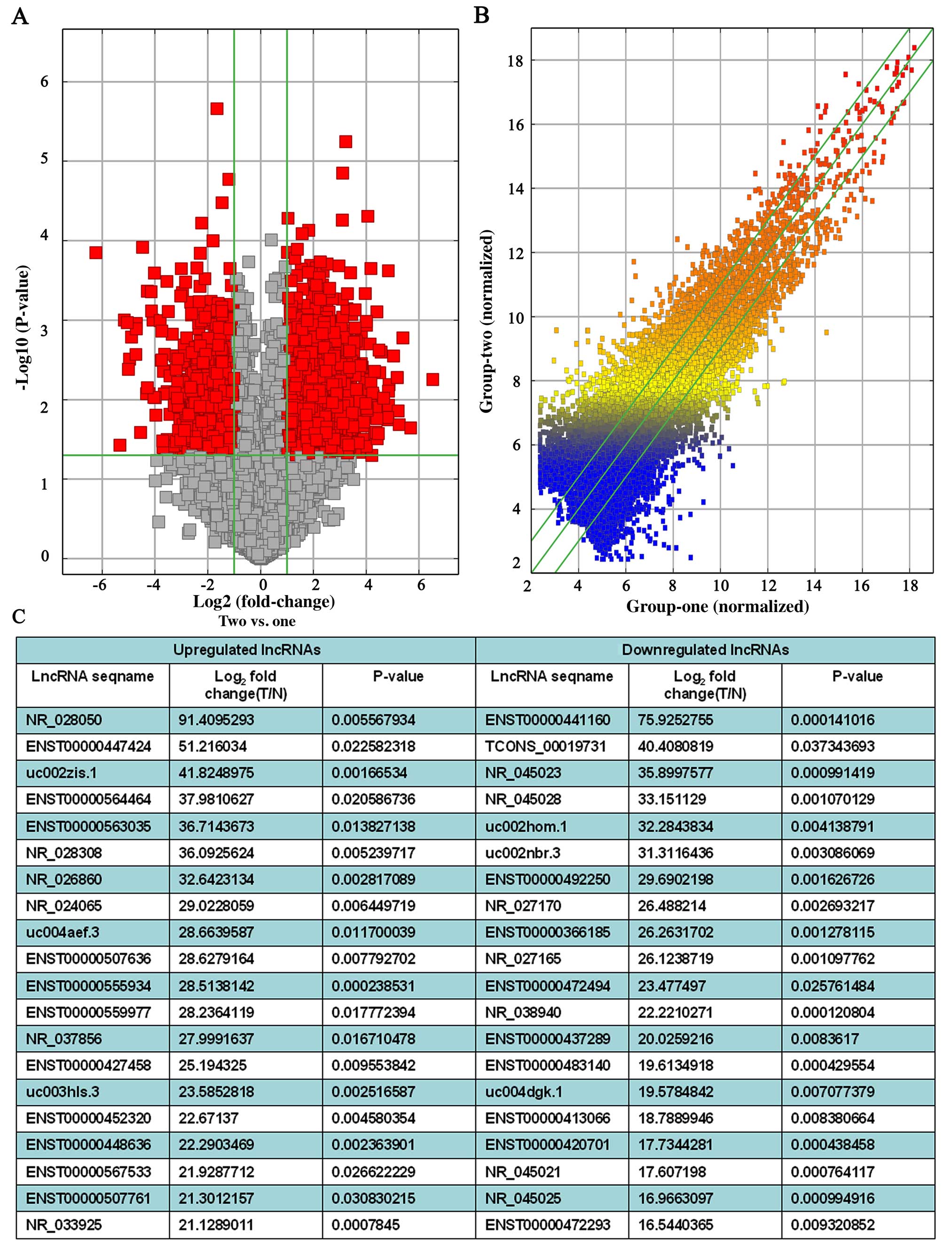
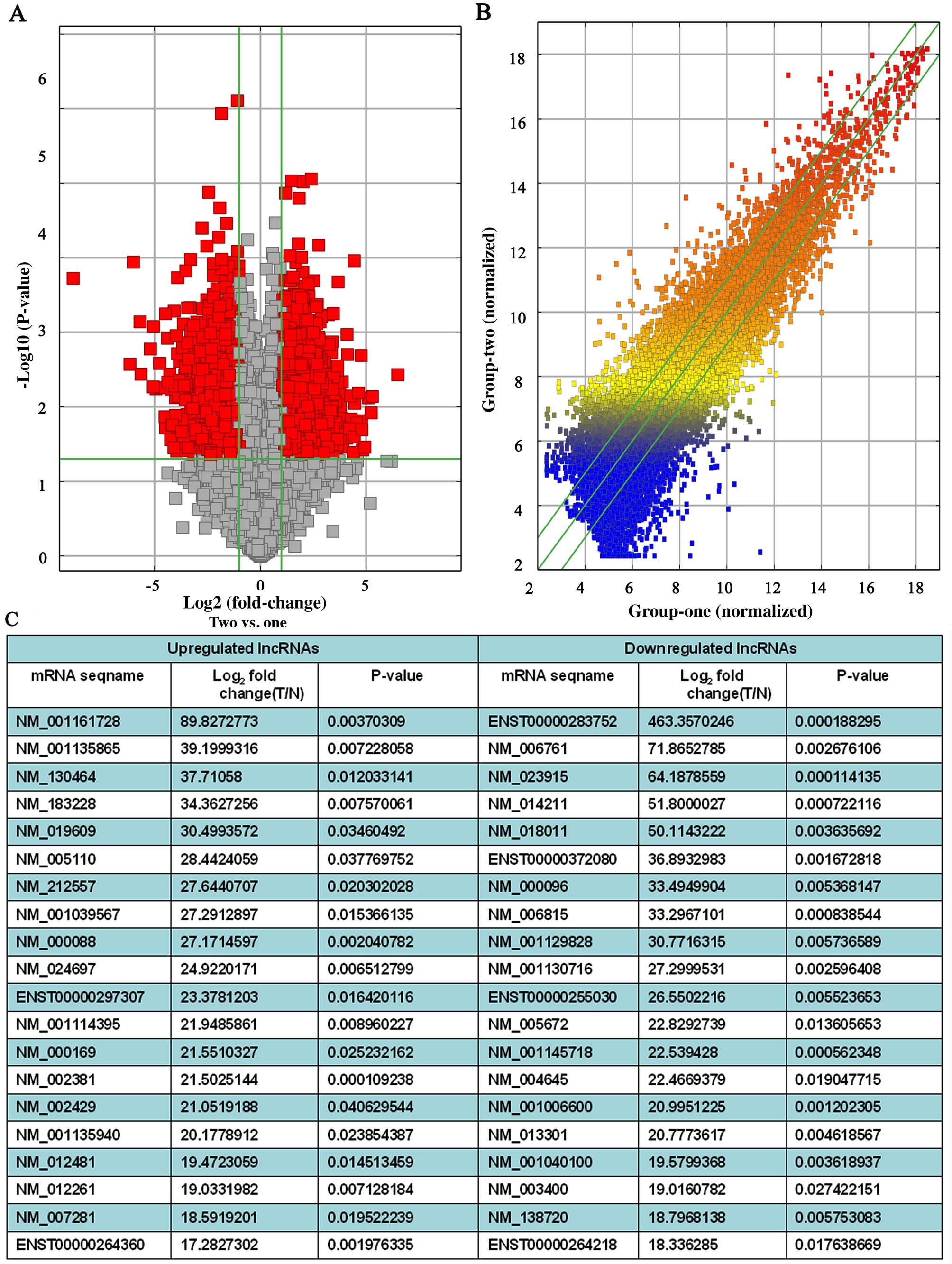
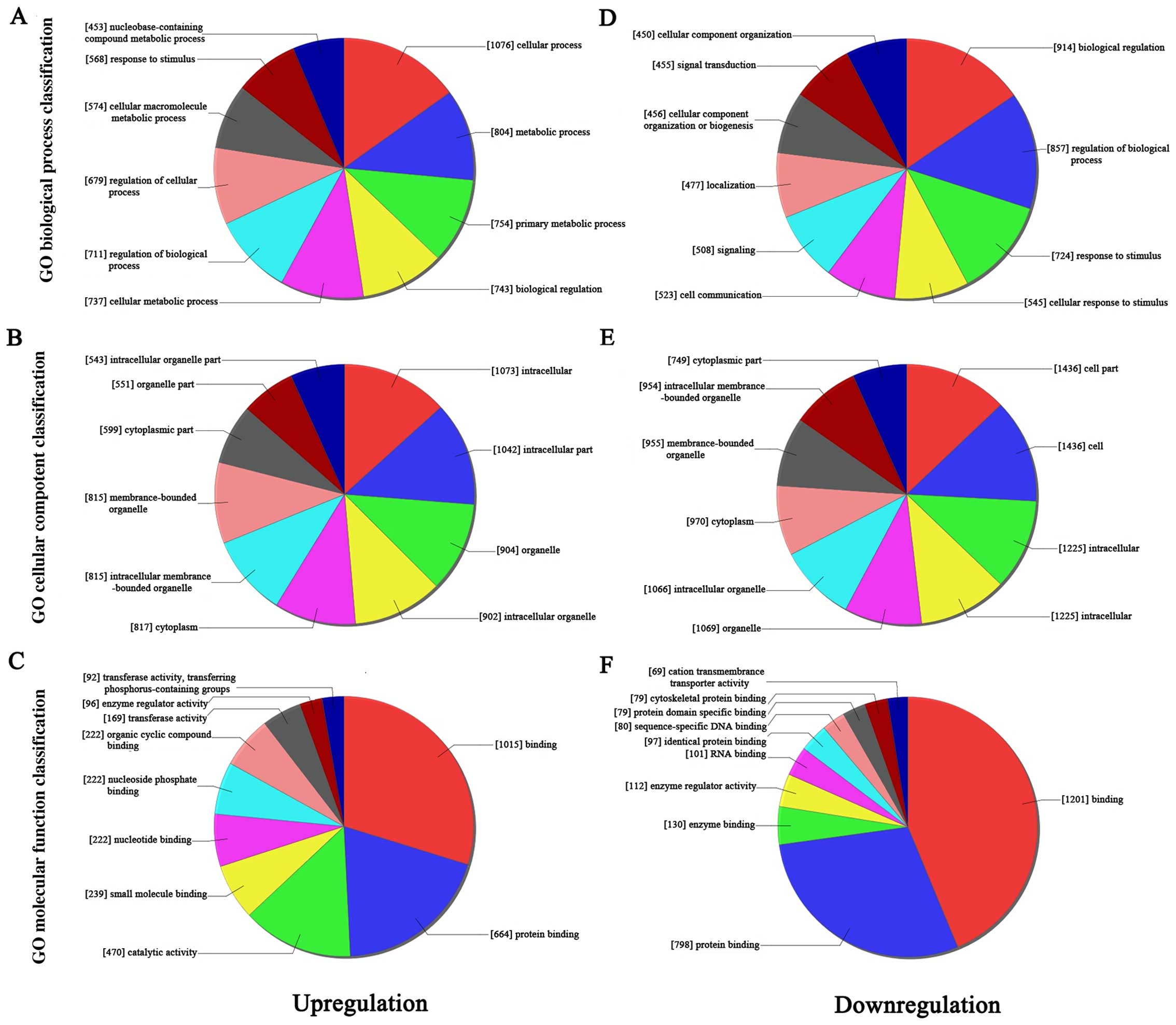
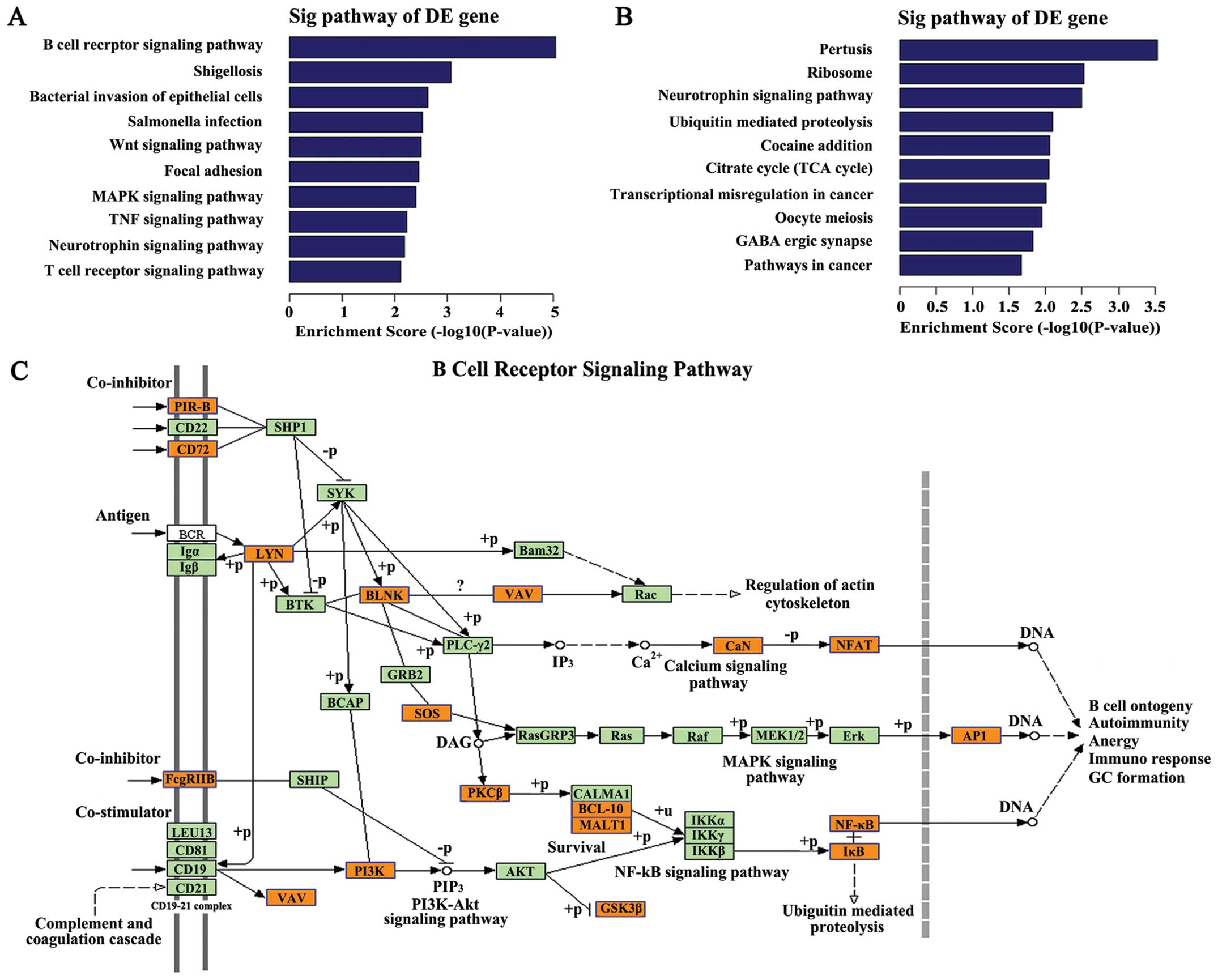
|
1
|
Kazanjian KK, Hines OJ, Duffy JP, Yoon DY,
Cortina G and Reber HA: Improved survival following
pancreaticoduodenectomy to treat adenocarcinoma of the pancreas:
The influence of operative blood loss. Arch Surg. 143:1166–1171.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Oettle H, Neuhaus P, Hochhaus A, Hartmann
JT, Gellert K, Ridwelski K, Niedergethmann M, Zülke C, Fahlke J,
Arning MB, et al: Adjuvant chemotherapy with gemcitabine and
long-term outcomes among patients with resected pancreatic cancer:
The CONKO-001 randomized trial. JAMA. 310:1473–1481. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Yang J, Li J, Zhu R, Zhang H, Zheng Y, Dai
W, Wang F, Shen M, Chen K, Cheng P, et al: K-ras mutational status
in cytohistological tissue as a molecular marker for the diagnosis
of pancreatic cancer: A systematic review and meta-analysis. Dis
Markers. 2014:5737832014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cowan RW and Maitra A: Genetic progression
of pancreatic cancer. Cancer J. 20:80–84. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dempe S, Stroh-Dege AY, Schwarz E,
Rommelaere J and Dinsart C: SMAD4: A predictive marker of PDAC cell
permissiveness for oncolytic infection with parvovirus H-1PV. Int J
Cancer. 126:2914–2927. 2010.
