Introduction
Breast cancer is the most commom malignancy in women
worldwide (1–3). Similar to many other solid tumors,
distant metastasis are responsible for >90% of breast
cancer-related mortality (4).
MicroRNAs (miRNAs or miRs) are a subclass of 19–25
nucleotides in length, non-coding RNAs that have received
increasingly attention in recent years. miRNAs play important
regulatory roles in a variety of biological processes, such as
cellular proliferation, differentiation, apoptosis and motility
(5). Growing evidence indicates
that the alteration of miRNA expression in tumors is associated
with tumor development and progression (6). miR-506 is a novel miRNA, and it has
been demonstrated that the expression pattern of miR-506 is
different in different types of malignant tumors, suggesting the
role of miR-506 is complex in cancer progression (7–10).
The meta-analysis revealed that miR-506 is related to the survival
of breast cancer patients (11).
However, the mechanism underlying miRNA-506 involvement in breast
carcinogenesis remains unclear.
In the present study, we investigated the expression
of miR-506 in different breast tissues and breast cancer cell
lines. In addition, gain-of-function and loss-of-function
experiments were performed in vitro to examine the role of
miR-506 in breast cancer cell proliferation, invasion and adhesion.
Furthermore, a novel target by which miR-506 exerts its effects on
breast carcinogenesis was identified.
Materials and methods
Clinical breast tissues
The present study was approved by the Ethics
Committee of the China-Japan Union Hospital of Jilin University.
The surgical specimens, including 48 normal, 42 fibroadenoma and 48
malignant breast tissues were obtained from patients in the Breast
Surgical Department of China-Japan Union Hospital of Jilin
University. Among these 48 patients with malignant breast cancer, 8
patients had stage 0, 13 patients had stage I, 10 patients had
stage II, 9 patients had stage III and the other 8 patient had
stage IV, at the time of the diagnosis. None of these patients had
received chemotherapy or radiation therapy treatment before
surgery. Written informed consent was obtained from each patient
prior to enrollment in this study.
Cell culture and cell transfection
Human breast cancer cell lines (T47D, MDA-MB-231,
MCF7, SK-BR-3 and HCC1937), normal human epithelial mammary cell
line MCF-10A and HEK293 cells were obtained from the American Type
Culture Collection (ATCC; Manassas, VA, USA) and grown in
Dulbecco's modified Eagle's medium (DMEM; Invitrogen, Carlsbad, CA,
USA) supplemented with 8% heat-inactivated fetal bovine serum (FBS;
Invitrogen). The cells were maintained at 37°C in 5% CO2
and were passaged every 2–3 days. The miR-negative control (NC),
miR-506 mimic, miR-506 inhibitor, IQGAP1-pcDNA3.1 and IQGAP1-shRNA
were transfected into the cells using Lipofectamine 2000
(Invitrogen) according to the manufacturer's instructions.
Luciferase assay
Luciferase constructs were made by ligating
fragments containing the wild-type (WT) and mutant-type (MUT) 3′
untranslated region (UTR) of IQ motif containing GTPase activating
protein 1 (IQGAP1) in pMIR-REPORT luciferase vector (Applied
Biosystems, Foster City, CA, USA). The luciferase constructs were
co-transfected with the miR-NC or the miR-506 mimic into the HEK293
cells. Firefly and Renilla luciferase activity were measured
using the Dual-Luciferase reporter assay system (Promega, Madison,
WI, USA).
MTT assay
Cells (5×103) were suspended and cultured
in the 96-well plates overnight. Following transfection, 10 μl MTT
solution (0.5 mg/ml; Sigma, St. Louis, MO, USA) was added to each
well, and the plates were incubated at 37°C for 4 h. The formazan
granules were dissolved using DMSO (Sigma), and then the absorbance
rates were measured at 570 nm using a microplate reader (Infinite
M200; Tecan Group Ltd., Männedorf, Switzerland).
Transwell-Matrigel invasion assay
The Transwell inserts (Corning Incorporated,
Corning, NY, USA) precoated with Matrigel (BD Biosciences, Franklin
Lakes, NJ, USA) was used to determine breast cancer cell invasion
ability. Briefly, the cells were suspended in the FBS-free medium
at the density of 5×104 cells/ml and placed in the upper
chambers. The lower chambers were filled with cell medium
containing 8% FBS. Following incubation at 37°C for 24 h, the cells
on the upper surface of the membrane were removed by a cotton swab,
and the cells on the lower surface of the insert were fixed in 95%
ethanol and stained with hematoxylin. The invaded cells were
counted under a light microscope (Nikon, Tokyo, Japan).
Cell adhesion assay
For adhesion assay, the 96-well plates were
precoated with fibronectin (Sigma), and then blocked with 1% bovine
serum albumin (BSA; Sigma) for 2 h. The cells were suspended in the
FBS-free medium at the density of 3×105 cells/ml and
seeded in the 96-well plates. Following incubation at 37°C for 2 h,
the adhesive cells were fixed in 4% paraformaldehyde and stained
with 0.5% crystal violet. The crystals were dissolved using SDS
(Amresco LLC, Solon, OH, USA) and then the absorbance rates were
measured at 570 nm using a microplate reader (Infinite M200; Tecan
Group Ltd.).
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA from the breast tissues and breast cancer
cell lines was extracted using the TRIzol reagent (Invitrogen). For
miR-506 amplification, mirVana RNA isolation kit (Ambion, Austin,
TX, USA) was used for RNA isolation following the manufacturer's
instructions. RNA samples were reverse transcribed to complementary
DNAs using the RevertAid First Strand cDNA Synthesis kit
(Fermentas, Vilnius, Lithuania). Subsequently, the cDNA was
amplified by real-time PCR on an ABI Prism 7500 Fast Real-Time PCR
system (Applied Biosystems) using the SYBR-Green PCR kit (Applied
Biosystems). The conditions for PCR amplification were as follows:
an initial 95°C for 5 min, followed by 40 cycles of 95°C for 15
sec, 58°C for 30 sec, and 72°C for 30 sec. The Ct value was
calculated using the ΔΔCt method.
Western blot analysis
The total protein was extracted using the Total
Protein Extraction kit (BioChain Institute Inc., Hayward, CA, USA).
Protein (30 μg) was resolved on 10% sodium dodecyl
sulphate-polyacrylamide gel electrophoresis (SDS-PAGE) and
transferred onto a nitrocellulose membrane (Millipore, Billerica,
MA, USA) by electroblotting. The membranes were then incubated with
5% BSA (Sigma) at 4°C overnight. After washing in Tris-buffered
saline with 0.1% Tween-20, the membranes were incubated with the
primary antibodies, including rabbit polyclonal to IQGAP1 (1:800;
cat. sc-10792; Santa Cruz Biotechnology, Santa Cruz, CA, USA),
rabbit polyclonal to B-Raf (1:400; cat. sc-9002; Santa Cruz
Biotechnology), rabbit monoclonal to Erk1/2 (Thr202/Tyr204) (1:800;
cat. 4695; Cell Signaling Technology, Beverly, MA, USA), rabbit
monoclonal to phospho-Erk1/2 (Thr202/Tyr204) (1:500; cat. 4376;
Cell Signaling Technology), and rabbit polyclonal to GAPDH
(1:1,000; cat. sc-25778; Santa Cruz Biotechnology) at 37°C for 2 h,
followed by the incubation of horseradish peroxidase-conjugated
goat anti-rabbit IgG (1:2,000; cat. sc-2004; Santa Cruz
Biotechnology) at 37°C for 1 h. Chemiluminescence detection was
carried out by using ECL Plus™ (GE Healthcare, Piscataway, NJ,
USA).
Statistical analysis
All data were expressed as the mean ± SD.
Differences between 2 groups were assessed by the Student's t-test.
Analysis was performed using SPSS 19.0 statistical software (SPSS,
Inc., Chicago, IL, USA). A P-value <0.05 was considered
significant.
