Introduction
Gliomas are the most common type of primary brain
tumors. The most malignant form, glioblastoma multiforme (GBM),
accounts for ~15% of all brain tumors, and >50% of all
astrocytomas (1). Current standard
treatment of GBM is surgical resection of the tumor, followed by
adjuvant radio- and chemotherapy. However, median survival time for
GBM patients is still poor, ~12–15 months despite multimodal
therapy (2), and currently there
are no effective long-term treatments for this malignancy. One of
the primary reasons for the poor prognosis is the development of
recurrence composed of highly proliferative and infiltrative tumor
cells which massively invade into the surrounding brain parenchyma
and contribute to the fatal progression (2).
Chemokines, small chemotactic cytokines, are known
to contribute to a broad spectrum of physiological and pathological
processes, including angiogenesis (3), haematopoiesis (4), development (5,6) and
also tumor initiation, survival and progression (7,8).
In particular, the chemokine CXCL12 (also termed
SDF-1, stromal cell-derived factor-1) and its receptors CXCR4 and
CXCR7 seem to play a pivotal role in tumor progression, as
described for different tumor types including GBM (9–11).
In GBM, CXCL12 and CXCR4 are overexpressed in tumor tissues when
compared to normal brain parenchyma and their expression level
correlates with tumor grade and poor prognosis (12). The CXCL12/CXCR4 activation in
glioma cells and specific cells of the surrounding microenvironment
(e.g., microglia, endothelial cells, and mesenchymal cells)
contributes to GBM proliferation, spreading, and chemoresistance as
reviewed for example by Würth et al (13). The long known receptor CXCR4 is
expressed on glioma cells with stem cell properties (14,15).
These cells are able to perform self-renewal, to recapitulate the
whole tumor and to differentiate into specific GBM subpopulations.
Thus, they are likely responsible for the development of
glioblastoma relapses and the poor prognosis of recurrent GBM
(16). Nevertheless, not only
CXCR4, but also CXCR7, which has been described in tumors, is a
regulator of GBM growth (13). For
example, CXCR7 is highly expressed in tumor endothelial, microglial
and GBM cells (15,17), controls tumor diffusion through
CXCL12 gradients and is frequently detected in GBM-associated
vasculature (18). Interestingly,
in contrast to CXCR4, CXCR7 was detected on more differentiated GBM
cells (15). However, a
significant correlation between CXCR4 and CXCR7 in GBM was observed
(19). These results are
fascinating and point to a pivotal role of CXCR4 and CXCR7 in
glioma progression including especially the development of
recurrences.
Materials and methods
Tumor specimens
GBM samples were surgically dissected tissues from
the Department of Neurosurgery (Kiel, Germany) and were obtained in
accordance with the Helsinki Declaration of 1975 with approval of
the ethics committee of the University of Kiel, Germany after
written informed consent of donors (file reference: D 536/15).
Tumors were classified according to the WHO criteria, and the
diagnosis was established by a pathologist. A total of 28 GBM (14
primary and 14 recurrent tumors, paired samples for each single
donor) was included. If enough material was available, matched
probes of individual tumor samples were used for different
investigations.
Reverse transcription and real-time PCR
(qRT-PCR)
RNA was isolated with the TRIzol reagent
(Invitrogen, Carlsbad, CA, USA), digested by DNase, cDNA was
synthesized, and quantitative reverse transcription real-time PCR
(qRT-PCR) was performed as described before (20) using TaqMan primer probes (Applied
Biosystems, Foster City, CA, USA):
glycerinaldehyde-3-phosphate-dehydrogenase (hGAPDH)
(Hs99999905_m1), hCXCR4 (Hs00237052_m1), hCXCR7
(Hs00664172_s1). Fluorescent data were converted into cycle
threshold (CT) measurements, and ΔCT values
of each sample were calculated as CTgene of interest −
CT GAPDH. Relative gene expression was calculated with
2(normalized CT non-stimulated − normalized CT
stimulated) = n-fold of control. A ΔCT value of
3.33 corresponds to one magnitude lower gene expression compared to
GAPDH. To visualize possible similarities in chemokine receptor
expression of individual primary-recurrent GBM pairs, relative gene
expression data were assigned to grey shades in Fig. 1. A relative gene expression value
of 1 (= equal expression in primary and recurrent GBM) was assigned
as 30% grey, lower n-fold expression values (lower expression in
recurrent compared to primary) were displayed with increasing
lighter shading with 0 corresponding to white. Relative expression
values >1 (higher expression in recurrence compared to primary)
were assigned with increasing darker grey shades until 3-fold
induction (or higher) which was assigned as maximum (black).
Afterwards, a ‘heatmap-like’ arrangement of individual
primary-recurrent GBM pairs was performed orientating to up- or
downregulation of CXCR4 and CXCR7 in recurrent samples.
Immunofluorescence
Cryostat sections of different primary and recurrent
GBM tissues were fixed in acetone/methanol, and sequentially
blocked with Sudan black and 0.1% bovine serum albumin as described
before (20). Primary antibodies
were applied overnight at 4°C, secondary antibodies were incubated
at 37°C for 1 h, nuclei were counterstained with
4′,6-diamidino-2-phenylindole (DAPI; Sigma-Aldrich, Hamburg,
Germany), slides were embedded with Immumount (ThermoShandon,
Pittsburgh, PA, USA) and digital photography was performed using a
Zeiss microscope and Zeiss camera (Zeiss, Oberkochen, Germany).
Primary antibodies were anti-OCT-4 (octamer binding transcription
factor 4; 1:150, rabbit; Cell Signaling, Danvers, MA, USA),
anti-SOX-2 (sex determining region Y-box 2; 1:200, rabbit; Santa
Cruz Biotechnology, CA, USA), anti-MUSASHI-1 [Musashi
(Drosophila) homolog 1; 1:100, mouse; R&D Systems,
Wiesbaden, Germany], anti-NANOG (‘Tir nan Og‘; 1:500, rabbit;
Thermo Fisher Scientific, Rockford, IL, USA), anti-KLF-4
(Krüppel-like factor 4; 1:250, mouse; Thermo Fisher Scientific),
anti-CXCR4 (1:200, rabbit; Imgenex IMG 125-2, San Diego, CA, USA)
and anti-CXCR7 (1:100, mouse, MAB42273; R&D Systems). If
primary antibodies were derived from the same species, unspecific
binding was blocked by F(ab) fragments derived from this species
[donkey anti-mouse and anti-rabbit F(ab) fragments, 1:100, from
Dianova, Hamburg, Germany]. Primary antibodies were omitted for
negative controls. As secondary antibodies donkey anti-mouse or
anti-rabbit IgGs labeled with Alexa Fluor 488 or Alexa Fluor 555
(1:1,000; Invitrogen) were used.