|
|
6
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ho AS, Huang X, Cao H, Christman-Skieller
C, Bennewith K, Le QT and Koong AC: Circulating miR-210 as a novel
hypoxia marker in pancreatic cancer. Transl Oncol. 3:109–113. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Kawaguchi T, Komatsu S, Ichikawa D,
Morimura R, Tsujiura M, Konishi H, Takeshita H, Nagata H, Arita T,
Hirajima S, et al: Clinical impact of circulating miR-221 in plasma
of patients with pancreatic cancer. Br J Cancer. 108:361–369. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ponting CP, Oliver PL and Reik W:
Evolution and functions of long noncoding RNAs. Cell. 136:629–641.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tay Y, Karreth FA and Pandolfi PP:
Aberrant ceRNA activity drives lung cancer. Cell Res. 24:259–260.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Beroukhim R, Mermel CH, Porter D, Wei G,
Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J,
Urashima M, et al: The landscape of somatic copy-number alteration
across human cancers. Nature. 463:899–905. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Futreal PA, Coin L, Marshall M, Down T,
Hubbard T, Wooster R, Rahman N and Stratton MR: A census of human
cancer genes. Nat Rev Cancer. 4:177–183. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Stratton MR, Campbell PJ and Futreal PA:
The cancer genome. Nature. 458:719–724. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Nagano T and Fraser P: No-nonsense
functions for long noncoding RNAs. Cell. 145:178–181. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Flynn RA and Chang HY: Active chromatin
and noncoding RNAs: An intimate relationship. Curr Opin Genet Dev.
22:172–178. 2012. View Article : Google Scholar :
|
|
17
|
Kung JT, Colognori D and Lee JT: Long
noncoding RNAs: Past, present, and future. Genetics. 193:651–669.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Hung T and Chang HY: Long noncoding RNA in
genome regulation: Prospects and mechanisms. RNA Biol. 7:582–585.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
20
|
Jiang J, Jing Y, Cost GJ, Chiang JC, Kolpa
HJ, Cotton AM, Carone DM, Carone BR, Shivak DA, Guschin DY, et al:
Translating dosage compensation to trisomy 21. Nature. 500:296–300.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Yildirim E, Kirby JE, Brown DE, Mercier
FE, Sadreyev RI, Scadden DT and Lee JT: Xist RNA is a potent
suppressor of hematologic cancer in mice. Cell. 152:727–742. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hajjari M and Salavaty A: HOTAIR: An
oncogenic long non-coding RNA in different cancers. Cancer Biol
Med. 12:1–9. 2015.PubMed/NCBI
|
|
23
|
Pang EJ, Yang R, Fu XB and Liu YF:
Overexpression of long non-coding RNA MALAT1 is correlated with
clinical progression and unfavorable prognosis in pancreatic
cancer. Tumour Biol. 36:2403–2407. 2015. View Article : Google Scholar
|
|
24
|
Ding C, Yang Z, Lv Z, Du C, Xiao H, Peng
C, Cheng S, Xie H, Zhou L, Wu J, et al: Long non-coding RNA PVT1 is
associated with tumor progression and predicts recurrence in
hepatocellular carcinoma patients. Oncol Lett. 9:955–963.
