Introduction
Head and neck squamous cell carcinoma (HNSCC)
originates from mucosal epithelial lining of the upper
aerodigestive tract. The incidence of the disease has gradually
increased over the last three decades, especially associated with
HPV infection (1). A typical case
presents at an advanced stage as a metastatic cancer, which
accounts for the poor mortality rate. Despite the progress in
methodology and clinical efficacy of radiotherapy, chemotherapy and
surgical approaches up to 50% of patients who die from HNSCC have
local or regional recurrence (2).
Detailed molecular characterization of HNSCC tumors might help to
identify subpopulations for appropriate therapeutic modalities and
lead to improvement of the prognosis.
Evidence from xenotransplantation studies revealed
that only a small subpopulation in cancer cells has the driving
force of cancer growth. Those cells responsible for tumor
initiation, carcinogenesis, progression, and metastasis defined to
be tumor-initiating cells or cancer stem cells (CSC) (3). In addition, CSCs also possess
significant resistance to chemotherapy and radiotherapy (4,5).
CSCs have been isolated in several types of epithelial cancers
including HNSCC (6). Thus, studies
on CSCs may provide clues for improvement of prognosis associated
with development of metastases and therapy-resistant local and
regional recurrences in HNSCC.
There are a number of methods for isolating and
characterizing cancer stem cells, mostly based on
fluorescence-activated cell sorting (FACS). Expression of cell
surface markers such as CD44, CD133 or drug-detoxifying surface
transporter proteins has been used commonly in CSC isolation.
However, xenotransplantation study is the gold standard for
verification of CSC property, as tumor initiation ability of the
isolated cells is evaluated in NOD/SCID mice through serial
dilutions (7). Moreover, Aldefluor
assay has been developed to isolate CSCs based on aldehyde
dehydrogenase 1 (ALDH1) activity (8). Although all these methods were also
validated in HNSCC, Aldefluor assay has been found to show higher
sensitivity in isolation of CSCs as compared to the expression of
cell surface marker, CD44 (9,10).
MicroRNAs (miRNAs) are small non-protein coding RNA
molecules, which act to regulate gene expression by
post-transcriptional interactions with mRNA. miRNAs are assumed to
regulate more than one-third of all genes related with various
physiological and pathological processes including carcinogenesis
(11). In our previous study, we
showed the role of miRNA-200 family in
epithelial-mesenchymal-transition, migration and invasion ability
of HNSCC (12). Moreover, recent
research has indicated the biological importance of miRNAs in CSC
dysregulation (13).
In human genome, so far >2,500 mature miRNAs are
described according to latest release of miRNA database, miRBase 21
(14). However, currently no data
exist concerning miRNA expression profiles of cancer stem cells in
HNSCC. Thus, the identification of cancer stem cell-related miRNAs
would provide valuable information for a better understanding of
cancer stem cell properties and the molecular pathways of
oncogenesis.
In the present study, we isolated a subpopulation of
HNSCC cells with increased ALDH1 activity and verified their CSC
properties in vitro and in vivo conditions. Moreover,
we performed microRNA profile analysis to further explore the
characteristics and to uncover microRNAs that may serve as novel
therapeutic targets in HNSCC.
Materials and methods
Ethics statement
Experimental mice were maintained in accordance with
the guidelines, and approval of the Institutional Animal Care and
Use Committee of Wakayama Medical University (permit number: 672).
Any animal found unhealthy or sick were promptly euthanized.
Cell lines and cultures
UTSCC-9 and UTSCC-90 cell lines (15,16)
were kindly provided by Dr R. Grenman (Department of
Otolaryngology, Turku University, Finland). UTSCC cells were grown
in RPMI-1640 medium supplemented with 10% fetal bovine serum, 1%
penicillin/streptomycin and 1 μl/ml amphotericin B (Gibco™,
Invitrogen, Japan), and all cell lines were cultured in a
humidified incubator with 5% CO2 at 37°C. UTSCC-9 and
UTSCC-90 cell lines were established from squamous cell carcinoma
of laryngeal carcinoma and tongue carcinoma, respectively.
Aldefluor assay and
fluorescence-activated cell sorting (FACS)
We used an Aldefluor assay kit (Stem Cell
Technologies™, Vancouver, Canada) to determine ALDH1 activity of
cells according to the manufacturer's protocol. Cells were
suspended in Aldefluor assay buffer containing 1 μmol/l per
1×106 cells of the ALDH substrate,
boron-dipyrromethene-aminoacetaldehyde (BAAA), and incubated for 45
min at 37°C. Each sample was treated with 45 mmol/l of an
ALDH-specific inhibitor, diethylaminobenzaldehyde (DEAB), as a
negative control. Stained cells were analyzed by BD FACSAria I™ (BD
Biosciences, San Jose, CA, USA). Cells were stained with 1 μg/ml of
propidium iodide to evaluate their viability prior to analysis. The
brightly fluorescent ALDH1-expressing cells (ALDH1high)
were detected in the green fluorescence channel (520–540 nm).
Xenograft transplantation
ALDH1high and ALDH1low cells
were isolated by FACS and resuspended at 5.0×103 cells
in 100 μl PBS and Matrigel (BD Biosciences) mixture (1:1). Then
each mixture was injected subcutaneously into the right/left middle
back areas of 6-week-old female non-obese diabetic/severe combined
immune-deficiency (NOD/SCID) mice (NOD/ShiJic-scid Jcl, Clea-Japan,
Tokyo, Japan) under inhalation anesthesia by isoflurane. Tumor
initiation and progression were observed weekly and external tumor
volume was calculated as 0.5 × Dmax × (Dmin)2
[mm3] (Dmax:long axis, Dmin:short axis of mass).
Sphere formation assay
ALDH1high and ALDH1low cells
were isolated by FACS and then cultured in 6-well ultra-low
attachment surface dishes (Corning, Tewksbury, MA, USA) at 5,000
cells per well. The cells were cultured in stem-cell medium,
serum-free DMEM/F12 (Life Technologies) supplemented with N-2
supplement (Life Technologies), 10 ng/ml recombinant human
epithelial growth factor (R&D Systems, Minneapolis, MN, USA),
10 ng/ml human basic fibroblast growth factor (R&D Systems).
Morphological change was observed daily under a light microscope
for 28 days. Round cell clusters >100 μm were judged as
spheres.
mRNA processing and quantitative
real-time PCR
Preparation of cDNA from mRNA was performed directly
from cultured cell lysate using the TaqMan® Gene
Expression Cells-to-CT™ kit (Ambion, Japan), according to the
manufacturer's instructions. Cell lysate were reverse transcribed
to cDNA using the Reverse Transcription (RT) Enzyme Mix and
appropriate RT buffer (Ambion). Finally the cDNA was amplified by
quantitative real-time PCR (qRT-PCR) using the included TaqMan Gene
Expression Master Mix and the specific TaqMan primer/probe assay
designed for the investigated genes: ALDH1A1
(Hs00946916_m1), NANOG (Hs02387400_g1), OCT4
(Hs03005111_g1), SOX2 (Hs01053049_s1) and GAPDH
(Hs99999905_m1), (Applied Biosystems, Tokyo, Japan).