Results
Expression of miR-506 in human breast
tissues and breast cancer cell lines
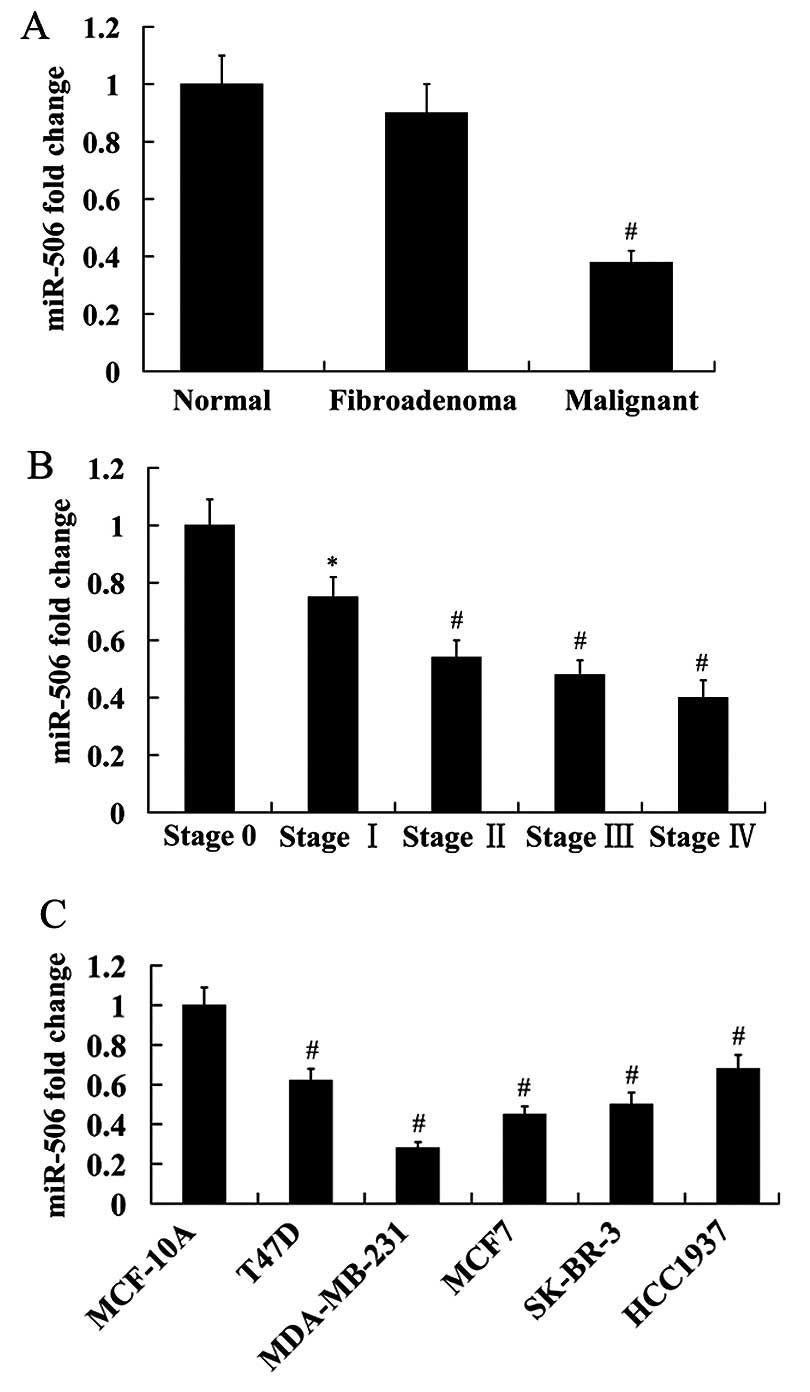
Expression of miR-506 in the normal, fibroadenoma
and malignant breast tissues was analyzed using RT-qPCR. The
results showed that miR-506 expression was not significantly
different between the normal and fibroadenoma breast tissues.
However, miR-506 expression was significantly reduced in the
malignant breast tissues compared with the normal and fibroadenoma
breast tissues (Fig. 1A).
Furthermore, we found that miR-506 expression was decreased with
the increasing of tumor stage (Fig.
1B).
We detected the expression of miR-506 in five breast
cancer cell lines (T47D, MDA-MB-231, MCF7, SK-BR-3 and HCC1937). A
normal human epithelial mammary cell line MCF-10A was used as
control. The results obtained from RT-qPCR analysis revealed that
compared with the control, miR-506 expression was significantly
decreased in the breast cancer cell lines. Among these breast
cancer cell lines, miR-506 had the lowest expression in MDA-MB-231
cells, and highest expression in HCC1937 cells (Fig. 1C).
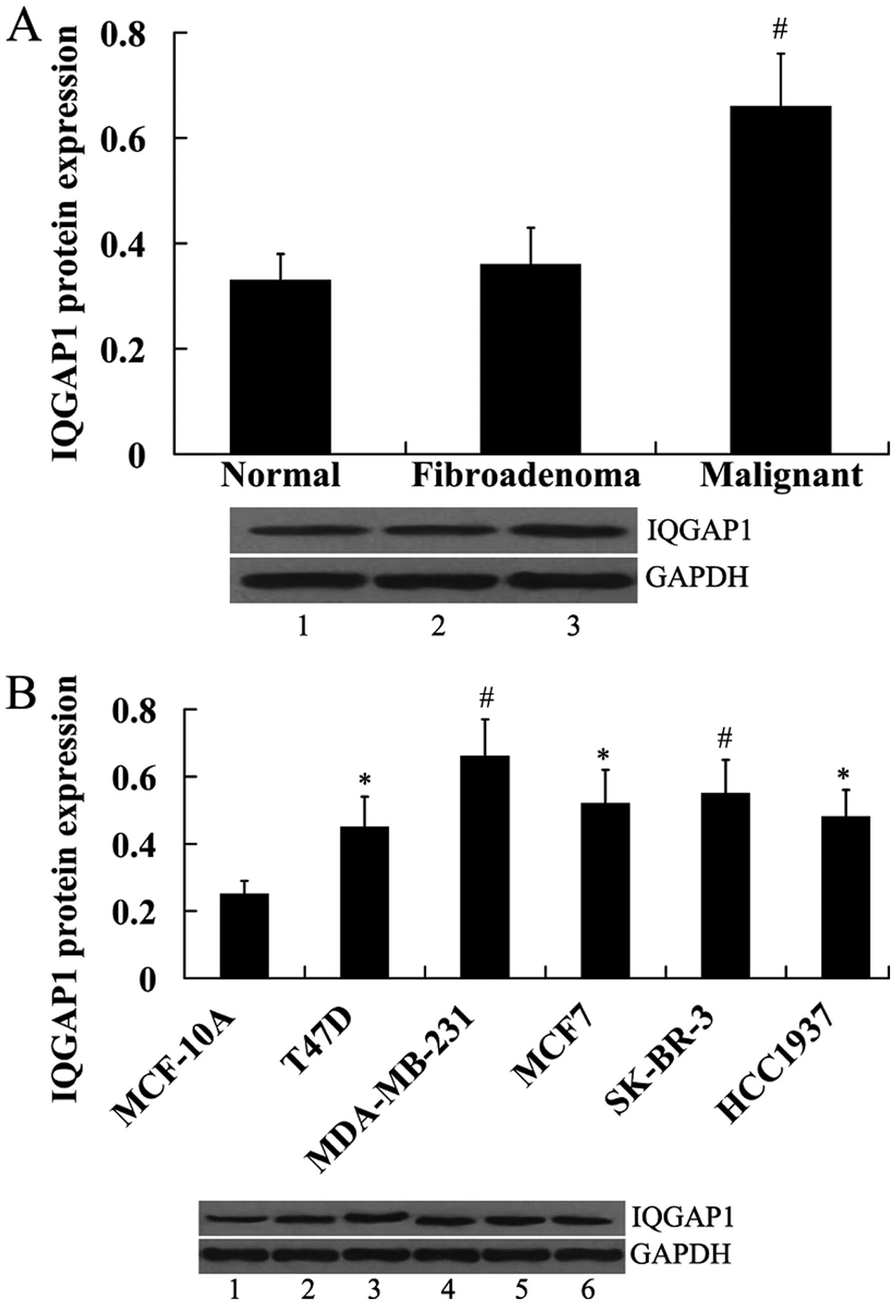
Expression of IQGAP1 in human breast
tissues and breast cancer cell lines
IQGAP1 expression was also examined in human breast
tissues and breast cancer cell lines. In contrast to miR-506
expression, IQGAP1 protein was significantly upregulated in the
malignant breast tissues compared with the normal and fibroadenoma
breast tissues (Fig. 2A). In
addition, IQGAP1 protein was significantly increased in the breast
cancer cell lines compared with the normal control MCF-10A cells
(Fig. 2B).