Statistical analysis
For statistical analyses a two-tailed Student's
t-test with matched samples was used. Significance level was
p<0.05 (indicated by an asterisk (*) in the figures).
Results
Expression of CXCR4 and CXCR7 in human
primary and recurrent GBM pairs
To evaluate mRNA expression levels of CXCR4 and
CXCR7, qRT-PCR analysis was performed using matched probes of solid
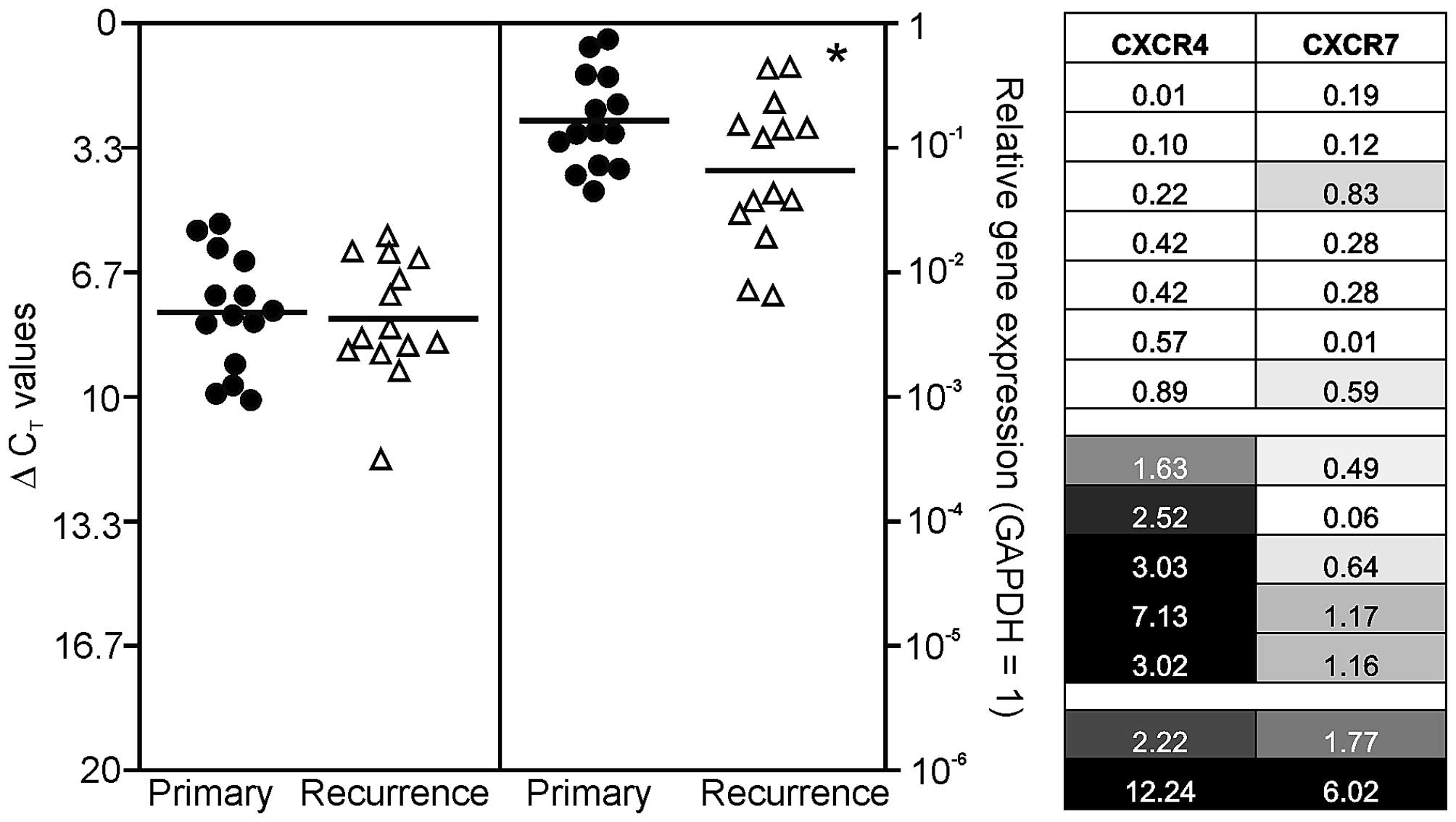
human primary and recurrent GBM. Results are shown in Fig. 1, where single ΔCT values
for primary GBM samples are demonstrated in black circles and
ΔCT values for recurrent ones in white triangles. It
should be kept in mind that a ΔCT value of 3.33
corresponds to one magnitude lower gene expression.
Irrespective of the tumor identity (primary versus
recurrent), both CXCR4 and CXCR7 were detected at considerable
levels in solid GBM tissues with CXCR7 clearly to higher extent.
Additionally, when comparing primary and recurrent GBM samples, the
chemokine receptor CXCR7 was expressed in lower amounts in
recurrences (mean reduction to ~40%; p<0.05), while this was not
observed for CXCR4. In detail, the normalized averaged
ΔCT values for investigated primary/recurrent GBM
samples were: 7.74/7.91 (CXCR4), and 2.61/3.95 (CXCR7),
respectively (Fig. 1, left).
To compare chemokine receptor expression differences
between individual primary and recurrent GBM pairs more in detail,
we arranged the n-fold expression changes of all individual
primary-recurrent GBM pairs as a heatmap: equal n-fold expression
in primary and recurrent GBM was assigned as 30% grey, lower n-fold
expression values were displayed with increasing lighter shading
with 0 corresponding to white, and relative n-fold expression
values >1 were assigned with increasing darker grey shades until
3-fold induction (or higher) which was assigned as maximum (black).