The gene expression levels were normalized to the
expression of the housekeeping gene GAPDH and were expressed as
fold changes relative to the expression of the untreated cells. The
amplification profile was initiated by 10-min incubation at 95°C,
followed by two-step amplification of 15 sec at 95°C and 60 sec at
60°C for 40 cycles. All experiments were performed including
non-template controls to exclude contamination of the reagents.
Quantification was done with the delta/delta Ct calculation method
(17). Each sample was analyzed in
triplicate on the 7500 Real-Time PCR system in a 96-well plate
(Applied Biosystems).
MicroRNA processing and qRT-PCR
Preparation of cDNA from microRNA was also performed
directly from cultured cell lysate using the TaqMan®
MicroRNA Cells-to-CT™ kits (Ambion) according to the manufacturer's
instructions. Cell lysate was reverse transcribed to cDNA using the
RT Enzyme Mix, RT buffer and appropriate RT primer (Applied
Biosystems). Finally the cDNA of each mature microRNA was amplified
separately by qRT-PCR using the TaqMan Universal Master Mix and the
specific primer and probe mix included in predesigned TaqMan
MicroRNA assays: hsa-miR-424; ID: 000604, hsa-miR-4730; ID:
462061_mat, hsa-miR-6836-5p; ID: 467207_mat, hsa-miR-6873-5p; ID:
467097_mat, hsa-miR-7152-3; ID: 466958_mat, hsa-hsa-let-7a; ID:
000377, hsa-miR-147b; ID: 002262, hsa-miR-3622a-3p; ID: 464955_mat,
hsa-miR- miR-1976; ID: 121207_mat (Applied Biosystems). Calculation
of microRNA expression was performed with the same method as
described above and for normalization RNU6B RNA (RNU6B; ID: 001093,
P/N: 4373381, Applied Biosystems) was used as internal control.
Each sample was analyzed in triplicate.
Total RNA extraction and microarray
profiling of miRNAs
Total RNA was isolated and purified from cell lines
using miRNeasy Mini kit (Qiagen, Tokyo, Japan) as recommended by
the manufacturer. RNA yield and quality was determined using the
Agilent RNA 6000 Nano kit and Agilent 2100 Bioanalyzer (Agilent
Technologies, Inc.). For all samples RNA integrity number (RIN) was
>7.
Affymetrix GeneChip miRNA 4.0 array were used for
miRNA profiling. Total RNA (1 μg per sample) was labeled using the
FlashTag™ Biotin HSR RNA Labeling kit (Affymetrix, Inc., USA) as
described by the manufacturer. The samples were hybridized on
GeneChip miRNA 4.0 arrays (Affymetrix, Inc.) for 18 h at 48°C.