Effect of miR-506 on cell proliferation,
invasion and adhesion of breast cancer cells
To reveal the effect of miR-506 on breast cancer
cell proliferation, invasion and adhesion, the miR-506 mimic was
transfected into the MDA-MB-231 cells, which have low endogenous
miR-506 expression, to overex-press miR-506. In addition, we
knocked down the expression of miR-506 by transfection of the
miR-506 inhibitor into the HCC1937 cells, which show high miR-506
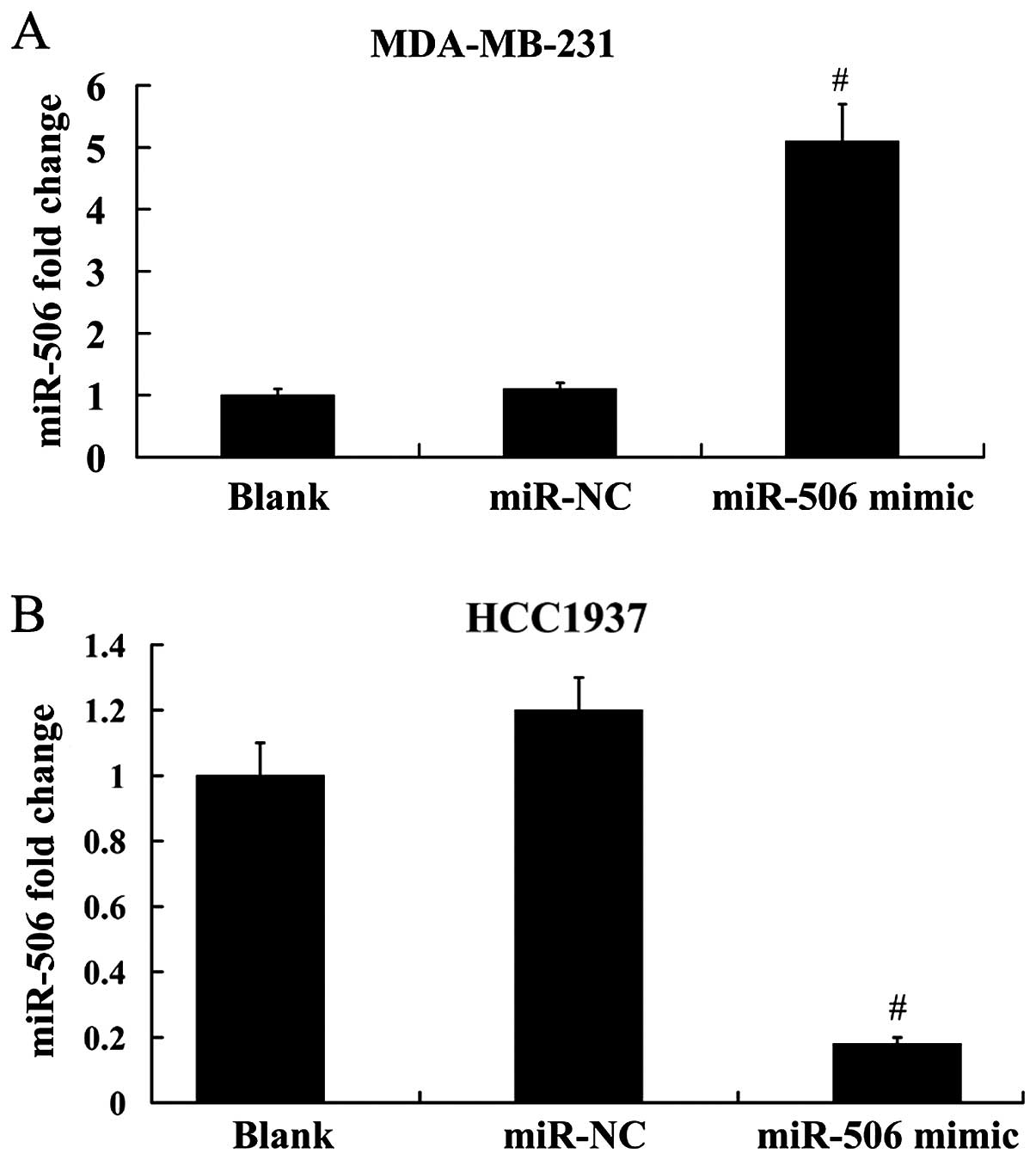
expression. As demonstrated in Fig.
3A, the expression level of miR-506 was increased ~4.6-fold in
MDA-MB-231 cells following transfection with the miR-506 mimic;
however, miR-506 expression was decreased ~85% in HCC1937 cells
following transfection with the miR-506 inhibitor (Fig. 3B).
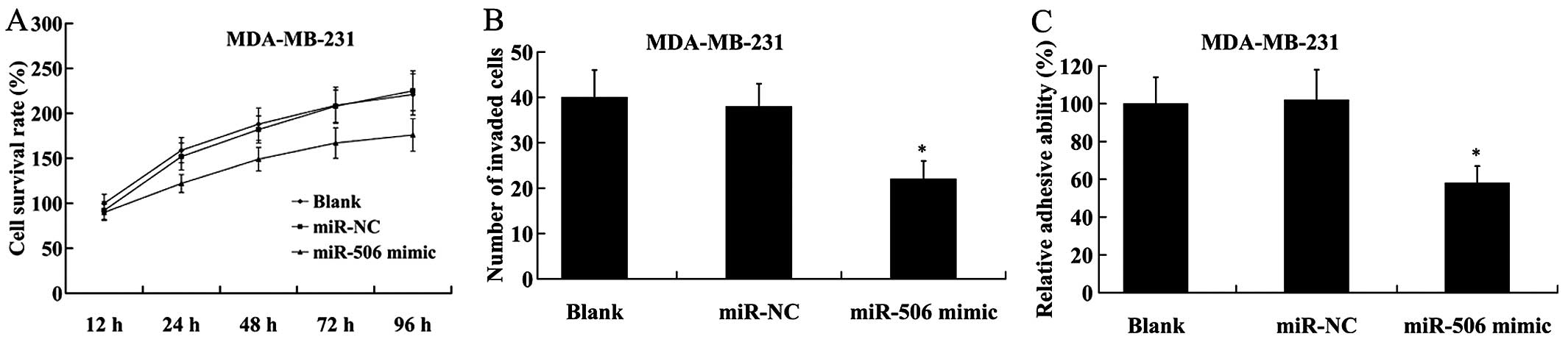
We found that compared with the control, MDA-MB-231
cells with ectopic expression of miR-506 showed a significant
decrease in cell proliferation by MTT assay (Fig. 4A). The number of invaded cells was
also decreased in MDA-MB-231 cells transfected with the miR-506
mimic in comparison to the cells transfected with the miR-NC
(Fig. 4B). Furthermore, cell
adhesion assay revealed that following transfection with the
miR-506 mimic, the adhesive ability of the MDA-MB-231 cells was
significantly reduced compared with that of the miR-NC (Fig. 4C).
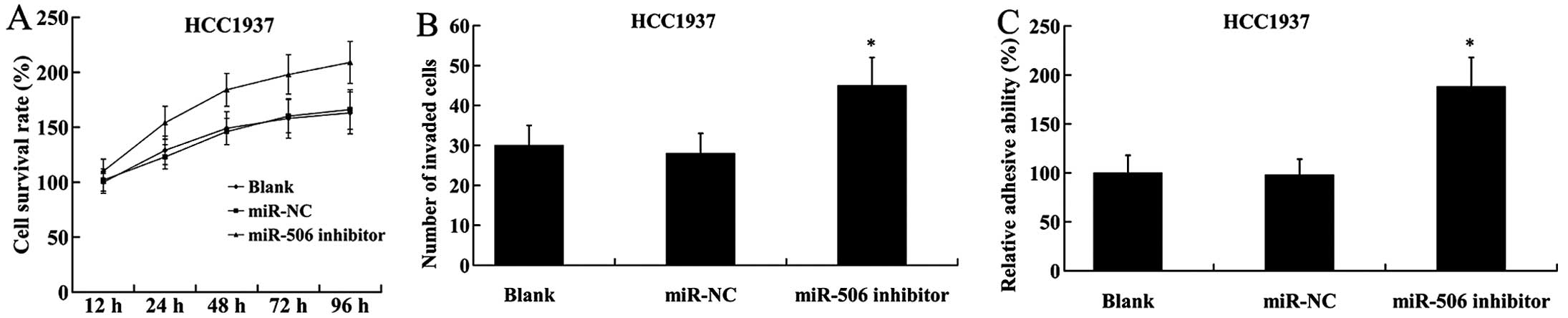
On the contrary, inhibition of miR-506 in HCC1937
cells led to the induction of cell proliferation, increased number
of invaded cells and adhesive ability (Fig. 5).