By this, three different GBM groups in our collective could be
identified (Fig. 1, right): the
first group, containing seven primary-recurrent GBM pairs, was
characterized by a general lower mRNA expression of CXCR4 and CXCR7
in recurrent samples. The second group, containing five
primary-recurrent GBM pairs, was characterized by higher expression
of CXCR4 in combination with lower expression of CXCR7. The third
group included two GBM pairs and was characterized by higher
expression of both chemokine receptors in recurrences.
Interestingly, a combination of increased CXCR7 and decreased CXCR4
expression was not observed in our cohort. Summarized, various
combinations of loss or gain of CXCR4 and CXCR7 mRNA expression are
apparently detectable in our collective of paired primary and
recurrent GBM samples, and it became clear that a more precise
evaluation of chemokine receptor expression during the progression
from primary to recurrent GBM is possible if each individual pair
is analyzed in detail.
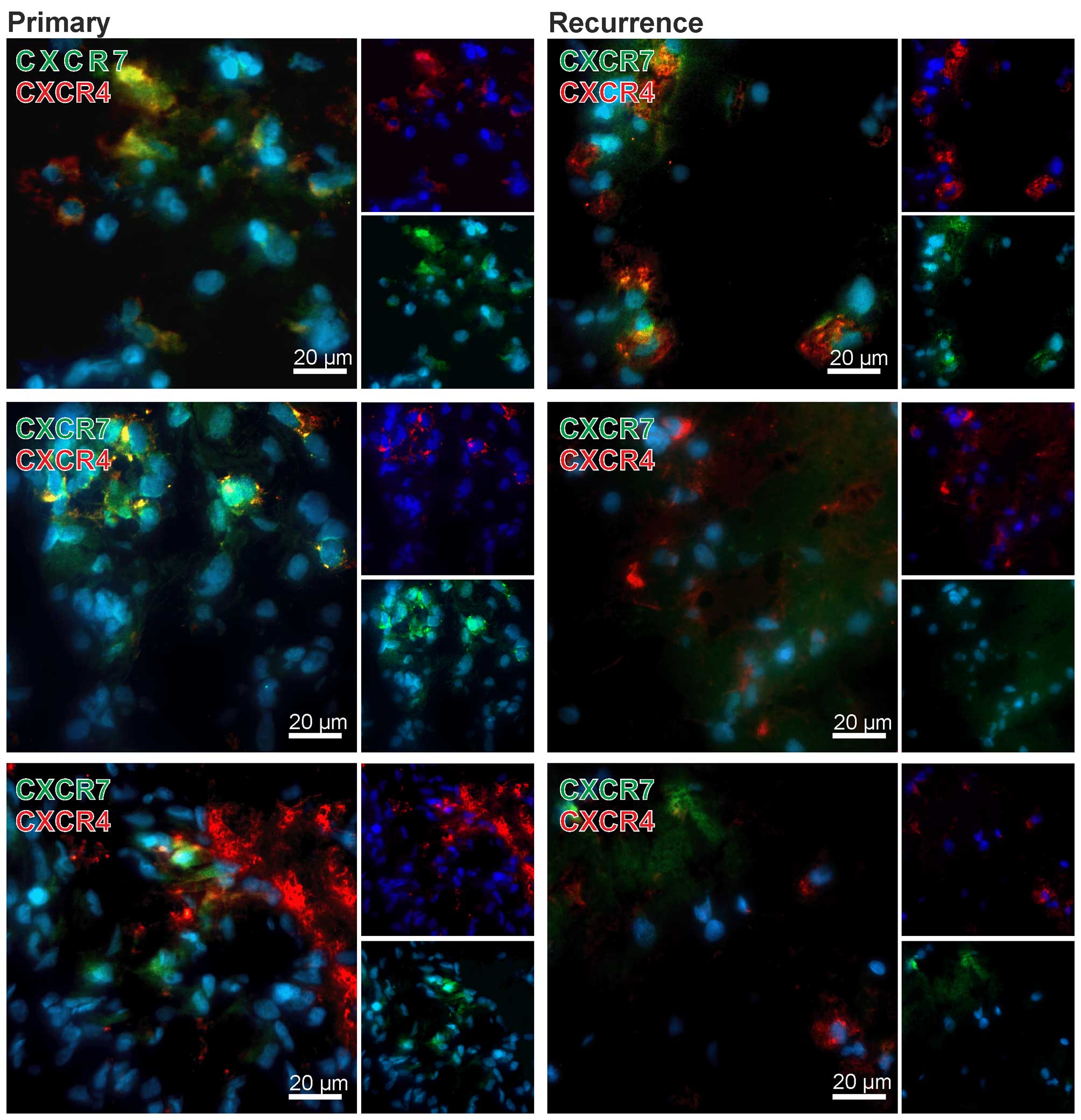
To confirm qRT-PCR results and to evaluate CXCR4 and
CXCR7 expression also on the protein level in primary-recurrent GBM
pairs, we performed double-immunofluorencence staining of both
chemokine receptors. Nuclei were stained with DAPI. Exemplary
results of stained matched primary and recurrent samples are shown
in Fig. 2. Although no clear
quantitative data could be obtained using fluorescence
immunostaining of cryo-sections the primary tumors showed high
amounts of CXCR4 or CXCR7 positively stained cells, whereas in
matched recurrent samples only few CXCR7-positive cells, but
considerable amounts of CXCR4-positive cells were visible. In
addition, the majority of tumor cells were solely positive for
CXCR4 or CXCR7, respectively (exemplified in Fig. 2), only single cells expressed CXCR4
and CXCR7 in the same cell regions [examples for merged regions
(yellow) are shown in Fig. 2].
Summarized, while both CXCR4 and CXCR7 are expressed
at considerable levels in primary and recurrent GBM samples, CXCR7
expression is apparently downregulated in relapsed cases. Regarding
CXCR4/CXCR7 expression differences between primary and recurrent
samples, our cohort included different GBM groups of combined
induction or reduction upon relapse. Expression of CXCR4 and CXCR7
was confirmed on protein level, and co-stainings revealed single as
well as double-positive cells.