Arrays were washed to remove non-specifically bound nucleic acids
and stained on Fluidics Station 450 (Affymetrix, Inc.) and then
scanned on GeneChip Scanner 3000 7G system (Affymetrix, Inc.).
Finally, Microarray Data Analysis Tool version 3.2 (Filgen, Inc.,
Japan) was used as described in the manufacturer's instructions for
all subsequent data analysis.
Statistical analysis
Data were analyzed with GraphPad Prism 5.0 (GraphPad
Software, San Diego, CA, USA). To evaluate significant differences
between two matched pair groups or between two independent groups
of samples, paired t-tests and non-paired t-tests were used,
respectively. Pearson's correlation coefficient (r) was used to
measure correlation, and logarithmic regression was used to
calculate the R2 and to create the equation of the slope.
Results
Isolation of ALDH1high cells
from HNSCC cell lines
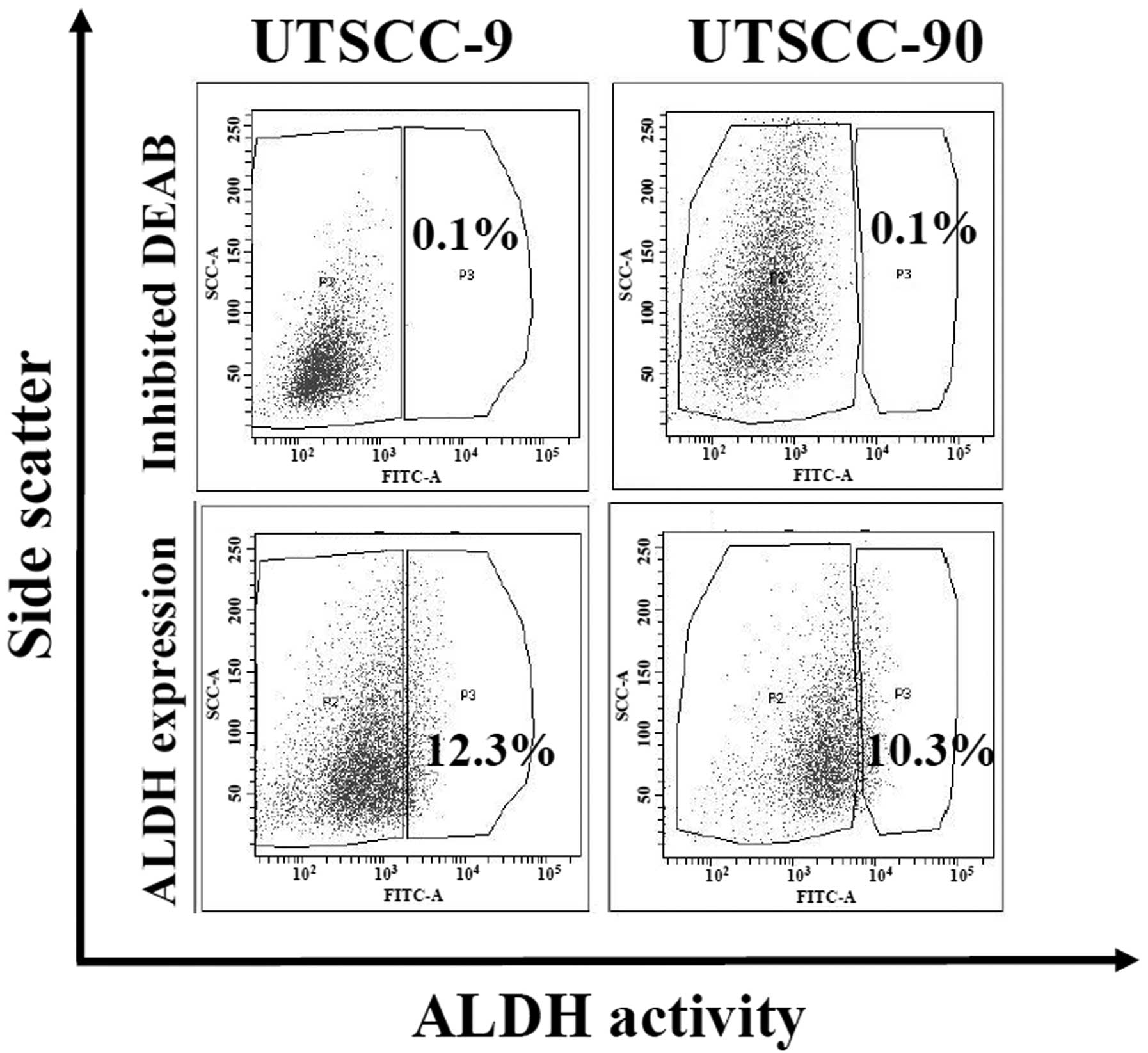
ALDH1 activity is demonstrated to be a reliable
marker for the isolation of cancer stem cells in several cancers
including HNSCC (10). We analyzed
ALDH activity by Aldefluor assay in two human HNSCC cell lines
(UTSCC-9 and UTSCC-90). Cancer cells were sorted and isolated into
two subpopulations as ALDH1 high (ALDH1high) and low
(ALDH1low) by flow cytometry. Proportion of
ALDH1high cells was found to be 12.3 and 10.3% for
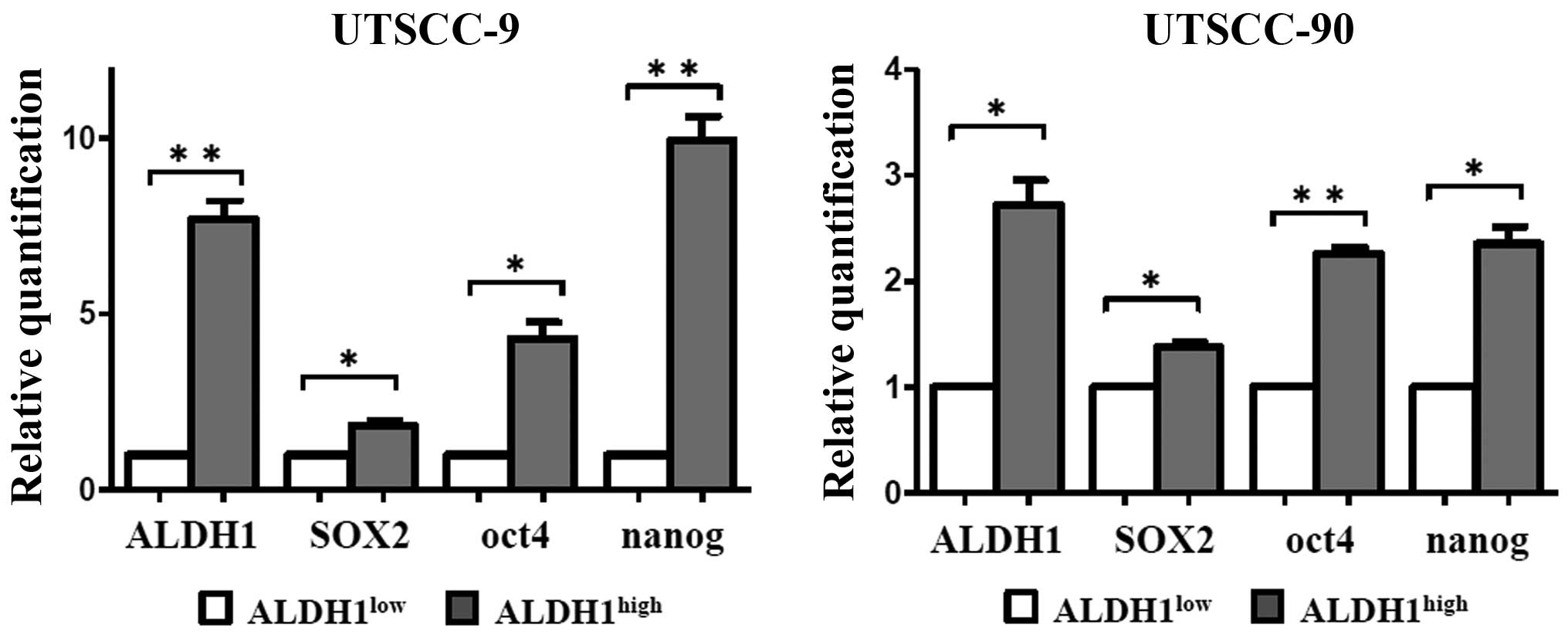
UTSCC-9 and UTSCC-90 cells, respectively (Fig. 1). These results were also confirmed
by quantitative real-time PCR (qPCR) analysis. mRNA expression of
ALDH1 was significantly higher in ALDH1high cells as
compared to ALDH1low in both UTSCC-9 and UTSCC-90 cells
(Fig. 2).
Characterization of ALDH1high
cells
To analyze whether ALDH1high cells
represent the CSC population of HNSCC, several characteristics
related to stem cell were evaluated and compared between
ALDH1high and ALDH1low portions both for
UTSCC-9 and UTSCC-90 cells.
First, the expression levels of stem cell markers
including SOX2, Nanog and Oct4 were investigated by qPCR. The
ALDH1high population of both HNSCC cell lines
demonstrated increased expression levels of SOX2, Nanog and Oct4 as
compared to ALDH1low cells with statistical significance
(Fig. 2). The results were
reproduced in three independent experiments. These results verified
that ALDH1high cells derived from each cell line has
similar genetic properties with CSCs.