Effect of miR-506 on IQGAP1, B-Raf and
phosphorylated (pho)-extracellular signal regulated kinase (Erk)
1/2 expression
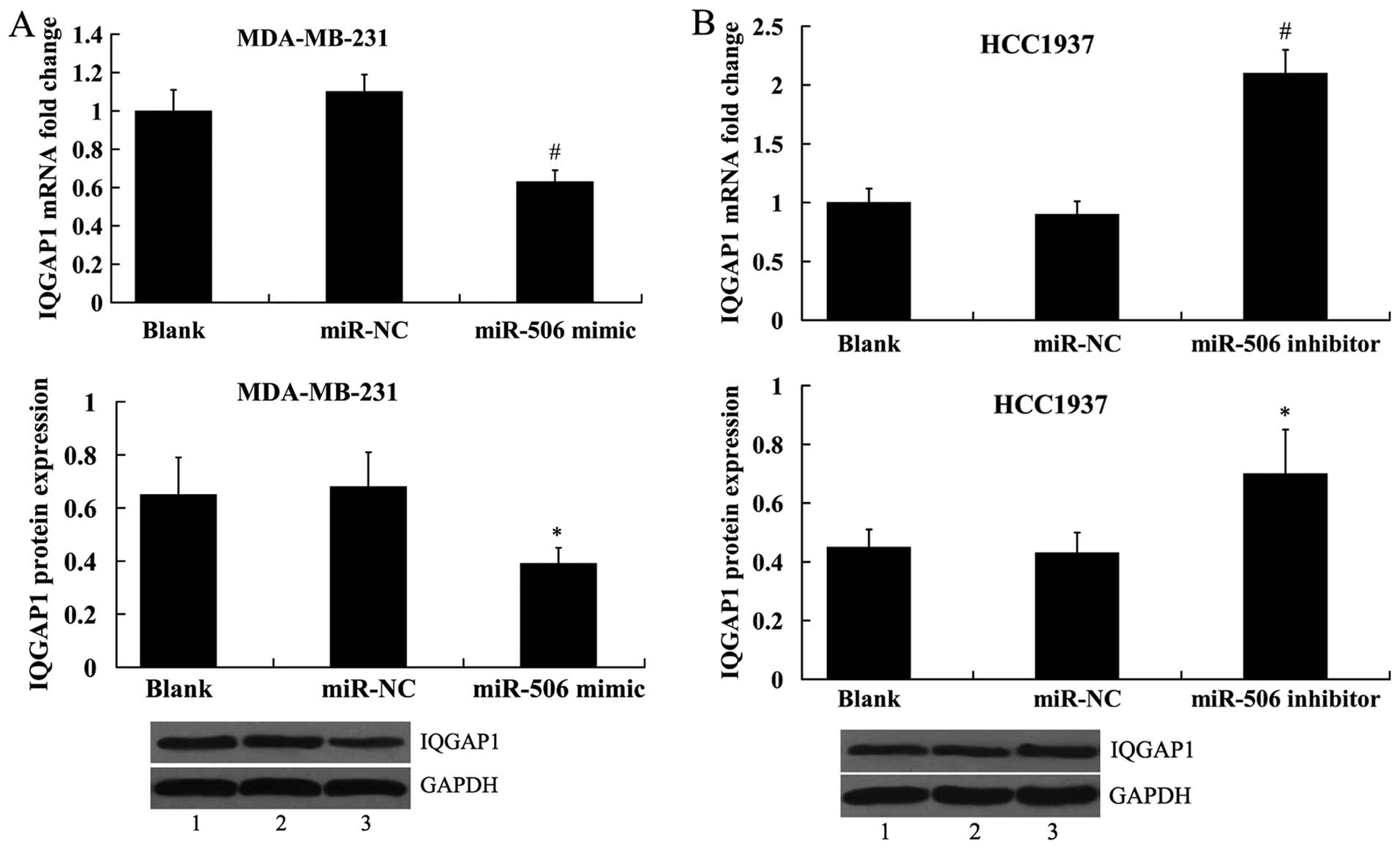
To investigate whether miR-506 regulates IQGAP1
expression, the miR-506 mimic and the miR-506 inhibitor was
transfected into the MDA-MB-231 and HCC1937 cells, respectively,
and IQGAP1 expression was analyzed both at the mRNA level and
protein level. It was shown that IQGAP1 expression was
significantly decreased in the MDA-MB-231 cells transfected with
the miR-506 mimic (Fig. 6A).
Transfection of miR-506 inhibitor into the HCC1937 cells led to
increased expression of IQGAP1 (Fig.
6B).
In addition, we found that the ERK MAPK pathway was
suppressed in MDA-MB-231 cells by transfection of the miR-506
mimic, as evidenced by the decreased expression of B-Raf and
pho-Erk1/2 (Fig. 7A). In HCC1937
cells, the expression of B-Raf and pho-Erk1/2 was significantly
increased by the miR-506 inhibitor (Fig. 7B).
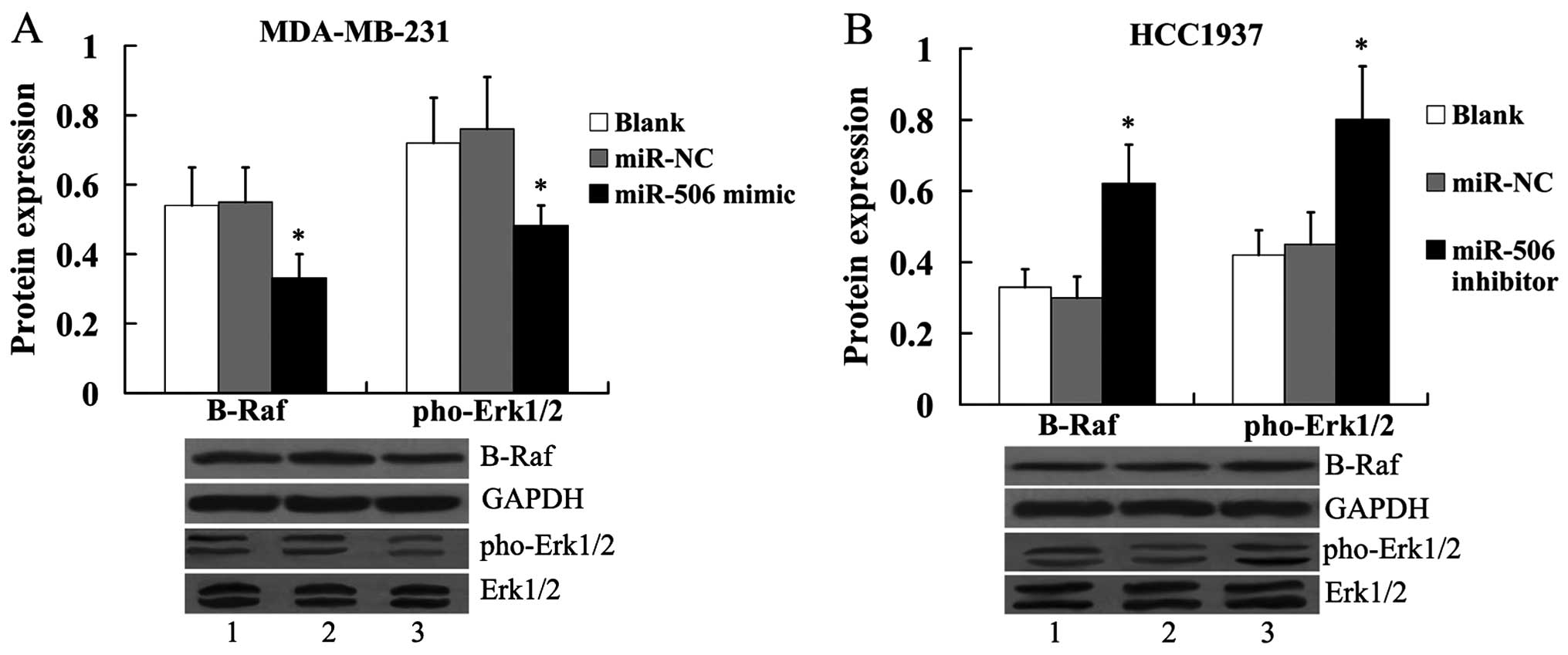 | Figure 7Effect of miR-506 on B-Raf and
pho-Erk1/2 protein expression in breast cancer cells. (A) Effect of
miR-506 overexpression on B-Raf and pho-Erk1/2 protein expression
in MDA-MB-231 cells. Lane 1, blank; lane 2, miR-NC; lane 3, miR-506
mimic. (B) Effect of miR-506 suppression on B-Raf and pho-Erk1/2
protein expression in HCC1937 cells. The expression of B-Raf was
normalized to GAPDH, while the expression of pho-Erk1/2 was
normalized to total Erk1/2. Lane 1, blank; lane 2, miR-NC; lane 3,
miR-506 inhibitor. *P<0.05 compared with the miR-NC.
