Introduction
Cancer stem cell (CSC) is a cell within a tumor that
possesses the capacity to self-renew and to cause the heterogeneous
lineages of cancer cells that comprise the tumor (1). Although it has been proven that
epithelial-mesenchymal transition (EMT) can generate cells with
stem cell properties (2), it is
still unknown which of the stem-like cancer cells are generated by
these phenotype transitions.
The function of Snail genes is best known for
induction of EMT. Both in development and during carcinoma
progression, Snail1 (Snail) is expressed at the onset of the
transition, whereas Snail2 (Slug), Zeb genes, E47 and Twist are
subsequently induced to maintain the migratory mesenchymal state
(3). As transcriptional
repressors, Snail and Slug can regulate the expression of genes
mediating therapeutic resistance and acquiring of stem-like
phenotype in ovarian cancer cells (4).
Snail and Slug have complicated interactions with
many key molecules. Snail can cause functional deficiency of p53 in
tumor cells with mutant KRAS (5).
We know GSK3β, the downstream molecule of Akt (6), can phosphorylate Snail to cause the
degradation or the nuclear export of Snail (7). However, KRAS can interact with the
PI3K/Akt pathway (8), and then
affect GSK3β. Slug can escape degradation when p53 is mutant in
lung cancer (9). In addition, the
experssion of Slug can be regulated by mutant KRAS in colon cancer
cells (10). Snail and Slug are
also connected to other key molecules (e.g., EGFR, ERCC1) (11–13).
Certain key molecules, including KRAS, p53, EGFR,
ERCC1, are all connected to EMT (11,14–16)
and stem cell biology (17–20).
This makes the SNAIL-related phenotype transitions much more
complicated (Fig. 1A). Notably,
these key molecules are also found to be of great value in
non-small cell lung cancer patients (21–24).
In addition, the great value of personalized therapy suggests the
important role of genetic background in lung cancer (25). Snail and Slug are important
regulators in the stemness of lung cancer (26,27).
Plenty of studies have already revealed SNAILs can induce
EMT-related phenotype transition of lung cancer. Accordingly, we
were interested in investigation of whether the in lung cancer
cells with wild-type of these key molecules can provide some
significant clarification.
We conducted the currently study to investigate the
EMT-inducing roles of SNAILs in selected cancer cells (lung cancer
cell line with relatively stable genome, selection criteria are
described in Materials and methods), in order to provide further
knowledge for the investigation of EMT-related phenotype
transitions in cancer.
Materials and methods
Antibodies and reagents
Primary antibodies were: anti-Snail, anti-Slug,
anti-γ-H2AX (Abcam), anti-Snail, anti-Slug, anti-Bax, anti-Bcl-2,
anti-Bcl-xl, anti-cytochrome c, anti-caspase 3 (Cell
Signaling Technology), anti-total (Ser473)-Akt, anti-phospho
(Ser473)-Akt, anti-total (Ser9)-GSK3β, anti-phospho (Ser9)-GSK3β,
anti-Myc (Signalway Antibody), anti-β-actin (Beijing Biosynthesis
Biotechnology Co., Ltd.), anti-p53, anti-p21, anti-ERCC1,
anti-E-cadherin, anti-Vimentin, anti-CK8/18 (Beijing Zhongshan
Golden Bridge Biotechnology Co., Ltd.). Secondary antibodies were:
HRP-conjugated goat anti-rabbit IgG, HRP-conjugated goat anti-mouse
IgG (Beijing Zhongshan Golden Bridge Biotechnology Co., Ltd.),
Alexa Fluor 594-conjugated anti-rabbit IgG (Molecular Probe). PI,
Hoechst 33342, cisplatin (DDP) and G418 were purchased from
Sigma.
Cell lines
Human cell lines A549, H460, H292, HUVEC and HBE
were purchased from and tested by the American Type Culture
Collection (ATCC; Manassas, VA, USA). All the cell lines were used
within 6 months after receipt or resuscitation. All the cell lines
were maintained in appropriate medium that contained 10% FBS (both
from Life Technologies), penicillin (100 U/ml) and streptomycin
(100 U/ml) (both from Thermo).
Selection of the cell line to establish
stable cell lines
In order to maximally avoid the interference caused
by genomic mutation (genomic instability), H292 cell line was used
to establish stable cell lines. In the known non-small cell lung
cancer cell lines, we found only H292 cells possess the relatively
stable genome (with wild-type
KRAS/TP53/EGFR/ERCC1/Keap1/Nrf2) (28–36).
Plasmids construction
RNAi sequences of Snail and Slug were previously
described (11). The shRNA
constructs were synthesized by Shanghai GeneChem Co., Ltd. The
plasmids carrying the full-length human cDNA of Snail or Slug were
purchased from OriGene. The target fragments were cut out and
inserted into the overexpression vector, pcDNA3.1 pre-inserted
fragments encoding EGFP. The two fragments shared the same promoter
and each of them had their own initiation and termination codons.
All plasmids were confirmed by PCR sequencing.
Cell transfection and construction of
stable cell lines
All clones presented in this report were generated
by stable transfection. We performed transfection by Lipofectamine
2000 reagent (Invitrogen) following the instructions. Selected by
G418 (400 μg/ml), mono-clones were picked up and confirmed by
western blotting. Stable clones were cultured with 15% FBS and 200
μg/ml G418. The medium described in cell cultures were used when
testing. Cells carrying pGCsil-vector were used as control after
stable cell lines were established.
DDP cytotoxicity and cell growth
For cytotoxicity, cells were seeded on 96-well
plates and viability was detected by Cell Counting Kit-8 (DojinDo).
There were 5–6-wells for each concentration of DDP. For the cell
growth curve, cells were seeded in 24-well plates and the cell
number was counted three times a day after subculture (for 6 days).
We counted 4-wells for each cell line every day. The difference
between day 5 and 6 after subculture is equal to day 6 minus day 5.
For EdU test, cells were seeded on 6-well plates and performed by
Cell-Light™ EdU Apollo®567 In Vitro Flow Cytometry kit
(Guangzhou RiboBio Co., Ltd.).
Irradiation, serum deprivation and
stimulation
Cells cultured in culture vessels at suitable times
were irradiated with 8 Gy by a biological irradiation instrument
(Rad Source, USA). For p53 and p21 detection, 8 h after
irradiation, cells were fixed in 4% paraformaldehyde for
immunofluorescence. For G1/S phase arrest after irradiation,
experimental conditions were the same as in p53 and p21 detection.
For γ-H2AX detection, cells were fixed 30 min after irradiation.
For serum deprivation, cells were seeded on 6 cm dishes and
cultured overnight. The day after seeding, culture medium was
switched into serum-free medium. For late apoptosis, cells were
cultured in serum-free medium for 54 h. For western blotting, cells
were cultured and harvested at 0, 2, 4 and 6 days after medium
switching. For serum stimulation, cells were seeded on 6 cm dishes
and cultured overnight. On the day after seeded, culture medium was
switched into serum-free medium. Following culture in serum-free
medium for 24 h, culture medium was switched into complete medium
with 10% serum. Cells were then harvested at 0, 4 and 8 h after
medium switching. Cells were washed with pre-warmed PBS three times
before medium switching.
Immunofluorescence
Cells were fixed in 4% paraformaldehyde for 1 h at
RT, and then blocked in 3% BSA/PBS. Primary antibodies were
incubated in a routine manner, and the secondary antibodies were
incubated for 30 min at RT. Images were collected with a microscope
(Axio Scope.A1; Carl Zeiss) using Plan-Apochromat objective lens
(10X, 0.45; 20X, 0.8; and 63X, 1.4, Oil) and a camera (AxioCam MRm;
Carl Zeiss) at 25°C, illumination with single wavelength LED
fluorescent light source (365, 470 and 590 nm). AxioVision Rel. 4.7
software (Carl Zeiss) was used to acquire images. Images were
auto-adjusted using the Office Picture Manager. All cell lines were
tested.
Scratch-test
For migration ability, cells were seeded on 6-well
plates and cultured until 100% confluent. Then, culture medium was
switched to serum-free medium and the cell culture continued for 24
h. Then, a straight scratch was made in the middle of the dish.
After washing by pre-warmed PBS three times, cells were cultured
with complete medium for another 24 h. Then images were collected
by inverted microscope (Olympus IX71-22FL/PH). DP controller
1.1.1.65 software (Olympus IX) was used to acquire images. All cell
lines were tested. The results with significant difference are
reported.
Flow cytometric analysis
For the cell cycle, cells were fixed in 70% ice-cold
alcohol and stained with PI (20 μg/ml) solution containing
DNase-free RNase (200 μg/ml). For evaluation of apoptosis, cells
were harvested and stained with Hoechst 33342 (25 μg/ml) and PI (1
μg/ml) solution. All samples were analyzed on a BD FACSAria flow
cytometry. For qualitative analysis of the cell cycle, the results
are shown in a pseudocolor form. All cell lines were tested. Only
the results with significant difference are shown.