2015.PubMed/NCBI
|
|
25
|
Yu M, Ting DT, Stott SL, Wittner BS,
Ozsolak F, Paul S, Ciciliano JC, Smas ME, Winokur D, Gilman AJ, et
al: RNA sequencing of pancreatic circulating tumour cells
implicates WNT signalling in metastasis. Nature. 487:510–513. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Miyabayashi K, Ijichi H, Mohri D, Tada M,
Yamamoto K, Asaoka Y, Ikenoue T, Tateishi K, Nakai Y, Isayama H, et
al: Erlotinib prolongs survival in pancreatic cancer by blocking
gemcitabine-induced MAPK signals. Cancer Res. 73:2221–2234. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Jun S, Lee S, Kim HC, Ng C, Schneider AM,
Ji H, Ying H, Wang H, DePinho RA and Park JI: PAF-mediated MAPK
signaling hyperactivation via LAMTOR3 induces pancreatic
tumorigenesis. Cell Rep. 5:314–322. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang Y, Yan W, Collins MA, Bednar F,
Rakshit S, Zetter BR, Stanger BZ, Chung I, Rhim AD and di Magliano
MP: Interleukin-6 is required for pancreatic cancer progression by
promoting MAPK signaling activation and oxidative stress
resistance. Cancer Res. 73:6359–6374. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Boreddy SR, Pramanik KC and Srivastava SK:
Pancreatic tumor suppression by benzyl isothiocyanate is associated
with inhibition of PI3K/AKT/FOXO pathway. Clin Cancer Res.
17:1784–1795. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Roy SK, Chen Q, Fu J, Shankar S and
Srivastava RK: Resveratrol inhibits growth of orthotopic pancreatic
tumors through activation of FOXO transcription factors. PLoS One.
6:e251662011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ma Y, Gu Y, Zhang Q, Han Y, Yu S, Lu Z and
Chen J: Targeted degradation of KRAS by an engineered ubiquitin
ligase suppresses pancreatic cancer cell growth in vitro and in
vivo. Mol Cancer Ther. 12:286–294. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Hajjari M and Khoshnevisan A: Potential
long non-coding RNAs to be considered as biomarkers or therapeutic
targets in gastric cancer. Front Genet. 4:2102013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Huang L, Liao LM, Liu AW, Wu JB, Cheng XL,
Lin JX and Zheng M: Overexpression of long noncoding RNA HOTAIR
predicts a poor prognosis in patients with cervical cancer. Arch
Gynecol Obstet. 290:717–723. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Panzitt K, Tschernatsch MM, Guelly C,
Moustafa T, Stradner M, Strohmaier HM, Buck CR, Denk H, Schroeder
R, Trauner M, et al: Characterization of HULC, a novel gene with
striking up-regulation in hepatocellular carcinoma, as noncoding
RNA. Gastroenterology. 132:330–342. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Matouk IJ, Abbasi I, Hochberg A, Galun E,
Dweik H and Akkawi M: Highly upregulated in liver cancer noncoding
RNA is overexpressed in hepatic colorectal metastasis. Eur J
Gastroenterol Hepatol. 21:688–692. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Liu Y, Pan S, Liu L, Zhai X, Liu J, Wen J,
Zhang Y, Chen J, Shen H and Hu Z: A genetic variant in long
non-coding RNA HULC contributes to risk of HBV-related
hepatocellular carcinoma in a Chinese population. PLoS One.
7:e351452012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sun M, Jin FY, Xia R, Kong R, Li JH, Xu
TP, Liu YW, Zhang EB, Liu XH and De W: Decreased expression of long
noncoding RNA GAS5 indicates a poor prognosis and promotes cell
proliferation in gastric cancer. BMC Cancer. 14:3192014. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Renganathan A, Kresoja-Rakic J, Echeverry
N, Ziltener G, Vrugt B, Opitz I, Stahel RA and Felley-Bosco E: GAS5
long non-coding RNA in malignant pleural mesothelioma. Mol Cancer.