Cellular allocation of CXCR4 and CXCR7 in
human primary and recurrent GBM pairs
We and others have shown that CXCR4 is predominately
expressed in (tumor) stem-like cells (14,15),
and GBM stem-like cells are supposed to give rise to the
recurrences. Therefore, we analyzed in a next step in more detail
which cells might account for expression of CXCR4 and CXCR7 in
primary and recurrent GBM. Thus, co-stainings with neural
stem/progenitor (RNA-binding protein MUSASHI-1) and different
embryonic stem cell markers [KLF-4, OCT-4, SOX-2 as embryonic stem
cell transcription factors, and NANOG which initially was used as
readout for efficient reprogramming of iPSCs (21)] were performed and analyzed by
fluorescence microscopy.
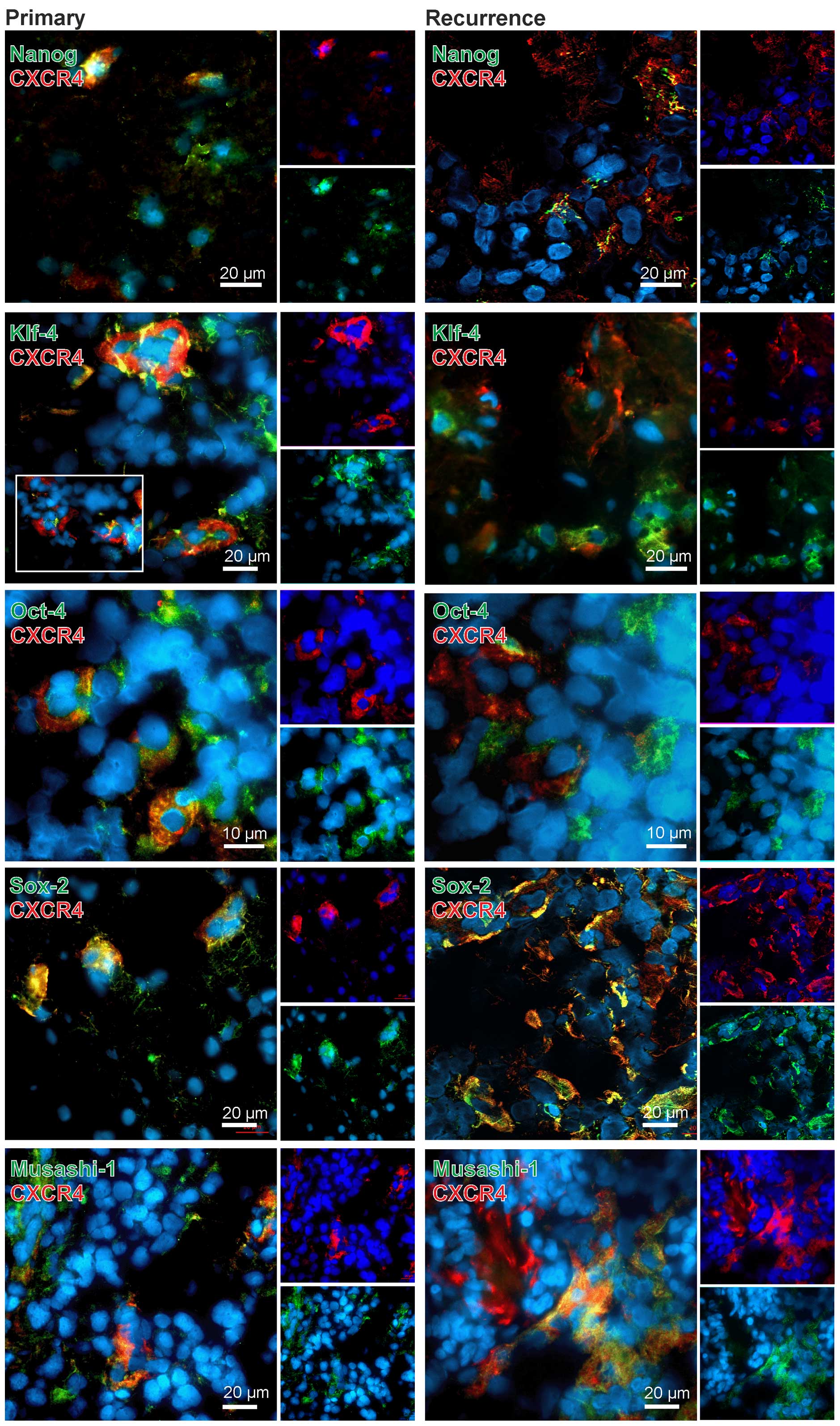
In general, CXCR4 yielded clear co-stainings with
different stem cells markers in both primary and recurrent samples
(exemplified in Fig. 3). However,
this method does not allow for a valid quantification of both
staining intensities and amounts of positively stained cells.
Additionally, different antigens are not localized in the same
cellular structures, thus signals do not merge in all cases, but
can be found in the same regions. Nevertheless, especially in the
co-staining CXCR4-SOX2 nearly all CXCR4-positive cells were also
SOX-2-positive. For KLF-4, OCT-4 and MUSASHI-1 we also observed
clear co-stainings with CXCR4 in both primary and recurrent GBM,
with OCT-4-CXCR4 double-positive cells being remarkably localized
in cluster-like structures. However, also CXCR4, KLF-4, OCT-4 and
MUSASHI-1 single-positive cells existed within the sections. For
NANOG, a co-staining with CXCR4 was also observed but not as
prominent as detected for the other stem cell markers (exemplified
in Fig. 3).