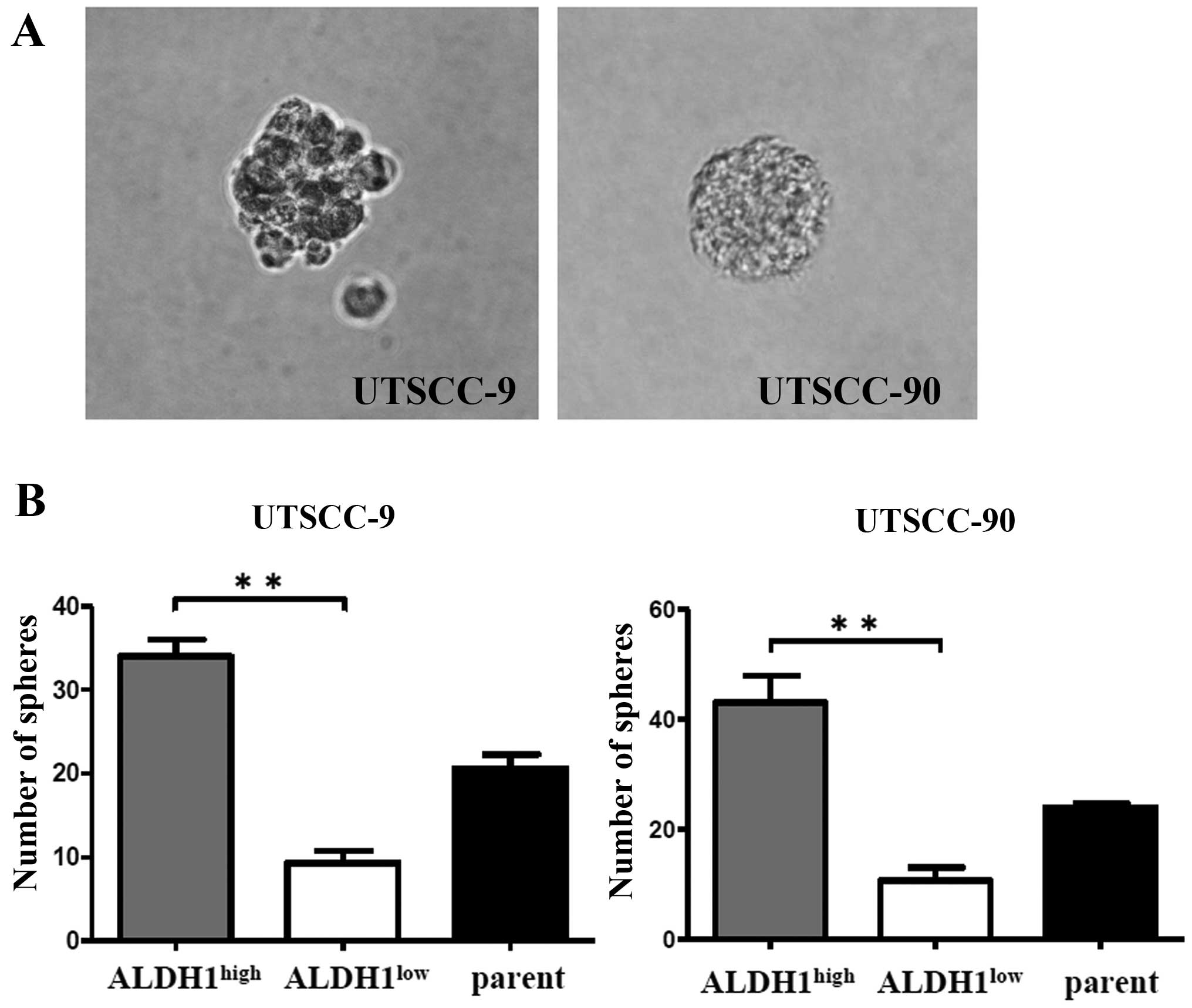
CSCs are known to be able to grow in floating
culture conditions as non-adherent, three-dimensional sphere
clusters (spheres) (18).
Therefore, sphere formation ability of ALDH1high
population was compared with ALDH1low and unsorted
parent cells of UTSCC-9 and UTSCC-90. Sorted 5×103 cells
per well were incubated into a 6-well plate in an
anchorage-independent environment. The sphere formation assays were
repeated in three independent experiments. Total number of spheres
per well were counted after 28 days, and used to calculate the mean
number of spheres.
ALDH1high cells derived from UTSCC-9
cells exhibited significantly increased sphere formation efficiency
compared to ALDH1low cells (P<0.01). Similar results
were obtained also for UTSCC-90 cells (P<0.01) (Fig. 3).
Increased tumorigenicity of
ALDH1high cells
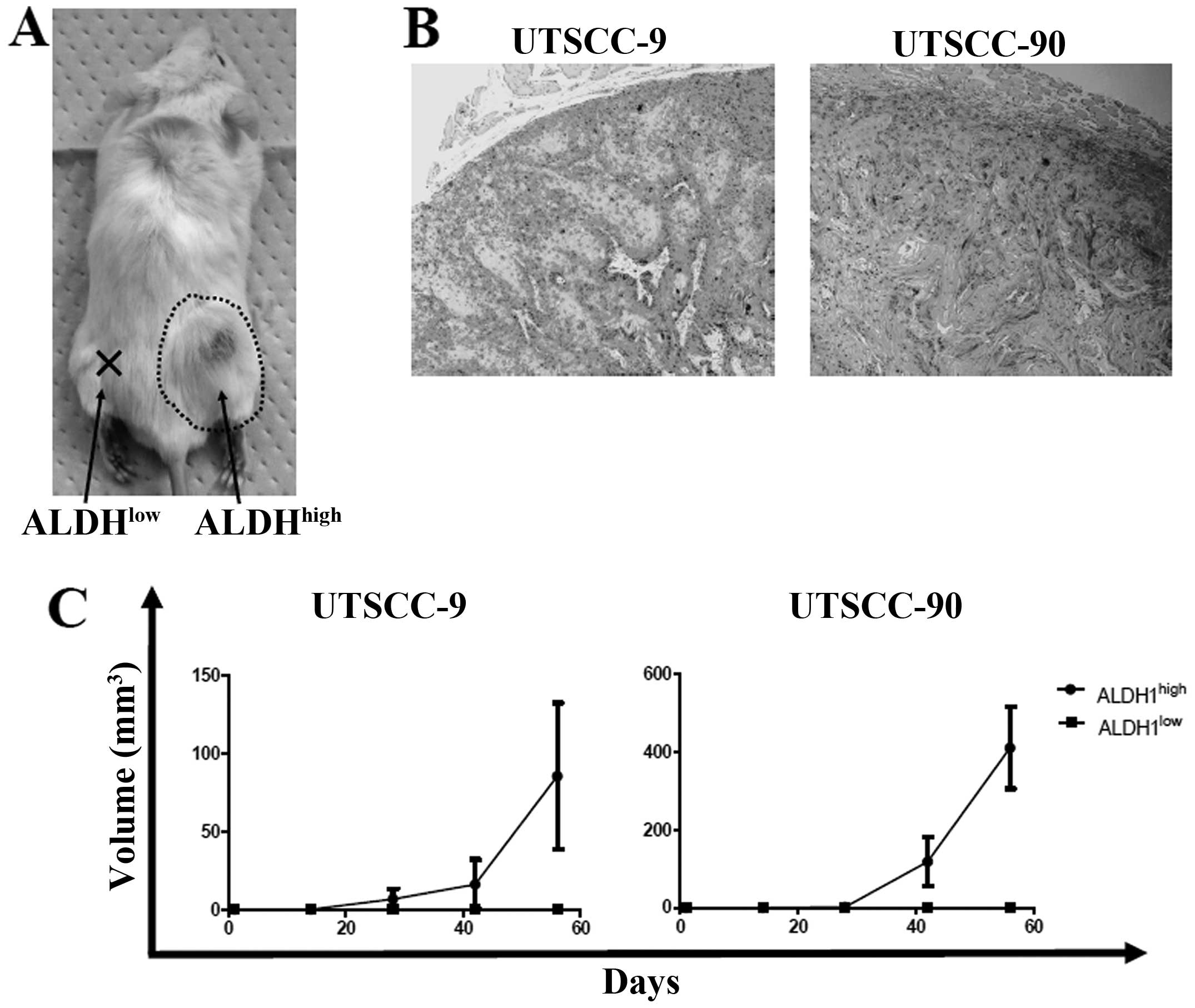
To address the in vivo tumorigenicity of
ALDH1high and ALDH1low cells, xenograft
transplantation was performed into four NOD/SCID mice for each cell
line group (UTSCC-9 and UTSCC-90). Sorted 5×103 cells
from ALDH1high and ALDH1low cells were
injected simultaneously into the subcutaneous region of right and
left back sites for each NOD/SCID mice, respectively. One mouse in
UTSCC-9 group died in the third week without any tumor detection.