NC, negative control; IQGAP1, IQ motif containing GTPase activating
protein 1; Erk, extracellular signal regulated kinase; pho,
phosphorylated. |
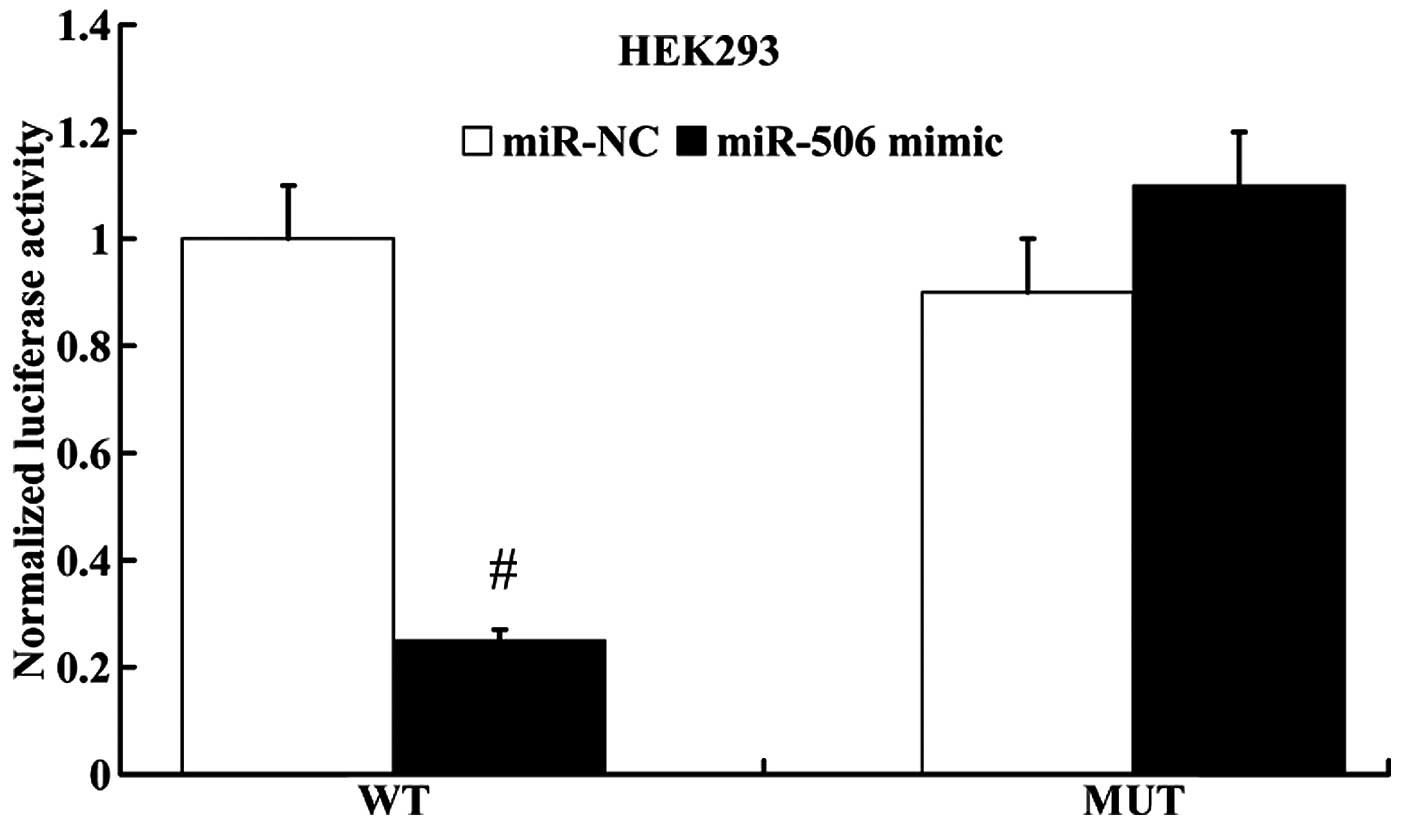
miR-506 regulates IQGAP1 expression by
directly targeting its 3′UTR
miRanda algorithms were employed to search for
putative gene targets of miR-506, and IQGAP1 was identified as a
potential target. Subsequently, luciferase reporter assay was
performed to determine whether IQGAP1 was a direct target of
miR-506. IQGAP1 3′UTR (wt 3′UTR or mut 3′UTR) luciferase reporter
vector was co-transfected with the miR-506 mimic or miR-NC into the
HEK293 cells. As shown in Fig. 8,
the luciferase activity of IQGAP1 wt 3′UTR was notably decreased in
the miR-506 mimic group compared with the miR-NC group. However,
the luciferase activity of IQGAP1 mut 3′UTR had no significant
difference between the miR-506 mimic group and the miR-NC
group.
IQGAP1 attenuates the effect of miR-506
on cell proliferation, invasion and adhesion of breast cancer
cells
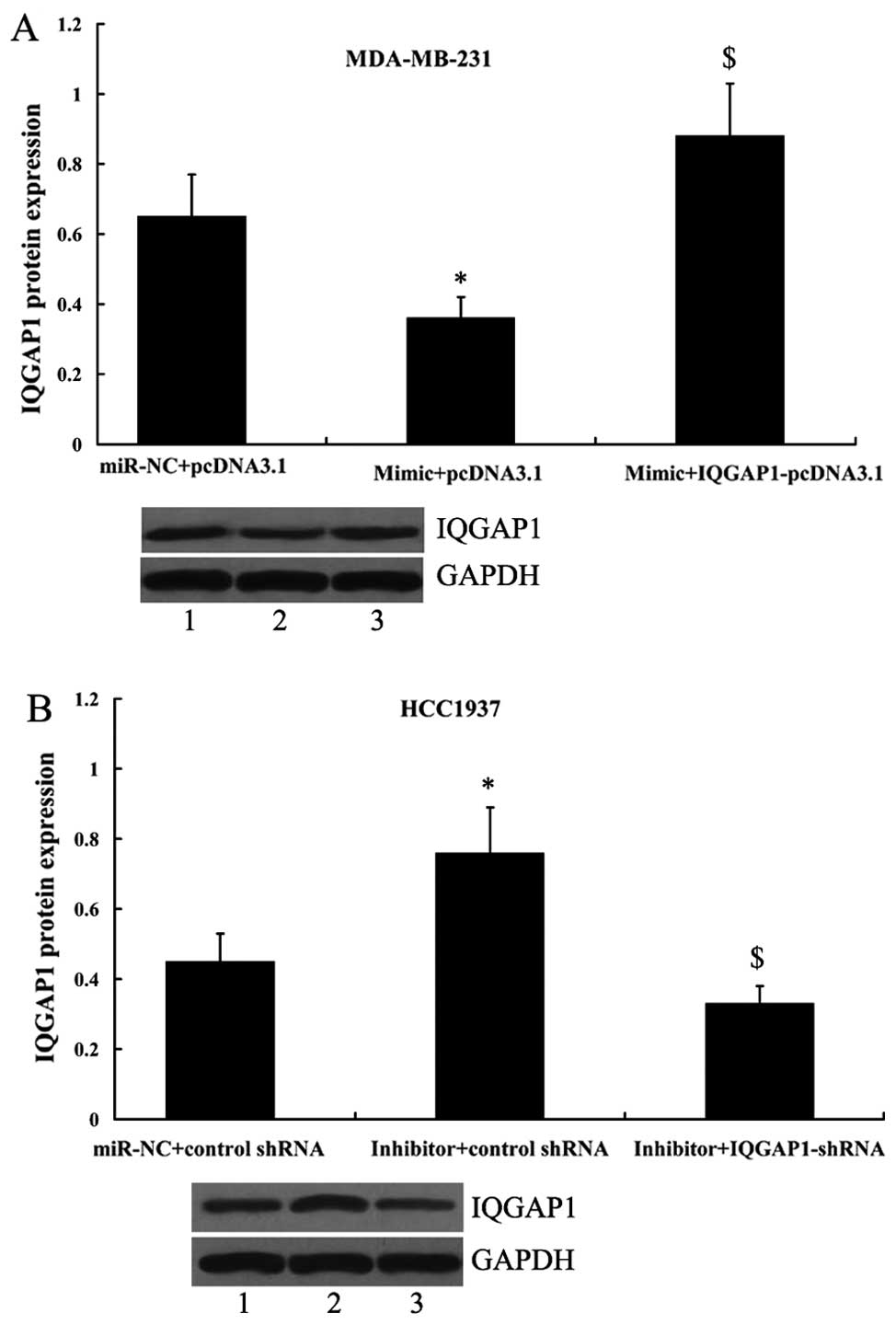
Since IQGAP1 was shown to be a direct target of
miR-506, its principle role was further investigated by its
overexpression or suppression in MDA-MB-231 and HCC1937 cells in
the presence of the miR-506 mimic or the miR-506 inhibitor. As
shown in Fig. 9A, the expression
of IQGAP1 was significantly increased in MDA-MB-231 cells following
transfection with the miR-506 mimic and IQGAP1-pcDNA3.1 compared
with the cells only transfected with the miR-506 mimic. In
addition, transfection of the IQGAP1 shRNA in HCC1937 cells led to
significantly decreased expression of IQGAP1 (Fig. 9B).
