Introduction
Loss of chromosome 3p in many tumors is well
established and it has been supposed for a long time that this
chromosomal region might carry several tumor suppressor genes
(1,2). In particular, sporadic clear-cell
renal cell carcinoma (ccRCC) is characterized by a high frequency
of allelic deletion or loss of heterozygosity (LOH) on chromosome
3p (>90%), causing biallelic inactivation of von Hippel-Lindau
tumor suppressor (VHL) gene (3–5). It
was reported that deletion of fragile histidine triad (FHIT)
gene and flanking loci might occur as an initiating event followed
by deletions at 3p12.2, 3p21.31-3p21.32, and 3p24.2-3p26.1 in the
early stages, then continuous large deletions of 3p21.3-3p26.1 and
3p14.1-3p26.1 (6). Next-generation
sequencing studies have identified frequent mutations in genes
involved in chromatin modification such as polybromo 1
(PBRM1), SET domain containing 2 (SETD2), and BRCA1
associated protein-1 (BAP1) (7–10),
all of which are located on 3p21. Moreover, BAP1 mutations
were mutually exclusive with PBRM1 mutations (9,11).
Germline BAP1 mutations are associated with
an increased risk of malignant mesothelioma (MM), atypical
melanocytic tumors, and other neoplasms (12–15).
They also predispose to RCC (16,17).
Moreover, somatic BAP1 mutation was reported in ~10% of
ccRCC patients of any ethnic background (9–11,18).
Recently, Wang et al reported that mice deficient for Vhl
together with one allele of Bap1 in nephron progenitor cells
developed ccRCC, but mice deficient in Vhl did not (19). Wang et al suggested that
BAP1 is a more potent tumor suppressor gene than VHL, which
they assessed as a weak tumor suppressor gene in the kidney. The
question remains as to why VHL mutations occur at a much higher
frequency than BAP1 mutations in both familial and sporadic
ccRCC (20).
The BAP1 protein is a nuclear protein that has been
considered to function as a deubiquitinase of histone H2A (21). Two types of BAP1 inactivation have
been reported: sequence-level mutation of BAP1 combined with
monoallelic loss of 3p, and biallelic deletion comprising broad
deletions of 3p21 and narrow deletions of several exons or the
entire BAP1 gene. In metastasized uveal melanoma, the former
mutation type occurs frequently (22,23).
In MM, we reported a high frequency of biallelic loss of
BAP1 (24,25), in addition to sequence-level
mutations in the BAP1 gene (26). However, the observed frequency of
BAP1 inactivation in MM has been variable among studies: low
frequencies of mutations found by sequencing alone (12,26,27),
and high frequencies of loss of nuclear staining, or genomic
mutations identified by sequencing and copy number analysis
(24,28,29).
Nasu et al suggested immunostaining as the most accessible
and reliable technique to detect BAP1 inactivation in MM biopsies
(29). It was also reported that
BAP1 expression in the nucleus was reduced in 44% of esophageal
squamous carcinoma, although the mutation rate of the gene was ~2%
(30). It is increasingly clear
that BAP1 protein nuclear translocation may play a key role in
tumorigenesis (31,32).
To clarify the status of inactivation of the
BAP1 gene in ccRCC development, we performed genomic
analysis of the BAP1 gene, assessed for 3p rearrangement,
and conducted immunostaining for the BAP1 protein in RCC
patients.
Materials and methods
Tumor specimens
We obtained 45 RCC samples from 45 patients who had
radical nephrectomy at the Hospital of Hyogo College of Medicine.
The patient summary is presented in Table I. Fresh frozen tumor tissues
dissected from the surgical specimen and unaffected kidney tissue
adjacent to the tumor regions for most patients (ID: RCC01-56), or
peripheral blood for 13 patients (ID: RCC57-69), as a matched
control were analyzed. Pathological examination showed that 42
tumors were diagnosed as ccRCC and three as chromophobe RCC
(chRCC). Our study was approved by the Ethics Committee of Hyogo
College of Medicine (permission no.: RINHI277), and performed in
accordance with the Declaration of Helsinki (1995) of the World
Medical Association (as revised in Fortalez, 2013). All patients
provided written informed consent.
 | Table ISummary of genomic alterations
detected in RCC tumors. |
Table I
Summary of genomic alterations
detected in RCC tumors.
| Patient
information | 3p | VHL | BAP1 | PBRM1 | SETD2 |
|---|
|
|
|
|
|
|
|
|---|
| RCC_ID | Sex | Age | Histological
type | TMN | Recurrencea | Survivalb | LOHc | Monoallelic
lossd | Somatic
mutation | Promoter
methylatione | Monoallelic
lossf | Somatic
mutation | Nuclear
staining | Somatic
mutation | Somatic
mutation |
|---|
| 2 | M | 56 | ccRCC | 3a | − | + | + | + | p.E70X | ND | + | Wild | (−) | p.I571V | Wild |
| 4 | M | 45 | ccRCC | 2a | − | + | + | + | p.N78S | ND | + | Wild | (−) | Wild | Wild |
| 5 | M | 52 | ccRCC | 2a | − | + | + | + | Wild | + | + | Wild | (−) | Wild | Wild |
| 6 | M | 77 | ccRCC | 1b | − | + | − | − | Wild | + | − | Wild | (+) | Wild | Wild |
| 7 | M | 45 | ccRCC | 1a | − | + | − | − | Wild | − | − | Wild | (+) | Wild | Wild |
| 8 | M | 52 | ccRCC | 1a | − | + | + | + | p.D126fs | ND | + | Wild | (+) | p.H24fs | Wild |
| 9 | M | 73 | ccRCC | 3a | − | −g | + | + | p.F136fs | ND | + | Wild | (+) | p.S987X | p.T220fs |
| 10 | M | 55 | ccRCC | 1b | − | + | + | + | p.L188P | ND | + | Wild | (−) | Wild | Wild |
| 11 | M | 54 | ccRCCh | 3a | + | − | + | + | p.L158fs | ND | + | Wild | (−) | p.I1345fs | Wild |
| 13 | M | 58 | ccRCC | 3a | − | + | + | + | p.F119L | ND | + | p.K286fs | (−) | Wild | Wild |
