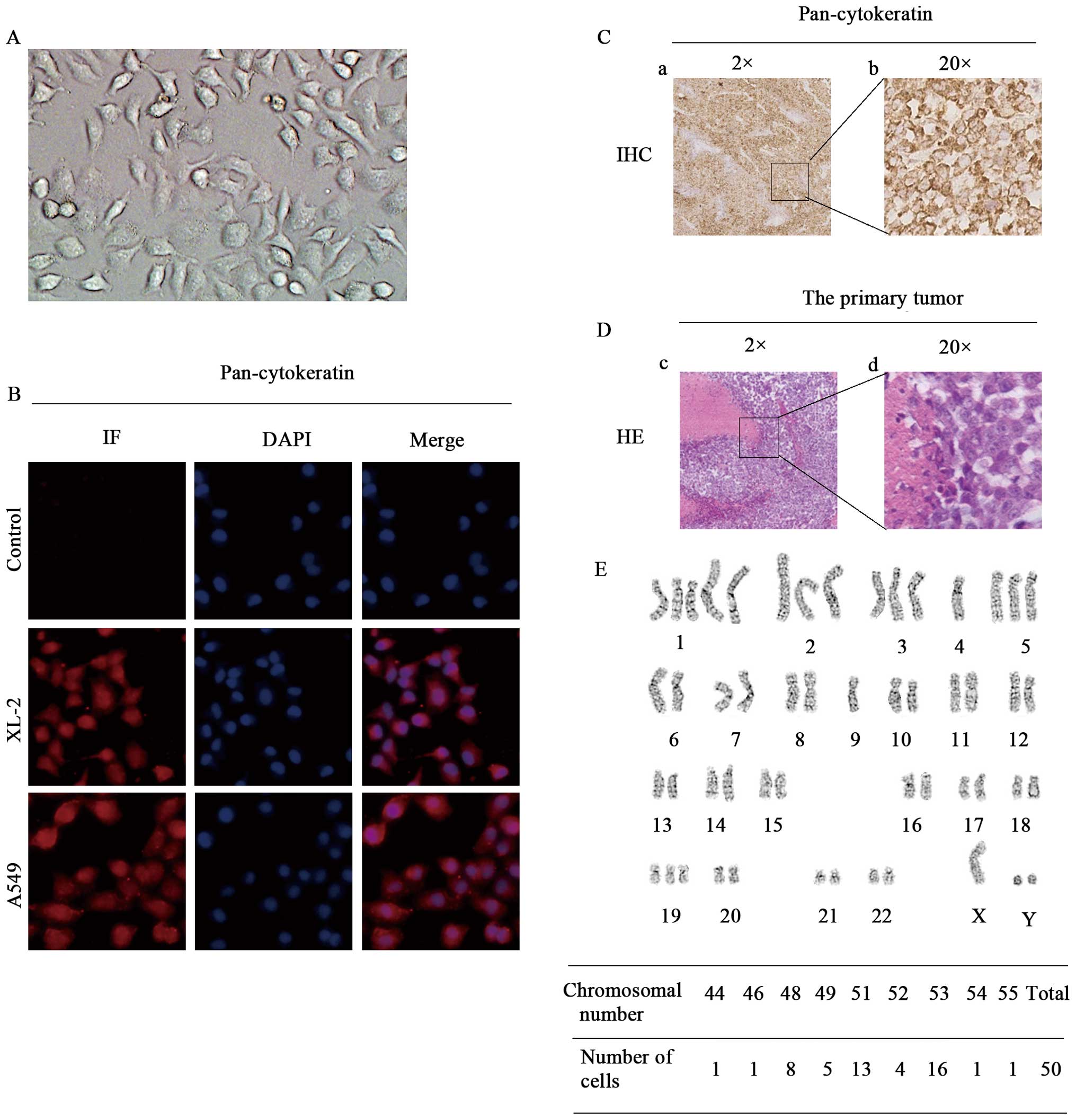
Cell morphology and phenotyping
A new lung cancer cell line designated XL-2 was
generated from ascites fluid. Cells were regularly certified as
free of bacteria or mycoplasma contamination. Determining the
morphology revealed that monolayer, polygonal, epithelial-like
cells adhered tightly to the bottom of the cell culture flasks
(Fig. 1A). The epithelial origin
was confirmed by positive immunoreactivity for epithelial marker
(pan-cytokeratin) in primary cells (Fig. 1B) and IHC analyses of the s.c.
tumor slides (Fig. 1C). XL-2 cell
line engrafted well and gave rise to growth in vivo.
Tumorigenicity rate was 100% in ten mice (data not shown). H&E
staining of the primary s.c. tumor is shown in Fig. 1D.
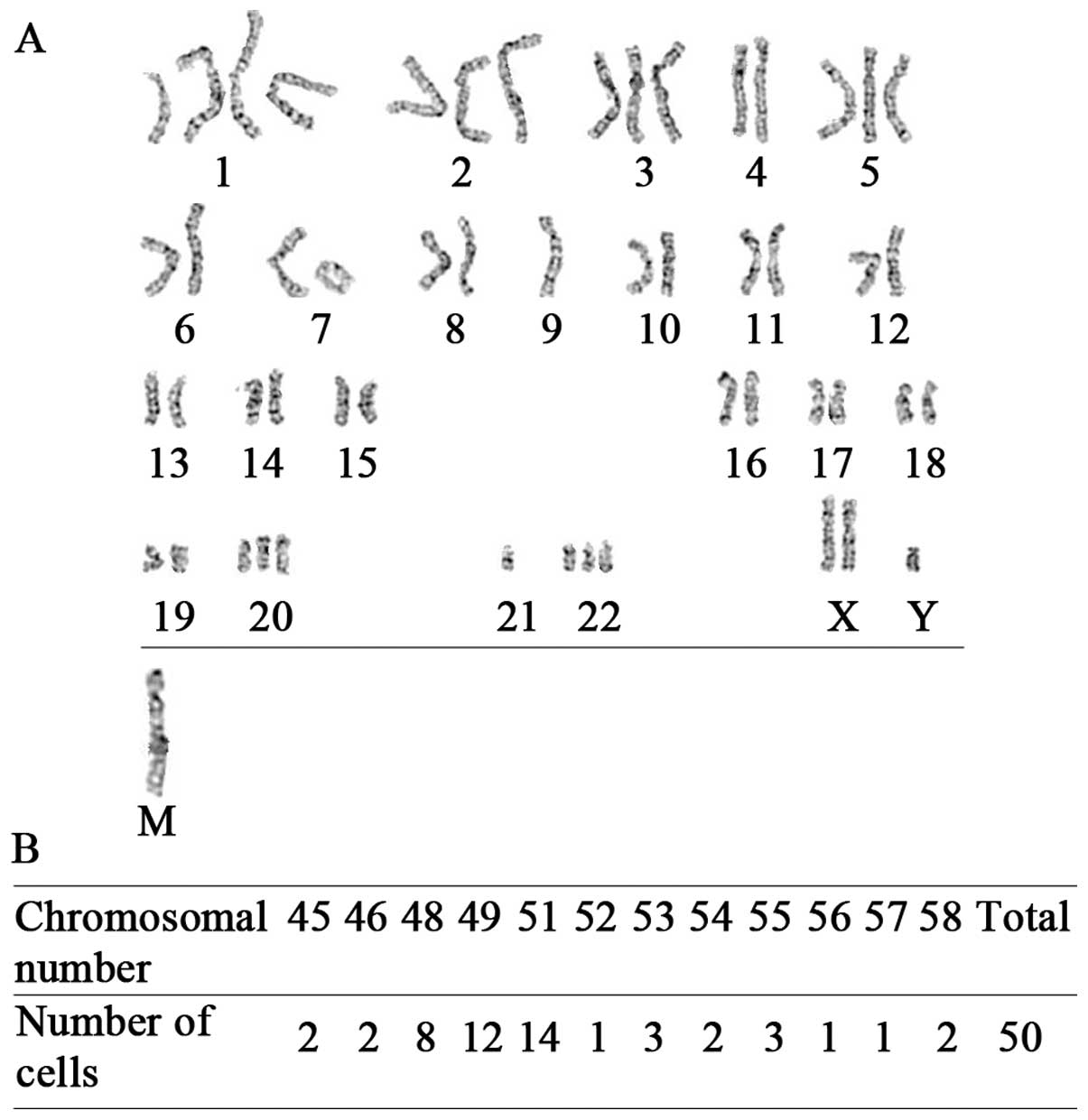
Chromosomal instability
Chromosome structural and numerical aberrations are
critical for the initiation of tumorigenesis (36) and are crucial features of cancer
cells versus normal ones. We therefore performed a karyotype
analysis of XL-2 cell line. As shown in Fig. 1E (lower panel), the 50 randomly
selected cells in metaphase contained between 44 and 55
chromosomes, with 51 and 53 being the predominant numbers. Both
gains and loss of chromosomes were frequent in this cell line
(Fig. 1E, upper panel).
der(1)(?::q11→q11::qter) and
der(14)(14pter→q11.2::1q12→qter)
were the two significant chromosomal translocations identified. In
summary, XL-2 cell line displayed chromosomal instability and
abnormal mitosis, which are hallmarks of cancer cells.