Protein extraction and western blot
analysis
Whole cell proteins were obtained by the Total
Protein Extraction kit (KeyGen Biotech). Western blotting was
performed with the Mini-PROTEAN Tetra Cell and Mini Trans-Blot Cell
systems (BioRad). Immunoblots were detected by chemiluminescence
using the ECL kit (Pierce).
Statistical analysis
The data are expressed as means ± SD from the number
of independent experiments as indicated. Statistical analysis was
performed using Student's t-test. P-values of <0.05 were
considered statistically significant.
Results
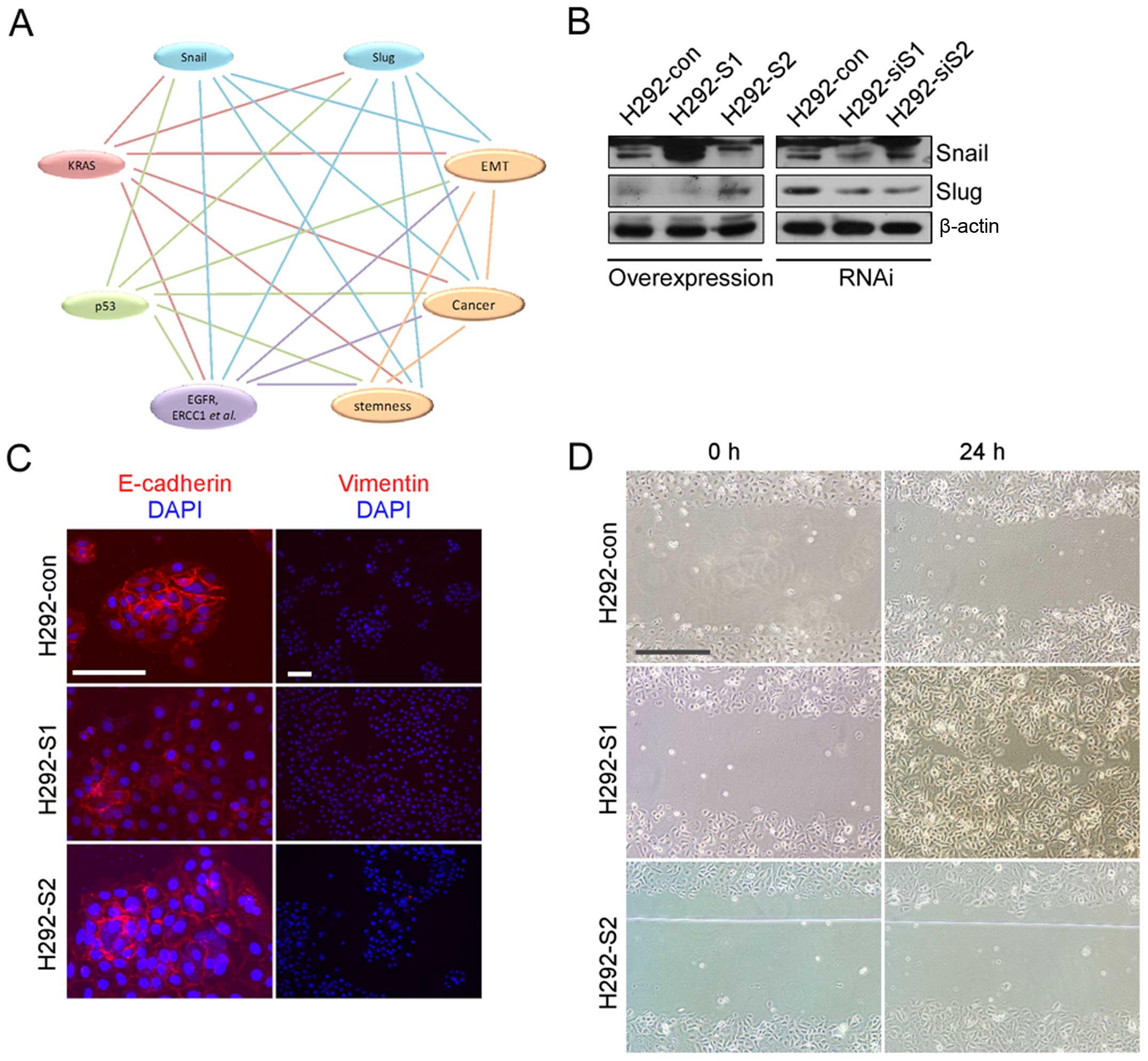
Establishment of stable cell lines
We established stable cell lines following the
description of Materials and methods. Because all the clones of the
same transfection possess the same phenotype, we chose one clone of
the transfection for further examinations (Fig. 1B).
SNAILs fail to induce completed EMT
Although expression of E-cadherin was weakened and
cell migration was enhanced in H292 overexpressing Snail (H292-S1),
Vimentin was still negative (Fig. 1C
and D). Overexpressing Snail or Slug did not induce completed
EMT. Obvious changes were shown in cell growth and DDP treatment.
We analyzed in detail the mechanism of changes of cell growth and
DDP treatment to investigate why SNAILs failed to induce completed
EMT.
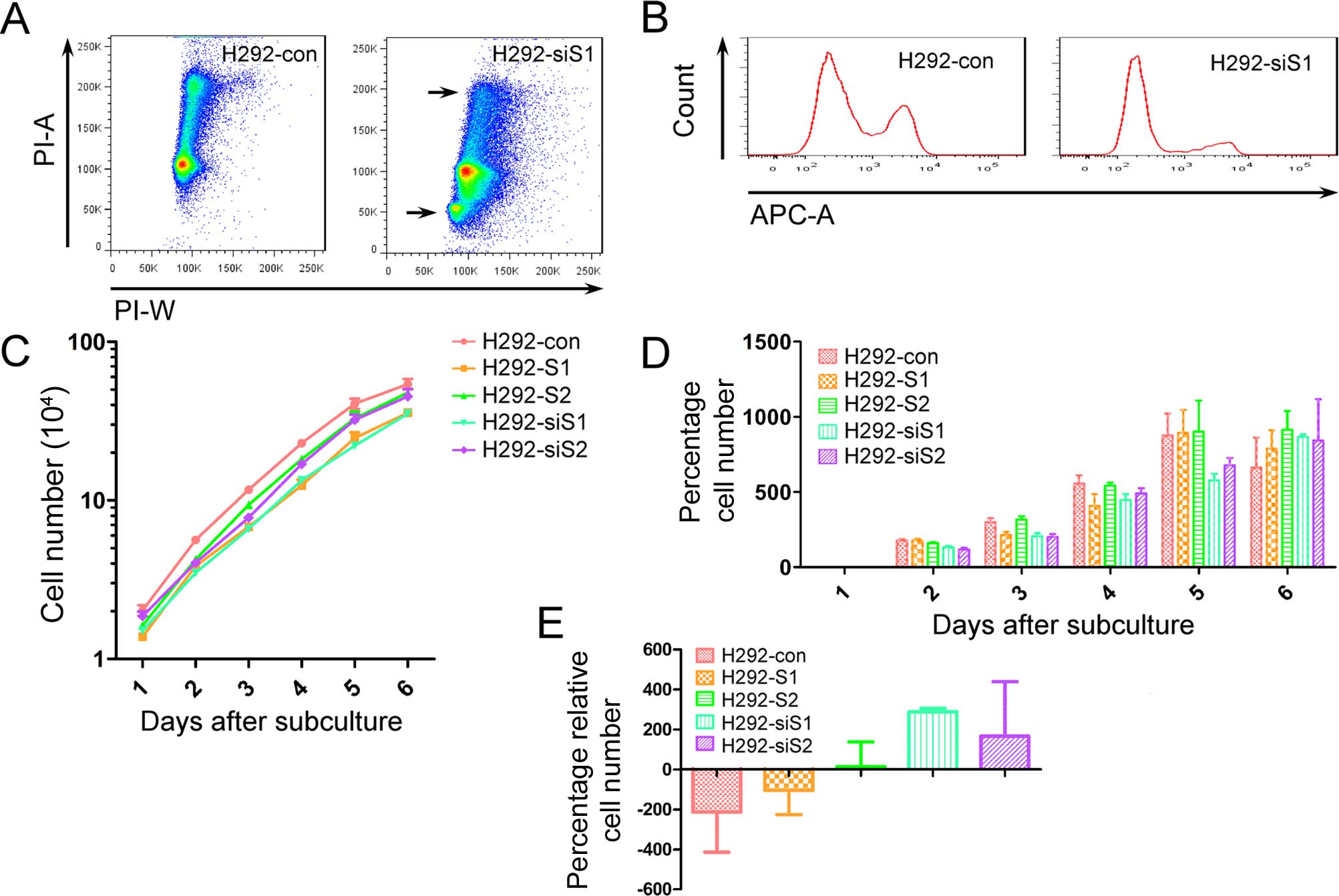
Better cell growth needs SNAILs
To confirm the effects of SNAILs on cell growth, we
utilized three methods: cell cycle, EdU test and growth curve. The
percentage of G2/M phase in H292 silencing Snail (H292-siS1)
obviously decreased compared with H292-con and an apoptotic subset
was detected in H292-siS1 (Fig.