| 14 | F | 59 | ccRCC | 1a | − | + | + | + | Splicing
acceptor | ND | + | Wild | (+) | Wild | Wild |
| 15 | M | 73 | ccRCC | 1a | − | + | + | + | p.Y185X | ND | + | Wild | (+) | p.S1253fs | Wild |
| 17 | F | 66 | ccRCC | 3a | − | + | + | + | p.E70X | ND | + | Wild | (−) | p.P703L | Wild |
| 18 | F | 58 | ccRCC | 1a | − | + | + | + | p.S65X | ND | + | Wild | (−) | p.F684Y | Wild |
| 20 | M | 56 | ccRCC | 1a | + | + | + | + | p.T133fs | ND | + | p.W202X | (−) | Wild | Wild |
| 21 | M | 81 | ccRCC | 1a | − | + | + | + | Wild | + | + | Wild | (+) | Wild | Wild |
| 24 | M | 74 | ccRCC | 3a | + | + | + | + | p.D179fs | ND | + | Wild | (+) | Wild | Wild |
| 25 | M | 71 | ccRCC | 1b | − | + | + | + | p.L135fs | ND | − | Wild | (+) | p.K553X | p.V1820E |
| 26 | M | 74 | ccRCC | 1a | − | + | + | + | p.S80R | ND | + | Wild | (+) | Splicing donor | Wild |
| 30 | M | 68 | ccRCC | 4 | + | + | + | + | p.N131X | ND | + | Wild | (+) | p.H712P | p.S204X |
| 31 | F | 67 | ccRCC | 3a | − | + | + | + | p.Q96fs | ND | + | Wild | ND | p.K1282fs | Wild |
| 32 | M | 71 | ccRCC | 2a | + | + | + | + | p.C162W | ND | + | Wild | ND | Wild | Wild |
| 33 | M | 50 | ccRCC | 2a | − | + | + | + | p.L169P | ND | + | Wild | (+) | Wild | Wild |
| 34 | M | 60 | ccRCC | 3a | + | + | + | + | p.L178fs | ND | + | Wild | (+) | p.E1501fs | Splicing donor |
| 38 | M | 75 | ccRCC | 1a | − | + | + | + | p.F136fs | ND | + | Wild | (−) | p.Y998X | Wild |
| 41 | M | 78 | ccRCC | 1a | − | + | + | + | p.V74D | ND | + | Wild | (−) | p.H573fs | Wild |
| 43 | F | 63 | ccRCC | 2b | − | + | + | + | Splicing donor | ND | + | Wild | (−) | Wild | Wild |
| 50 | M | 63 | ccRCC | 3a | + | + | + | + | p.G144fs | ND | − | p.C91F | (−) | Wild | p.K2525X |
| 52 | F | 46 | ccRCC | 1b | − | + | + | + | p.L163P | ND | + | Wild | (+) | p.E532X | Wild |
| 53 | M | 71 | ccRCC | 1b | − | + | + | + | p.G114fs | ND | + | Wild | (+) | p.S1253fs | Wild |
| 55 | F | 72 | ccRCC | 3a | + | + | − | − | Wild | − | − | Wild | ND | Wild | Wild |
| 57 | M | 84 | ccRCC | 3b | − | + | + | − | p.R120fs | ND | − | Wild | (+) | Wild | Wild |
| 58 | F | 57 | ccRCC | 3a | + | + | + | + | p.A149fs | ND | + | Wild | ND | Splicing
acceptor | Wild |
| 61 | M | 72 | ccRCC | 1a | − | + | + | + | p.R161P | ND | + | Wild | (+) | p.K1373fs | Wild |
| 62 | M | 63 | ccRCC | 1a | − | + | + | + | p.W88X | ND | + | p.I586fs | (−) | Wild | Wild |
| 63 | F | 53 | ccRCC | 1b | − | + | + | + | p.N90Y | ND | + | Wild | (+) | Wild | p.T1761fs |
| 64 | F | 62 | ccRCC | 3a | + | + | + | + | p.L135fs | ND | + | p.R701fs | (−) | Wild | Wild |
| 65 | M | 74 | ccRCC | 1b | − | + | + | + | Splicing
acceptor | ND | + | Wild | (+) | p.V964del | Wild |
| 66 | M | 83 | ccRCC | 3a | − | + | + | + | p.S111R | ND | + | Wild | (+) | Wild | p.A1591fs |
| 67 | F | 77 | ccRCC | 1a | − | + | + | − | Wild | + | − | Wild | (+) | Wild | Wild |
| 68 | F | 72 | ccRCC | 3a | − | + | + | + | p.S111R | ND | + | Wild | (+) | Splicing donor | Wild |
| 69 | M | 44 | ccRCC | 1b | − | + | + | + | p.T157fs | ND | + | Wild | (+) | p.Q238X | Wild |
| 23 | F | 65 | chRCC | 1a | − | + | − | − | Wild | − | − | Wild | (−) | Wild | Wild |
| 36 | F | 73 | chRCC | 1b | − | + | + | − | Wild | − | − | Wild | ND | Wild | Wild |
| 44 | F | 46 | chRCC | 2b | − | + | + | + | Wild | − | − | Wild | (−) | Wild | Wild |
Microsatellite (MS) genotyping of
chromosome 3p
We analyzed 13 polymorphic MS markers, identified
from the ‘Heterozygosity of MS Markers in the Japanese Population’
database (https://dbarchive.biosciencedbc.jp/jp/heterozygosity-jp/desc.html),
using a modified version of the method described by Dietrich et
al (33). We examined the
following markers: D3S1263 (3p25.3); D3S1266 (3p24.1); D3S1611
(3p22.2); D3S3687 (3p22.1); D3S3678 (3p22.1); D3S2420 (3p21.31);
D3S1568 (3p21.31); D3S1573 (3p21.31); D3S3561 (3p21.1); D3S3648
(3p21.1); D3S1289 (3p14.3); D3S1300 (3p14.2); and D3S1285 (3p14.1).
When the locus was heterozygous in the matched control sample, we
calculated the ratio of the peak area of allele 1 compared with
that of allele 2, and determined the allelic imbalance between the
allele ratio of the tumor and that of the matched control sample.
We judged LOH and deduced the rate of contaminating non-tumor cells
included in RCC tumors based on the value of allelic imbalance. The
rates of tumor content were lower than the rates estimated by
pathological examination.
Multiplex ligation probe amplification
(MLPA) analysis
We carried out MLPA analysis of genomic DNA for each
of the 17 exons of the BAP1 gene using a SALSA MLPA BAP1 kit
according to the manufacturer's instructions (MRC Holland,
Amsterdam, The Netherlands). This kit detects copy number changes
in each exon of BAP1 plus an additional ten genes on
chromosome 3: MLH1, RBM5, RASSF1, ZMYND10, HESX1, FHIT, MITF,
ROBO1, PROS1 and CPOX. The peak area obtained using
gene-specific probes in each sample was first normalized to the
average peak areas obtained using control probes specific for other
chromosomal locations. A final ratio was then calculated by
dividing by the value for genomic DNA isolated from the matched
control. The sample was scored to have loss or gain of one copy
number (monoallelic loss or amplification) by comparing the MLPA
ratio and the rate of tumor content in the RCC sample. For example,
if the rate of tumor content calculated by MS analysis and the MLPA
ratio were 0.5 and 0.75, respectively, this would indicate
monoallelic loss of BAP1 in an RCC sample that consisted of
≤50% non-tumor cells.