13:1192014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Arita T, Ichikawa D, Konishi H, Komatsu S,
Shiozaki A, Shoda K, Kawaguchi T, Hirajima S, Nagata H, Kubota T,
et al: Circulating long non-coding RNAs in plasma of patients with
gastric cancer. Anticancer Res. 33:3185–3193. 2013.PubMed/NCBI
|
|
40
|
Xie H, Ma H and Zhou D: Plasma HULC as a
promising novel biomarker for the detection of hepatocellular
carcinoma. BioMed Res Int. 2013:1361062013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Rinn JL, Kertesz M, Wang JK, Squazzo SL,
Xu X, Brugmann SA, Goodnough LH, Helms JA, Farnham PJ, Segal E, et
al: Functional demarcation of active and silent chromatin domains
in human HOX loci by noncoding RNAs. Cell. 129:1311–1323. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gupta RA, Shah N, Wang KC, Kim J, Horlings
HM, Wong DJ, Tsai MC, Hung T, Argani P, Rinn JL, et al: Long
non-coding RNA HOTAIR reprograms chromatin state to promote cancer
metastasis. Nature. 464:1071–1076. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Tsai MC, Manor O, Wan Y, Mosammaparast N,
Wang JK, Lan F, Shi Y, Segal E and Chang HY: Long noncoding RNA as
modular scaffold of histone modification complexes. Science.
329:689–693. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Yang Z, Zhou L, Wu LM, Lai MC, Xie HY,
Zhang F and Zheng SS: Overexpression of long non-coding RNA HOTAIR
predicts tumor recurrence in hepatocellular carcinoma patients
following liver transplantation. Ann Surg Oncol. 18:1243–1250.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Kogo R, Shimamura T, Mimori K, Kawahara K,
Imoto S, Sudo T, Tanaka F, Shibata K, Suzuki A, Komune S, et al:
Long noncoding RNA HOTAIR regulates polycomb-dependent chromatin
modification and is associated with poor prognosis in colorectal
cancers. Cancer Res. 71:6320–6326. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Kim K, Jutooru I, Chadalapaka G, Johnson
G, Frank J, Burghardt R, Kim S and Safe S: HOTAIR is a negative
prognostic factor and exhibits pro-oncogenic activity in pancreatic
cancer. Oncogene. 32:1616–1625. 2013. View Article : Google Scholar
|
|
47
|
Lin R, Maeda S, Liu C, Karin M and
Edgington TS: A large noncoding RNA is a marker for murine
hepatocellular carcinomas and a spectrum of human carcinomas.
Oncogene. 26:851–858. 2007. View Article : Google Scholar
|
|
48
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA, and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Schorderet P and Duboule D: Structural and
functional differences in the long non-coding RNA hotair in mouse
and human. PLoS Genet. 7:e10020712011. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Liu JH, Chen G, Dang YW, Li CJ and Luo DZ:
Expression and prognostic significance of lncRNA MALAT1 in
pancreatic cancer tissues. Asian Pac J Cancer Prev. 15:2971–2977.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wang KC, Yang YW, Liu B, Sanyal A,
Corces-Zimmerman R, Chen Y, Lajoie BR, Protacio A, Flynn RA, Gupta
RA, et al: A long noncoding RNA maintains active chromatin to
coordinate homeotic gene expression. Nature. 472:120–124. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Li Z, Zhao X, Zhou Y, Liu Y, Zhou Q, Ye H,
Wang Y, Zeng J, Song Y, Gao W, et al: The long non-coding RNA
HOTTIP promotes progression and gemcitabine resistance by
regulating HOXA13 in pancreatic cancer. J Transl Med. 13:842015.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ma C, Nong K, Zhu H, Wang W, Huang X, Yuan
Z and Ai K: H19 promotes pancreatic cancer metastasis by
derepressing let-7's suppression on its target HMGA2-mediated EMT.
Tumour Biol. 35:9163–9169. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Peng W, Gao W and Feng J: Long noncoding
RNA HULC is a novel biomarker of poor prognosis in patients with
pancreatic cancer. Med Oncol. 31:3462014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
You L, Chang D, Du HZ and Zhao YP:
Genome-wide screen identifies PVT1 as a regulator of Gemcitabine
sensitivity in human pancreatic cancer cells. Biochem Biophys Res
Commun. 407:1–6. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Tahira AC, Kubrusly MS, Faria MF, Dazzani
B, Fonseca RS, Maracaja-Coutinho V, Verjovski-Almeida S, Machado MC
and Reis EM: Long noncoding intronic RNAs are differentially
expressed in primary and metastatic pancreatic cancer. Mol Cancer.