2A). EdU-positive cells also obviously decreased in H292-siS1
(Fig. 2B). Although there were no
significant differences on the cell cycle and EdU test compared to
the others, downregulation of Snail or Slug was not conducive to
cell culture. For better understanding of growth differences, we
drew cell growth curves for all stable cell lines (Fig. 2C). To avoid the potential different
effects of chemo-biological reaction between each stable cell line,
CCK-8 test was not used for the cell growth curve. Through
comparing the percentage of increased cell number, we found cell
growth was limited more in H292-siS1 and H292-siS2 (Fig. 2D and E). During the late stage of
subculture, cell growth was limited less in H292-S1 and H292-S2.
This can be the result of weakened contact inhibition. The results
indicate better cell culture needs for SNAILs.
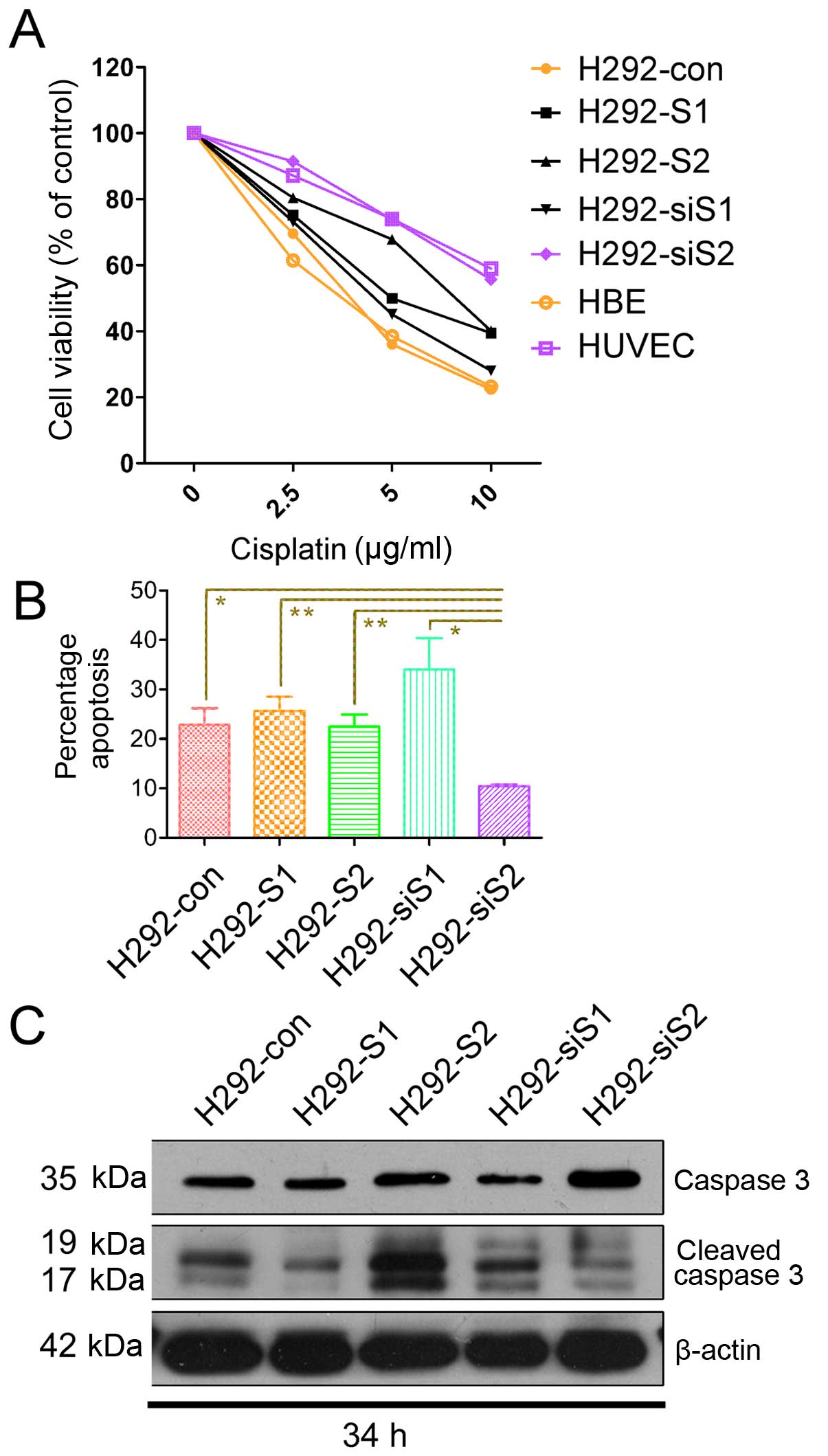
SNAILs respond differently to DDP
treatment
To confirm the effects of SNAILs on DDP treatment,
we investigated three aspects: cell viability, late apoptosis and
cleaving of caspase 3. The cell viability of H292-siS2 reduced to a
maximum of 60% when the concentration of DDP was up to 10 μg/ml
(Fig. 3A). We utilized double
stain of Hoechst 33342/PI to detect late apoptosis, in order to
avoid false-positive Annexin V causing membrane damage during cell
digestion (digestion is time-consuming in H292 compared with other
lung cancer cell lines). The rate of late apoptosis also
significantly reduced in H292-siS2 compared with the others
(Fig. 3B). There were no
statistical differences in the rate of late apoptosis of H292-siS1
compared with H292-con, H292-S1 and H292-S2, but we observed
increasing tendency of late apoptosis in H292-siS1. Through
analyzing the cleaved caspase 3, we confirmed DDP-related apoptosis
significantly increased in H292-S2 and H292-siS1, and decreased in
H292-S1 and H292-siS2 (Fig.
3C).
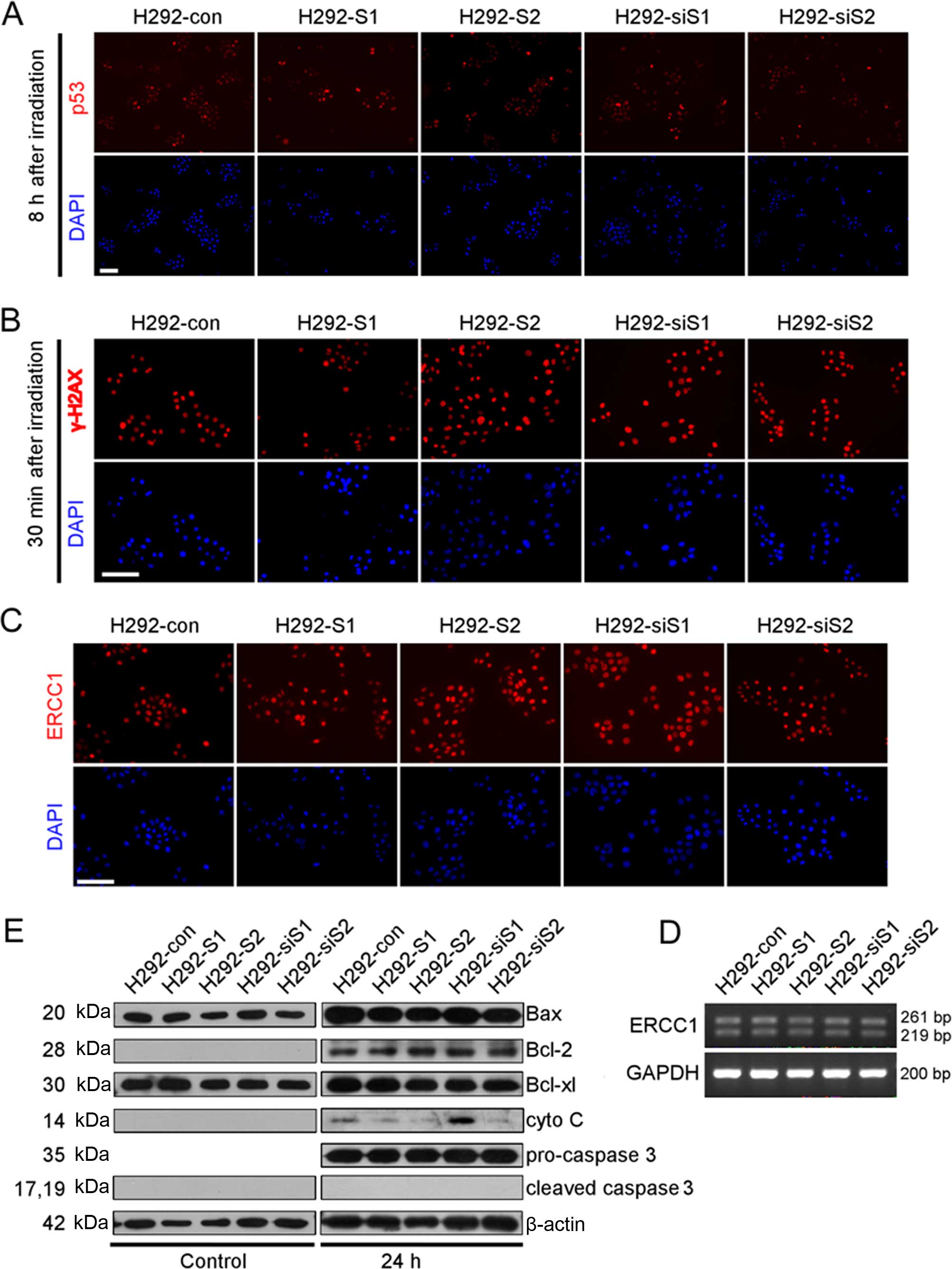
SNAILs affect the apoptotic stage after
DDP treatment
Surprised by the responses of SNAILs to DDP
treatment, we first checked the response process after DDP
interaction with DNA. We did not check the intracellular
concentration of DDP because it may not be appropriate in our
situation (the drug has passive entry and ATP-dependent efflux).