Target sequencing
We isolated DNA using an AllPrep DNA/RNA mini kit
(Qiagen, Germantown, MD, USA) according to the manufacturer's
instructions, and sequenced the coding regions of VHL, BAP1,
SETD2 and PBRM1. For VHL, direct sequencing was
conducted using a BigDye Terminator v3.1 kit on an Applied
Biosystems 3130xl Genetic Analyzer (Foster City, CA, USA); primer
sequences are presented in Table
II. By consulting the contamination rate of non-tumor cells in
each tumor sample, a barely imperceptible variant in the tumor
sample could be detected by sequencing on both strands. For the
other genes, next-generation sequencing was performed on an
Illumina MiSeq (San Diego, CA, USA) using paired-end 250-bp runs.
Libraries were prepared from 250 ng of genomic DNA from 45
independent RCC tumors using a Haloplex Custom kit (Agilent
Technologies, Santa Clara, CA, USA) according to the manufacturer's
instructions. Illumina paired-end reads were each aligned to the
human NCBI Build 37 reference sequence using bwa software
(bio-bwa.sourceforge.net/, version 0.6.0). The aligned sequence
files were sorted and merged using SAMtools (samtools.
sourceforge.net/, version 0.1.18). GATK (http://www.broadinstitute.org/gatk/) was used for
realignment, base quality score recalibration, single nucleotide
variant or indel (small insertions and deletions) variant calling,
and variant quality recalibration. SnpEff was used to categorize
the effects of variants by impact (34). Functional annotation of genetic
variants provided by ANNOVAR (www.openbioinformatics.org/annovar/, version 2013, Feb
11) and dbNSFP (35) were used to
identify protein-damaging mutations. Somatic mutations or germline
variants were judged by comparison between Sanger sequence data
from the ccRCC tumor and its matched control.
 | Table IISequences of the primers used for the
direct sequencing of the VHL gene. |
Table II
Sequences of the primers used for the
direct sequencing of the VHL gene.
| Exon | Uses for PCR or
sequencing | Forward or
reverse | Sequence |
|---|
| 1 | PCR and
sequencing | Forward |
TGGAAATACAGTAACGAGTTGGC |
| PCR and
sequencing | Reverse |
GCTTCAGACCGTGCTATCGT |
| 2 | PCR | Forward |
GGAGAAAATAGGTGCCCTGAC |
| PCR and
sequencing | Reverse |
GGCAAAAATTGAGAACTGGGCT |
| Sequencing | Forward |
CCAAAGTGCTGGGATTACAGG |
| 3 | PCR | Forward |
GGGGGCCATCAGCATAACAC |
| PCR and
sequencing | Reverse |
TACTTCTCTAATGGGCAGGCA |
| Sequencing | Forward |
GTAGTTGTTGGCAAAGCCTC |
Promoter methylation of VHL
Bisulfite sequencing was performed for the patients
who did not have a mutation in the VHL gene: ID 5, 6, 7, 21,
23, 36, 44, 55 and 67. After bisulfite modification of 100 ng tumor
or normal DNA using an EpiTect Fast DNA Bisulfite kit (Qiagen), the
regions containing both methylated and unmethylated alleles across
26 CpG dinucleotides in the VHL promoter were amplified
using a touchdown PCR program with an annealing temperature of
50°C: 1st PCR primers: 5′-TTAYGGAGGTYGATTYGGGAG-3′ and
5′-ACRATTACAAAAAATAACCTA-3′ (amplicon: 335 bp); nested PCR primers:
5′-YGGGTGGTTTGGATYG-3′ and 5′-AATTCACCRAACRCAACA-3′ (amplicon: 226
bp), as previously reported (36).
The cytosine positions in CpGs were inspected for thymine or
cytosine signals on the chromatograms. Promoter methylation was not
detected with normal samples; tumor samples with ≥30% methylated
CpGs were judged as methylated.
Nuclear staining
RCC tissues embedded in paraffin were cut into
3-μm-thick sections and heated in antigen retrieval solution (Bond
Epitope Retrieval Solution 2, pH 9.0; Leica Biosystems, Nussloch,
Germany) at 98°C for 10 min. Then, the sections were incubated with
mouse monoclonal antibody against human BAP1 (C-4; sc-28383; Santa
Cruz Biotechnology, Santa Cruz, CA, USA; 1:150 dilution). Bond
Polymer Refine Detection (Leica Biosystems) was used for
visualization with 3, 3′-diaminobenzidine tetrahydrochloride.
Substantial tumors had heterogeneous nuclear staining for BAP1,
with some cells staining positively and some cells staining
negatively. Negativity was evaluated when the majority of tumor
cell nuclei were stained negatively while there was obvious
staining in the nuclei of normal renal tubule cells on the same
slide. A pathologist reviewed all slides and categorized tumors as
positive, negative, or weak-positive. After samples with
weak-positive expression were excluded, 40 samples were available
for analysis (Table I).
Statistical analysis
Survival curves were calculated by the Kaplan-Meier
method and compared by log-rank test. Two patients died of cancer,
therefore the analysis for overall survival was not done. All
p-values were two-tailed, and differences were considered
significant at p<0.05.