Additionally, the distinction between the karyotype of XL-2 and
that of a mouse, which includes 40 acrocentric chromosomes,
confirmed its human cell origin at the same time.
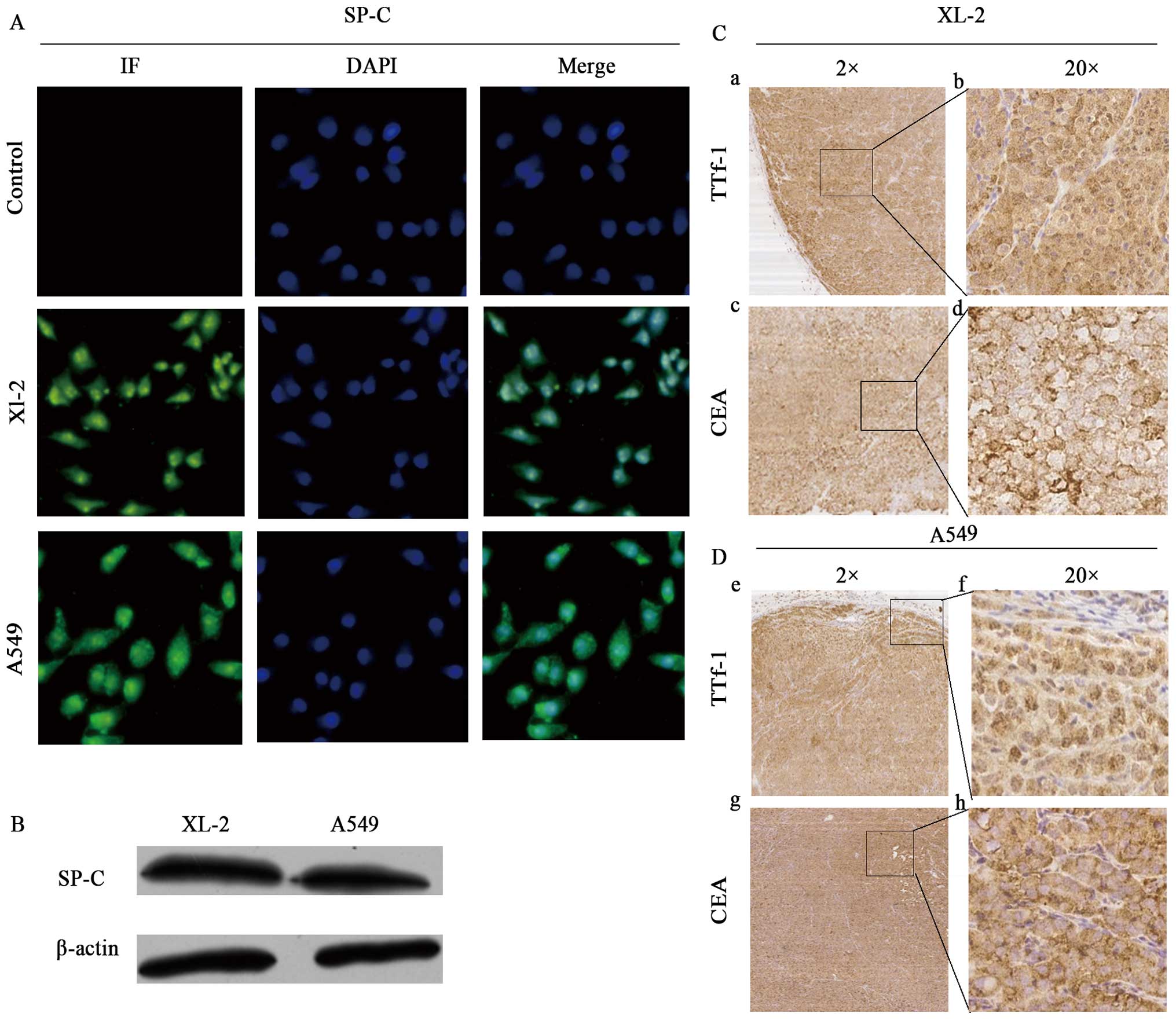
The characteristics of lung
adenocarcinoma
Because the clinicopathologic type of the tumor is
lung adenocarcinoma, to determine whether XL-2 possesses the
corresponding biological features, we first needed to prove that
XL-2 is a lung cancer cell line. Immunofluorescence staining and
western blotting were used to detect the expression of SP-C, which
is produced by AEC-II. As shown in Fig. 2A, the cells were positive for SP-C
by immunofluorescence detection. Western blot analysis (Fig. 2B) indicated that the XL-2 cells had
a similar secretion level to that of the A549 cells (derived from
AEC-II, a positive control). As CEA (37,38)
and TTf-1 (39–41) are common markers used for
identification of lung adeno-carcinoma, we further performed IHC
analysis of these two proteins using slides collected from the s.c.
tumors of mice in the tumorigenicity assay. Samples derived from
the A549 s.c. tumors was used as positive control. Remarkably, the
tissues were positive for both CEA and TTf-1, with cytoplasmic and
nuclear immunostaining, respectively (Fig. 2C), just as the control group
(Fig. 2D). This immunoprofile was
compatible with the features of adenocarcinoma. Thus, the XL-2 cell
line showed consistency with the clinicopathologic diagnosis.