In UTSCC-90 group, ALDH1high cells produced tumors in
all four mice, while ALDH1low cells did not produce any
tumor. Similarly, tumor formation was observed in 2 of the 3 mice
for ALDH1high cells, whereas no tumor occurred in sites
injected with ALDH1low cells in UTSCC-9 group (Fig. 4A and C). The histology of tumors
formed in NOD/SCID mice was verified to be squamous cell carcinoma
(Fig. 4B). These results confirmed
higher tumor-initiating ability of ALDH1high cells
compared to ALDH1low cells in vivo.
MicroRNA expression profiles of
ALDH1high vs ALDH1low cells
In order to characterize the miRNA expression
profile that regulates genes involved in cancer stem cells of
HNSCC, we performed a microarray analysis in UTSCC-9 and UTSCC-90
cells. Total RNA samples were prepared from ALDH1high vs
ALDH1low subpopulations of each cell type. GeneChip
miRNA 4.0 array (Affymetrix, Inc.) was used which contains 2,578
human mature miRNA probe sets annotated in the miRBase 20 database.
One hybridization reaction was performed for each cell type.
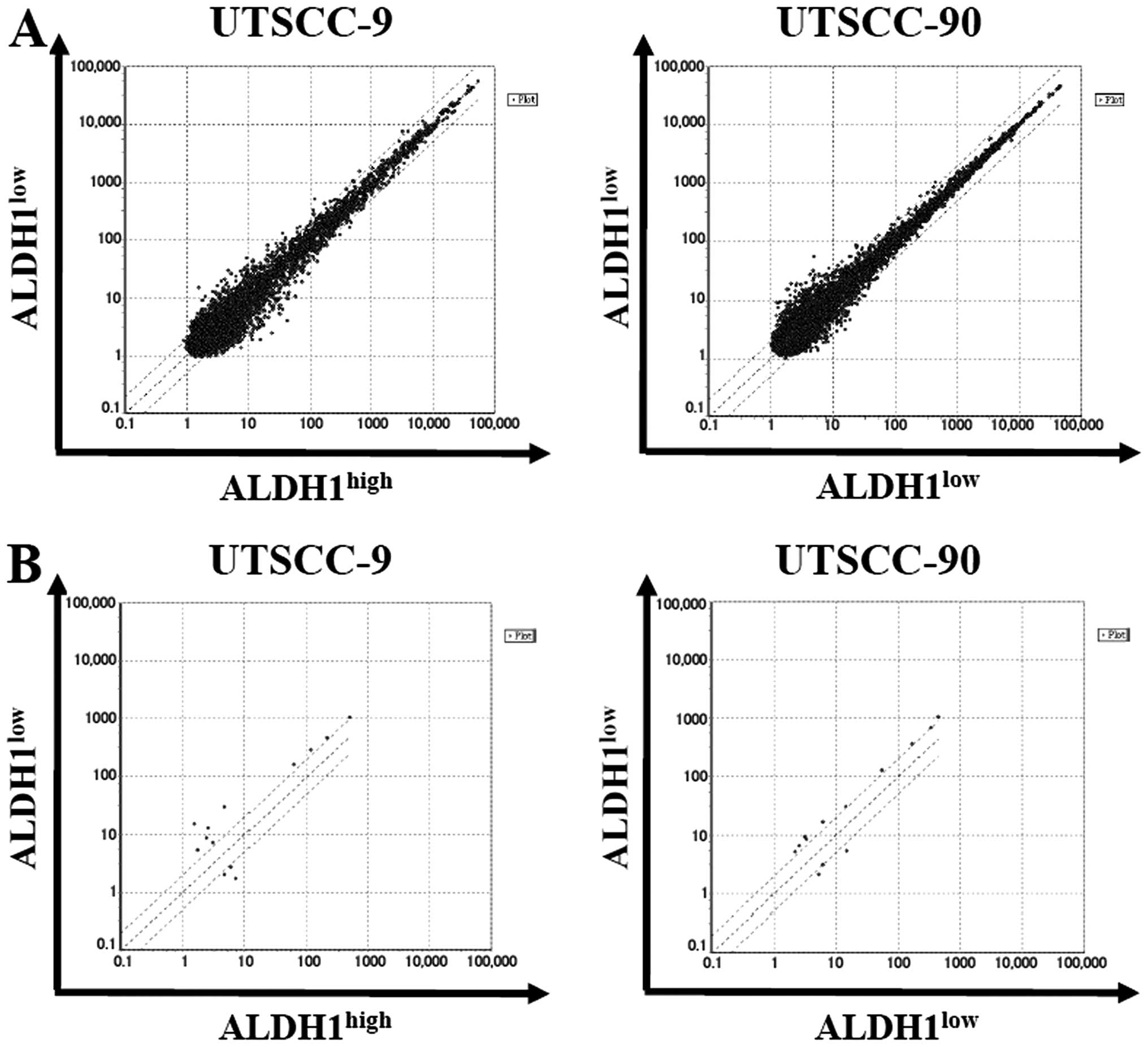
By using stringent significance criteria of a
≥2-fold difference in expression level and P<0.05, we identified
that nine miRNAs were differentially expressed between
ALDH1high and ALDH1low subpopulations
(Fig. 5). In ALDH1high
cells, six out of nine miRNAs, including miR-424, let-7a, miR-4730,
miR-6836, miR-6873, miR-7152 were significantly downregulated,
while three miRNAs including miR-147b, miR-3622a-3p and miR-1976
were significantly upregulated.