The process of cellular response mainly includes three parts:
damage perception, repairing, and outcome (survival or apoptosis)
(37). The expression of p53 and
γ-H2AX, regulated by ATM/ATR (38), the main molecules conducting DNA
damage signal during DDP treatment (39), were upregulated after irradiation,
but had no significant differences among the stable cell lines
(Fig. 4A and B). Thus, the ability
of DNA damage exists, but there was no significant differences
among the stable cell lines. The expression of ERCC1, the main
repair factor in DDP-related DNA damage, was also similar at mRNA
and protein levels among each stable cell line (Fig. 4C and D). This indicates the
capacity of DDP-related DNA damage repair, but again no significant
difference was observed in stable cell line.
At the early stage of DPP-induced apoptosis, the
expression of apoptotic signal molecules were increased in each
stable cell line (Fig. 4E). Except
cytochrome c (cyto c), manipulating either Snail or
Slug did not change the expression of Bax, Bcl-2, Bcl-xl,
pro-caspase 3 and cleaved-caspase 3. The expression of cyto
c, reflecting the coordinating effect of pro-apoptotic
(e.g., Bax) and pro-survival (e.g., Bcl-2, Bcl-xl) signals, was
significantly increased in H292-siS1 compared with H292-con, but
significantly decreased in H292-S1, H292-S2 and H292-siS2. The
higher level of cyto c in H292-siS1 indicates the absence of
Snail enhancing the activation of apoptotic signal. The lower level
of cyto c in both H292-S2 and H292-siS2 suggests the effect
of Slug on the DDP-related apoptosis is not dominated by
DDP-induced apoptotic signal.
SNAILs affect DDP-related apoptosis via
different approaches
Survival pathways can affect DDP sensitivity
(37). In addition, Snail can
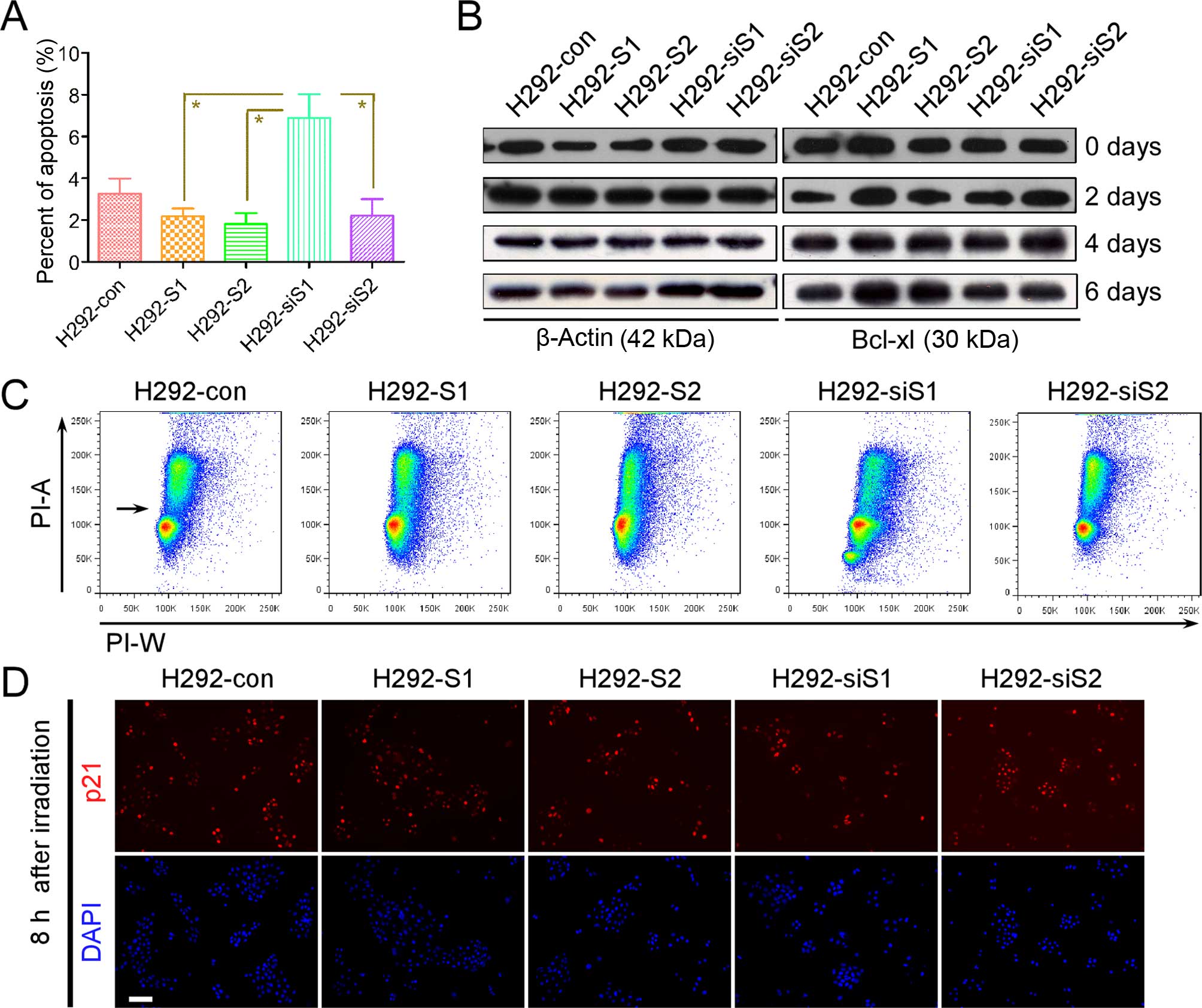
activate survival pathways in serum deprivation (SD) (40). Our results show the absence of
Snail enhanced activation of the apoptotic signal. Hence, we
speculated Snail enhances the pro-survival signal (e.g., Bcl-xl, of
which the function can be enhanced by survival pathways) to affect
the apoptotic signal. We treated cells with SD, and the rate of
late apoptosis after SD was higher in H292-siS1 (Fig. 5A). Then, we analyzed the expression
alteration of Bcl-xl after SD (Fig.
5B). At each check point, the level of Bcl-xl did not increase
in H292-siS1, but was always higher in H292-S1. These results
support that Snail utilizes pro-survival signal to decrease
DDP-related apoptosis. Moreover, the rate of late apoptosis was not
significantly different between H292-S2 and H292-siS2 (Fig. 5A). This also suggests that Slug
utilizes a different approach to affect DDP-related apoptosis.
DDP sensitivity can also be affected by the function
of G1 phase monitoring point. Knockout of TP53 in MCF-7
cells greatly improved DDP sensitivity through functional
deficiency of G1 phase arrest or nucleotide excision repair (NER)
(41). We wondered whether the
effect of Slug to DDP sensitivity is related to G1 phase monitoring
point. Cells, with normal function of G1 phase monitoring point,
will show a G1/S phase arrest after irradiation (42). Thus, we investigated G1/S phase
arrest after irradiation. H292-S2 did not show the arrest after
irradiation (Fig. 5C). Since we
studied changes at only one time point, it only supports that
overexpressing Slug does delay the generation of G1/S phase
arrest.
p21 is a key effector molecule guiding G1/S phase
arrest. Hence, we suspected Slug affects the function of p21, and
weakens the function of G1 phase monitoring point. We used
immunofluorescence to detect changes of p21 expression before and
after irradiation. The fluorescence expression of p21 had no
significant differences between each irradiated stable cell lines
(Fig. 5D). Thus Slug has no direct
effects on the monitoring point, considering no significant
alteration of expression of p53 was seen after irradiation. The
generation of G1/S phase arrest is the result of interaction
between the forward momentum of cell cycle and the resistance of
monitoring point in G1 phase. It was clear that monitoring point
has no significant functional deficiency. Slug enhances the forward
momentum of the cell cycle, and delays the emergence of G1/S phase
arrest. i.e., Slug promotes G1/S phase transition. This causes more
DNA damage to enter G2/M phase when DNA damage stimuli (e.g., DDP,
irradiation) exist. As a consequence, cell death will be more
easily triggered in mitotic phase.