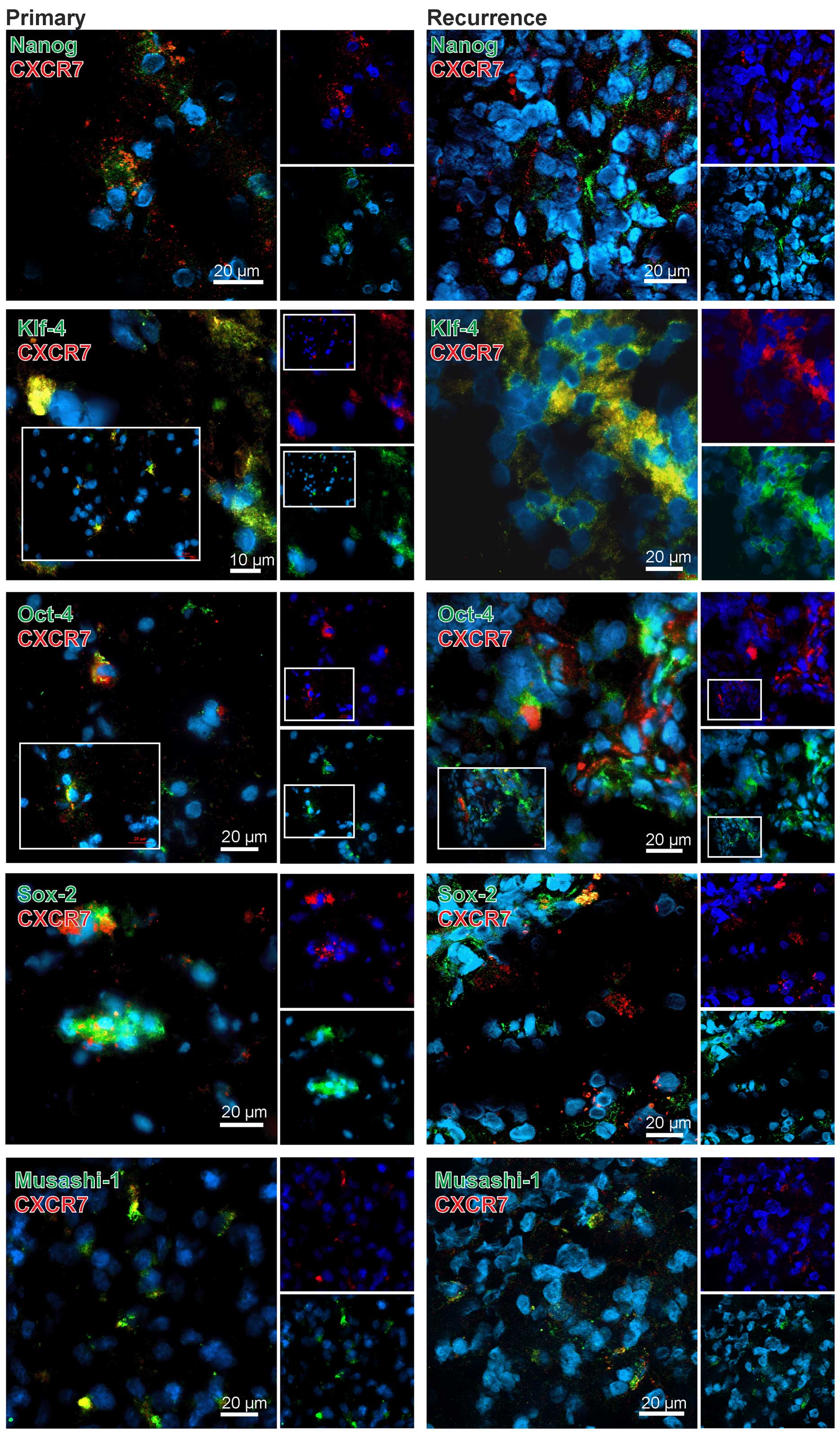
In comparison to CXCR4 the results for CXCR7 were
clearly different. With exception of the combination CXCR7-KLF-4
only very few CXCR7-stem cell marker double-positive cells were
found in the GBM specimens (exemplified in Fig. 4). In detail, when looking at OCT-4
and SOX-2 in CXCR7-positive regions, some OCT-4 or SOX-2-positive
cells (and here more obvious in recurrences) were found,
respectively, but it seems that in the majority of the cases CXCR7
was expressed in different, OCT-4 or SOX-2 negative cells. In line
with these results, a clear co-staining of CXCR7 with NANOG or
MUSASHI-1 was not detectable although positive cells were found in
CXCR7-positive cell regions. In contrast, KLF-4 and CXCR7 were
found co-expressed in the same cells and, although this method does
not allow a valid quantification, this was more prominent in
recurrent samples (exemplified in Fig.
4). Nevertheless, also KLF-4 and CXCR7 single-positive cells
were found in the tumor sections, respectively.
Summarized, CXCR4 was clearly expressed in stem cell
marker positive cells in both primary and recurrent GBM samples,
whereas CXCR7, with the exception of a clear co-staining with
KLF-4-positive cells especially in recurrences, was predominantly
found in stem cell marker negative cells in the investigated
samples.
Discussion
The chemokine CXCL12/SDF-1 is involved in glioma
progression (10,13,15,17).
In GBM, CXCL12 and its receptor, CXCR4, are localized in regions of
necrosis and angiogenesis (22),
and are supposed to mediate proliferation of GBM progenitor cells
(14). In contrast, the CXCL12
receptor CXCR7, which was initially regarded as a decoy receptor
scavenging CXCL12 to prevent CXCR4 signaling and effects (23), mediates apoptosis resistance in
human and rat glioma cells (15,17).
Previous results focusing on the expression of both receptors in
human GBM indeed yielded an expression of CXCR4 by a subpopulation
of GBM cells with stem cell properties (14), whereas the bulk of more
differentiated GBM cells express CXCR7 (15,24).
Conversely, other studies reported the existence of CXCR4-CXCR7
double-positive cells and showed that this subpopulation might
regulate the stem cell phenotype (25), and was able to initiate
intracranial tumors in vivo (26). Nevertheless, also CXCR4 and CXCR7
single populations exist and a high level of heterogeneity in both
the surface expression and functions of CXCR4 and CXCR7 in primary
human GBM cells of the proliferative subclass was determined
(26). In addition, GBM primary
cell cultures show a heterogeneous cell surface expression of CXCR4
and CXCR7 despite similar levels of corresponding mRNAs (25), and the expression of CXCR4 and
CXCR7 is significantly correlated (19).