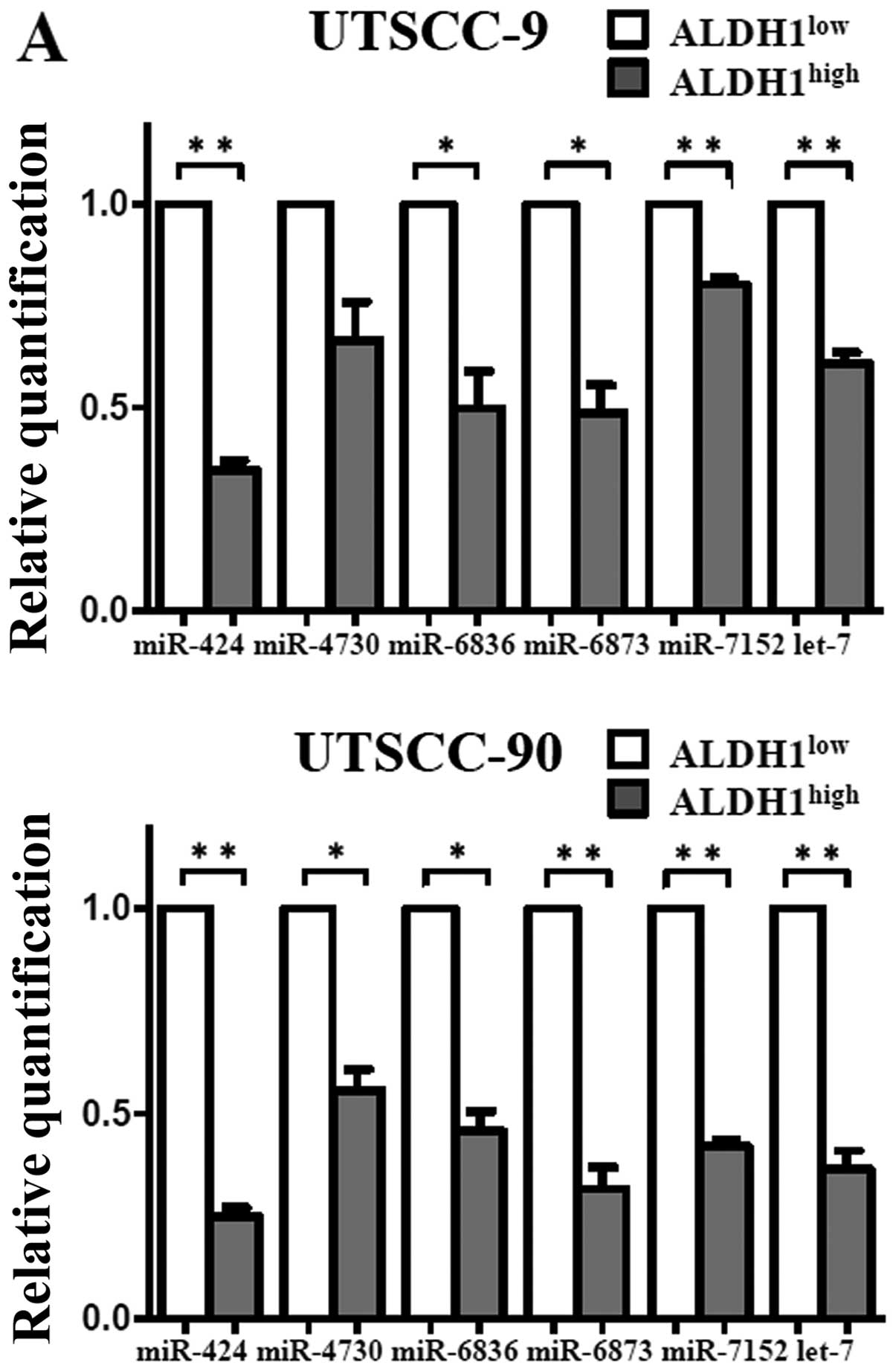
Validation of microarray results by
qPCR
To validate the microarray results, qPCR was
performed with nine miRNAs that exhibited >2-fold change in
expression. In agreement with the microarray results, expression of
miR-424, let-7a, miR-6836, miR-6873 and miR-7152 were
downregulated, whereas miR-147b was upregulated with statistical
significance in the ALDH1high subpopulation (Fig. 6). Thus, the miRNA expression
profiles of the cancer stem cells in HNSCC were confirmed by
qPCR.
Discussion
In the present study, we found that the
subpopulation of HNSCC cells with increased ALDH1 activity
(ALDH1high) represent cancer stem cells of HNSCC.
Furthermore, we also revealed that ALDH1high HNSCC cells
had a distinct microRNA expression profile as compared to
ALDH1low cells. These findings might contribute to
understanding miRNA-driven pathways related to the tumor-initiating
capability of HNSCC.
Initially, we tested whether ALDH1high
subpopulations of two HNSCC cell lines, UTSCC-9 and UTSCC-90, have
cancer stemness properties. We isolated ALDH1high
subpopulation by FACS and evaluated their stem cell features under
in vitro and in vivo conditions. Results of our
experiments including expression of stem cell markers
(stemness-related genes), sphere formation capacity and xenograft
transplantation into NOD/SCID mice supplied evidence that increased
ALDH1 activity in HNSCC presents a subpopulation with high
tumor-initiating ability.
Several types of approaches have been reported to
isolate CSC fraction of cancer cells (19). CD44 is the first and most widely
used cell surface biomarker to identify CSC in HNSCC (6). However, validity of CD44 has been
questioned recently, since it is abundantly expressed in carcinoma
cells and normal epithelium of head and neck region (20,21).
Furthermore, Clay et al proposed a new hypothesis on the
relation of CD44 and ALDH1 with the cancer stem cell compartment in
HNSCC. They suggest that ALDH-high subpopulation of HNSCC cells
comprise only a minor fraction among the CD44+ cells,
whereas more efficiently represent the CSC subpopulation itself
(10). Recently, other reports
also demonstrated ALDH1 expression as a reliable method to identify
CSC in various epithelial cancers and HNSCC (9,22).
Our observations are consistent with previous data on the role of
ALDH1 as a promising marker for identification of CSC.
Recently several studies have pointed out the
essential role of miRNAs in regulation of CSCs in etiology and
progression of various cancers including HNSCC (26–35).
Total number of human mature microRNAs has increased remarkably
from 1,105 to 2,578 according to the miRBase database in the last 5
years (14). However, only a
limited number of studies have been reported on microRNA expression
profiles of cancer stem cells in a few types of cancers (23,24).