Results
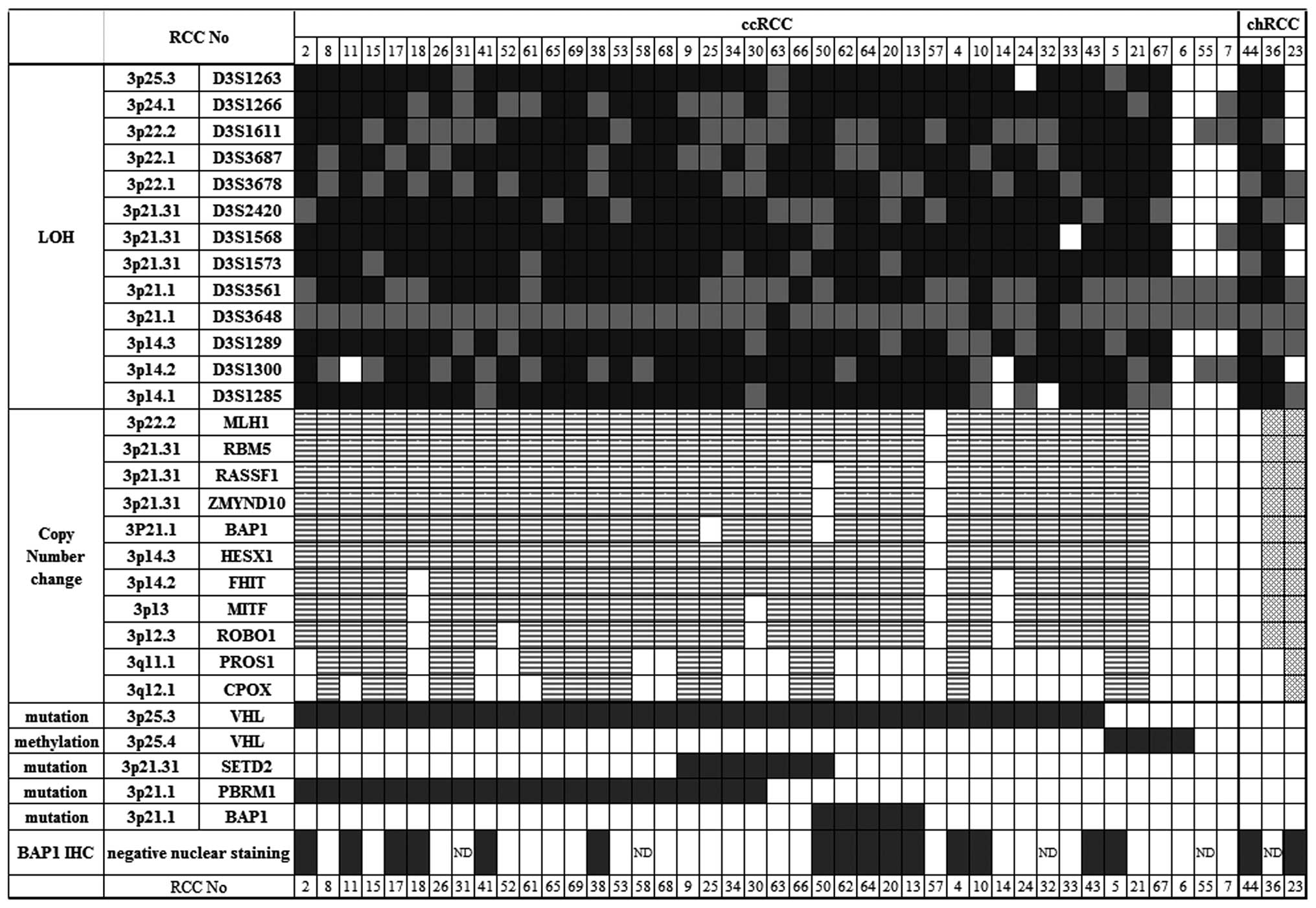
Ninety-five percent of ccRCC tumors had
either biallelic mutation or promoter methylation of VHL, while
chRCCs did not
A high frequency, 36/45 (80%), of VHL
mutation was detected in RCC tumors by Sanger sequencing. The
mutation type was: nonsense in six, frameshift in 15, splice site
in three, and missense in 12 (Table
I). The RCC tumors with VHL mutation showed LOH on 3p
(Fig. 1). No patients had germline
mutation of this gene (data not shown). Promoter methylation of
VHL was detected in four tumors: three (RCC05, 21 and 67)
with LOH and RCC06 without LOH. Especially prominent
hypermethylation was detected in RCC06 and RCC67 (data not shown).
By pathological examination, three (RCC23, 36, and 44) out of five
tumors without VHL mutation were noted to be chRCC. All
tumors with either somatic mutation or promoter methylation in
VHL were from patients with ccRCC.
Seventy-eight percent of RCC tumors had
monoallelic loss of BAP1, but none showed biallelic loss of
BAP1
All selected MS markers had a high frequency of
heterozygosity in the Japanese population except for D3S3648;
because this marker is close to the BAP1 locus, the
homozygosity rate was high [42 of our 45 subjects (93.3%)], and LOH
was mostly not determined for the D3S3648 region. Instead, we
deduced LOH of the BAP1 region by combining the LOH data of
flanking loci with copy number data determined by comparison of RCC
tumor and matched control samples.
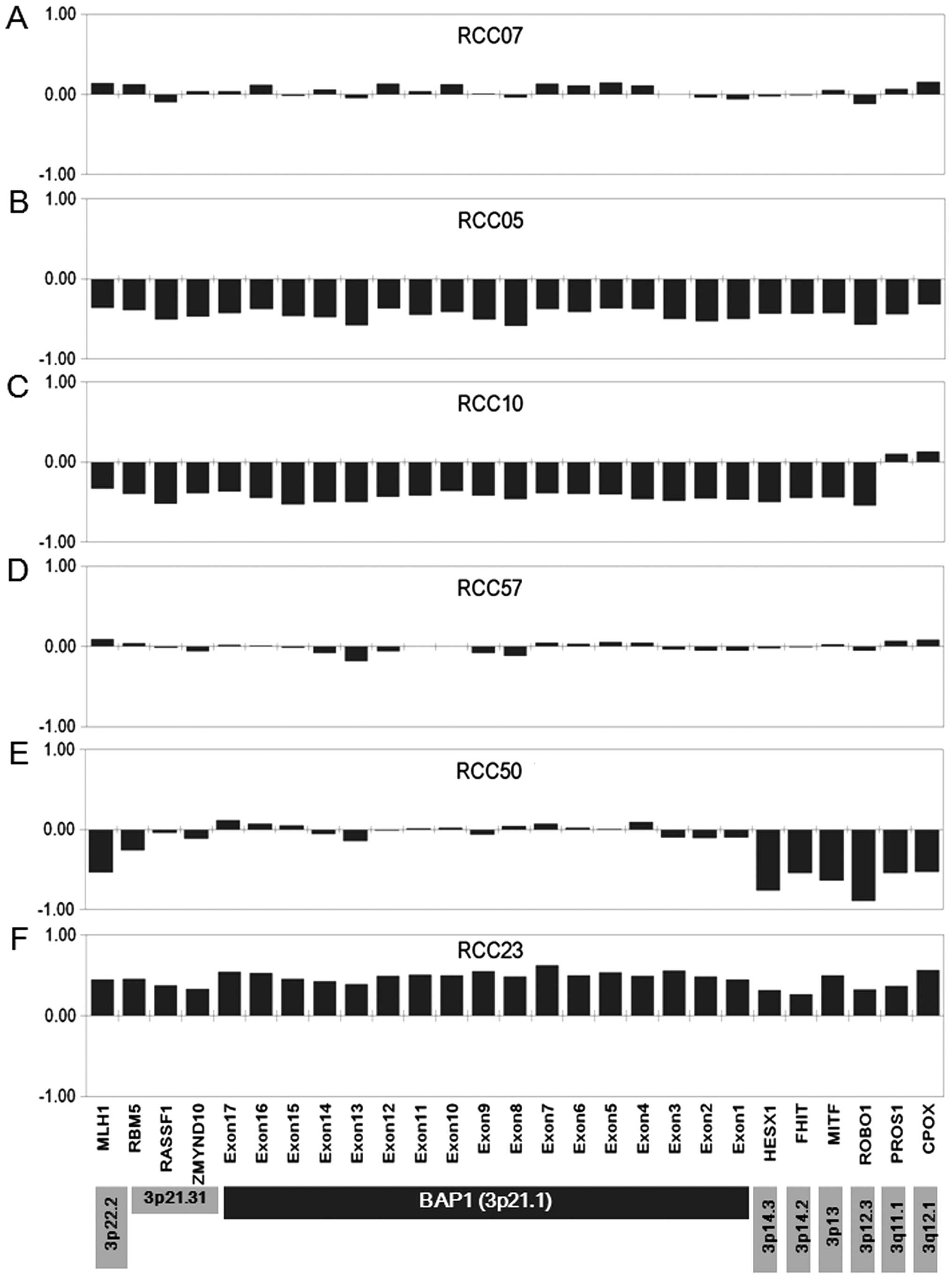
We did not find biallelic loss of BAP1 in RCC
samples. LOH in 3p was detected at a frequency of 41/45 (91%;
Table I and Fig. 1), and using MLPA analysis we
identified monoallelic loss of all exons of BAP1 together
with genes on 3p22.2-3p14.3 in 35 of 45 tumors (77.8%; Figs. 1, 2B
and 2C). Two ccRCCs (RCC14 and 18) demonstrated loss of
BAP1, but not FHIT nor genes on 3p13-12. Although
showing 3p LOH, three tumors that had no changes in copy number to
any BAP1 exons and any genes analyzed on 3p were judged to
show uniparental disomy: RCC44, 57 and 67 (Figs. 1 and 2D). Two tumors (RCC25 and 50) retained
two copies of BAP1 although other genes on 3p had lost one
copy (Fig. 2E). In two chRCCs
(RCC23 and 36), MLPA indicated amplification of all exons of
BAP1 together with other genes on 3p (Fig. 2F), although gross copy number
changes among the genome regions designed as normalization probes
(13 control probes specific to chromosomes other than chromosome 3)
were also detected (data not shown).