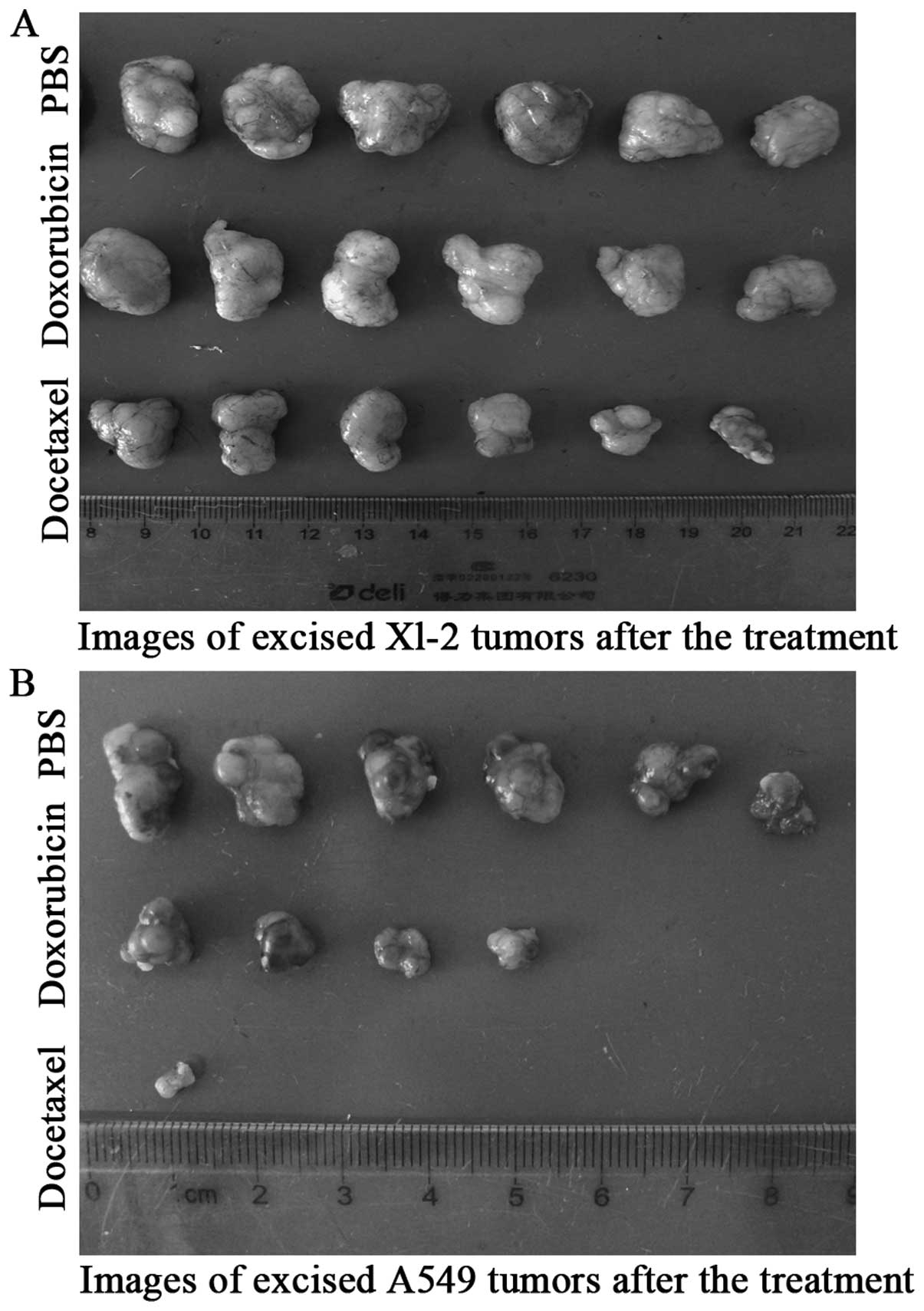
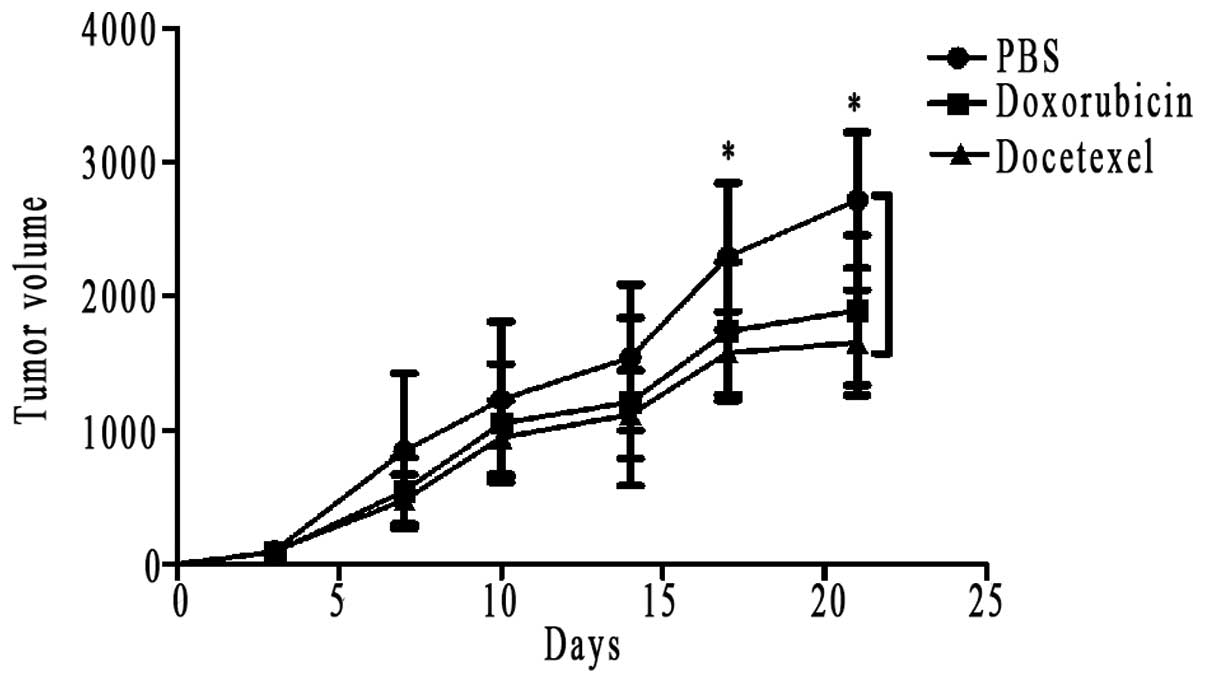
Drug susceptibility
Doxorubicin (42,43)
and docetaxel (44–47), which are commonly used as
chemotherapy regimens for advanced non-small cell lung cancer
(NSCLC), were introduced in the drug-sensitive of Xl-2 cells. As
shown in Fig. 3A, in the three
subgroups of XL-2 cells, docetaxel was apparently effective in
inhibiting tumor volume, compared with the rapid tumor growth of
PBS subgroup at day 21, and the difference was statistically
significant (Fig. 4). Same
conclusion could also be drawn in the A549 positive group, with
docetaxel almost completely inhibiting the tumor growth (Fig. 3B). Because some tumors disappeared
in A549 group, we can not depict its tumor growth curves. With
regard to the sensitivity to doxorubicin, XL-2 and A549 cells
exhibited different responses. In the subgroup of A549, the
majority of s.c. tumors exhibited a smaller volume than the PBS
subgroup, and the tumors of two mice even disappeared (Fig. 3B). However, the antitumor
efficiency of doxorubicin in XL-2 subgroup was not so evident, and
there was not a statistical difference between the tumor volume of
doxorubicin subgroup and the PBS subgroup (Fig. 4).
Highly metastatic XL-2sci cell line
obtained from in vivo selection
At the fifth round of screening, the lung metastases
were most obvious, and we obtained a highly metastatic subline of
XL-2 (designated XL-2sci). Additionally, we representatively
present the data in the first, third and fifth rounds of selection
to prove the escalating trend of the lung metastatic rate (Table I).
 | Table ILung metastatic rate and selection
periods in the first, third and fifth round of in vivo
metastasis selection. |
Table I
Lung metastatic rate and selection
periods in the first, third and fifth round of in vivo
metastasis selection.
| Round | Lung metastatic
ratea | Selection periods
(days)b |
|---|
| First | 3/10 | 99 |
| Third | 6/10 | 78 |
| Fifth | 9/10 | 63 |
Karyotype analysis of XL-2sci was also performed,
and results showed that in the 50 randomly selected cells in
meta-phase, the number of chromosomes ranged from 45 to 58, with 49
and 51 being the predominant numbers. The two chromosomal
translocations identified in XL-2 cells were also observed in
XL-2sci cells. However, der(9)(9pter→p11::1q11→qter) was the third
translocation unique to this cell line. Furthermore, there was a
marker chromosome in XL-2sci cells (data shown in Fig. 5).