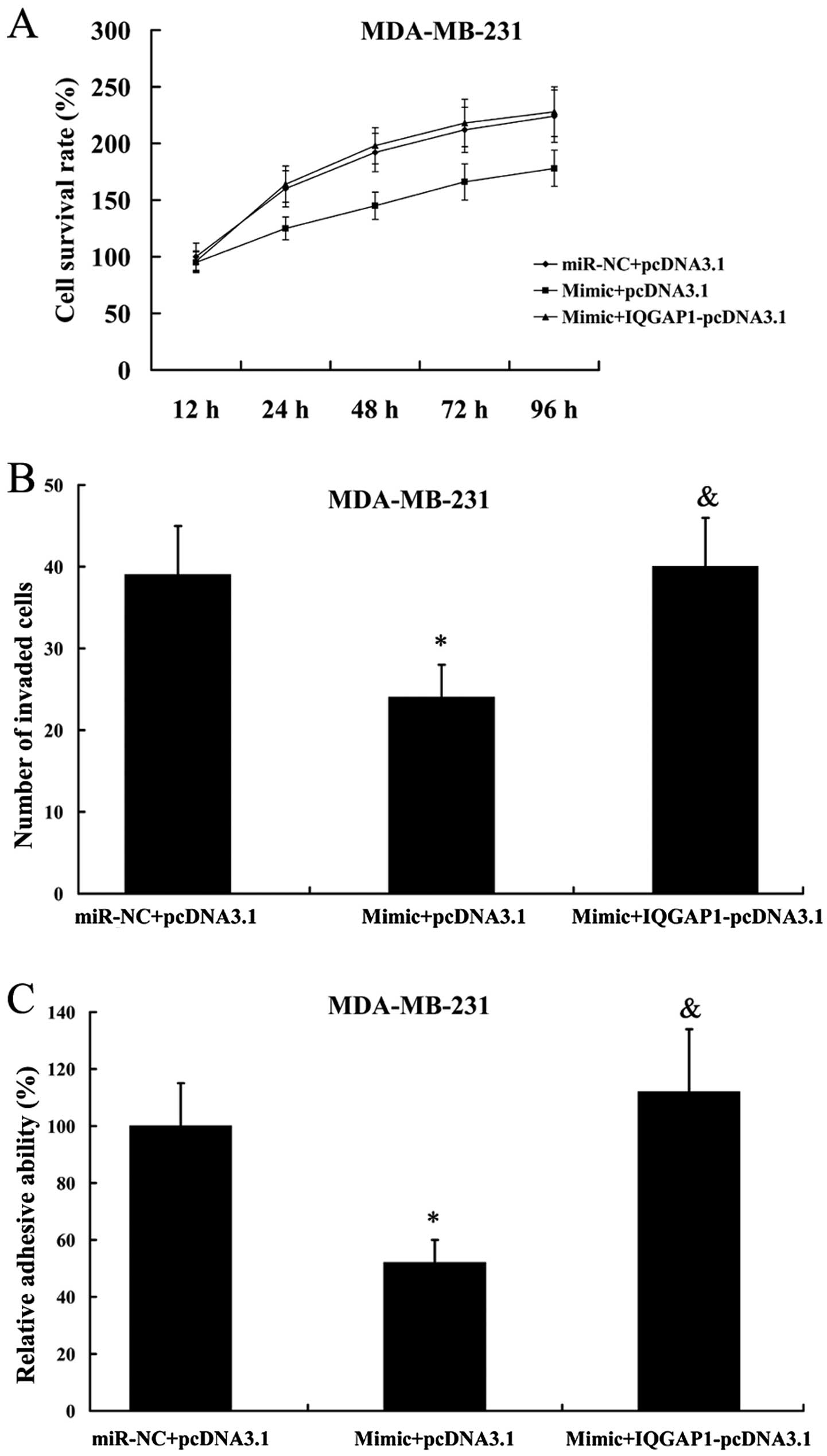
We found that the inhibition of cell proliferation,
invasion and adhesion as a consequence of miR-506 mimic
transfection in MDA-MB-231 cells was attenuated by the
overexpression of IQGAP1 (Fig.
10).
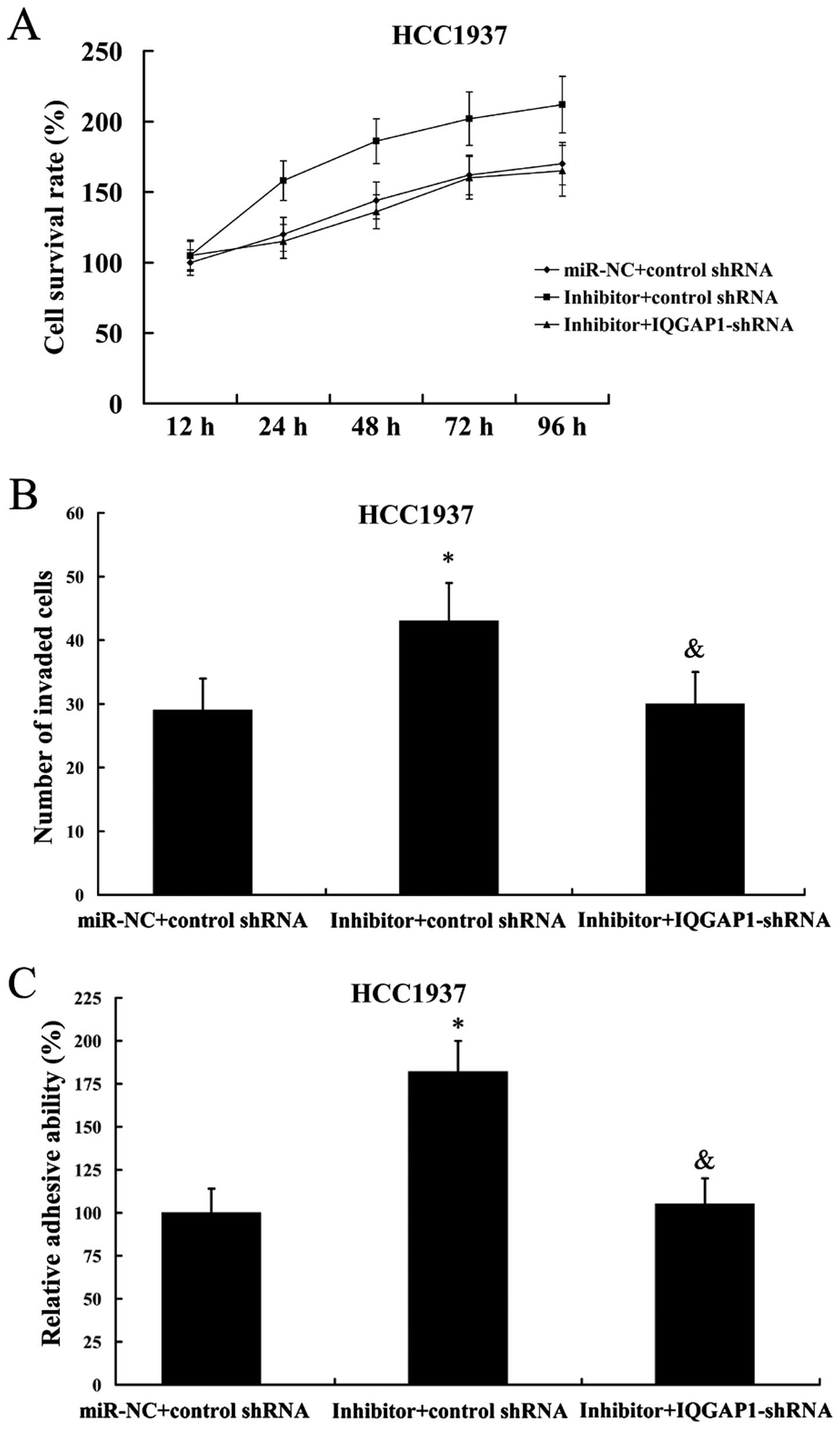
As expected, IQGAP1 suppression also reversed the
effect of miR-506 inhibitor on cell proliferation, invasion and
adhesion in HCC1937 cells (Fig.