Since comparative investigations regarding the
expression of CXCR4 and CXCR7 in matched samples of primary and
recurrent GBM are still lacking, we analysed this point in 14
different human primary-recurrent GBM pairs. We were able to show
that irrespective of used materials (primary/recurrent GBM) both
CXCR4 and CXCR7 were expressed at considerable amounts on the mRNA
level with CXCR7 clearly to higher extent. Whereas CXCR4 expression
was nearly equal in primary and recurrent samples, CXCR7 was found
in lower amounts in recurrent GBM samples (~40%) in comparison to
primary ones. Nevertheless when analyzing each individual pair in
detail, two primary-recurrent pairs were observed in our GBM
collective with higher CXCR7 mRNA level in the recurrent samples.
Although immunofluorescence staining of cryo-sections cannot
clearly deliver quantitative data, the CXCR7 expression appeared
lower in recurrent samples on the protein level, which is in
accordance with the results obtained by qRT-PCR.
Interestingly, Razmkhah et al (27) reported that CXCR7 was undetectable
in secondary brain tumors (sarcoma and breast cancer), whereas
CXCR4 was expressed. Conversely to our observations, in that glioma
cohort, CXCR4 expression was ~110-fold higher than its counterpart
CXCR7. However, only 7 samples of glioma (4 high-grade gliomas and
3 low-grade gliomas without any histopathological specification)
were included.
Further, when investigating CXCR4 and CXCR7
expression in primary and recurrent GBM on the protein level by
double-immunofluorescence technique, we found that the chemokine
receptors were mainly detectable on different cell types, only very
few CXCR4-CXCR7 double-positive cells existed in the samples. In
accordance with this, heterogeneity of CXCR4 and CXCR7 expression
was reported before for primary GBM cells pointing to the existence
of different subpopulations measurable by variable percentages of
CXCR4+CXCR7−,
CXCR4−CXCR7+ and
CXCR4+CXCR7+ cells in glioma samples
(26). In addition, since for
GBM-positive correlations not only for the expression of CXCR7 and
CXCR4 but also for the expression of CXCR7 and HIF1α and CXCR7 and
IDH1 were reported, these results also indicate the existence of
different chemokine receptor expressing subclones (19). By co-staining of CXCR4 and CXCR7
with stem cell markers we could show in our primary-recurrent GBM
cohort that CXCR4 was mostly found on stem-cell marker positive GBM
cells, whereas CXCR7 was usually expressed on stem-cell marker
negative cells, with exception of the combination CXCR7-KLF-4. An
embryonic stem cell gene signature is well known to correlate with
a more undifferentiated phenotype in various cancers (28), and a large scale tissue microarray
analysis including 80 low-grade and 98 high-grade gliomas showed an
upregulated protein level of NANOG, KLF-4, OCT-4 and SOX-2 in
high-grade gliomas (29). Several
other studies also point to the relevance of neural and embryonic
stem cell markers in GBM progression (30–36).
In the present study we showed that both neural and embryonic stem
cells markers are preferentially co-expressed with CXCR4 in matched
samples of primary and recurrent GBM pairs, whereas CXCR7 was
mostly found on stem cell marker negative cells. In line with these
results, CXCR4 expression in neuroblastoma is associated with
highly aggressive undifferentiated tumors, while CXCR7 expression
was detected in more differentiated and mature neuroblastic tumors
(37). In addition, CXCR4 is
essential for the self-renewal of GBM stem-like cells since
disruption of the CXCL12/CXCR4 pathway resulted in reduced
expression of stem-like markers like Nestin and MUSASHI-1 or genes
which are involved in regulating stem cell properties such as OCT-4
and NANOG (38). The miR-137
inhibited glioma stem-like cell self-renewal and promoted their
differentiation by targeting RTVP-1, which downregulated CXCR4
(39). The inhibition of CXCR4 in
glioma-initiating cells disrupted the SHH-GLI-NANOG network, which
is involved in self-renewal and expression of the embryonic stem
cell-like signature (40).
Interestingly, Chen et al (41) reported that CXCR7 can mediate
neural progenitor cell migration to CXCL12 independently of CXCR4.
In line with this, our primary-recurrent GBM cohort showed a
distinct subpopulation of CXCR7-KLF-4 double-positive cells, which
was more prominent in the recurrences. Since it has been shown that
the miR-152 targets KLF-4 in GBM cells and thereby influences cell
proliferation, invasion, apoptosis and cell migration processes
(42), the CXCR7-KLF-4
double-positive GBM cells in our cohort could be a subpopulation of
highly migratory tumor cells. However, the paracrine PGI signaling
initiated by mesenchymal glioma cells also induces self-renewal and
tumorigenic potential of glioma stem cells through induction of
KLF-4 (43). The functional role
of CXCR7 in KLF-4-positive glioma-stem cells remains still highly
speculative.
Summarized, we were able to show that in a cohort of
matched primary-recurrent GBM samples the CXCL12 receptors CXCR4
and CXCR7 are expressed both on the mRNA and protein levels in
large amounts, with CXCR7 mRNA being statistically significantly
downregulated in recurrent samples. A co-expression of both
receptors was rare. In accordance with this, CXCR4 was co-expressed
with all investigated neural and embryonic stem cell markers in
both primary and recurrent tissues, whereas CXCR7 was mostly found
on stem cell marker-negative cells but was co-expressed with KLF-4
on a distinct GBM cell subpopulation. These results point to an
individual role of CXCR4 and CXCR7 in the contribution of stem cell
marker-positive GBM cells in glioma progression and underline the
potential of chemokine receptors as targets for new therapeutic
approaches in GBM intervention.
Acknowledgements
We thank Brigitte Rehmke, Fereshteh Ebrahim and Jörg
Krause for expert technical assistance. This study was supported by
the University of Kiel and by the popgen 2.0 network [(P2N;
supported by a grant from the German Ministry for Education and
Research (01EY1103)].