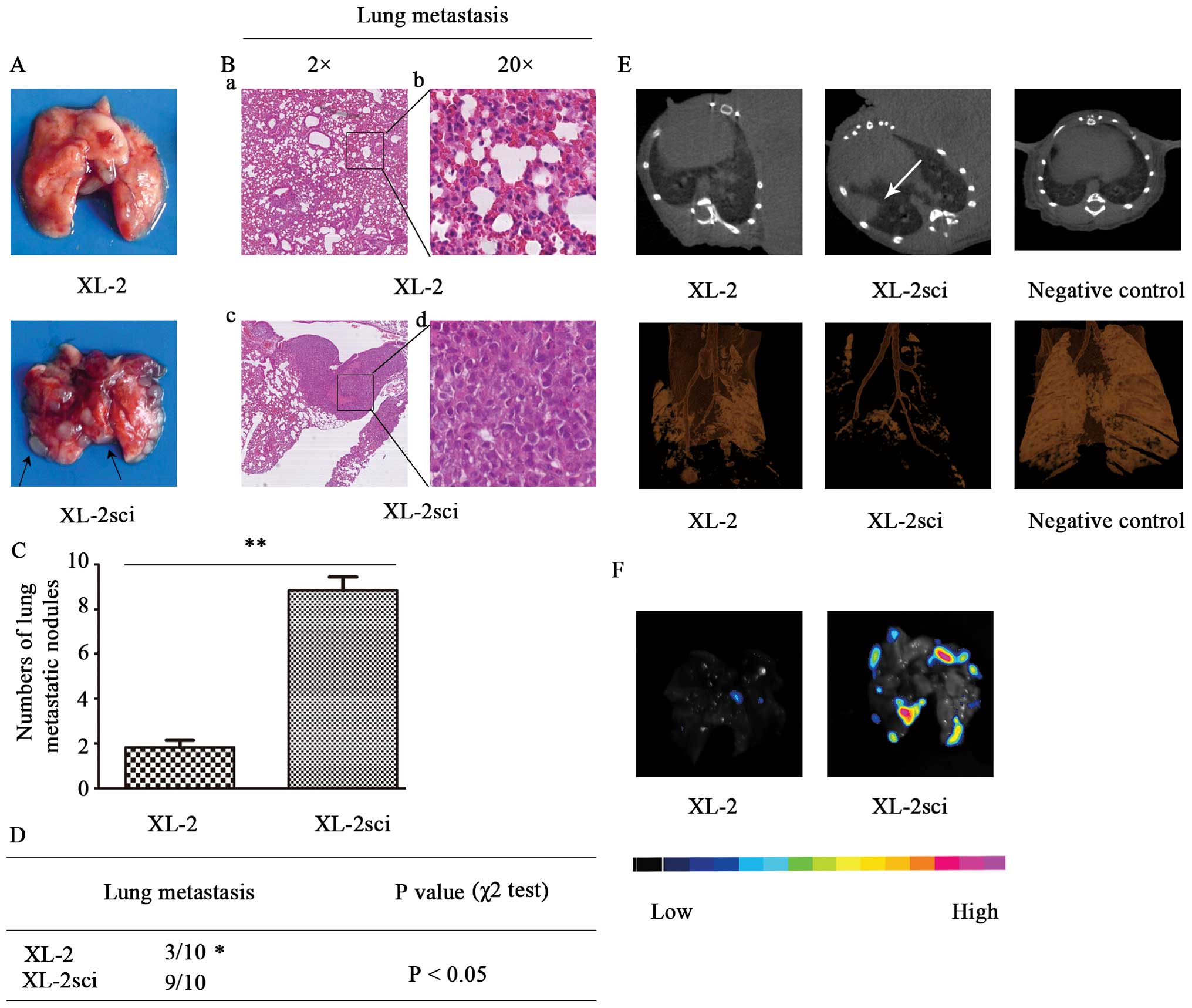
Metastatic lesions were more apparent in mice
injected with XL-2sci cells (Fig.
6A), which were further identified by histological analysis
(Fig. 6B). Mice in XL-2sci group
had significantly more pulmonary metastatic spots than those in
XL-2 group (P<0.01) (Fig. 6C).
The metastasis rates were 9/10 and 3/10 (P<0.05) in XL-2sci and
XL-2 tumor-bearing mice, respectively (Fig. 6D). In brief, the two cell lines
differed in metastatic ability, with XL-2sci being superior to
XL-2.
Comparison of the metastatic ability in
vivo with optical imaging and micro-CT scanning
To provide a more intuitive way to estimate the
in vivo characteristics of these two cell lines, we
subcutaneously injected GFP- and luciferase-labeled XL-2 and
XL-2sci cells as described above, which permitted us to monitor
tumor growth and metastasis non-invasively and semi-quantitatively.
When mice displayed signs of being moribund (9 weeks later), lung
metastases were monitored by in vivo BLI. However, the
positive BLI signals from primary s.c. tumors were so strong that
they interfered with the identification of pulmonary nodules.
Micro-CT imaging was then introduced. As shown in Fig. 6E, obvious tumor nodules were noted
in XL-2sci group. Changes in XL-2 group also appeared but were
minor. Significant differences with regard to tumor burden were
observed between the two groups. In the subsequent disposal, ex
vivo BFI imaging was applied to the evaluation of pulmonary
metastasis. A good accordance was demonstrated between ex
vivo BFI and micro-CT data from the same time-point. As shown
in Fig. 6F, light-emitting foci
were mostly localized in the XL-2sci group because only a low level
of photons was emitted from the recipients in the XL-2 group. In
short, XL-2sci cells were more prone to metastasize in
vivo.
Comparison of cellular migration and
invasive capability in vitro
Considerable differences were observed in the
migration and invasive properties of these two cell lines, as
evaluated by in vitro Transwell migration and invasion
assays (Fig. 7A). Regarding the
invasive potential, significantly more XL-2sci cells passed through
the Matrigel than did XL-2 cells. The parallel migration assay
yielded similar results. These data suggest that the XL-2sci cells
are markedly more aggressive than their parental cells.
Consequently, we developed a subpopulation of XL-2 cells (XL-2sci)
with enhanced metastatic competence.
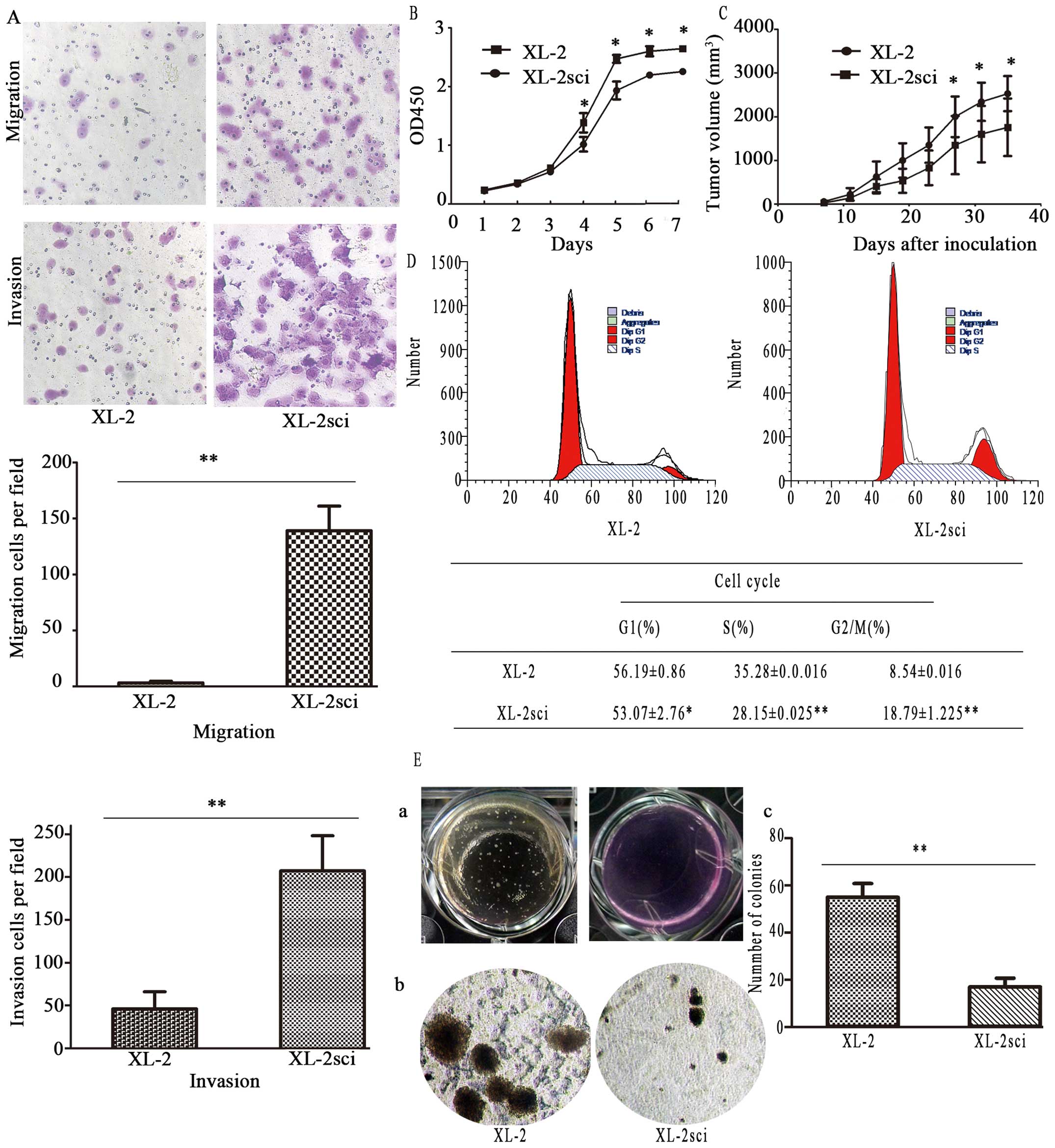 | Figure 7The comparison of migration, invasive
and proliferation potential. (A) Comparison of migration and
invasive potential of XL-2 and XL-2sci cells. The phase-contrast
images (upper panels) show that more XL-2sci cells penetrated
through the underside of the inserts than did XL-2 cells in both
migration and invasion assays (magnification, ×100). The lower
panels show histograms of the quantification. The results are
represented as the mean ± SD from at least three independent
experiments, **P<0.01. (B) In vitro growth
curves of XL-2 and XL-2sci cells. Cells were seeded into the
corresponding 96-well plates, after which cells were counted at the
indicated times. Results are expressed as the mean ± SD;
*P<0.05. (C) In vivo growth curve of XL-2 and
XL-2sci cells. Cells (2.0×106 per mouse) were
subcutaneously implanted into BALB/c-nu/nu mice, after which the
tumor volume was measured every four days. The results are
expressed as the mean ± SD, *P<0.05. (D) Cell cycle
analysis. Upper panel, representative images of the cell cycle
assays. Lower panel, a table depicting the results of the cell
cycle assays. The percentage of XL-2sci cells in S phase was lower
than that of XL-2 cells, *P<0.05,
**P<0.01. (E) Soft agar colony formation assay
(clonogenic assay) of XL-2 and XL-2sci cells. (a) Large-scale image
of the soft agar colony formation assay. (b) Representative image
of the microscopic colonies. (c) Quantification of the colonies
expressed as the mean ± SD; **P<0.01. |
Comparison of proliferation ability
The proliferation ability of XL-2 and XL-2sci cells
was compared using growth curves and soft agar colony formation
assay. The doubling time of XL-2 and XL-2sci cells was 31.92±2.73
and 37.10±4.90 h, respectively (P<0.05, Fig. 7B), indicating that the
proliferation rate of XL-2sci was significantly lower than that of
XL-2. We achieved similar results as to the growth rate of s.c.