11).
IQGAP1 attenuates the effect of miR-506
on B-Raf and pho-Erk1/2 expression
In addition, we found that the inhibitory effect of
miR-506 mimic on the expression of B-Raf and pho-Erk1/2 in
MDA-MB-231 cells could be reversed in the presence of IQGAP1
overexpression (Fig. 12A).
However, the increased expression of B-Raf and pho-Erk1/2 by the
transfection of miR-506 inhibitor was abolished by IQGAP1-shRNA
(Fig. 12B).
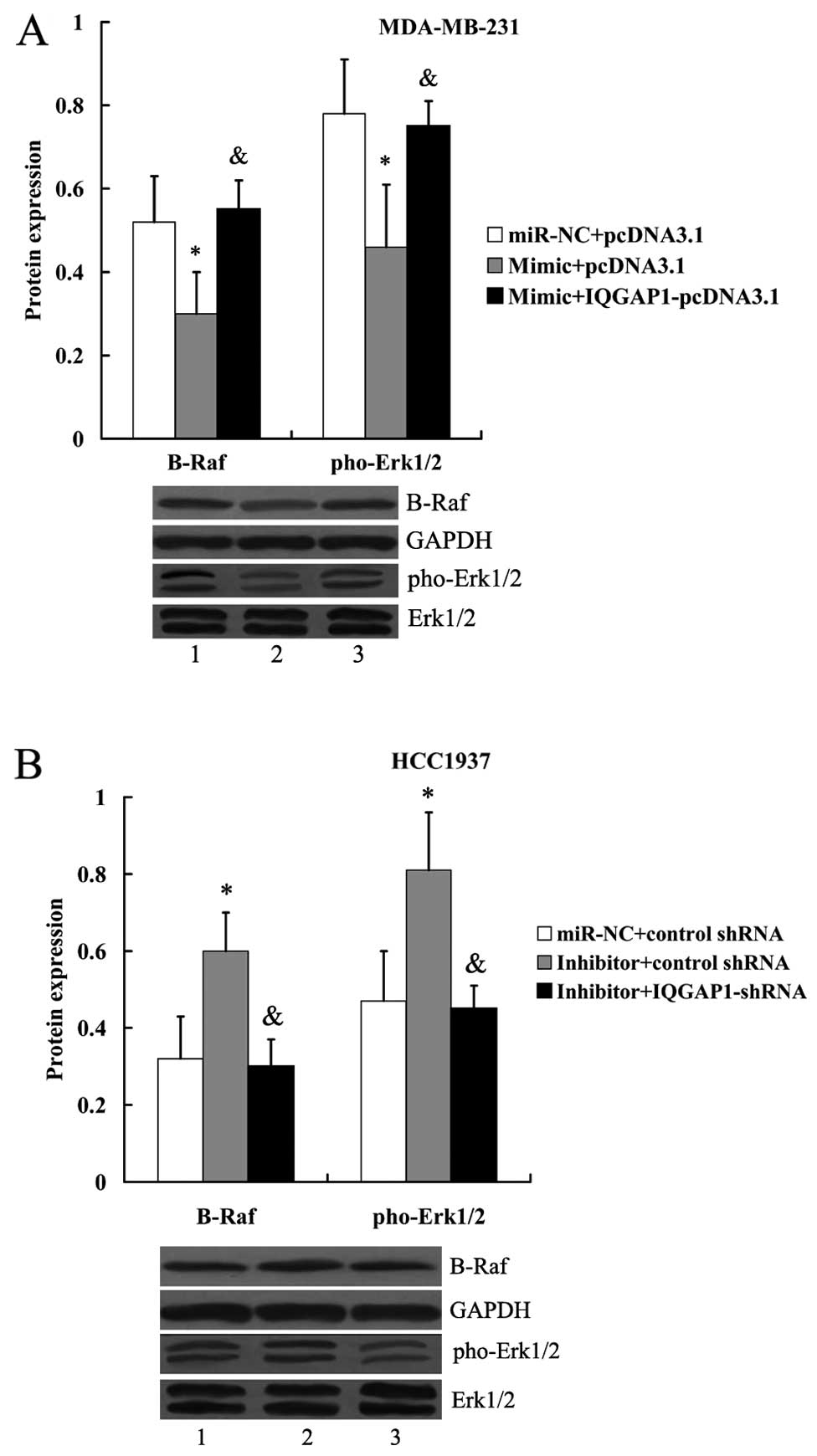 | Figure 12IQGAP1 attenuates the effect of
miR-506 on B-Raf and pho-Erk1/2 expression. (A) Expression of B-Raf
and pho-Erk1/2 protein in MDA-MB-231 cells following transfection
with the miR-506 mimic and IQGAP1-pcDNA3.1. Lane 1,
miR-NC+pcDNA3.1; lane 2, mimic+pcDNA3.1; lane 3,
mimic+IQGAP1-pcDNA3.1. *P<0.05 compared with
miR-NC+pcDNA3.1; &P<0.05 compared with
mimic+pcDNA3.1. (B) Expression of B-Raf and pho-Erk1/2 protein in
HCC1937 cells following transfection with the miR-506 inhibitor and
IQGAP1 shRNA. Lane 1, miR-NC+pcDNA3.1; lane 2, inhibitor+control
shRNA; lane 3, inhibitor+ IQGAP1-shRNA. *P<0.05
compared with miR-NC+control shRNA; &P<0.05
compared with inhibitor+control shRNA. The expression of B-Raf was
normalized to GAPDH, while the expression of pho-Erk1/2 was
normalized to total Erk1/2. NC, negative control; IQGAP1, IQ motif
containing GTPase activating protein 1; Erk, extracellular signal
regulated kinase; pho, phos-phorylated. |
Discussion
In the present study, we demonstrated downregulation
of miR-506 in both the breast malignant tissues and human breast
cancer cell lines. In addition, expression level of miR-506 was
decreased with the increasing of tumor stage. These results
indicate that miR-506 is clearly involved in the development of
human breast cancer. Subsequently, in vitro experiments were
performed to investigate the role of miR-506 on breast cancer cell
proliferation, invasion and adhesion.
miR-506 was cloned relatively recently, and it acts
as a tumor suppressive miRNA in various cancers and malignantly
transformed cells (7–9,12,13).
Downregulation of miR-506 has been identified in various tumors,
and overexpres sion of miR-506 shows inhibitory effect on the
development and progression of hepatocellular carcinoma, cervical
cancer and ovarian cancer (7,9,13).
However, Streicher et al (10) reported that miR-506 was upregulated
and acts as an oncogene in melanomas. Taken together, these
findings suggested that the function of miR-506 appears to be cell
type-specific. In the present study, we clearly demonstrated the
anti-oncogenic role of miR-506 in breast cancer cells. Both
gain-of-function and loss-of-function experiments revealed that
miR-506 suppresses cell proliferation, invasion and adhesion of
breast cancer cells. These findings are consistent with the reports
by Arora et al (8).
To date, there has been no report on the molecular
mechanisms underlying the role of miR-506 in breast cancer. In this
study, we first identified IQGAP1 as a direct target of miR-506.
IQGAP1 is a scaffold protein which has ubiquitous expression
(14,15). It contains multiple
protein-interacting domains, including one calponin homology
domain, one Ras-GAP-related domain, a polyproline binding domain
and four calmodulin-binding motifs. IQGAP1 interacts with
components of the cytoskeleton, the intercellular adhesion complex,
and several signaling molecules, thus, having roles in many
different aspects of cell physiology (16).