Additionally, almost all of them were designed according to the
previous and narrower version of probe entries. Accordingly, any
miRNA signature has not been evaluated in HNSCC yet. Only one
recent report, evaluated the relation of miRNAs with the
radiosensitivity in laryngeal cancer stem cells represented by
CD133+ sphere forming subpopulations (25). They studied the alterations of
miRNAs before and after radiotherapy exposure in only one cell
line, whereas expression profile of miRNAs specific to cancer stem
cell itself was not investigated.
In the next step, we evaluated microRNA expression
profile of ALDH1high HNSCC cells. The results of
microarray analysis in the cell lines of HNSCC showed that nine
miRNAs were differentially expressed in ALDH1high HNSCC
cells compared to ALDH1low cells. Among them, decreased
expression of miR-424, let-7a, miR-4730, miR-6836, miR-6873,
miR-7152 and overexpression of miR-147 were confirmed by qPCR
assay, confirming the array results. This miRNA expression profile
indicates stemness characteristics of HNSCC. Thus, miRNA regulation
is intricately related to distinctive features and the biological
performance of HNSCC cancer stem cells. By defining miRNAs related
to the characteristics of HNSCC stem cells, we will be able to
further clarify the regulatory pathway and gain insight into the
features of these cancers.
Concerning miRNAs differentially expressed in our
ALDH1high cells, there is no data currently in the
literature on the function of four miRNAs (miR-4730, miR-6836,
miR-6873 and miR-7152). This is due to the fact that these are
novel miRNAs revealed by just the recent version of miRBase 20
(14). On the other hand, the role
of miR-424, let-7a and miR-147 in carcinogenesis has been described
by several studies.
miR-424 has been found to be downregulated in
various type of cancers and it is proposed to act as a tumor
suppressor in carcinogenesis (26,27).
Our data are the first on the expression status of miR-424 in
HNSCC, while its downregulation was also observed in squamous cell
carcinoma of the cervical region (28). miR-424 is predicted, by TargetScan,
to target many mRNAs and recent studies have already validated
important targets in cancer cells. miR-424 inhibits Wee1 and Chk1
related with chemo-resistance and radio-resistance in cancer cells
(29). It also targets inhibitor
of beta-catenin and TCF-4 (ICAT) and leads to inhibition of
epithelial-mesenchymal transition (EMT) in hepatocellular carcinoma
cells (30). Based on these
findings, it is essential to clarify the role of miR-424 in
regulation of CSC of HNSCC.
The let-7 family has been widely studied to have an
essential function in stem cell differentiation and
tumor-suppressive activity. Recently, let-7a was shown to
negatively modulate the expression of stemness genes, and inhibit
chemo-resistance and tumor initiating ability of ALDH1 (+) cell
population in HNSCC (31). Our
findings supported the tumor suppressive role of let-7a through
cancer stem cells in HNSCC.
Profiling studies demonstrated that miR-147 was
significantly overexpressed in gastric cancers and tongue cancers,
consistent with our findings (32,33),
contrarily it was shown to be underexpressed in colon carcinomas
(34). Interestingly, miR-147 was
also found significantly associated with chemo-resistance in small
cell lung cancer (35).
The limitation of this study is the lack of
functional studies to verify the potential role of those miRNAs
differentially expressed in ALDH1high HNSCC cells. As a
further step, designs of loss-of-function and gain-of-function
experiments are required for each miRNA to evaluate their effect on
cancer stem-like phenotypes such as drug resistance and cell
invasion.
In conclusion, we validated cancer stem cell
properties of ALDH1high subpopulation in HNSCC through
in vivo and in vitro experiments. Furthermore, we
identified a subset of miRNAs that were differentially expressed in
ALDH1high subpopulation of HNSCC. These findings may
provide new microRNA targets to study dysregulation of
HNSCC-initiating cells and develop therapeutic strategies aimed at
eradicating the tumorigenic subpopulation of cells in HNSCC. Thus,
further investigation is required to validate the role of these
microRNA candidates in HNSCC.
Acknowledgements
This study was supported in part by Doctorate
Program Research Grant of Wakayama Medical University.
References
|
1
|
Dufour X, Beby-Defaux A, Agius G and Lacau
St Guily J: HPV and head and neck cancer. Eur Ann Otorhinolaryngol
Head Neck Dis. 129:26–31. 2012. View Article : Google Scholar
|
|
2
|
Marur S and Forastiere AA: Head and neck
cancer: Changing epidemiology, diagnosis, and treatment. Mayo Clin
Proc. 83:489–501. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Nguyen LV, Vanner R, Dirks P and Eaves CJ:
Cancer stem cells: An evolving concept. Nat Rev Cancer. 12:133–143.
2012.PubMed/NCBI
|
|
4
|
Bertolini G, Roz L, Perego P, Tortoreto M,
Fontanella E, Gatti L, Pratesi G, Fabbri A, Andriani F, Tinelli S,
et al: Highly tumorigenic lung cancer CD133+ cells
display stem-like features and are spared by cisplatin treatment.
Proc Natl Acad Sci USA. 106:16281–16286. 2009. View Article : Google Scholar
|
|
5
|
Baumann M, Krause M and Hill R: Exploring
the role of cancer stem cells in radioresistance. Nat Rev Cancer.
8:545–554. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Prince ME, Sivanandan R, Kaczorowski A,
Wolf GT, Kaplan MJ, Dalerba P, Weissman IL, Clarke MF and Ailles
LE: Identification of a subpopulation of cells with cancer stem
cell properties in head and neck squamous cell carcinoma. Proc Natl
Acad Sci USA. 104:973–978. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
O’Brien CA, Kreso A and Jamieson CH:
Cancer stem cells and self-renewal. Clin Cancer Res. 16:3113–3120.
2010. View Article : Google Scholar
|
|
8
|
Ginestier C, Hur MH, Charafe-Jauffret E,
Monville F, Dutcher J, Brown M, Jacquemier J, Viens P, Kleer CG,
Liu S, et al: ALDH1 is a marker of normal and malignant human
mammary stem cells and a predictor of poor clinical outcome. Cell
Stem Cell. 1:555–567. 2007. View Article : Google Scholar
|
|
9
|
Chen YC, Chen YW, Hsu HS, Tseng LM, Huang
PI, Lu KH, Chen DT, Tai LK, Yung MC, Chang SC, et al: Aldehyde
dehydrogenase 1 is a putative marker for cancer stem cells in head
and neck squamous cancer. Biochem Biophys Res Commun. 385:307–313.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Clay MR, Tabor M, Owen JH, Carey TE,
Bradford CR, Wolf GT, Wicha MS and Prince ME: Single-marker
identification of head and neck squamous cell carcinoma cancer stem
cells with aldehyde dehydrogenase. Head Neck. 32:1195–1201. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shcherbata HR, Hatfield S, Ward EJ,
Reynolds S, Fischer KA and Ruohola-Baker H: The MicroRNA pathway
plays a regulatory role in stem cell division. Cell Cycle.