tumors (Fig. 7C). In subsequent
cell cycle analysis (Fig. 7D),
data showed that 35.28% of XL-2 cells were in S phase, in contrast
to 28.15% of XL-2sci cells. The number of XL-2sci cells in G2/M
phase (18.79%) was approximately two times higher than that of XL-2
cells, indicating a more proliferative phenotype in XL-2 compared
with XL-2sci. In the soft agar colony formation assay, both cell
lines produced colonies but had significantly different
colony-formation ability (Fig.
7E-a). Both the number and size of XL-2 colonies increased when
compared with those of the XL-2sci colonies (Fig. 7E-b). The two cell lines showed a
significant difference (P<0.01) in the colony formation rate
(Fig. 7E-c).
Differentially expressed proteins between
XL-2sci and Xl-2
After one technical replicate, we respectively
identified a total of 3347/3330 proteins (global FDR <1%) using
ProteinPilot 4.1. Limited by the screening condition of unused
protein score >1.3 and a number of peptides ≥2, 2148/2373
proteins were identified. There were 18 upregulated (iTRAQ ratio
115:114 >1.5) and 32 downregulated (iTRAQ ratio 115:114
<0.67) proteins in XL-2sci cells, compared with XL-2 cells
(Tables II and III).
 | Table IIList of upregulated proteins. |
Table II
List of upregulated proteins.
| Accession
no.a | Protein name | Unused score (mean
± SD)b | % cov (95) (mean ±
SD)c | Peptides (95%)
(mean ± SD)d | iTRAQ rate (mean ±
SD)e |
|---|
| P00918 | CAH2 |
7.8±0.282842712 |
28.45±8.697413409 | 6±1 |
2.49565±0.259578899 |
| P02787 | TRFE |
2.34±0.890954544 | 12.5±0 | 3±1 |
1.9729±0.332481609 |
| P04083 | ANXA1 |
47.845±3.217335854 |
65.6±2.828427125 | 59±4 |
1.933±0.088105505 |
| P05787 | K2C8 |
83.885±4.207285348 |
71.7±0.141421356 | 75±4 |
1.6534±0.096873629 |
| P09525 | ANXA4 |
18.435±8.195367594 |
52.2±6.929646456 | 12±4 |
2.67675±0.485428805 |
| P10768 | ESTD |
7.16±1.060660172 |
28.2±0.282842712 | 6±1 |
2.4864±1.081166268 |
| Q04446 | GLGB |
119.205±6.75286976 |
74.55±1.060660172 | 123±1 |
1.7236±0.257245447 |
| Q16658 | FSCN1 |
19.53±1.824335495 |
35.7±7.778174593 | 14±0 |
2.2537±0.205202388 |
| Q16719 | KYNU |
30.67±1.555634919 |
47.95±2.757716447 | 19±1 |
1.83485±0.332693741 |
| Q32MZ4 | LRRF1 |
29.275±0.360624458 | 46±0 | 22±1 |
1.89915±0.135976634 |
| Q92597 | NDRG1 |
17.18±1.187939392 |
28.9±0.424264069 | 10±0 |
2.2508±0.747977553 |
| Q96FQ6 | S10AG |
4.02±0.028284271 |
8.65±2.899137803 | 2±0 |
2.0106±0.222172951 |
| Q99584 | S10AD |
4.88±3.577960313 | 42.7±0 | 3±1 |
3.37905±1.226335291 |
| Q9H2J7 | S6A15 |
3.365±1.859690835 | 25±6.505382387 | 3±1 |
1.6524±0.053740115 |
| Q9H2J7 | S6A15 |
2.84±1.48492424 | 8±1.272792206 | 3±1 |
1.91475±0.062296107 |
| B2WTI4 | B2WTI4 |
7.135±1.364716088 |
23.55±1.626345597 | 6±1 |
1.60065±0.08336789 |
| E7EU05 | E7EU05 |
9.18±0.339411255 |
21.25±2.61629509 | 7±0 |
3.11955±1.429699201 |
| G3V1S6 | G3V1S6 |
17.255±0.091923882 |
32.75±1.48492424 | 10±2 |
3.9979±1.49949064 |
 | Table IIIList of downregulated proteins. |
Table III
List of downregulated proteins.