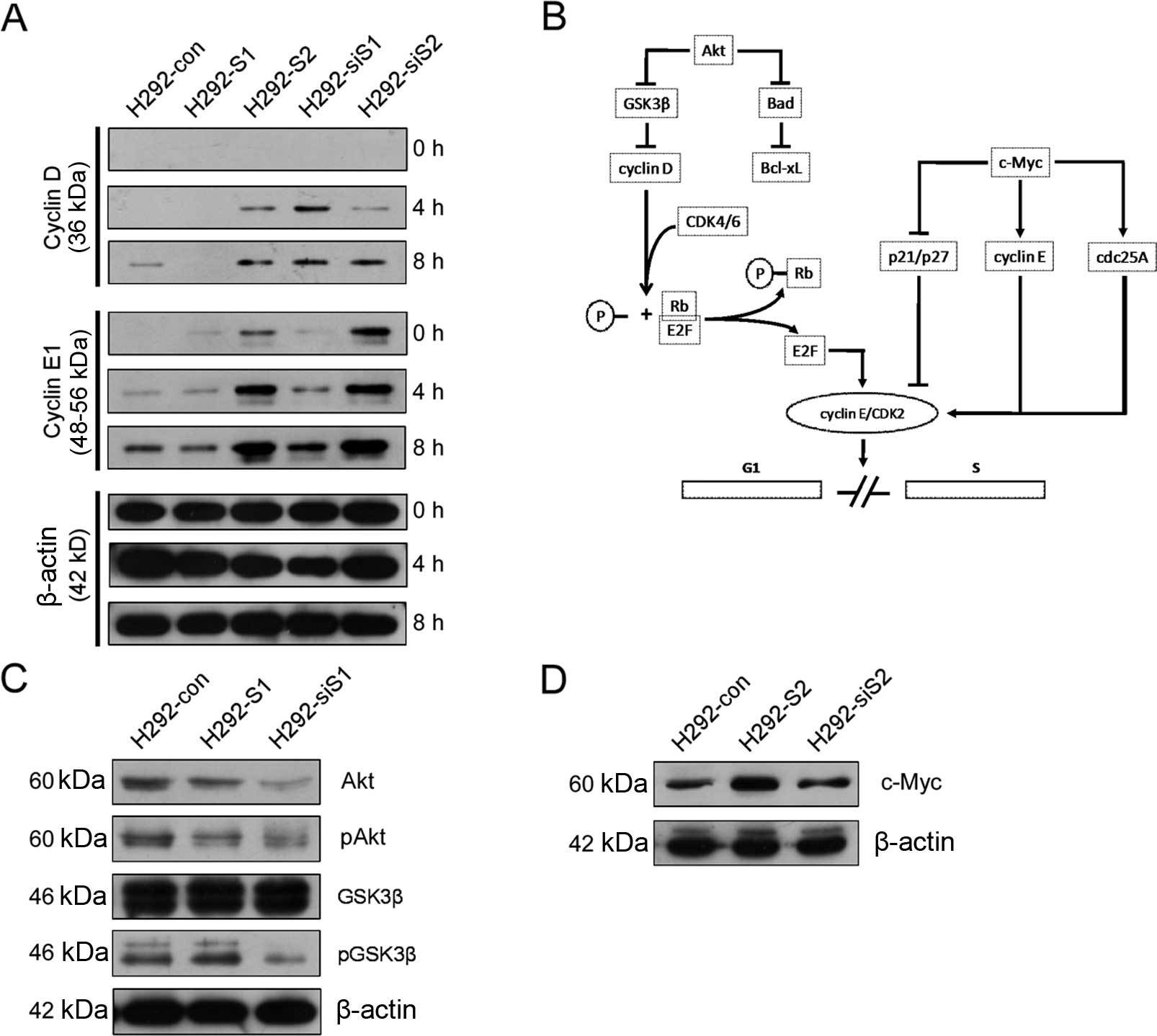
SNAILs promote different stages of G1
phase via different pathway
The aforementioned results have shown that SNAILs
differently affects DDP-related apoptosis via different molecular
mechanisms. Considering the impact on cell growth, we speculated
that the mechanism of integrated effects of SNAILs will be a change
of cell cycle kinetics in G1 phase. To prove this hypothesis, we
treated cells with serum stimulation and detected the dynamic
alteration of expression of cyclin D1 and E1. The expression
alterations of cyclin D1 and E1 were combined to reveal the
progression kinetics of G1 phase (both early and late stages), and
also to partly reveal the functional status of each other.
After serum stimulation (Fig. 6A), the expression of cyclin D1 was
significantly lower in H292-S1, but higher in H292-siS1. The
expression of cyclin E1 did not significantly differ between the
two cell lines. The results indicate H292-S1 more easily enter into
the late stage of G1 phase compared with H292-siS1. This supports
that the progression of early G1 phase is faster in H292-S1, while
slower in H292-siS1. It does not indicate whether the function of
cyclin D1 is enhanced or weakened in H292-S1 and H292-siS1. The
expression of cyclin D1 had no significant differences between
H292-S2 and H292-siS2. However, the expression of cyclin E1 had
significant differences in the two sets compared with the control
group. Cyclin E1 was increasing faster in H292-S2 than in H292-con
and H292-siS2. Although the baseline expression of cyclin E1 in
H292-siS2 was significantly increased, the speed was slower than
H292-con and H292-S2. These results illustrate that the late G1
phase progression is faster in H292-S2 than in H292-siS2. The
expression alteration of cyclins indicates the function of cyclin
E1 is weakened in H292-siS2.
In conclusion, both Snail and Slug promote G1 phase.
We speculated the process of promoting G1 phase may involve Akt or
c-Myc pathway (Fig. 6B), and
detected relevant molecules. The expression of Akt, phospho-Akt,
and phospho-GSK3β significantly reduced in H292-siS1 (Fig. 6C). This indicates Snail is
essential for the Akt/GSK3β pathway. The expression of c-Myc was
significantly upregulated in H292-S2 (Fig. 6D), supporting that Slug can
upregulate c-Myc. These results indicate Snail and Slug can utilize
Akt or c-Myc to promote G1 phase.
Although upregulation of Akt/GSK3β pathway was not
detected in H292-S1 (Fig. 6C),
overexpressing Snail can still facilitate G1 phase progression
after cell cycle synchronization by SD, via the ability of
activating Akt in the absence of serum (40). Besides, downregulation of c-Myc was
not detected in H292-siS2 either (Fig.
6D). Knockdown of Slug can downregulate c-Myc in Xenopus
laevis embryos (43). Because
KRAS can activate c-Myc through MAPK (44), no-decline of c-Myc expression in
H292-siS2 can result by the neutralizing effect of KRAS activated
by culture medium containing serum. Moreover, combined with the
weakened function of cyclin E1 in H292-siS2 (Fig. 6A), the expression of c-Myc in
H292-S2 and H292-siS2 indicates Slug enhances not only the
expression but also the function of c-Myc. Hence, downregulation of
Slug can still block G1 phase progression after cell cycle
synchronization by SD.
Discussion
The most important finding of this investigation is
the promoting action of SNAILs to G1 phase. It is contrary to the
acknowledged blocking effect (40,45).
Drug-induced downregulation of Snail was connected with
upregulation of p21 and G1 phase arrest (46). However, those who first found the
repression effect of Snail on p21 (47), already indicate it may be dependend
on the type of the cell line. In an epithelial cell line (MDCK
cells), Snail induced G0/G1 arrest through increased expression of
p21. In a mesenchymal cell line (MG63 cells), Snail suppressed
E2A-dependent activation of the p21 promoter. In contrast, the G1
phase-promoting effect reported by us is inherent in an epithelial
cell line and not related to the drug. Our results revealed a novel
value of the heredity alteration during phenotype transition.
The G1 phase-blocking effect is one of the basic
roles that make Snail genes the regulators of epithelial phenotype
and of cell adhesion and movement (3,48).