10:1412011. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Ding YC, Yu W, Ma C, Wang Q, Huang CS and
Huang T: Expression of long non-coding RNA LOC285194 and its
prognostic significance in human pancreatic ductal adenocarcinoma.
Int J Clin Exp Pathol. 7:8065–8070. 2014.
|
|
58
|
Katayama S, Tomaru Y, Kasukawa T, Waki K,
Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J, et
al: FANTOM Consortium: Antisense transcription in the mammalian
transcriptome. Science. 309:1564–1566. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Wang D, Garcia-Bassets I, Benner C, Li W,
Su X, Zhou Y, Qiu J, Liu W, Kaikkonen MU, Ohgi KA, et al:
Reprogramming transcription by distinct classes of enhancers
functionally defined by eRNA. Nature. 474:390–394. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Li W, Notani D, Ma Q, Tanasa B, Nunez E,
Chen AY, Merkurjev D, Zhang J, Ohgi K, Song X, et al: Functional
roles of enhancer RNAs for oestrogen-dependent transcriptional
activation. Nature. 498:516–520. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Hah N, Danko CG, Core L, Waterfall JJ,
Siepel A, Lis JT and Kraus WL: A rapid, extensive, and transient
transcriptional response to estrogen signaling in breast cancer
cells. Cell. 145:622–634. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Melo CA, Drost J, Wijchers PJ, van de
Werken H, de Wit E, Oude Vrielink JA, Elkon R, Melo SA, Léveillé N,
Kalluri R, et al: eRNAs are required for p53-dependent enhancer
activity and gene transcription. Mol Cell. 49:524–535. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Huarte M and Rinn JL: Large non-coding
RNAs: Missing links in cancer? Hum Mol Genet. 19R:R152–R161. 2010.
View Article : Google Scholar
|
|
64
|
Gibb EA, Brown CJ and Lam WL: The
functional role of long non-coding RNA in human carcinomas. Mol
Cancer. 10:382011. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Tsai MC, Spitale RC and Chang HY: Long
intergenic noncoding RNAs: New links in cancer progression. Cancer
Res. 71:3–7. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Maruyama R and Suzuki H: Long noncoding
RNA involvement in cancer. BMB Rep. 45:604–611. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Spizzo R, Almeida MI, Colombatti A and
Calin GA: Long non-coding RNAs and cancer: A new frontier of
translational research? Oncogene. 31:4577–4587. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Reference Genome Group of the Gene
Ontology Consortium. The Gene Ontology's Reference Genome Project:
A unified framework for functional annotation across species. PLoS
Comput Biol. 5:e10004312009. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Mizuma M, Rasheed ZA, Yabuuchi S, Omura N,
Campbell NR, de Wilde RF, De Oliveira E, Zhang Q, Puig O, Matsui W,
et al: The gamma secretase inhibitor MRK-003 attenuates pancreatic
cancer growth in preclinical models. Mol Cancer Ther. 11:1999–2009.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Feng J, Ma T, Ge Z, Lin J, Ding W, Chen H,
Zhu W, Zhou S and Tan Y: PKM2 gene regulates the behavior of
pancreatic cancer cells via mitogen-activated protein kinase
pathways. Mol Med Rep. 11:2111–2117. 2015.
|
|
71
|
Zhang X, Lian Z, Padden C, Gerstein MB,
Rozowsky J, Snyder M, Gingeras TR, Kapranov P, Weissman SM and
Newburger PE: A myelopoiesis-associated regulatory intergenic
noncoding RNA transcript within the human HOXA cluster. Blood.
113:2526–2534. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
McCarthy DJ and Smyth GK: Testing
significance relative to a fold-change threshold is a TREAT.
Bioinformatics. 25:765–771. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Su X, Malouf GG, Chen Y, Zhang J, Yao H,
Valero V, Weinstein JN, Spano JP, Meric-Bernstam F, Khayat D, et
al: Comprehensive analysis of long non-coding RNAs in human breast
cancer clinical subtypes. Oncotarget. 5:9864–9876. 2014. View Article : Google Scholar : PubMed/NCBI
|