| Accession
no.a | Protein name | Unused score (mean
± SD)b | % cov (95) (mean ±
SD)c | Peptides (95%)
(mean ± SD)d | iTRAQ rate (mean ±
SD)e |
|---|
| O14965 | AURKA |
4.415±2.057680733 |
23.3±9.475230868 | 3±1 |
0.4295±0.107904495 |
| O43175 | SERA |
20.53±1.569777054 |
32.65±4.030508653 | 12±0 |
0.31645±0.016475588 |
| O75643 | U520 |
62.62±2.884995667 |
31.9±4.101219331 | 41±7 |
0.63135±0.032880465 |
| O95433 | AHSA1 |
20.09±1.329360749 |
53.25±2.050609665 | 16±3 |
0.538±0.045537677 |
| P04818 | TYSY |
11.505±0.728319985 |
30.2±3.818376618 | 6.5±1 |
0.42345±0.25335636 |
| P05120 | PAI2 |
23.4±0.353553391 |
55.15±6.151828996 | 14.5±1 |
0.34675±0.047305444 |
| P09874 | PARP1 |
32.07±5.246732316 |
32.9±0.282842712 | 21.5±5 |
0.4633±0.086974134 |
| P11216 | PYGB |
51.085±0.091923882 |
52.1±1.414213562 | 36±1 |
0.52305±0.160866793 |
| P12277 | KCRB |
21.635±2.312239174 |
46.05±7.283199846 | 14±3 |
0.24005±0.01251579 |
| P17931 | LEG3 |
16.34±5.628569978 |
45.6±7.919595949 | 16.5±1 |
0.23475±0.095105862 |
| P18669 | PGAM1 | 31±5.868986284 |
63.15±4.737615434 | 22.5±2 |
0.5916±0.080610173 |
| P23246 | SFPQ |
16.28±1.796051224 |
29.95±0.494974747 | 10.5±1 |
0.421±0.02192031 |
| P23381 | SYWC |
19.145±2.877924599 |
36.95±1.767766953 | 14±0 |
0.34825±0.065407377 |
| P23526 | SAHH |
28.685±0.16263456 |
42.85±0.636396103 | 22.5±1 |
0.2939±0.01145513 |
| P25815 | S100P |
8.58±2.008183259 |
44.75±3.74766594 | 5±0 |
0.4637±0.066185195 |
| P27695 | APEX1 |
18.65±0.707106781 |
43.25±2.899137803 | 13±0 |
0.4232±0.03026417 |
| P43490 | NAMPT |
70.375±2.679934701 |
73.65±2.474873734 | 58±4 |
0.60645±0.051265242 |
| P46821 | MAP1B |
57.255±6.130615793 |
28.5±2.545584412 | 34±3 |
0.54755±0.109813683 |
| P62805 | H4 |
19.71±2.870853532 |
60.2±1.414213562 | 18.5±2 |
0.037±0.010040916 |
| Q05639 | EF1A2 |
10.905±3.712310601 |
59.55±2.899137803 | 33.5±1 |
0.2535±0.159664711 |
| Q14432 | PDE3A |
3.575±0.827314934 |
10.15±1.767766953 | 2±0 |
0.61095±0.004030509 |
| Q15020 | SART3 |
11.395±1.803122292 |
20.95±2.333452378 | 7±1 |
0.44965±0.040941483 |
| Q16555 | DPYL2 |
46.505±6.130615793 | 58.4±0 | 31.5±2 |
0.48235±0.124238661 |
| Q6EMK4 | VASN |
3.53±0.381837662 |
10.65±5.161879503 | 2.5±1 |
0.47635±0.110662211 |
| Q6FI13 | H2A2A | 2±0 |
51.9±3.818376618 | 12.5±1 |
0.3077±0.09277241 |
| Q9NR30 | DDX21 |
29.19±0.537401154 |
37.65±8.838834765 | 17.5±1 |
0.38045±0.019869701 |
| Q9Y2J2-2 | E41L3 |
12.555±2.453660531 |
21.35±0.494974747 | 8.5±1 |
0.2993±0.090226825 |
| D6RD18 | D6RD18 |
10.985±0.784888527 |
36.9±3.252691193 | 8±1 |
0.42375±0.071347074 |
| F8W1S7 | F8W1S7 |
35.69±2.771858582 |
70.75±1.060660172 | 41±6 |
0.55985±0.15125014 |
| F8W6L6 | F8W6L6 |
37.705±8.20950973 |
32.5±1.555634919 | 32±10 |
0.2693±0.012303658 |
| J3QSV6 | J3QSV6 |
8.94±0.721248917 |
29.05±0.636396103 | 4.5±1 |
0.46375±0.236527218 |
| Q4JM47 | Q4JM47 |
11.21±3.012274888 |
44.65±2.192031022 | 8±0 |
0.1281±0.108894444 |
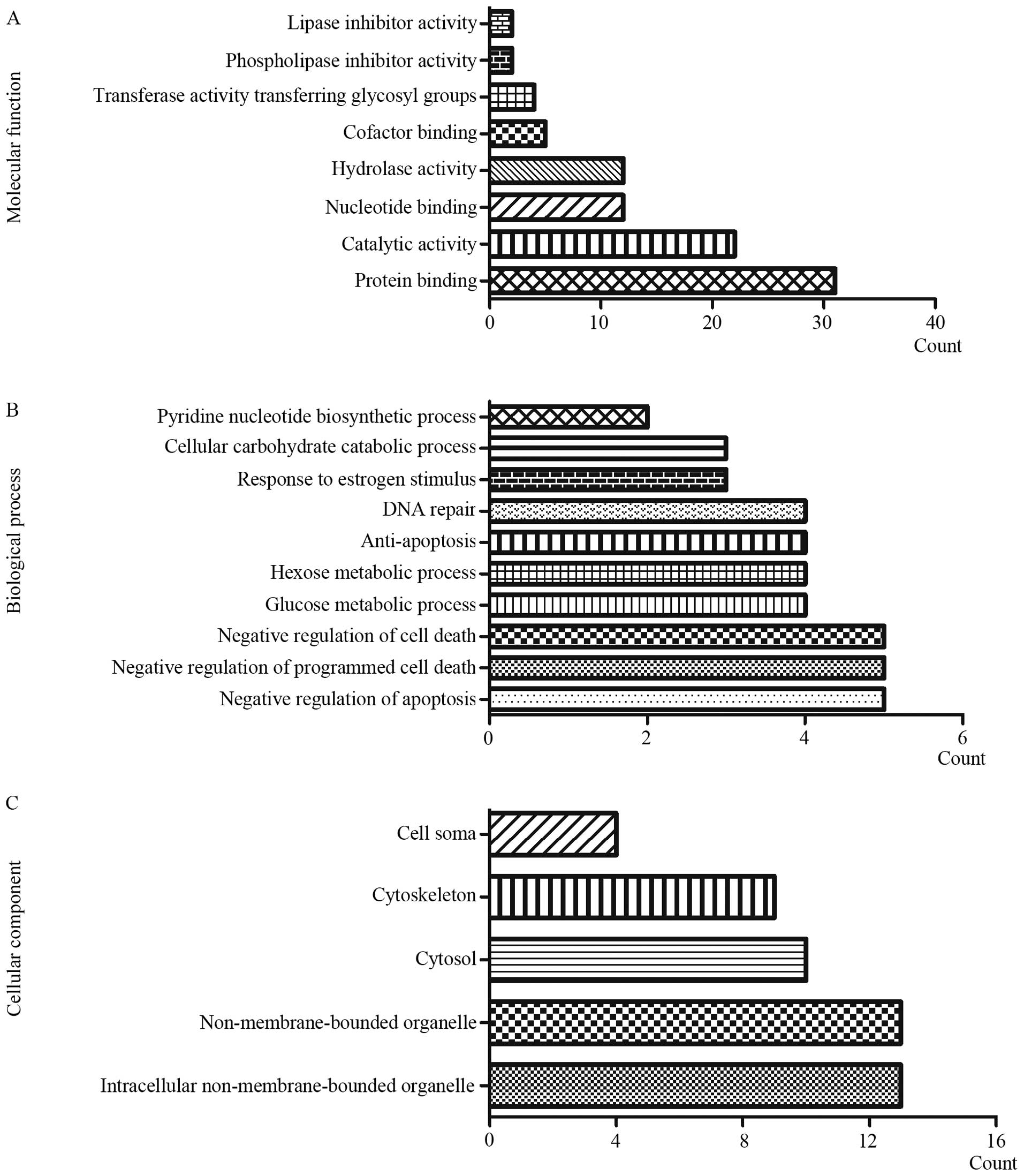
GO analysis
GO analysis of the dysregulated proteins was
performed via CapitalBio MAS 3.0. In terms of cellular component
(CC), the differentially expressed proteins mainly focused on
intracellular non-membrane-bound organelle, non-membrane-bound
organelle and cytosol; regarding molecular functions (MF), the
dysregulated proteins mainly centered on protein binding, catalytic
activity and nucleotide binding; as for biological processes (BP),
the top three of the focused aspects are negative regulation of
apoptosis, negative regulation of programmed cell death and
negative regulation of cell death (Fig. 8).
Discussion
Well-characterized primary tumor cell lines can be
invaluable tools for better understanding cancer biology (48) and exploring new treatment
modalities. Nevertheless, many cell lines have lost their original
biological characteristics during long-term culture in vitro
(49,50). Thus, the establishment of novel
cell lines that reliably reflect clinical features is apparently
needed. In general, the availability of newly established lung
cancer cell lines is relatively more limited compared with other
types of epithelia-originating cell lines. Not direct ascites fluid
incubation, but gradient centrifugation was introduced in our
study. The advances in cell culture methods enabled us to develop a
permanently growing lung cancer cell line.
Through cellular and histological techniques, XL-2
was finally identified as a lung adenocarcinoma cell line. The
structural and numerical aberrations in karyotype analysis
illustrated its genomic abnormalities. Drug trail showed that it
was sensitive to docetaxel and resistant to doxorubicin, which
might be attributed to the patient's multiple chemotherapy with
doxorubicin. Hence, XL-2 cell line may serve as a cell line model
of drug resistance for identifying mechanisms involved in the
resistance to doxorubicin. To sum up, this newly isolated cell line
should provide a beneficial addition to the cell lines currently
available for investigating the biological characteristics of lung
cancer.