Biallelic inactivation of BAP1 was
detected in five ccRCCs, but not combined mutation of both BAP1 and
PBRM1
Somatic mutation in BAP1 was detected in five
ccRCCs with LOH (mutation frequency 11.1%); three ccRCCs had a
frameshift mutation (RCC13, 62, and 64), one had a nonsense
mutation (RCC20), and one had a missense change at amino acid
Cys91, which is essential for ubiquitin C-terminal hydrolase
activity (RCC50) (http://www.uniprot.org/uniprot/Q92560). Thus, all
mutations detected in BAP1 were predicted to cause loss-of-
function. The mutation frequencies of PBRM1 and SETD2
in our RCCs were 21/45 (46.7%), and 7/45 (15.6%), respectively
(Table I and Fig. 1). Only RCC50 had a mutation in both
BAP1 and SETD2. Somatic mutations in both BAP1
and PBRM1 were not detected; this combined mutation has been
reported to be rare.
Approximately 1/3 of ccRCCs with
hemizygous normal BAP1 showed negative nuclear staining
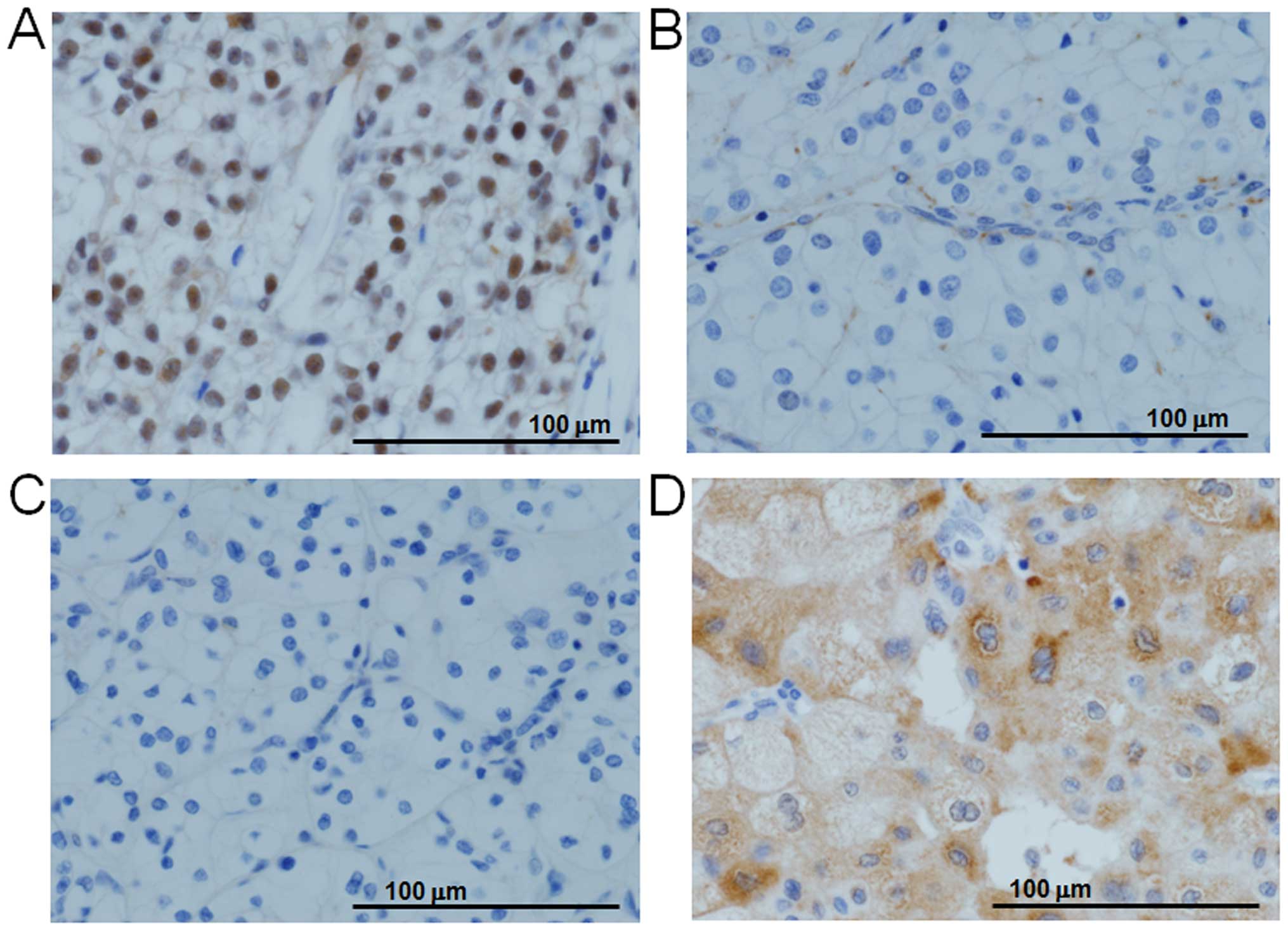
BAP1 nuclear staining was negative in the tumors
with biallelic mutation of BAP1 (Fig. 3B). In addition, BAP1 nuclear
staining was negative in 10 ccRCCs with hemizygous normal
BAP1 (10/31, 32.3%) (Figs.