IQGAP1 has attracted attention because IQGAP1 plays
important roles in the control of cell adhesion, polarization and
migration (17). Overexpression of
IQGAP1 has been shown in glioblastoma, hepatocellular carcinoma,
lung and pancreatic cancer (18–22).
IQGAP1 expression is also associated with the development of breast
cancer (23,24). In the present study, we
demonstrated that IQGAP1 expression is regulated by miR-506.
Furthermore, IQGAP1 could attenuate the effect of miR-506 on breast
cancer cell proliferation, invasion and adhesion.
The ERK mitogen-activated protein kinase (MAPK)
pathway signaling is downstream of IQGAP1 (25), and this pathway was found to be
repressed by miR-506 in the present study. There is a direct
interaction between IQGAP1 and B-Raf, which modulates the
activation of B-Raf (26). Raf was
able to phosphorylate and activate the dual specificity protein
kinases MEK1 and MEK2, thus leading to the phosphorylation of Erk1
and Erk2 (27). In addition, it
has been demonstrated that IQGAP1 contributes to the regulation of
epidermal growth factor (EGF)-stimulated ERK activity (28,29).
The Ras/Raf/MEK/ERK cascade is suggested to play vital roles in
several fundamental cellular activities (30). In the present study, we first
proved that miR-506 represses the activation of B-Raf and Erk1/2,
at least partially, by downregulation of IQGAP1.
Collectively, these findings revealed that miR-506
expression is downregulated in breast cancer and associated with
the tumor stage. miR-506 inhibits breast cancer cell proliferation,
invasion and adhesion, as well as the ERK MAPK pathway, at least
partially, by directly downregulating IQGAP1. The
miR-506/IQGAP1/ERK pathway may be a novel therapeutic target in
breast cancer.
Acknowledgements
The present study is supported by the Jilin Province
Key Scientific and Technical Project (no. 20150204081SF),
China.
References
|
1
|
Badar F, Faruqui ZS, Ashraf A and Uddin N:
Third world issues in breast cancer detection. J Pak Med Assoc.
57:137–140. 2007.PubMed/NCBI
|
|
2
|
Jamal S, Mamoon N, Moghal S, Mushtaq S and
Luqman M: Carcinoma breast: A histopathological audit. J Coll
Physicians Surg Pak. 16:117–119. 2006.PubMed/NCBI
|
|
3
|
Jemal A, Bray F, Center MM, Ferlay J, Ward
E and Forman D: Global cancer statistics. CA Cancer J Clin.
61:69–90. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chaffer CL and Weinberg RA: A perspective
on cancer cell metastasis. Science. 331:1559–1564. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Nelson KM and Weiss GJ: MicroRNAs and
cancer: Past, present, and potential future. Mol Cancer Ther.
7:3655–3660. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu G, Sun Y, Ji P, Li X, Cogdell D, Yang
D, Parker Kerrigan BC, Shmulevich I, Chen K, Sood AK, et al:
MiR-506 suppresses proliferation and induces senescence by directly
targeting the CDK4/6-FOXM1 axis in ovarian cancer. J Pathol.
233:308–318. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Arora H, Qureshi R and Park WY: miR-506
regulates epithelial mesenchymal transition in breast cancer cell
lines. PLoS One. 8:e642732013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wen SY, Lin Y, Yu YQ, Cao SJ, Zhang R,
Yang XM, Li J, Zhang YL, Wang YH, Ma MZ, et al: miR-506 acts as a
tumor suppressor by directly targeting the hedgehog pathway
transcription factor Gli3 in human cervical cancer. Oncogene.
34:717–725. 2015. View Article : Google Scholar
|
|
10
|
Streicher KL, Zhu W, Lehmann KP,
Georgantas RW, Morehouse CA, Brohawn P, Carrasco RA, Xiao Z, Tice
DA, Higgs BW, et al: A novel oncogenic role for the miRNA-506-514
cluster in initiating melanocyte transformation and promoting
melanoma growth. Oncogene. 31:1558–1570. 2012. View Article : Google Scholar
|
|
11
|
Buffa FM, Camps C, Winchester L, Snell CE,
Gee HE, Sheldon H, Taylor M, Harris AL and Ragoussis J:
microRNA-associated progression pathways and potential therapeutic
targets identified by integrated mRNA and microRNA expression
profiling in breast cancer. Cancer Res. 71:5635–5645. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhao Y, Liu H, Li Y, Wu J, Greenlee AR,
Yang C and Jiang Y: The role of miR-506 in transformed 16HBE cells
induced by anti-benzo[a]pyrene-trans-7,8-dihydrodiol-9,10-epoxide.
Toxicol Lett. 205:320–326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang Y, Cui M, Sun BD, Liu FB, Zhang XD
and Ye LH: MiR-506 suppresses proliferation of hepatoma cells
through targeting YAP mRNA 3′UTR. Acta Pharmacol Sin. 35:1207–1214.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Weissbach L, Settleman J, Kalady MF,
Snijders AJ, Murthy AE, Yan YX and Bernards A: Identification of a
human rasGAP-related protein containing calmodulin-binding motifs.
J Biol Chem. 269:20517–20521. 1994.PubMed/NCBI
|
|
15
|
Hart MJ, Callow MG, Souza B and Polakis P:
IQGAP1, a calmodulin-binding protein with a rasGAP-related domain,
is a potential effector for cdc42Hs. EMBO J. 15:2997–3005.
1996.PubMed/NCBI
|
|
16
|
Noritake J, Watanabe T, Sato K, Wang S and
Kaibuchi K: IQGAP1: A key regulator of adhesion and migration. J
Cell Sci. 118:2085–2092. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
White CD, Brown MD and Sacks DB: IQGAPs in
cancer: A family of scaffold proteins underlying tumorigenesis.
FEBS Lett. 583:1817–1824. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Johnson M, Sharma M and Henderson BR:
IQGAP1 regulation and roles in cancer. Cell Signal. 21:1471–1478.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Lu SH, Jiang XJ, Xiao GL, Liu DY and Yuan
XR: miR-124a restoration inhibits glioma cell proliferation and
invasion by suppressing IQGAP1 and β-catenin. Oncol Rep.
32:2104–2110. 2014.PubMed/NCBI
|
|
20
|
Chen F, Zhu HH, Zhou LF, Wu SS, Wang J and
Chen Z: IQGAP1 is overexpressed in hepatocellular carcinoma and
promotes cell proliferation by Akt activation. Exp Mol Med.
42:477–483. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nakamura H, Fujita K, Nakagawa H, Kishi F,
Takeuchi A, Aute I and Kato H: Expression pattern of the scaffold
protein IQGAP1 in lung cancer. Oncol Rep. 13:427–431.
2005.PubMed/NCBI
|
|
22
|
Wang XX, Li XZ, Zhai LQ, Liu ZR, Chen XJ
and Pei Y: Overexpression of IQGAP1 in human pancreatic cancer.
Hepatobiliary Pancreat Dis Int. 12:540–545. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Erdemir HH, Li Z and Sacks DB: IQGAP1
binds to estrogen receptor-α and modulates its function. J Biol
Chem. 289:9100–9112. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Jadeski L, Mataraza JM, Jeong HW, Li Z and
Sacks DB: IQGAP1 stimulates proliferation and enhances
tumorigenesis of human breast epithelial cells. J Biol Chem.
283:1008–1017. 2008. View Article : Google Scholar
|
|
25
|
Brown MD and Sacks DB: IQGAP1 in cellular
signaling: Bridging the GAP. Trends Cell Biol. 16:242–249. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ren JG, Li Z and Sacks DB: IQGAP1
modulates activation of B-Raf. Proc Natl Acad Sci USA.
104:10465–10469. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Schaeffer HJ and Weber MJ:
Mitogen-activated protein kinases: Specific messages from
ubiquitous messengers. Mol Cell Biol. 19:2435–2444. 1999.PubMed/NCBI
|
|
28
|
Roy M, Li Z and Sacks DB: IQGAP1 is a
scaffold for mitogen-activated protein kinase signaling. Mol Cell
Biol. 25:7940–7952. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Roy M, Li Z and Sacks DB: IQGAP1 binds
ERK2 and modulates its activity. J Biol Chem. 279:17329–17337.
2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
McCubrey JA, Steelman LS, Chappell WH,
Abrams SL, Wong EW, Chang F, Lehmann B, Terrian DM, Milella M,
Tafuri A, et al: Roles of the Raf/MEK/ERK pathway in cell growth,
malignant transformation and drug resistance. Biochim Biophys Acta.
1773:1263–1284. 2007. View Article : Google Scholar
|