Although metastasis is the foremost cause of lung
cancer recurrence and mortality, we have limited access to the
mechanisms. Searching for metastasis-associated genes and proteins
will contribute greatly to the inhibition of tumor development and
progression. This spontaneous metastatic model derived from s.c.
injection provided us with a new cell line with much increased
metastatic properties, XL-2sci. Its enhanced metastatic capacity
versus XL-2 was identified by optical imaging, micro-CT scanning
in vivo and Transwell assays in vitro. Besides, by
means of ITRAQ labeled proteomics profiling study, we also found 50
dysregulated proteins in XL-2sci, some of which played an important
role in tumor metastasis. For example, FSCN1 has been reported to
be relevant with NSCLC metastasis, and G3V1S6 was said to be
involved in the distant metastasis of breast cancer (51), gastric cancer (52) plus liver cancer (53). In addition, NDRG1 was connected
with colorectal cancer and liver cancer, whereas, it was highly
expressed in lung cancer. Hence, in a subsequent study, we can
choose some of these metastasis-associated proteins having been
reported in other types of tumors to further explore their role in
lung cancer metastasis.
Proliferation ability of the two cell lines were
also evaluated in our study. Not only the doubling time and soft
agar colony formation assays in vitro but also the growth
rate of s.c. tumors in vivo suggested that XL-2sci cell line
had a decreased proliferation ability, indicating its enhanced
invasion and metastatic activity was not facilitated by a faster
growth rate. An inverse correlation between proliferation and
metastasis was demonstrated in our study, which was consistent with
previous reports (31,54).
In conclusion, we triumphantly established a lung
adenocarcinoma cell line (XL-2) from clinical specimens and
characterized it at cellular and histological levels. By means of
XL-2 cells, we developed a metastatic model via s.c. injection and
harvested a novel cell line with enhanced metastatic competence
(XL-2sci). Retaining the clinical features of original tumor, this
metastatic model will offer a reliable platform for in-depth
investigation of lung cancer.
Acknowledgements
This study was supported by Natural Science
Foundation of China (81472176), Shanghai Science and Technology
Developing Program (13140900502), State Key Laboratory of Oncogenes
and Related Genes Research Fund (91-14-15). We would like to thank
Li-Xing Zhang, Li-Xia Wu and Yu Zhang for their kind suggestions
and excellent technical assistance. We also thank the Nature
Publishing Group Language Editing for revising the English used in
this manuscript.
References
|
1
|
Siegel R, Naishadham D and Jemal A: Cancer
statistics, 2012. CA Cancer J Clin. 62:10–29. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hanahan D and Weinberg RA: The hallmarks
of cancer. Cell. 100:57–70. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Crinò L, Weder W, van Meerbeeck J and
Felip E; ESMO Guidelines Working Group. Early stage and locally
advanced (non-metastatic) non-small-cell lung cancer: ESMO Clinical
Practice Guidelines for diagnosis, treatment and follow-up. Ann
Oncol. 21(Suppl 5): V103–V115. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sekhon HS, London CA, Sekhon M, Iversen PL
and Devi GR: c-MYC antisense phosphosphorodiamidate morpholino
oligomer inhibits lung metastasis in a murine tumor model. Lung
Cancer. 60:347–354. 2008. View Article : Google Scholar
|
|
5
|
Fork MA, Murua Escobar H, Soller JT,
Sterenczak KA, Willenbrock S, Winkler S, Dorsch M, Reimann-Berg N,
Hedrich HJ, Bullerdiek J, et al: Establishing an in vivo model of
canine prostate carcinoma using the new cell line CT1258. BMC
Cancer. 8:2402008. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Capellá G, Farré L, Villanueva A, Reyes G,
García C, Tarafa G and Lluís F: Orthotopic models of human
pancreatic cancer. Ann NY Acad Sci. 880:103–109. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Talmadge JE, Singh RK, Fidler IJ and Raz
A: Murine models to evaluate novel and conventional therapeutic
strategies for cancer. Am J Pathol. 170:793–804. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Minn AJ, Gupta GP, Siegel PM, Bos PD, Shu
W, Giri DD, Viale A, Olshen AB, Gerald WL and Massagué J: Genes
that mediate breast cancer metastasis to lung. Nature. 436:518–524.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Daniel VC, Marchionni L, Hierman JS,
Rhodes JT, Devereux WL, Rudin CM, Yung R, Parmigiani G, Dorsch M,
Peacock CD, et al: A primary xenograft model of small-cell lung
cancer reveals irreversible changes in gene expression imposed by
culture in vitro. Cancer Res. 69:3364–3373. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Frese KK and Tuveson DA: Maximizing mouse
cancer models. Nat Rev Cancer. 7:645–658. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Voskoglou-Nomikos T, Pater JL and Seymour
L: Clinical predictive value of the in vitro cell line, human
xenograft, and mouse allograft preclinical cancer models. Clin
Cancer Res. 9:4227–4239. 2003.PubMed/NCBI
|
|
12
|
Hammer S, Sommer A, Fichtner I, Becker M,
Rolff J, Merk J, Klar U and Hoffmann J: Comparative profiling of
the novel epothilone, sagopilone, in xenografts derived from
primary non-small cell lung cancer. Clin Cancer Res. 16:1452–1465.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Masui O, White NM, DeSouza LV, Krakovska
O, Matta A, Metias S, Khalil B, Romaschin AD, Honey RJ, Stewart R,
et al: Quantitative proteomic analysis in metastatic renal cell
carcinoma reveals a unique set of proteins with potential
prognostic significance. Mol Cell Proteomics. 12:132–144. 2013.
View Article : Google Scholar :
|
|
14
|
Hou Q, Tan HT, Lim KH, Lim TK, Khoo A, Tan
IB, Yeoh KG and Chung MC: Identification and functional validation
of caldesmon as a potential gastric cancer metastasis-associated
protein. J Proteome Res. 12:980–990. 2013. View Article : Google Scholar
|
|
15
|
Qin X, Chen Q, Sun C, Wang C, Peng Q, Xie
L, Liu Y and Li S: High-throughput screening of tumor
metastatic-related differential glycoprotein in hepatocellular
carcinoma by iTRAQ combines lectin-related techniques. Med Oncol.
30:4202013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yu T, Li J, Yan M, Liu L, Lin H, Zhao F,
Sun L, Zhang Y, Cui Y, Zhang F, et al: MicroRNA-193a-3p and -5p
suppress the metastasis of human non-small-cell lung cancer by
downregulating the ERBB4/PIK3R3/mTOR/S6K2 signaling pathway.
Oncogene. 34:413–423. 2015. View Article : Google Scholar
|
|
17
|
Lin HC, Zhang FL, Geng Q, Yu T, Cui YQ,
Liu XH, Li J, Yan MX, Liu L, He XH, et al: Quantitative proteomic
analysis identifies CPNE3 as a novel metastasis-promoting gene in
NSCLC. J Proteome Res. 12:3423–3433. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Dunkley PR, Jarvie PE and Robinson PJ: A
rapid Percoll gradient procedure for preparation of synaptosomes.
Nat Protoc. 3:1718–1728. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Zhao Q, Feng Y, Jia X, Yin L, Zheng Y,
Ouyang D, Zhou H and Zhang L: Proteome analysis of hepatic
non-parenchymal cells of immune liver fibrosis rats. Sci China Life
Sci. 57:303–314. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bartnitzke S, Motzko H, Caselitz J,
Kornberg M, Bullerdiek J and Schloot W: A recurrent marker
chromosome involving chromosome 1 in two mammary tumors of the dog.
Cytogenet Cell Genet. 60:135–137. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chambers JK, Uchida K, Ise K and Nakayama
H: Cystic rete ovarii and uterine tube adenoma in a rabbit. J Vet
Med Sci. 76:909–912. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Cousins FL, Murray A, Esnal A, Gibson DA,
Critchley HO and Saunders PT: Evidence from a mouse model that
epithelial cell migration and mesenchymal-epithelial transition
contribute to rapid restoration of uterine tissue integrity during
menstruation. PLoS One. 9:e863782014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kamalidehghan B, Houshmand M,
Kamalidehghan F, Jafarzadeh N, Azari S, Akmal SN and Rosli R:
Establishment and characterization of two human breast carcinoma
cell lines by spontaneous immortalization: Discordance between
estrogen, progesterone and HER2/neu receptors of breast carcinoma
tissues with derived cell lines. Cancer Cell Int. 12:432012.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sozo F, Wallace MJ, Hanna MR, Flecknoe SJ,
Cock ML, Maritz GS, Harding R and Hooper SB: Alveolar epithelial
cell differentiation and surfactant protein expression after mild
preterm birth in sheep. Pediatr Res. 59:151–156. 2006. View Article : Google Scholar
|
|
25
|
Woodcock-Mitchell J, Mitchell JJ, Reynolds
SE, Leslie KO and Low RB: Alveolar epithelial cell keratin
expression during lung development. Am J Respir Cell Mol Biol.