Snail genes as regulators of phenotype transition in development,
are also very important to many pathological processes (e.g., tumor
metastasis, tissue repair/regeneration). Considering the
connections of EMT, tumor metastasis and tissue repair/regeneration
(49), Mani et al further
proved that EMT can generate cells with stem cell properties
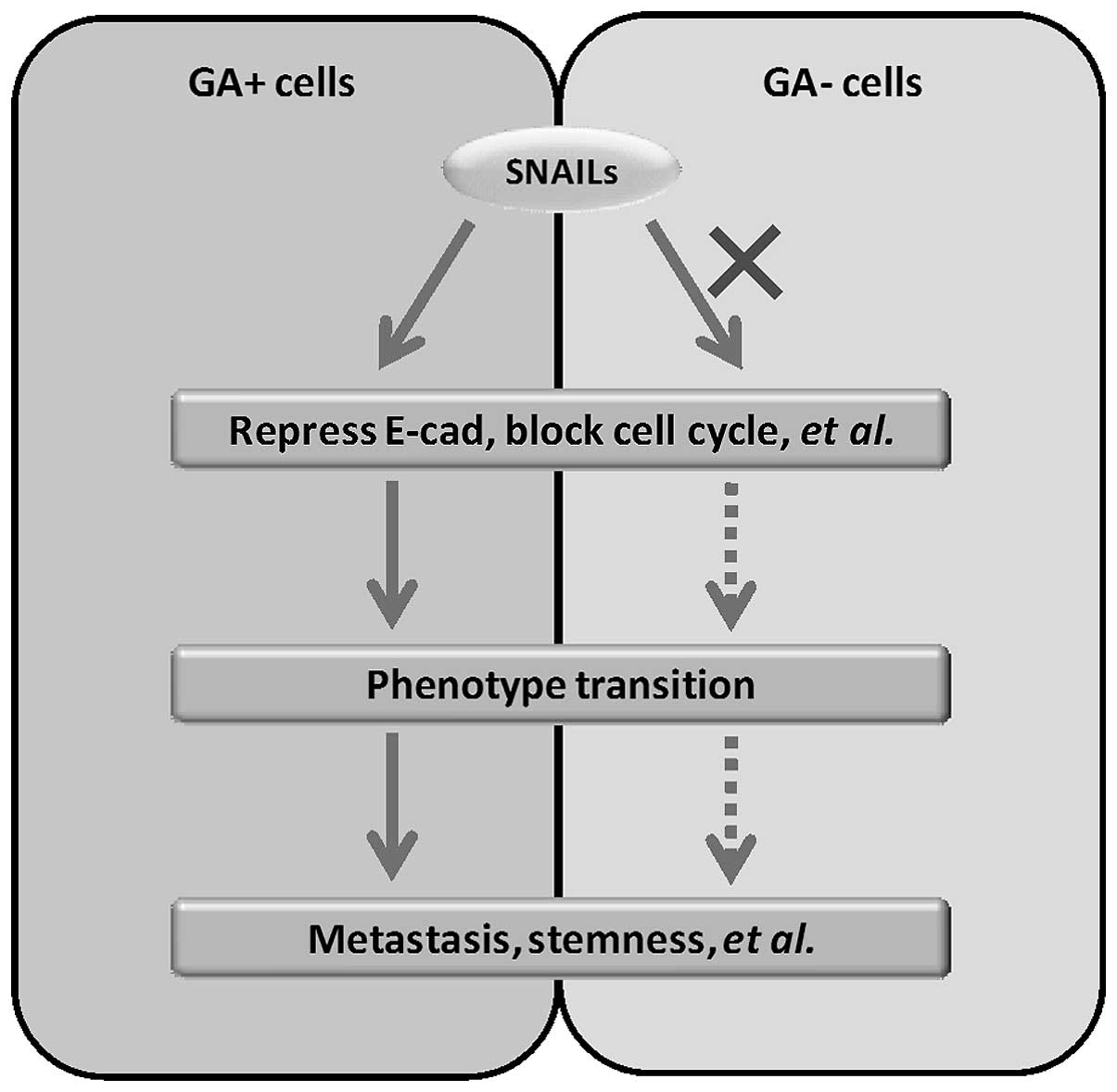
(2). Hence, blocking G1 phase is
one of the basic conditions for the onset of EMT-related phenotype
transitions. However, SNAILs fail to block G1 phase in deliberately
selected cancer cells. The discovery of this unexpected phenomenon
typically reveals the heterogeneity of cancer cells. In fact,
heterogeneity is caused by heredity alteration (genetic or
epigenetic). Hence, our findings reveal the onset of EMT-related
phenotype transitions in cancer needs not only the induction and
activation of SNAILs, but also some particular heredity alterations
(Fig. 8).
The unexpected phenomenon is very rare. We cannot
deny its existence, even though the results mainly came from single
cell clonal populations derived from a single cell line. After all,
the results of each stable cell line can be supported by each
other. In addition, all clones of the same transfection possess the
same phenotype (data not show).
According to our results, we cannot say that the
genotype of interest (wild-type
KRAS/TP53/EGFR/ERCC1/Keap1/Nrf2) is the right heredity
alteration. In addition, the results of studies involving phenotype
and these molecules also indicate the situation is much more
complicated than first thought. We discovered the unexpected
phenomenon, but we do not know which hereditary factors caused the
phenomenon. The unknown heredity alterations can be genetic or
epigenetic. To identify the actually relevant heredity alterations
in human body, comparison of alteration between primary and matched
metastatic tumor tissue (50,51)
may be an efficient approach. However, the results of published
studies are inconsistent. Hence, further studies need to be
conducted to establish the most efficient and feasible
approach.
Our study uncovered a new connection between
heredity alteration and phenotype transition. This connection will
offer new clues and rationales for distinguishing particular cancer
cell populations (e.g., metastatic and non-metastatic cancer cells,
‘innate’ and ‘acquired’ CSCs). Heredity alteration-based
distinguishing will identify the right target cells, no matter when
or whether they begin phenotype transitions. If the correct
heredity alterations can be identified, heredity
alteration-directed specific target therapy would be an efficient
approach to prevent tumor recurrence and metastasis.
Via EMT, cancer cells can acquire a stem cell
phenotype and capacity of tumorigenesis, metastasis and therapeutic
resistance (3). Hence, the origin
of CSCs is being questioned (52).
According to our results, the generation of ‘acquired’ CSCs
(induced by EMT) at least needs some particular heredity
alterations. In fact, genetic heterogeneity was already found to
exist in lung CSC populations, and related to intrinsical cellular
diversity (53,54). Notably, Kreso et al proposed
their own hypothesis regarding genetic alteration and CSC (two
mutually exclusive models for tumor hererogeneity) (55). They speculated tumor-initiating
cells (T-ICs) can evolve and acquire additional genetic mutations.
However, studies cited to support their speculation are mainly the
description of correlation, and importantly, there is no direct
evidence indicating that non-T-ICs do not generate T-ICs after
acquiring aggressive mutations. Taken together, more attention to
heredity alteration is required, whether investigating the origin
of CSCs or the causality of mutually exclusive models.
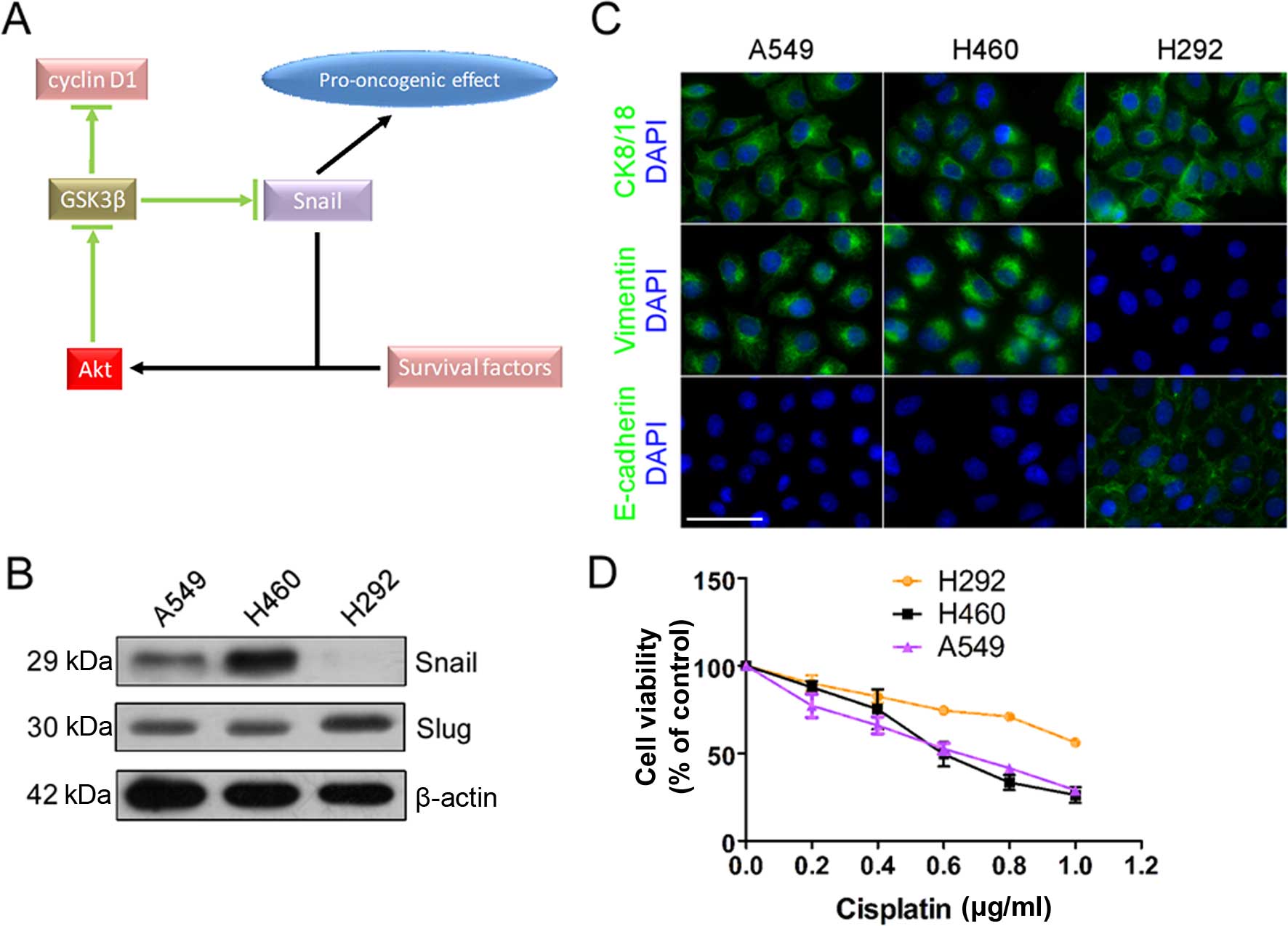
Overexpressing Snail activated Akt signaling in the
absence of serum (Fig. 5A and B)
(31). When serum existed,
overexpressing Snail did not enhance the Akt/GSK3β pathway, but
Snail was essential for pathway activation (Fig. 6C). GSK3β can inhibit Snail and be
inhibited by Akt. So, we proposed Snail/Akt/GSK3β constitutes a
signal loop (Fig. 7A). In cells
with minimal amount of Snail (e.g., H292 cells, compared with A549
and H460, Fig. 7B), survival
factors (e.g., some kind of growth factors in serum) activate Akt
and inhibit GSK3β, and then induce upregulation of Snail and
stabilization of cyclin D1. However, the high level of Snail no
longer enhances the activity of Akt. KRAS can interact with
PI3K/Akt pathway and mutant KRAS is needed for the pathological
roles of Snail in pancreatic fibrotic disease (56). In A549 and H460 cells with mutant
KRAS and wild-type TP53/EGFR/ERCC1 (H460 also has
PI3K mutation), we observed high level of Snail (Fig. 7B), and a mesenchymal phenotype
(Fig. 7C). DDP sensitivity was
increased in A549 and H460 (Fig.