Abbreviations:
|
CT
|
cycle of threshold
|
|
DAPI
|
4′,6-diamidino-2-phenylindole
|
|
DMEM
|
Dulbecco's modified Eagle's medium
|
|
FCS
|
fetal calf serum
|
|
GAPDH
|
glycerinaldehyde-3-phosphate-dehydrogenase
|
|
GBM
|
glioblastoma multiforme
|
|
KLF-4
|
Krüppel-like factor 4
|
|
MUSASHI-1
|
Musashi (Drosophila) homolog
1
|
|
NANOG
|
‘Tir nan Og’
|
|
OCT-4
|
octamer binding transcription
factor
|
|
qRT-PCR
|
quantitative reverse transcription
polymerase chain reaction
|
|
SOX-2
|
sex determining region Y-box 2
|
|
WHO
|
World Health Organization
|
References
|
1
|
Ohgaki H and Kleihues P: Epidemiology and
etiology of gliomas. Acta Neuropathol. 109:93–108. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Stupp R, Mason WP, van den Bent MJ, Weller
M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn
U, et al; European Organisation for Research and Treatment of
Cancer Brain Tumor and Radiotherapy Groups; National Cancer
Institute of Canada Clinical Trials Group. Radiotherapy plus
concomitant and adjuvant temozolomide for glioblastoma. N Engl J
Med. 352:987–996. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Strieter RM, Polverini PJ, Kunkel SL,
Arenberg DA, Burdick MD, Kasper J, Dzuiba J, Van Damme J, Walz A,
Marriott D, et al: The functional role of the ELR motif in CXC
chemokine-mediated angiogenesis. J Biol Chem. 270:27348–27357.
1995. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Broxmeyer HE and Kim CH: Regulation of
hematopoiesis in a sea of chemokine family members with a plethora
of redundant activities. Exp Hematol. 27:1113–1123. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Dambly-Chaudière C, Cubedo N and Ghysen A:
Control of cell migration in the development of the posterior
lateral line: Antagonistic interactions between the chemokine
receptors CXCR4 and CXCR7/RDC1. BMC Dev Biol. 7:232007. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hattermann K, Ludwig A, Gieselmann V,
Held-Feindt J and Mentlein R: The chemokine CXCL16 induces
migration and invasion of glial precursor cells via its receptor
CXCR6. Mol Cell Neurosci. 39:133–141. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Vandercappellen J, Van Damme J and Struyf
S: The role of CXC chemokines and their receptors in cancer. Cancer
Lett. 267:226–244. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
O'Hayre M, Salanga CL, Handel TM and Allen
SJ: Chemokines and cancer: Migration, intracellular signalling and
intercellular communication in the microenvironment. Biochem J.
409:635–649. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Müller A, Homey B, Soto H, Ge N, Catron D,
Buchanan ME, McClanahan T, Murphy E, Yuan W, Wagner SN, et al:
Involvement of chemokine receptors in breast cancer metastasis.
Nature. 410:50–56. 2001. View
Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hattermann K and Mentlein R: An infernal
trio: The chemokine CXCL12 and its receptors CXCR4 and CXCR7 in
tumor biology. Ann Anat. 195:103–110. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zlotnik A, Burkhardt AM and Homey B:
Homeostatic chemokine receptors and organ-specific metastasis. Nat
Rev Immunol. 11:597–606. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bian XW, Yang SX, Chen JH, Ping YF, Zhou
XD, Wang QL, Jiang XF, Gong W, Xiao HL, Du LL, et al: Preferential
expression of chemokine receptor CXCR4 by highly malignant human
gliomas and its association with poor patient survival.
Neurosurgery. 61:570–579. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Würth R, Bajetto A, Harrison JK, Barbieri
F and Florio T: CXCL12 modulation of CXCR4 and CXCR7 activity in
human glioblastoma stem-like cells and regulation of the tumor
microenvironment. Front Cell Neurosci. 8:144eCollection.
2014.PubMed/NCBI
|
|
14
|
Ehtesham M, Mapara KY, Stevenson CB and
Thompson RC: CXCR4 mediates the proliferation of glioblastoma
progenitor cells. Cancer Lett. 274:305–312. 2009. View Article : Google Scholar :
|
|
15
|
Hattermann K, Held-Feindt J, Lucius R,
Müerköster SS, Penfold ME, Schall TJ and Mentlein R: The chemokine
receptor CXCR7 is highly expressed in human glioma cells and
mediates antiapoptotic effects. Cancer Res. 70:3299–3308. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Auffinger B, Spencer D, Pytel P, Ahmed AU
and Lesniak MS: The role of glioma stem cells in chemotherapy
resistance and glioblastoma multiforme recurrence. Expert Rev
Neurother. 15:741–752. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hattermann K, Mentlein R and Held-Feindt
J: CXCL12 mediates apoptosis resistance in rat C6 glioma cells.
Oncol Rep. 27:1348–1352. 2012.PubMed/NCBI
|
|
18
|
Liu Y, Carson-Walter EB, Cooper A, Winans
BN, Johnson MD and Walter KA: Vascular gene expression patterns are
conserved in primary and metastatic brain tumors. J Neurooncol.
99:13–24. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bianco AM, Uno M, Oba-Shinjo SM, Clara CA,
de Almeida Galatro TF, Rosemberg S, Teixeira MJ and Nagahashi Marie
SK: CXCR7 and CXCR4 expression in infiltrative astrocytomas and
their interactions with HIF1α expression and IDH1 mutation. Pathol
Oncol Res. 21:229–240. 2015. View Article : Google Scholar
|
|
20
|
Kubelt C, Hattermann K, Sebens S, Mehdorn
HM and Held-Feindt J: Epithelial-to-mesenchymal transition in
paired human primary and recurrent glioblastomas. Int J Oncol.
46:2515–2525. 2015.PubMed/NCBI
|
|
21
|
Takahashi K and Yamanaka S: Induction of
pluripotent stem cells from mouse embryonic and adult fibroblast
cultures by defined factors. Cell. 126:663–676. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Rempel SA, Dudas S, Ge S and Gutiérrez JA:
Identification and localization of the cytokine SDF1 and its
receptor, CXC chemokine receptor 4, to regions of necrosis and
angiogenesis in human glioblastoma. Clin Cancer Res. 6:102–111.
2000.PubMed/NCBI
|
|
23
|
Naumann U, Cameroni E, Pruenster M,
Mahabaleshwar H, Raz E, Zerwes HG, Rot A and Thelen M:
CXCR/functions as a scavenger for CXCR12 and CXCL11. Plos One.
5:e91752010. View Article : Google Scholar
|
|
24
|
Gatti M, Pattarozzi A, Bajetto A, Würth R,
Daga A, Fiaschi P, Zona G, Florio T and Barbieri F: Inhibition of
CXCL12/CXCR4 autocrine/paracrine loop reduces viability of human
glioblastoma stem-like cells affecting self-renewal activity.