2:503–514. 1990. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ito Y, Ahmad A, Kewley E and Mason RJ:
Hypoxia-inducible factor regulates expression of surfactant protein
in alveolar type II cells in vitro. Am J Respir Cell Mol Biol.
45:938–945. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ito Y and Mason RJ: The effect of
interleukin-13 (IL-13) and interferon-γ (IFN-γ) on expression of
surfactant proteins in adult human alveolar type II cells in vitro.
Respir Res. 11:1572010. View Article : Google Scholar
|
|
28
|
Peng T, Zhang P, Liu J, Nguyen T,
Bolshakov S, Belousov R, Young ED, Wang X, Brewer K, López-Terrada
DH, et al: An experimental model for the study of
well-differentiated and dedifferentiated liposarcoma; deregulation
of targetable tyrosine kinase receptors. Lab Invest. 91:392–403.
2011. View Article : Google Scholar :
|
|
29
|
Lv S, Tang Z, Li M, Lin J, Song W, Liu H,
Huang Y, Zhang Y and Chen X: Co-delivery of doxorubicin and
paclitaxel by PEG-polypeptide nanovehicle for the treatment of
non-small cell lung cancer. Biomaterials. 35:6118–6129. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Munoz R, Man S, Shaked Y, Lee CR, Wong J,
Francia G and Kerbel RS: Highly efficacious nontoxic preclinical
treatment for advanced metastatic breast cancer using combination
oral UFT-cyclophosphamide metronomic chemotherapy. Cancer Res.
66:3386–3391. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jia D, Yan M, Wang X, Hao X, Liang L, Liu
L, Kong H, He X, Li J and Yao M: Development of a highly metastatic
model that reveals a crucial role of fibronectin in lung cancer
cell migration and invasion. BMC Cancer. 10:3642010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Caceres G, Zhu XY, Jiao JA, Zankina R,
Aller A and Andreotti P: Imaging of luciferase and GFP-transfected
human tumours in nude mice. Luminescence. 18:218–223. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ciana P, Brena A, Sparaciari P, Bonetti E,
Di Lorenzo D and Maggi A: Estrogenic activities in rodent
estrogen-free diets. Endocrinology. 146:5144–5150. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Takahashi M, Furihata M, Akimitsu N,
Watanabe M, Kaul S, Yumoto N and Okada T: A highly bone marrow
metastatic murine breast cancer model established through in vivo
selection exhibits enhanced anchorage-independent growth and cell
migration mediated by ICAM-1. Clin Exp Metastasis. 25:517–529.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang F, Lin H, Gu A, Li J, Liu L, Yu T,
Cui Y, Deng W, Yan M, Li J, et al: SWATH™- and iTRAQ-based
quantitative proteomic analyses reveal an overexpression and
biological relevance of CD109 in advanced NSCLC. J Proteomics.
102:125–136. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Conway AE, Lindgren A, Galic Z, Pyle AD,
Wu H, Zack JA, Pelligrini M, Teitell MA and Clark AT: A
self-renewal program controls the expansion of genetically unstable
cancer stem cells in pluripotent stem cell-derived tumors. Stem
Cells. 27:18–28. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mimura T, Ito A, Sakuma T, Ohbayashi C,
Yoshimura M, Tsubota N, Okita Y and Okada M: Novel marker D2-40,
combined with calretinin, CEA, and TTF-1: An optimal set of
immunodiagnostic markers for pleural mesothelioma. Cancer.
109:933–938. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Ordóñez NG: The immunohistochemical
diagnosis of mesothelioma: A comparative study of epithelioid
mesothelioma and lung adenocarcinoma. Am J Surg Pathol.
27:1031–1051. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yokouchi H, Yamazaki K, Asahina H,
Shigemura M, Moriyama T, Takaoka K, Moriya J, Itoh T, Kinoshita I,
Dosaka-Akita H, et al: Establishment and characterization of
amylase-producing lung adenocarcinoma cell line, IMEC-2. Anticancer
Res. 26B:2821–2827. 2006.
|
|
40
|
Moldvay J, Jackel M, Bogos K, Soltész I,
Agócs L, Kovács G and Schaff Z: The role of TTF-1 in
differentiating primary and metastatic lung adenocarcinomas. Pathol
Oncol Res. 10:85–88. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bejarano PA, Baughman RP, Biddinger PW,
Miller MA, Fenoglio-Preiser C, al-Kafaji B, Di Lauro R and Whitsett
JA: Surfactant proteins and thyroid transcription factor-1 in
pulmonary and breast carcinomas. Mod Pathol. 9:445–452.
1996.PubMed/NCBI
|
|
42
|
Minotti G, Menna P, Salvatorelli E, Cairo
G and Gianni L: Anthracyclines: Molecular advances and
pharmacologic developments in antitumor activity and
cardiotoxicity. Pharmacol Rev. 56:185–229. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Tacar O, Sriamornsak P and Dass CR:
Doxorubicin: An update on anticancer molecular action, toxicity and
novel drug delivery systems. J Pharm Pharmacol. 65:157–170. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ramalingam SS and Khuri FR: The role of
the taxanes in the treatment of older patients with advanced stage
non-small cell lung cancer. Oncologist. 14:412–424. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Pan B, Chen D, Huang J, Wang R, Feng B,
Song H and Chen L: HMGB1-mediated autophagy promotes docetaxel
resistance in human lung adenocarcinoma. Mol Cancer. 13:1652014.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Wang R, Huang J, Feng B, De W and Chen L:
Identification of ING4 (inhibitor of growth 4) as a modulator of
docetaxel sensitivity in human lung adenocarcinoma. Mol Med.
18:874–886. 2012.PubMed/NCBI
|
|
47
|
Miller VA and Kris MG: Docetaxel
(Taxotere) as a single agent and in combination chemotherapy for
the treatment of patients with advanced non-small cell lung cancer.
Semin Oncol. 27(Suppl 3): 3–10. 2000.PubMed/NCBI
|
|
48
|
Ku JL, Yoon KA, Kim IJ, Kim WH, Jang JY,
Suh KS, Kim SW, Park YH, Hwang JH, Yoon YB, et al: Establishment
and characterisation of six human biliary tract cancer cell lines.
Br J Cancer. 87:187–193. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Burdall SE, Hanby AM, Lansdown MR and
Speirs V: Breast cancer cell lines: Friend or foe? Breast Cancer
Res. 5:89–95. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
50
|
Osborne CK, Hobbs K and Trent JM:
Biological differences among MCF-7 human breast cancer cell lines
from different laboratories. Breast Cancer Res Treat. 9:111–121.
1987. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Gill DJ, Tham KM, Chia J, Wang SC,
Steentoft C, Clausen H, Bard-Chapeau EA and Bard FA: Initiation of
GalNAc-type O-glycosylation in the endoplasmic reticulum promotes
cancer cell invasiveness. Proc Natl Acad Sci USA. 110:E3152–E3161.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Hua D, Shen L, Xu L, Jiang Z, Zhou Y, Yue
A, Zou S, Cheng Z and Wu S: Polypeptide
N-acetylgalactosaminyltransferase 2 regulates cellular
metastasis-associated behavior in gastric cancer. Int J Mol Med.
30:1267–1274. 2012.PubMed/NCBI
|
|
53
|
Wu YM, Liu CH, Hu RH, Huang MJ, Lee JJ,
Chen CH, Huang J, Lai HS, Lee PH, Hsu WM, et al: Mucin
glycosylating enzyme GALNT2 regulates the malignant character of
hepatocellular carcinoma by modifying the EGF receptor. Cancer Res.
71:7270–7279. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Zhu LF, Hu Y, Yang CC, Xu XH, Ning TY,
Wang ZL, Ye JH and Liu LK: Snail overexpression induces an
epithelial to mesenchymal transition and cancer stem cell-like
properties in SCC9 cells. Lab Invest. 92:744–752. 2012. View Article : Google Scholar : PubMed/NCBI
|