7D), accompanying with significantly increased proliferation
capacity. We speculate that KRAS participates in the
Snail/Akt/GSK3β signal loop and is one of the guarantors of the
canonical roles of Snail, but more study to confirm this is
required.
In conclusion, we uncovered an unexpected role of
SNAILs in selected cancer cells, and provided significant knowledge
to the investigation of EMT-related phenotype transitions. However,
more questions are raised than answered. More study is necessary in
order to achieve better effect of personalized therapy in cancer
patients.
Acknowledgements
We thank Wen-Tong Meng, Qiao-Rong Huang, Tie Chen,
and Sheng-Liang Zhang for scientific discussions. This study was
supported by the National Major Project of China (no.
2011ZX09302-001-01), and the Natural Science Fund of China (no.
81172131).
References
|
1
|
Clarke MF, Dick JE, Dirks PB, Eaves CJ,
Jamieson CH, Jones DL, Visvader J, Weissman IL and Wahl GM: Cancer
stem cells - perspectives on current status and future directions:
AACR Workshop on cancer stem cells. Cancer Res. 66:9339–9344. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan
A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al: The
epithelial-mesenchymal transition generates cells with properties
of stem cells. Cell. 133:704–715. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Thiery JP, Acloque H, Huang RY and Nieto
MA: Epithelial-mesenchymal transitions in development and disease.
Cell. 139:871–890. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kurrey NK, Jalgaonkar SP, Joglekar AV,
Ghanate AD, Chaskar PD, Doiphode RY and Bapat SA: Snail and slug
mediate radioresistance and chemoresistance by antagonizing
p53-mediated apoptosis and acquiring a stem-like phenotype in
ovarian cancer cells. Stem Cells. 27:2059–2068. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lee SH, Lee SJ, Chung JY, Jung YS, Choi
SY, Hwang SH, Choi D, Ha NC and Park BJ: p53, secreted by
K-Ras-Snail pathway, is endocytosed by K-Ras-mutated cells;
implication of target-specific drug delivery and early diagnostic
marker. Oncogene. 28:2005–2014. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cross DA, Alessi DR, Cohen P, Andjelkovich
M and Hemmings BA: Inhibition of glycogen synthase kinase-3 by
insulin mediated by protein kinase B. Nature. 378:785–789. 1995.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhou BP, Deng J, Xia W, Xu J, Li YM,
Gunduz M and Hung MC: Dual regulation of Snail by
GSK-3beta-mediated phosphorylation in control of
epithelial-mesenchymal transition. Nat Cell Biol. 6:931–940. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Castellano E and Downward J: RAS
interaction with PI3K: More than just another effector pathway.
Genes Cancer. 2:261–274. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang SP, Wang WL, Chang YL, Wu CT, Chao
YC, Kao SH, Yuan A, Lin CW, Yang SC, Chan WK, et al: p53 controls
cancer cell invasion by inducing the MDM2-mediated degradation of
Slug. Nat Cell Biol. 11:694–704. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang Y, Ngo VN, Marani M, Yang Y, Wright
G, Staudt LM and Downward J: Critical role for transcriptional
repressor Snail2 in transformation by oncogenic RAS in colorectal
carcinoma cells. Oncogene. 29:4658–4670. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hsu DS, Lan HY, Huang CH, Tai SK, Chang
SY, Tsai TL, Chang CC, Tzeng CH, Wu KJ, Kao JY, et al: Regulation
of excision repair cross-complementation group 1 by Snail
contributes to cisplatin resistance in head and neck cancer. Clin
Cancer Res. 16:4561–4571. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Gan Y, Shi C, Inge L, Hibner M, Balducci J
and Huang Y: Differential roles of ERK and Akt pathways in
regulation of EGFR-mediated signaling and motility in prostate
cancer cells. Oncogene. 29:4947–4958. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chang TH, Tsai MF, Su KY, Wu SG, Huang CP,
Yu SL, Yu YL, Lan CC, Yang CH, Lin SB, et al: Slug confers
resistance to the epidermal growth factor receptor tyrosine kinase
inhibitor. Am J Respir Crit Care Med. 183:1071–1079. 2011.
View Article : Google Scholar
|
|
14
|
Horiguchi K, Shirakihara T, Nakano A,
Imamura T, Miyazono K and Saitoh M: Role of Ras signaling in the
induction of snail by transforming growth factor-beta. J Biol Chem.
284:245–253. 2009. View Article : Google Scholar
|
|
15
|
Chang CJ, Chao CH, Xia W, Yang JY, Xiong
Y, Li CW, Yu WH, Rehman SK, Hsu JL, Lee HH, et al: p53 regulates
epithelial-mesenchymal transition and stem cell properties through
modulating miRNAs. Nat Cell Biol. 13:317–323. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lo HW, Hsu SC, Xia W, Cao X, Shih JY, Wei
Y, Abbruzzese JL, Hortobagyi GN and Hung MC: Epidermal growth
factor receptor cooperates with signal transducer and activator of
transcription 3 to induce epithelial-mesenchymal transition in
cancer cells via up-regulation of TWIST gene expression. Cancer
Res. 67:9066–9076. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Van Meter ME, Díaz-Flores E, Archard JA,
Passegué E, Irish JM, Kotecha N, Nolan GP, Shannon K and Braun BS:
K-RasG12D expression induces hyperproliferation and
aberrant signaling in primary hematopoietic stem/progenitor cells.
Blood. 109:3945–3952. 2007. View Article : Google Scholar
|
|
18
|
Lin T, Chao C, Saito S, Mazur SJ, Murphy
ME, Appella E and Xu Y: p53 induces differentiation of mouse
embryonic stem cells by suppressing Nanog expression. Nat Cell
Biol. 7:165–171. 2005. View
Article : Google Scholar
|
|
19
|
Aguirre A, Rubio ME and Gallo V: Notch and
EGFR pathway interaction regulates neural stem cell number and
self-renewal. Nature. 467:323–327. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Niedernhofer LJ, Essers J, Weeda G,
Beverloo B, de Wit J, Muijtjens M, Odijk H, Hoeijmakers JH and
Kanaar R: The structure-specific endonuclease Ercc1-Xpf is required
for targeted gene replacement in embryonic stem cells. EMBO J.
20:6540–6549. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Riely GJ, Marks J and Pao W: KRAS
mutations in non-small cell lung cancer. Proc Am Thorac Soc.
6:201–205. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Mogi A and Kuwano H: TP53 mutations in
nonsmall cell lung cancer. J Biomed Biotechnol. 2011:5839292011.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lynch TJ, Bell DW, Sordella R,
Gurubhagavatula S, Okimoto RA, Brannigan BW, Harris PL, Haserlat
SM, Supko JG, Haluska FG, et al: Activating mutations in the
epidermal growth factor receptor underlying responsiveness of
non-small-cell lung cancer to gefitinib. N Engl J Med.