1 and 3C). Three chRCCs had
strong BAP1 staining in the cytosol (Fig. 3D), although two of them were
negative and one was excluded from analysis with nuclear
staining.
Biallelic inactivation of BAP1 led to
worse outcome
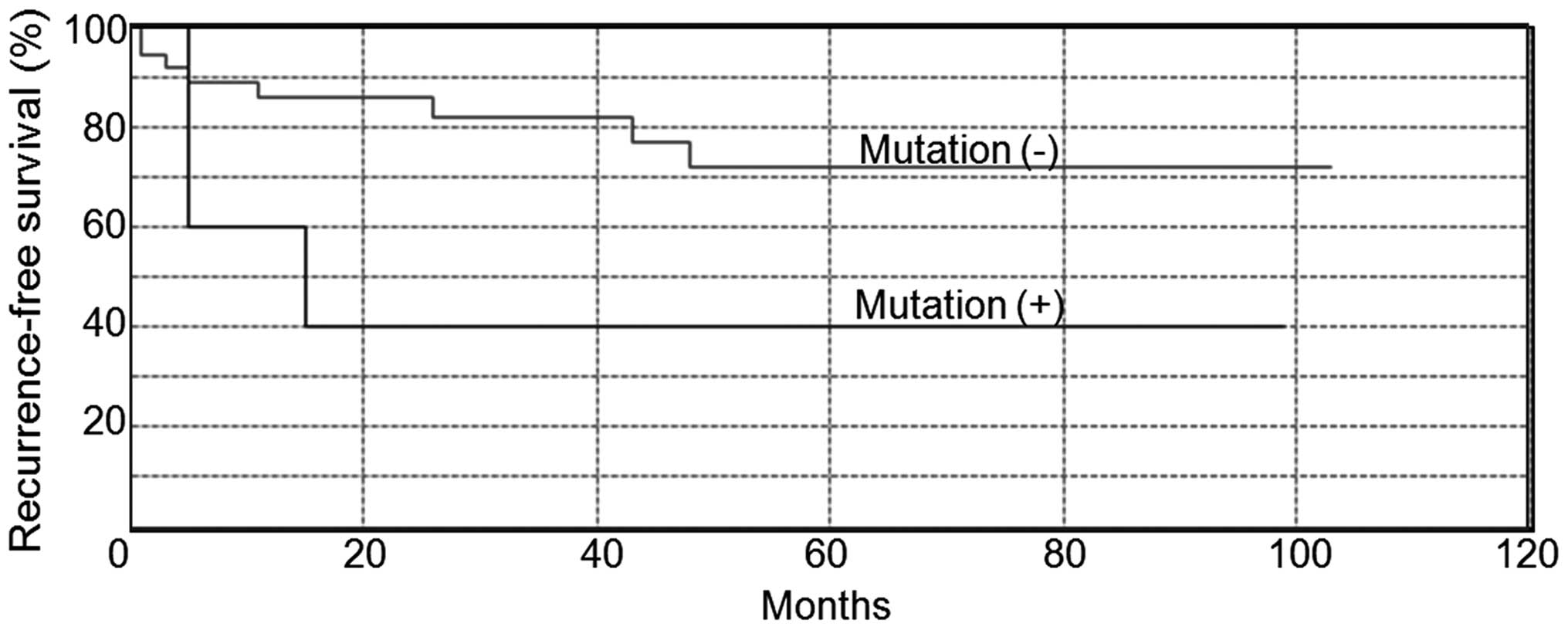
We investigated the relationship between BAP1
mutation and tumor recurrence. The ccRCC tumors with biallelic
loss-of-function mutation in BAP1 correlated with
recurrence-free survival time (p=0.046, Fig. 4), but not by multivariate analysis
combined with age, T stage, histological subtype, infiltration and
vascular invasion (data not shown). SETD2 mutations
significantly correlated with recurrence-free survival time
(p=0.007), but PBRM1 mutations did not (p=0.761) (data not
shown). The ccRCC tumors with negative BAP1 nuclear staining did
not associate with tumor recurrence (p=0.143, data not shown).
Discussion
We confirmed that 95% of our ccRCCs had features of
the VHL-dependent pathway of tumorigenesis. VHL inactivation
(mutation by base substitution or indel, or promoter methylation)
perfectly paralleled 3p LOH in ccRCC tumors. It is known that point
mutations in VHL, and loss of 3p, which has been suggested
to associate with the deletion of FHIT located on 3p14.2
(5,6), are early events during tumorigenesis
and are thought to initiate ccRCC development. Tumors from two
stage I ccRCC patients (RCC14 and 18) had monoallelic loss of
BAP1 together with genes on 3p22.2-3p14.3, but not
FHIT. Deletion of FHIT might not be essential for a
large loss of 3p, but mutations of several tumor suppressor genes
on 3p might trigger a large loss of 3p by cooperating with
VHL mutation.
Mutations in BAP1, PBRM1 and SETD2
were detected in ccRCCs with VHL mutation. The rate of
hemizygous truncation mutation of BAP1 in ccRCC was 4/42,
resulting in negative nuclear expression of the encoded protein.
RCC50, which showed uniparental disomy on chr3p21.31-3p21.1, had a
biallelic BAP1 missense mutation that would cause functional
loss, and also a biallelic SETD2 mutation. In addition, this
tumor was negative for BAP1 nuclear staining, although the mutation
did not occur in the nuclear-localization signal. Biallelic
deletion of BAP1 was not detected. The frequency of
BAP1 mutation (11.1%) is close to that in other studies
(9–11,18).
Even though the mutation rate of BAP1 was low, 1/3 of our
tumors showed negative nuclear staining of BAP1 by
immunohistochemistry. Peña-Llopis et al have also reported
this difference, but the difference was small because 25 out of 176
samples were negative for nuclear staining, and 22 out of 25 tumors
without expression had a BAP1 mutation (9). A subsequent multi-institutional
cohort study using immunohistochemistry without genetic testing
indicated that BAP1 protein was negative for nuclear staining in 82
of 559 ccRCC tumors (14.7%) (37).
Because of the genetic heterogeneity of ccRCC (38), it was difficult to validate
heterogeneous staining for BAP1 to obtain uniformity in scoring,
without differences among institutions.