Toxicology. 314:209–220. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Walters MJ, Ebsworth K, Berahovich RD,
Penfold ME, Liu SC, Al Omran R, Kioi M, Chernikova SB, Tseng D,
Mulkearns-Hubert EE, et al: Inhibition of CXCR7 extends survival
following irradiation of brain tumours in mice and rats. Br J
Cancer. 110:1179–1188. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu C, Pham K, Luo D, Reynolds BA, Hothi
P, Foltz G and Harrison JK: Expression and functional heterogeneity
of chemokine receptors CXCR4 and CXCR7 in primary patient-derived
glioblastoma cells. PLoS One. 8:e597502013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Razmkhah M, Arabpour F, Taghipour M,
Mehrafshan A, Chenari N and Ghaderi A: Expression of chemokines and
chemokine receptors in brain tumor tissue derived cells. Asian Pac
J Cancer Prev. 15:7201–7205. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ben-Porath I, Thomson MW, Carey VJ, Ge R,
Bell GW, Regev A and Weinberg RA: An embryonic stem cell-like gene
expression signature in poorly differentiated aggressive human
tumors. Nat Genet. 40:499–507. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
29
|
Elsir T, Edqvist P-H, Carlson J, Ribom D,
Bergqvist M, Ekman S, Popova SN, Alafuzoff I, Ponten F, Nistér M,
et al: A study of embryonic stem cell-related proteins in human
astrocytomas: Identification of Nanog as a predictor of survival.
Int J Cancer. 134:1123–1131. 2014. View Article : Google Scholar
|
|
30
|
Berezovsky AD, Poisson LM, Cherba D, Webb
CP, Transou AD, Lemke NW, Hong X, Hasselbach LA, Irtenkauf SM,
Mikkelsen T, et al: Sox2 promotes malignancy in glioblastoma by
regulating plasticity and astrocytic differentiation. Neoplasia.
16:193–206. 206.e19–206.e25. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Du Z, Jia D, Liu S, Wang F, Li G, Zhang Y,
Cao X, Ling EA and Hao A: Oct4 is expressed in human gliomas and
promotes colony formation in glioma cells. Glia. 57:724–733. 2009.
View Article : Google Scholar
|
|
32
|
Galatro TF, Uno M, Oba-Shinjo SM, Almeida
AN, Teixeira MJ, Rosemberg S and Marie SK: Differential expression
of ID4 and its association with TP53 mutation, SOX2, SOX4 and OCT-4
expression levels. PLoS One. 8:e616052013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Guo Y, Liu S, Wang P, Zhao S, Wang F, Bing
L, Zhang Y, Ling EA, Gao J and Hao A: Expression profile of
embryonic stem cell-associated genes Oct4, Sox2 and Nanog in human
gliomas. Histopathology. 59:763–775. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Holmberg J, He X, Peredo I, Orrego A,
Hesselager G, Ericsson C, Hovatta O, Oba-Shinjo SM, Marie SK,
Nistér M, et al: Activation of neural and pluripotent stem cell
signatures correlates with increased malignancy in human glioma.
PLoS One. 6:e184542011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ikushima H, Todo T, Ino Y, Takahashi M,
Saito N, Miyazawa K and Miyazono K: Glioma-initiating cells retain
their tumorigenicity through integration of the Sox axis and Oct4
protein. J Biol Chem. 286:41434–41441. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Muto J, Imai T, Ogawa D, Nishimoto Y,
Okada Y, Mabuchi Y, Kawase T, Iwanami A, Mischel PS, Saya H, et al:
RNA-binding protein Musashi1 modulates glioma cell growth through
the post-transcriptional regulation of Notch and PI3 kinase/Akt
signaling pathways. PLoS One. 7:e334312012. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Mühlethaler-Mottet A, Liberman J, Ascenção
K, Flahaut M, Balmas Bourloud K, Yan P, Jauquier N, Gross N and
Joseph JM: The CXCR4/CXCR7/CXCL12 axis is involved in a secondary
but complex control of neuroblastoma metastatic cell homing. PLoS
One. 10:e01256162015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lee CC, Lai JH, Hueng DY, Ma HI, Chung Y,
Sun YY, Tsai YJ, Wu WB and Chen CL: Disrupting the CXCL12/CXCR4
axis disturbs the characteristics of glioblastoma stem-like cells
of rat RG2 glioblastoma. Cancer Cell Int. 13:852013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Bier A, Giladi N, Kronfeld N, Lee HK,
Cazacu S, Finniss S, Xiang C, Poisson L, deCarvalho AC, Slavin S,
et al: MicroRNA-137 is downregulated in glioblastoma and inhibits
the stemness of glioma stem cells by targeting RTVP-1. Oncotarget.
4:665–676. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Fareh M, Turchi L, Virolle V, Debruyne D,
Almairac F, de-la- Forest Divonne S, Paquis P, Preynat-Seauve O,
Krause K-H, Chneiweiss H, et al: The miR 302–367 cluster
drastically affects self-renewal and infiltration properties of
glioma-initiating cells through CXCR4 repression and consequent
disruption of the SHH-GLI-NANOG network. Cell Death Differ.
19:232–244. 2012. View Article : Google Scholar :
|
|
41
|
Chen Q, Zhang M, Li Y, Xu D, Wang Y, Song
A, Zhu B, Huang Y and Zheng JC: CXCR7 mediates neural progenitor
cells migration to CXCL12 independent of CXCR4. Stem Cells.
33:2574–2585. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Ma J, Yao Y, Wang P, Liu Y, Zhao L, Li Z,
Li Z and Xue Y: MiR-152 functions as a tumor suppressor in
glioblastoma stem cells by targeting Krüppel-like factor 4. Cancer
Lett. 355:85–95. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhu XY, Wang L, Luan SH, Zhang HS, Huang
WT and Wang NH: The PGI-KLF4 pathway regulates self-renewal of
glioma stem cells residing in the mesenchymal niches in human
gliomas. Neoplasma. 61:401–410. 2014. View Article : Google Scholar : PubMed/NCBI
|