350:2129–2139. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Olaussen KA, Dunant A, Fouret P, Brambilla
E, André F, Haddad V, Taranchon E, Filipits M, Pirker R, Popper HH,
et al; IALT Bio Investigators. DNA repair by ERCC1 in
non-small-cell lung cancer and cisplatin-based adjuvant
chemotherapy. N Engl J Med. 355:983–991. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Mok TS: Personalized medicine in lung
cancer: What we need to know. Nat Rev Clin Oncol. 8:661–668. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tellez CS, Juri DE, Do K, Bernauer AM,
Thomas CL, Damiani LA, Tessema M, Leng S and Belinsky SA: EMT and
stem cell-like properties associated with miR-205 and miR-200
epigenetic silencing are early manifestations during
carcinogen-induced transformation of human lung epithelial cells.
Cancer Res. 71:3087–3097. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chiou SH, Wang ML, Chou YT, Chen CJ, Hong
CF, Hsieh WJ, Chang HT, Chen YS, Lin TW, Hsu HS, et al:
Coexpression of Oct4 and Nanog enhances malignancy in lung
adenocarcinoma by inducing cancer stem cell-like properties and
epithelial-mesenchymal transdifferentiation. Cancer Res.
70:10433–10444. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gandhi J, Zhang J, Xie Y, Soh J,
Shigematsu H, Zhang W, Yamamoto H, Peyton M, Girard L, Lockwood WW,
et al: Alterations in genes of the EGFR signaling pathway and their
relationship to EGFR tyrosine kinase inhibitor sensitivity in lung
cancer cell lines. PLoS One. 4:e45762009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Takezawa K, Okamoto I, Yonesaka K,
Hatashita E, Yamada Y, Fukuoka M and Nakagawa K: Sorafenib inhibits
non-small cell lung cancer cell growth by targeting B-RAF in KRAS
wild-type cells and C-RAF in KRAS mutant cells. Cancer Res.
69:6515–6521. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhou J, Oliveira P, Li X, Chen Z and
Bepler G: Modulation of the ribonucleotide reductase-antimetabolite
drug interaction in cancer cell lines. J Nucleic Acids.
2010:5970982010. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Mitsudomi T, Steinberg SM, Nau MM, Carbone
D, D'Amico D, Bodner S, Oie HK, Linnoila RI, Mulshine JL, Minna JD,
et al: p53 gene mutations in non-small-cell lung cancer cell lines
and their correlation with the presence of ras mutations and
clinical features. Oncogene. 7:171–180. 1992.PubMed/NCBI
|
|
32
|
O'Connor PM, Jackman J, Bae I, Myers TG,
Fan S, Mutoh M, Scudiero DA, Monks A, Sausville EA, Weinstein JN,
et al: Characterization of the p53 tumor suppressor pathway in cell
lines of the National Cancer Institute anticancer drug screen and
correlations with the growth-inhibitory potency of 123 anticancer
agents. Cancer Res. 57:4285–4300. 1997.PubMed/NCBI
|
|
33
|
You L, Yang CT and Jablons DM: ONYX-015
works synergistically with chemotherapy in lung cancer cell lines
and primary cultures freshly made from lung cancer patients. Cancer
Res. 60:1009–1013. 2000.PubMed/NCBI
|
|
34
|
McDermott U, Ames RY, Iafrate AJ,
Maheswaran S, Stubbs H, Greninger P, McCutcheon K, Milano R, Tam A,
Lee DY, et al: Ligand-dependent platelet-derived growth factor
receptor (PDGFR)-alpha activation sensitizes rare lung cancer and
sarcoma cells to PDGFR kinase inhibitors. Cancer Res. 69:3937–3946.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Jensen DE, Proctor M, Marquis ST, Gardner
HP, Ha SI, Chodosh LA, Ishov AM, Tommerup N, Vissing H, Sekido Y,
et al: BAP1: A novel ubiquitin hydrolase which binds to the BRCA1
RING finger and enhances BRCA1-mediated cell growth suppression.
Oncogene. 16:1097–1112. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yamadori T, Ishii Y, Homma S, Morishima Y,
Kurishima K, Itoh K, Yamamoto M, Minami Y, Noguchi M and Hizawa N:
Molecular mechanisms for the regulation of Nrf2-mediated cell
proliferation in non-small-cell lung cancers. Oncogene.
31:4768–4777. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Basu A and Krishnamurthy S: Cellular
responses to cisplatin-induced DNA damage. J Nucleic Acids.
2010:2013.672010. View Article : Google Scholar
|
|
38
|
Kuo LJ and Yang LX: Gamma-H2AX - a novel
biomarker for DNA double-strand breaks. In Vivo. 22:305–309.
2008.PubMed/NCBI
|
|
39
|
Yang J, Xu ZP, Huang Y, Hamrick HE,
Duerksen-Hughes PJ and Yu YN: ATM and ATR: Sensing DNA damage.
World J Gastroenterol. 10:155–160. 2004.PubMed/NCBI
|
|
40
|
Vega S, Morales AV, Ocaña OH, Valdés F,
Fabregat I and Nieto MA: Snail blocks the cell cycle and confers
resistance to cell death. Genes Dev. 18:1131–1143. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Fan S, Smith ML, Rivet DJ II, Duba D, Zhan
Q, Kohn KW, Fornace AJ Jr and O'Connor PM: Disruption of p53
function sensitizes breast cancer MCF-7 cells to cisplatin and
pentoxifylline. Cancer Res. 55:1649–1654. 1995.PubMed/NCBI
|
|
42
|
Lodish H, Berk A, Zipursky SL, Matsudaira
P, Baltimore D and Darnell J: Molecular Cell Biology. Molecular
Cell Biology. W. H. Freeman; New York: 2000
|
|
43
|
Rodrigues CO, Nerlick ST, White EL,
Cleveland JL and King ML: A Myc-Slug (Snail2)/Twist regulatory
circuit directs vascular development. Development. 135:1903–1911.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Seger R and Krebs EG: The MAPK signaling
cascade. FASEB J. 9:726–735. 1995.PubMed/NCBI
|
|
45
|
Liu J, Uygur B, Zhang Z, Shao L, Romero D,
Vary C, Ding Q and Wu WS: Slug inhibits proliferation of human
prostate cancer cells via downregulation of cyclin D1 expression.
Prostate. 70:1768–1777. 2010.PubMed/NCBI
|
|
46
|
Cheon MG, Kim W, Choi M and Kim JE: AK-1,
a specific SIRT2 inhibitor, induces cell cycle arrest by
downregulating Snail in HCT116 human colon carcinoma cells. Cancer
Lett. 356:637–645. 2015. View Article : Google Scholar
|
|
47
|
Takahashi E, Funato N, Higashihori N, Hata
Y, Gridley T and Nakamura M: Snail regulates p21WAF/CIP1
expression in cooperation with E2A and Twist. Biochem Biophys Res
Commun. 325:1136–1144. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Barrallo-Gimeno A and Nieto MA: The Snail
genes as inducers of cell movement and survival: Implications in
development and cancer. Development. 132:3151–3161. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Kondo M, Wagers AJ, Manz MG, Prohaska SS,
Scherer DC, Beilhack GF, Shizuru JA and Weissman IL: Biology of
hematopoietic stem cells and progenitors: Implications for clinical
application. Annu Rev Immunol. 21:759–806. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Vignot S, Frampton GM, Soria JC, Yelensky
R, Commo F, Brambilla C, Palmer G, Moro-Sibilot D, Ross JS, Cronin
MT, et al: Next-generation sequencing reveals high concordance of
recurrent somatic alterations between primary tumor and metastases
from patients with non-small-cell lung cancer. J Clin Oncol.
31:2167–2172. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Vakiani E, Janakiraman M, Shen R, Sinha R,
Zeng Z, Shia J, Cercek A, Kemeny N, D'Angelica M, Viale A, et al:
Comparative genomic analysis of primary versus metastatic
colorectal carcinomas. J Clin Oncol. 30:2956–2962. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Gupta PB, Chaffer CL and Weinberg RA:
Cancer stem cells: Mirage or reality? Nat Med. 15:1010–1012. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Curtis SJ, Sinkevicius KW, Li D, Lau AN,
Roach RR, Zamponi R, Woolfenden AE, Kirsch DG, Wong KK and Kim CF:
Primary tumor genotype is an important determinant in
identification of lung cancer propagating cells. Cell Stem Cell.
7:127–133. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Sutherland KD, Song JY, Kwon MC, Proost N,
Zevenhoven J and Berns A: Multiple cells-of-origin of mutant
K-Ras-induced mouse lung adenocarcinoma. Proc Natl Acad Sci USA.
111:4952–4957. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Kreso A and Dick JE: Evolution of the
cancer stem cell model. Cell Stem Cell. 14:275–291. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Shields MA, Khan MW, Grippo PJ and Munshi
HG: Concurrent Snail expression and Kras mutation promote
pancreatic fibrosis. Cancer Res. 72:13362012. View Article : Google Scholar
|