There are several ways to explain negative nuclear
expression of BAP1 protein without biallelic mutation or deletion:
epigenetic regulation of gene expression by DNA methylation or
histone modification, excess degradation of mRNA or protein,
repressed translation by miRNA, or dysregulation of nuclear
localization. Promoter methylation of BAP1 was not detected
in ccRCC (39) nor in MM (29). Nuclear localization is important
for the function of this protein (31). Recently a ubiquitin-conjugating
enzyme, UBE2O, has been noted to multi-monoubiquitinate the nuclear
localization signal of BAP1, resulting in its cytoplasmic
sequestration. UBE2O activity is counteracted by BAP1
autodeubiquitination through intramolecular interactions (32).
Three chRCC tumors showed strong cytosolic staining
of BAP1 that might result from copy number amplification of this
gene or aberrant transportation by BAP1 ubiquitinated by UBE2O. We
could not test the expression level of the BAP1 gene because
our RCC samples contained a substantial proportion of non-tumor
cells. Because ~30% of ccRCC tumors with hemizygous normal
BAP1 showed negative nuclear staining (Fig. 1), haploinsufficiency might be
related to RCC development.
BAP1 mutations have been tightly linked to worse
clinical outcome in multiple studies (9,11,40).
Our data showed that patients with biallelic BAP1 mutation
had worse recurrence-free survival than the patients without
biallelic mutation (p=0.046), although multivariate analyses did
not show the association between BAP1 mutation and tumor
recurrence probably due to small sample size. We did not find an
association between negative nuclear BAP1 staining and tumor
recurrence. This shows that genomic analysis of BAP1 gene is
useful in interpreting the prognosis of RCC.
Acknowledgements
This study was supported in part by a Grant-in-Aid
for Researchers from Hyogo College of Medicine, 2013. We are
grateful to Atsuko Iemoto and Daiki Haruguchi of Hyogo College of
Medicine for their technical assistance.
References
|
1
|
Angeloni D: Molecular analysis of
deletions in human chromosome 3p21 and the role of resident cancer
genes in disease. Brief Funct Genomics Proteomics. 6:19–39. 2007.
View Article : Google Scholar
|
|
2
|
Kok K, Naylor SL and Buys CH: Deletions of
the short arm of chromosome 3 in solid tumors and the search for
suppressor genes. Adv Cancer Res. 71:27–92. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gnarra JR, Tory K, Weng Y, Schmidt L, Wei
MH, Li H, Latif F, Liu S, Chen F, Duh FM, et al: Mutations of the
VHL tumour suppressor gene in renal carcinoma. Nat Genet. 7:85–90.
1994. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Clifford SC, Prowse AH, Affara NA, Buys CH
and Maher ER: Inactivation of the von Hippel-Lindau (VHL) tumour
suppressor gene and allelic losses at chromosome arm 3p in primary
renal cell carcinoma: Evidence for a VHL-independent pathway in
clear cell renal tumourigenesis. Genes Chromosomes Cancer.
22:200–209. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sükösd F, Kuroda N, Beothe T, Kaur AP and
Kovacs G: Deletion of chromosome 3p14.2-p25 involving the VHL and
FHIT genes in conventional renal cell carcinoma. Cancer Res.
63:455–457. 2003.PubMed/NCBI
|
|
6
|
Singh RB and Amare Kadam PS: Investigation
of tumor suppressor genes apart from VHL on 3p by deletion mapping
in sporadic clear cell renal cell carcinoma (cRCC). Urol Oncol.
31:1333–1342. 2013. View Article : Google Scholar
|
|
7
|
Varela I, Tarpey P, Raine K, Huang D, Ong
CK, Stephens P, Davies H, Jones D, Lin ML, Teague J, et al: Exome
sequencing identifies frequent mutation of the SWI/SNF complex gene
PBRM1 in renal carcinoma. Nature. 469:539–542. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dalgliesh GL, Furge K, Greenman C, Chen L,
Bignell G, Butler A, Davies H, Edkins S, Hardy C, Latimer C, et al:
Systematic sequencing of renal carcinoma reveals inactivation of
histone modifying genes. Nature. 463:360–363. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Peña-Llopis S, Vega-Rubín-de-Celis S, Liao
A, Leng N, Pavía-Jiménez A, Wang S, Yamasaki T, Zhrebker L,
Sivanand S, Spence P, et al: BAP1 loss defines a new class of renal
cell carcinoma. Nat Genet. 44:751–759. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Guo G, Gui Y, Gao S, Tang A, Hu X, Huang
Y, Jia W, Li Z, He M, Sun L, et al: Frequent mutations of genes
encoding ubiquitin-mediated proteolysis pathway components in clear
cell renal cell carcinoma. Nat Genet. 44:17–19. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sato Y, Yoshizato T, Shiraishi Y, Maekawa
S, Okuno Y, Kamura T, Shimamura T, Sato-Otsubo A, Nagae G, Suzuki
H, et al: Integrated molecular analysis of clear-cell renal cell
carcinoma. Nat Genet. 45:860–867. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Testa JR, Cheung M, Pei J, Below JE, Tan
Y, Sementino E, Cox NJ, Dogan AU, Pass HI, Trusa S, et al: Germline
BAP1 mutations predispose to malignant mesothelioma. Nat Genet.
43:1022–1025. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wiesner T, Obenauf AC, Murali R, Fried I,
Griewank KG, Ulz P, Windpassinger C, Wackernagel W, Loy S, Wolf I,
et al: Germline mutations in BAP1 predispose to melanocytic tumors.
Nat Genet. 43:1018–1021. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Abdel-Rahman MH, Pilarski R, Cebulla CM,
Massengill JB, Christopher BN, Boru G, Hovland P and Davidorf FH:
Germline BAP1 mutation predisposes to uveal melanoma, lung
adenocarcinoma, meningioma, and other cancers. J Med Genet.
48:856–859. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Carbone M, Ferris LK, Baumann F,
Napolitano A, Lum CA, Flores EG, Gaudino G, Powers A,
Bryant-Greenwood P, Krausz T, et al: BAP1 cancer syndrome:
Malignant mesothelioma, uveal and cutaneous melanoma, and MBAITs. J
Transl Med. 10:1792012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Popova T, Hebert L, Jacquemin V, Gad S,
Caux-Moncoutier V, Dubois-d'Enghien C, Richaudeau B, Renaudin X,
Sellers J, Nicolas A, et al: Germline BAP1 mutations predispose to
renal cell carcinomas. Am J Hum Genet. 92:974–980. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Farley MN, Schmidt LS, Mester JL,
Peña-Llopis S, Pavia-Jimenez A, Christie A, Vocke CD, Ricketts CJ,
Peterson J, Middelton L, et al: A novel germline mutation in BAP1
predisposes to familial clear-cell renal cell carcinoma. Mol Cancer
Res. 11:1061–1071. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Liao L, Testa JR and Yang H: The roles of
chromatin-remodelers and epigenetic modifiers in kidney cancer.