5:172–175. 2006. View Article : Google Scholar
|
|
12
|
Tamagawa S, Beder LB, Hotomi M, Gunduz M,
Yata K, Grenman R and Yamanaka N: Role of miR-200c/miR-141 in the
regulation of epithelial-mesenchymal transition and migration in
head and neck squamous cell carcinoma. Int J Mol Med. 33:879–886.
2014.PubMed/NCBI
|
|
13
|
DeSano JT and Xu L: MicroRNA regulation of
cancer stem cells and therapeutic implications. AAPS J. 11:682–692.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kozomara A and Griffiths-Jones S: miRBase:
Annotating high confidence microRNAs using deep sequencing data.
Nucleic Acids Res. 42(D1): D68–D73. 2014. View Article : Google Scholar :
|
|
15
|
Minn H, Clavo AC, Grénman R and Wahl RL:
In vitro comparison of cell proliferation kinetics and uptake of
tritiated fluorode-oxyglucose and L-methionine in squamous-cell
carcinoma of the head and neck. J Nucl Med. 36:252–258.
1995.PubMed/NCBI
|
|
16
|
Lansford CD, Grenman R, Bier H, Somers KD,
Kim S-Y, Whiteside TL, Clayman GL, Welkoborsky H-J and Carey TE:
Head and neck cancers. Human Cell Culture, Vol. 2, Cancer Cell
Lines Part 2. Masters JR and Palsson B: Norwell: Kluwer Academic
Publishers; pp. 185–255. 1999
|
|
17
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2 (−Delta Delta C (T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
18
|
Pastrana E, Silva-Vargas V and Doetsch F:
Eyes wide open: A critical review of sphere-formation as an assay
for stem cells. Cell Stem Cell. 8:486–498. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Major AG, Pitty LP and Farah CS: Cancer
stem cell markers in head and neck squamous cell carcinoma. Stem
Cells Int. 2013:3194892013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen C, Wei Y, Hummel M, Hoffmann TK,
Gross M, Kaufmann AM and Albers AE: Evidence for
epithelial-mesenchymal transition in cancer stem cells of head and
neck squamous cell carcinoma. PLoS One. 6:e164662011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Mack B and Gires O: CD44s and CD44v6
expression in head and neck epithelia. PLoS One. 3:e33602008.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Deng S, Yang X, Lassus H, Liang S, Kaur S,
Ye Q, Li C, Wang LP, Roby KF, Orsulic S, et al: Distinct expression
levels and patterns of stem cell marker, aldehyde dehydrogenase
isoform 1 (ALDH1), in human epithelial cancers. PLoS One.
5:e102772010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Nam EJ, Lee M, Yim GW, Kim JH, Kim S, Kim
SW and Kim YT: MicroRNA profiling of a CD133 (+) spheroid-forming
subpopulation of the OVCAR3 human ovarian cancer cell line. BMC Med
Genomics. 5:182012. View Article : Google Scholar
|
|
24
|
Sun JG, Liao RX, Qiu J, Jin JY, Wang XX,
Duan YZ, Chen FL, Hao P, Xie QC, Wang ZX, et al: Microarray-based
analysis of microRNA expression in breast cancer stem cells. J Exp
Clin Cancer Res. 29:1742010. View Article : Google Scholar
|
|
25
|
Huang CX, Zhu Y, Duan GL, Yao JF, Li ZY,
Li D and Wang QQ: Screening for MiRNAs related to laryngeal
squamous carcinoma stem cell radiation. Asian Pac J Cancer Prev.
14:4533–4537. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Oneyama C, Kito Y, Asai R, Ikeda J,
Yoshida T, Okuzaki D, Kokuda R, Kakumoto K, Takayama K, Inoue S, et
al: MiR-424/503-mediated Rictor upregulation promotes tumor
progression. PLoS One. 8:e803002013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xu J, Li Y, Wang F, Wang X, Cheng B, Ye F,
Xie X, Zhou C and Lu W: Suppressed miR-424 expression via
upregulation of target gene Chk1 contributes to the progression of
cervical cancer. Oncogene. 32:976–987. 2013. View Article : Google Scholar
|
|
28
|
Cheung TH, Man KN, Yu MY, Yim SF, Siu NS,
Lo KW, Doran G, Wong RR, Wang VW, Smith DI, et al: Dysregulated
microRNAs in the pathogenesis and progression of cervical neoplasm.
Cell Cycle. 11:2876–2884. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Pouliot LM, Chen YC, Bai J, Guha R, Martin
SE, Gottesman MM and Hall MD: Cisplatin sensitivity mediated by
WEE1 and CHK1 is mediated by miR-155 and the miR-15 family. Cancer
Res. 72:5945–5955. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhang Y, Li T, Guo P, Kang J, Wei Q, Jia
X, Zhao W, Huai W, Qiu Y, Sun L, et al: MiR-424-5p reversed
epithelial-mesenchymal transition of anchorage-independent HCC
cells by directly targeting ICAT and suppressed HCC progression.
Sci Rep. 4:62482014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yu CC, Chen YW, Chiou GY, Tsai LL, Huang
PI, Chang CY, Tseng LM, Chiou SH, Yen SH, Chou MY, et al: MicroRNA
let-7a represses chemoresistance and tumourigenicity in head and
neck cancer via stem-like properties ablation. Oral Oncol.
47:202–210. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Yao Y, Suo AL, Li ZF, Liu LY, Tian T, Ni
L, Zhang WG, Nan KJ, Song TS and Huang C: MicroRNA profiling of
human gastric cancer. Mol Med Rep. 2:963–970. 2009.PubMed/NCBI
|
|
33
|
Wong TS, Liu XB, Wong BY, Ng RW, Yuen AP
and Wei WI: Mature miR-184 as potential oncogenic microRNA of
squamous cell carcinoma of tongue. Clin Cancer Res. 14:2588–2592.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gaedcke J, Grade M, Camps J, Søkilde R,
Kaczkowski B, Schetter AJ, Difilippantonio MJ, Harris CC, Ghadimi
BM, Møller S, et al: The rectal cancer microRNAome - microRNA
expression in rectal cancer and matched normal mucosa. Clin Cancer
Res. 18:4919–4930. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ranade AR, Cherba D, Sridhar S, Richardson
P, Webb C, Paripati A, Bowles B and Weiss GJ: MicroRNA
92a-2*: A biomarker predictive for chemoresistance and
prognostic for survival in patients with small cell lung cancer. J
Thorac Oncol. 5:1273–1278. 2010. View Article : Google Scholar : PubMed/NCBI
|