Cancer Genet. 208:206–214. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang SS, Gu YF, Wolff N, Stefanius K,
Christie A, Dey A, Hammer RE, Xie XJ, Rakheja D, Pedrosa I, et al:
Bap1 is essential for kidney function and cooperates with Vhl in
renal tumorigenesis. Proc Natl Acad Sci USA. 111:16538–16543. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Peña-Llopis S, Christie A, Xie XJ and
Brugarolas J: Cooperation and antagonism among cancer genes: The
renal cancer paradigm. Cancer Res. 73:4173–4179. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Scheuermann JC, de Ayala Alonso AG, Oktaba
K, Ly-Hartig N, McGinty RK, Fraterman S, Wilm M, Muir TW and Müller
J: Histone H2A deubiquitinase activity of the Polycomb repressive
complex PR-DUB. Nature. 465:243–247. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Harbour JW, Onken MD, Roberson ED, Duan S,
Cao L, Worley LA, Council ML, Matatall KA, Helms C and Bowcock AM:
Frequent mutation of BAP1 in metastasizing uveal melanomas.
Science. 330:1410–1413. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ewens KG, Kanetsky PA, Richards-Yutz J,
Purrazzella J, Shields CL, Ganguly T and Ganguly A: Chromosome 3
status combined with BAP1 and EIF1AX mutation profiles are
associated with metastasis in uveal melanoma. Invest Ophthalmol Vis
Sci. 55:5160–5167. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Yoshikawa Y, Sato A, Tsujimura T, Emi M,
Morinaga T, Fukuoka K, Yamada S, Murakami A, Kondo N, Matsumoto S,
et al: Frequent inactivation of the BAP1 gene in epithelioid-type
malignant mesothelioma. Cancer Sci. 103:868–874. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Emi M, Yoshikawa Y, Sato C, Sato A, Sato
H, Kato T, Tsujimura T, Hasegawa S, Nakano T and Hashimoto-Tamaoki
T: Frequent genomic rearrangements of BRCA1 associated protein-1
(BAP1) gene in Japanese malignant mesothelioma-characterization of
deletions at exon level. J Hum Genet. 60:647–649. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bott M, Brevet M, Taylor BS, Shimizu S,
Ito T, Wang L, Creaney J, Lake RA, Zakowski MF, Reva B, et al: The
nuclear deubiquitinase BAP1 is commonly inactivated by somatic
mutations and 3p21.1 losses in malignant pleural mesothelioma. Nat
Genet. 43:668–672. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zauderer MG, Bott M, McMillan R, Sima CS,
Rusch V, Krug LM and Ladanyi M: Clinical characteristics of
patients with malignant pleural mesothelioma harboring somatic BAP1
mutations. J Thorac Oncol. 8:1430–1433. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Arzt L, Quehenberger F, Halbwedl I,
Mairinger T and Popper HH: BAP1 protein is a progression factor in
malignant pleural mesothelioma. Pathol Oncol Res. 20:145–151. 2014.
View Article : Google Scholar
|
|
29
|
Nasu M, Emi M, Pastorino S, Tanji M,
Powers A, Luk H, Baumann F, Zhang YA, Gazdar A, et al: High
incidence of somatic BAP1 alterations in sporadic malignant
mesothelioma. J Thorac Oncol. 10:565–576. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Mori T, Sumii M, Fujishima F, Ueno K, Emi
M, Nagasaki M, Ishioka C and Chiba N: Somatic alteration and
depleted nuclear expression of BAP1 in human esophageal squamous
cell carcinoma. Cancer Sci. 106:1118–1129. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yu H, Mashtalir N, Daou S, Hammond-Martel
I, Ross J, Sui G, Hart GW, Rauscher FJ III, Drobetsky E, Milot E,
et al: The ubiquitin carboxyl hydrolase BAP1 forms a ternary
complex with YY1 and HCF-1 and is a critical regulator of gene
expression. Mol Cell Biol. 30:5071–5085. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Mashtalir N, Daou S, Barbour H, Sen NN,
Gagnon J, Hammond-Martel I, Dar HH, Therrien M and Affar B:
Autodeubiquitination protects the tumor suppressor BAP1 from
cytoplasmic sequestration mediated by the atypical ubiquitin ligase
UBE2O. Mol Cell. 54:392–406. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Dietrich W, Katz H, Lincoln SE, Shin HS,
Friedman J, Dracopoli NC and Lander ES: A genetic map of the mouse
suitable for typing intraspecific crosses. Genetics. 131:423–447.
1992.PubMed/NCBI
|
|
34
|
Cingolani P, Platts A, Wang L, Coon M,
Nguyen T, Wang L, Land SJ, Lu X and Ruden DM: A program for
annotating and predicting the effects of single nucleotide
polymorphisms, SnpEff: SNPs in the genome of Drosophila
melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 6:80–92.
2012. View Article : Google Scholar
|
|
35
|
Liu X, Jian X and Boerwinkle E: dbNSFP
v2.0: A database of human non-synonymous SNVs and their functional
predictions and annotations. Hum Mutat. 34:E2393–E2402. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Herman JG, Graff JR, Myöhänen S, Nelkin BD
and Baylin SB: Methylation-specific PCR: A novel PCR assay for
methylation status of CpG islands. Proc Natl Acad Sci USA.
93:9821–9826. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Kapur P, Christie A, Raman JD, Then MT,
Nuhn P, Buchner A, Bastian P, Seitz C, Shariat SF, Bensalah K, et
al: BAP1 immunohistochemistry predicts outcomes in a
multi-institutional cohort with clear cell renal cell carcinoma. J
Urol. 191:603–610. 2014. View Article : Google Scholar
|
|
38
|
Gerlinger M, Horswell S, Larkin J, Rowan
AJ, Salm MP, Varela I, Fisher R, McGranahan N, Matthews N, Santos
CR, et al: Genomic architecture and evolution of clear cell renal
cell carcinomas defined by multiregion sequencing. Nat Genet.
46:225–233. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ibragimova I, Maradeo ME, Dulaimi E and
Cairns P: Aberrant promoter hypermethylation of PBRM1, BAP1, SETD2,
KDM6A and other chromatin-modifying genes is absent or rare in
clear cell RCC. Epigenetics. 8:486–493. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Hakimi AA, Ostrovnaya I, Reva B, Schultz
N, Chen YB, Gonen M, Liu H, Takeda S, Voss MH, Tickoo SK, et al;
ccRCC Cancer Genome Atlas (KIRC TCGA) Research Network
investigators. Adverse outcomes in clear cell renal cell carcinoma
with mutations of 3p21 epigenetic regulators BAP1 and SETD2: A
report by MSKCC and the KIRC TCGA research network. Clin Cancer
Res. 19:3259–3267. 2013. View Article : Google Scholar : PubMed/NCBI
|